Abstract
Background and Aims
Large-scale ploidy surveys using flow cytometry have become an essential tool to study plant genome dynamics and to gain insight into the mechanisms and genetic barriers framing ploidy diversity. As an ideal complement to traditional techniques such as chromosome counting, the analysis of cytotype diversity in plant systems such as Sorbus provides primary investigation into the potential patterns and evolutionary implications of hybrid speciation.
Methods
Ploidy was assessed by means of relative nuclear DNA content using propidium iodide flow cytometry in 474 Sorbus samples collected from 65 populations in southern Wales and South-West England. Statistical tests were applied to evaluate the utility of this technique to confidently discriminate ploidy in the genus.
Key Results
Flow cytometric profiles revealed the presence of four cytotypes (2x, 3x, 4x and 5x), confirming in many cases chromosome counts previously reported and demonstrating cytotype heterogeneity within specific Sorbus aggregates. Diploid cytotypes were restricted to the potential parental species and homoploid hybrids. Most of the samples processed were polyploid. The occurrence of the pentaploid cytotype had previously only been reported from a single specimen; it is now confirmed for two taxa occurring at different sites.
Conclusions
Flow cytometry results obtained have proved useful in shedding light on the taxonomy of several controversial taxa and in confirming the presence of cytoypes which occur at very low frequencies. Notably, the coexistence of several cytotypes in Sorbus populations has probably been facilitated by the overlapping distribution of many of the species studied, which might also explain the high incidence of potential hybrid apomictic polyploids. These results will provide a solid baseline for molecular research aiming to better understand the genetic pathways controlling the formation and establishment of polyploid Sorbus.
Keywords: Apomixis, cytotype mixture, flow cytometry, genome size, hybridization, ploidy, polyploidy, whitebeam
INTRODUCTION
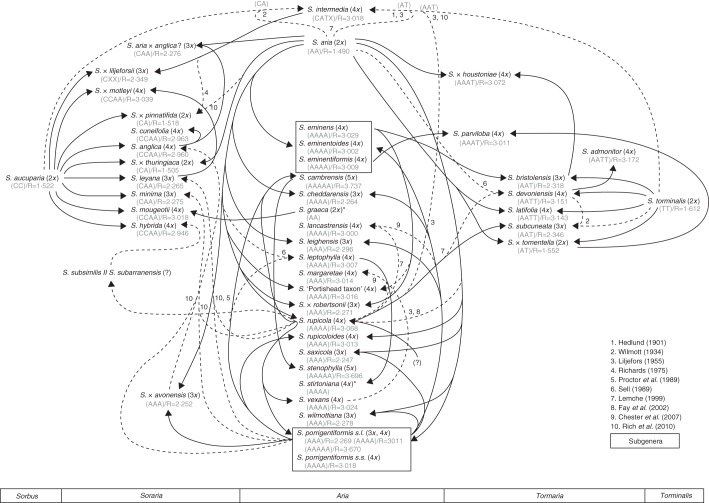
The genus Sorbus in Britain is represented by 45 native and seven introduced taxa (Rich et al., 2010). The origin of polyploid endemics in the genus is an ongoing question, but early studies (e.g. Liljefors, 1955) suggested that the sexual diploids S. aria, S. aucuparia and S. torminalis, with the apomictic tetraploid S. rupicola, probably form the backbone of this complex. In the last decade, new molecular markers for investigating population genetics have become increasingly useful in disentangling relationships in the genus, revealing a complex evolutionary history (Fig. 1) in which hybridization and recurrent episodes of polyploidization have played a crucial role (Nelson-Jones et al., 2002; Chester et al., 2007). The general interest that polyploidy has raised among researchers due to its frequent occurrence in plant systems has been essential in gaining insight into the dynamics of this evolutionary force and its implications in plant speciation (Soltis et al., 2009). Despite these advances, the potential advantages of apomixis in plant diversification and colonization (Hörandl et al., 2008), as well as the reproductive mechanisms of apomixis relating to polyploidy (Dickinson et al., 2007), are not fully understood and are still areas of active study.
Fig. 1.
Hypothesized network of relationships in Sorbus based on the results compiled by Rich et al. (2010) including diploids and the intermediate polyploid taxa. Proposed genome compositions are also included with the relative DNA content and ploidy inferred using FCM. Dotted lines indicate either multiple or dubious origins. *Ploidy based on published chromosome numbers (Warburg and Kárpáti, 1968; Bailey et al., 2008).
The number of species recognized in Sorbus varies between authors depending on the way that the numerous polyploid apomicts are treated, and the classification has been frequently revised (Proctor, 1999; Aldasoro et al., 2004; Rich and Proctor, 2009). Taxa such as the so-called ‘No Parking’ whitebeam, now known as S. admonitor, and Sorbus ‘Taxon D’, now known as S. margaretae, were unnamed for a long time (Proctor et al., 1989). Given that many of these polyploids originated via hybridization, the question arises as to whether they should be considered as hybrids or as new independent species, a question that might be related to the ability of these hybrid genotypes to produce viable offspring and perpetuate themselves. Nearly all taxa in the genus are relatively long-lived trees, mostly needing high light levels and are intolerant of grazing. For that reason many species have a preference for open cliffs where they have easy access to sunlight. As exceptions, S. aucuparia and representatives of the S. eminens aggregate are shade-tolerant, and S. torminalis is the only woodland species of Sorbus in Britain.
From a karyological point of view, chromosome counts have only been reported for 24 native Sorbus taxa. Estimates of ploidy inferred from the number of microsatellite alleles are also available for nine taxa, but chromosome numbers in 12 of the most recently described taxa are still unknown (Bailey et al., 2008; Houston et al., 2009; Robertson et al., 2010). Karyological information is available for all the introduced taxa. Three common ploidies (diploid, triploid and tetraploid; 2n = 34, 51 and 68, respectively) occur in Britain, and a single ‘pentaploid’ count (2n = approx. 87 chromosomes) was reported for a seedling of S. porrigentiformis sensu lato (s.l.) (Bailey et al., 2008). Most taxa have only been reported to occur at one ploidy, but three of the native taxa have been recorded to have more than one cytotype; S. anglica and S. subcuneata have both been reported to include triploids and tetraploids, and S. porrigentiformis s.l. includes triploids and tetraploids. The single pentaploid count was obtained from a cultivated seedling and, in the absence of information regarding the parentage of the seed, this count might not be representative of that for the parent. The introduced S. latifolia has been reported as both diploid and tetraploid, although the diploid counts probably refer to S. × tomentella (the primary hybrid between S. aria and S. torminalis).
The difficulties in obtaining suitable tissue for chromosome counts and the small size and relatively high numbers of chromosomes clearly indicate a need to find alternative techniques to estimate ploidy easily. The availability of knowledge of ploidy in addition to karyological data (see compilation of Bailey et al., 2008) could become a source of extra information to help in inferring species relationships and in hypothesizing the origins in controversial cases. Flow cytometry (hereafter FCM) has become widely used for estimating nuclear DNA contents in either absolute or relative (i.e. DNA ploidy) units, with many applications described in fields such as plant genome size diversity, biosystematics, ecology and population biology (Kron et al., 2007; Loureiro et al., 2010; Pellicer et al., 2010).
Recently, understanding of the patterns of occurrence and distribution of polyploids across plant populations has been facilitated by easy access to FCM, enabling us to explore the evolutionary implications of hybrid speciation (Leitch et al., 2008; Siljak-Yakovlev et al., 2008; Bennert et al., 2011; Marques et al., 2012). In this sense, FCM has become an ideal tool for tracking the dynamics of ploidy variation in many diverse plant groups and assessing the processes that frame cytotype distribution on a temporal scale (Ebihara et al., 2005; Suda et al., 2007; Marhold et al., 2010; Šafářová and Duchoslav, 2010; Trávníček et al., 2010, 2011). This technique is also especially useful in providing a more robust understanding of genetic data at the population level. An illustrative example can be found in the genus Gymnadenia (Orchidaceae). While chromosome studies in the complex (Marhold et al., 2005) reported the existence of 2x and 4x cytotypes, and nuclear microsatellite data (Stark et al., 2011) were useful only in differentiating between diploid and polyploid populations sensu lato, the ploidy screening carried out by Trávníček et al. (2011) using FCM revealed the coexistence of multiple cytotypes (2x, 3x, 4x, 5x, 6x) in the genus. Scenarios such as this not only show the importance of large-scale surveys, but also highlight the difficulties in interpreting genetic data for markers such as nuclear microsatellites for which allele number is dependent on ploidy (e.g. Sorbus; M. F. Fay et al., unpubl. res.). This illustrates the need to explore potential cytotype coexistence under environmental conditions by using complementary tools that provide data for gaining insight into the mechanisms of polyploidy and speciation, and assessing the genetic boundaries preserving such diversity and its consequences at the genome size level.
Given the previous evidence suggesting a frequent co-occurrence of cytotypes in British Sorbus (Bailey et al., 2008), we designed a population survey: (1) to ascertain the diversity of cytotypes in a range of species to help in clarifying species relationships; (2) to assess the coexistence of multiple ploidies within species and aggregates; and (3) to provide a general picture of the most prevalent cytotypes in British Sorbus populations.
MATERIALS AND METHODS
Plant material
Sixty-five populations of Sorbus, including 31 recognized species, seven known interspecific hybrids and 15 taxonomically unconfirmed specimens, were collected during summer 2011 covering (in many cases) the ranges of the known distribution of the rarer taxa (see Supplementary Data Table S1, available online, for locality and herbarium voucher information). In this study, the field sampling aimed to cover as wide a range of species in South-West England and Wales as possible with a good coverage of diploids to provide background data and information on known taxonomically difficult individuals. Additional collections from the National Botanic Garden of Wales, the Finnish Museum of Natural History and the Pyrenees were also included. In total, 474 samples were cytotyped. Herbarium vouchers are deposited in the Welsh National Herbarium (NMW).
Chromosome counts
Karyological information based on chromosome numbers compiled in the literature (Bailey et al., 2008; Rich et al., 2010) was used to confirm whether these previous reports correlated with our DNA ploidy estimations using FCM.
Flow cytometry
Samples were processed by FCM using propidium iodide (PI)-stained nuclei to unravel DNA ploidy levels (Suda et al., 2006). Fully expanded leaf tissue from each specimen collected in the field was preserved in plastic bags at 4 °C for up to 5 d. About 1 cm2 of the target sample was co-chopped with the selected internal standard (Oryza sativa ‘IR36’, 1C = 0·5 pg) using a new razor blade in a Petri dish containing 1 mL of ‘general purpose isolation buffer’ (GPB; Loureiro et al., 2007) with 3 % PVP-40 following the one-step procedure described by Doležel et al. (2007). The nuclear suspension was then filtered through a nylon mesh (30 µm) to remove debris, stained with PI (1 mg mL−1; Sigma) at a final concentration of 60 µg mL−1 and supplemented with 17 µL of a solution of 3 mg mL−1 ribonuclease A (RNase A; Sigma). After incubation for 10 min on ice, the relative nuclear DNA content was estimated by recording at least 3000 particles using a Partec Cyflow SL3 (Partec GmbH, Münster, Germany) flow cytometer fitted with a 100-mW green solid state laser (Cobolt Samba). The resulting histograms were analysed with the FlowMax software (v. 2·4, Partec GmbH). The absolute DNA content of the diploid species S. aria, S. aucuparia and S. torminalis was calculated following the same procedure described above but adjusting the acquisition to 5000 particles and analysing three independent specimens (three replicates each). Ploidy was determined on the basis of the sample/standard ratio, taking into account the FCM profiles of the diploid taxa. Given that only Oryza sativa was used as standard and its 2C-value is 1·0 pg, ratios calculated are equivalent to the relative DNA content.
Statistical analysis
Data were analysed with Statgraphics Plus v. 5·1 (Statistical Graphics Corp., Warrenton, VA, USA). The normality of the data distributions was tested using the Kolmogorov–Smirnov test and the homogeneity of variances by Levene's test. Differences between selected groups were tested using the non-parametric Kruskal–Wallis test. A linear regression analysis was also conducted to test for any correlation between relative DNA content and ploidy.
RESULTS
Accuracy of measurements: evaluation of flow histograms
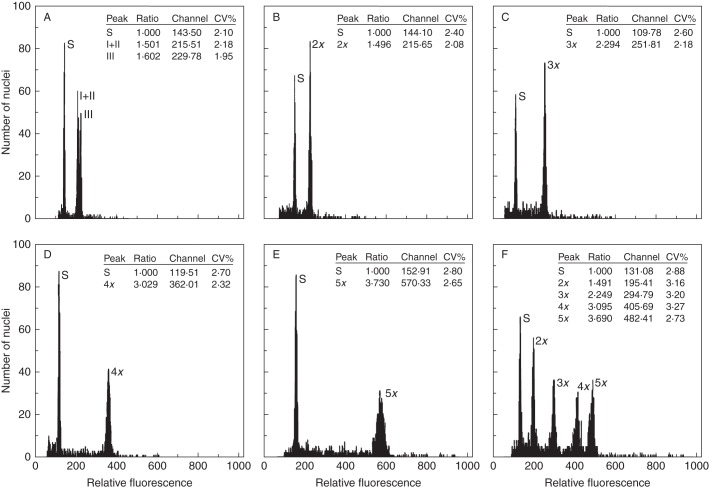
The Sorbus samples analysed produced high-resolution flow histograms with peaks of both the target and standard samples yielding low coefficients of variation (CV%): 1·80–3·50 (mean = 2·39 ± 0·52) and 1·22–3·65 (mean = 2·23 ± 0·45) (Fig. 2). Little variation (<2·8 %) was found between samples from the same species measured on different days. In most cases, only peaks with nuclei in phase G0/G1 of mitosis (2C peak) were recorded; those for G2 nuclei were almost imperceptible and did not interfere with interpretation of the results when present. The genome sizes of the diploid parental taxa were similar (Table 1, Fig. 2A), although that of S. torminalis was slightly larger (also reflected in the hybridogenous taxa with S. torminalis as one parent providing useful information on origins). Four easily recognizable distinct groups of fluorescence intensities were observed in taxa with chromosome counts available, therefore allowing us to infer reliable DNA ploidy (Fig. 2B–F).
Fig. 2.
Representative flow cytometric histograms illustrating all DNA ploidies found within the Sorbus complex investigated [internal standard Oryza sativa (S)]. Nuclei of both the target samples and the standard were isolated together and stained with PI. (A) combined sample of the diploid species S. aria (I), S. aucuparia (II) and S. torminalis (III); (B) diploid S. aria; (C) triploid S. leyana; (D) tetraploid S. eminens; (E) pentaploid S. cambrensis; (F) combined run including all cytotypes and the internal standard.
Table 1.
Nuclear DNA amounts estimated for the parental diploid Sorbus
| Taxon | 2n | Ploidy | 2C (pg, mean ± s.d.) | 1C (pg) | 1C (Mbp)* |
|---|---|---|---|---|---|
| S. aria (L.) Crantz | 34 | 2x | 1·484 ± 0·023 | 0·742 | 752·676 |
| S. aucuparia (L.) | 34 | 2x | 1·525 ± 0·011 | 0·762 | 745·236 |
| S. torminalis (L.) Crantz | 34 | 2x | 1·612 ± 0·002 | 0·810 | 792·180 |
* 1 pg = 978 Mbp (Doležel et al., 2003).
Ploidy variation in the Sorbus complex
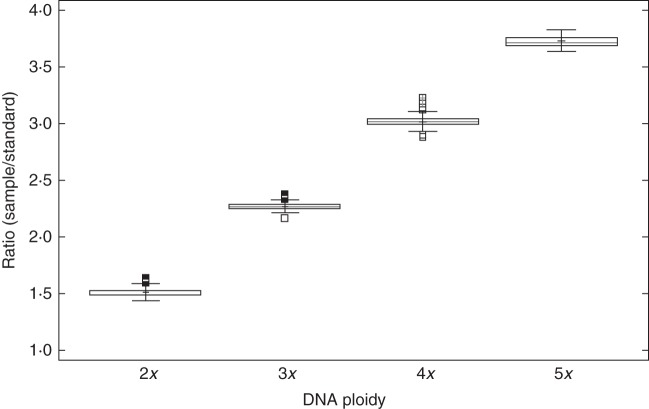
As expected, DNA peak ratios identified ploidy (Fig. 3; K = 409·83, P < 0·0001) based on the assumption that ratios ranging from 1·434 to 1·631 represented DNA-diploid individuals, from 2·286 to 2·370 triploids, 2·882 to 3·226 tetraploids and 3·647 to 3·833 pentaploids. Thus, four different cytotypes (2x, 3x, 4x and 5x) were found to occur among the samples analysed (Table 2, Supplementary Data Table S1). An illustrative flow cytometric histogram combining taxa of all different ploidies reported is presented in Fig. 2F.
Fig. 3.
Box and whisker plot representation of the relative DNA content according to ploidy in Sorbus.
Table 2.
Karyological data and population cytotypes for the studied Sorbus taxa (for more detailed information about specimen localities and FCM results, see Supplementary Data Table S1)
| Taxon | 2n* | Ploidy | DNA ploidy† | No. of specimens | No. of populations | Ratio‡ (mean ± s.d.) |
|---|---|---|---|---|---|---|
| S. admonitor M.Proctor | approx. 68 | 4x | 4x | 4 | 1 | 3·172 ± 0·038 |
| S. anglica Hedl. | 51, 68 | 3x, 4x | 4x | 16 | 10 | 2·960 ± 0·038 |
| S. aria (L.) Crantz | 34 | 2x | 2x | 82 | 25 | 1·490 ± 0·017 |
| S. cf. aria | – | – | 3x | 14 | 7 | 2·270 ± 0·028 |
| S. aucuparia L. | 34 | 2x | 2x | 13 | 10 | 1·522 ± 0·011 |
| S. bristoliensis Wilmott | 51 | 3x | 3x | 1 | 1 | 2·318 |
| S. cambrensis M.Proctor | 68 | 4x | 5x | 13 | 3 | 3·737 ± 0·050 |
| S. cheddarensis L.Houston & Ashley Robertson | – | 3x§ | 3x | 5 | 2 | 2·264 ± 0·018 |
| S. cf. cheddarensis | – | – | 3x | 15 | 4 | 2·265 ± 0·022 |
| S. cuneifolia T.C.G.Rich | – | – | 4x | 1 | 1 | 2·963 |
| S. devoniensis E.F.Warb. | 68 | 4x | 4x | 6 | 3 | 3·151 ± 0·033 |
| S. eminens E.F.Warb. | 68 | 4x | 4x | 32 | 8 | 3·029 ± 0·018 |
| S. cf. eminens | – | – | 4x | 3 | 2 | 3·028 ± 0·016 |
| S. eminentiformis T.C.G.Rich | 68 | 4x | 4x | 10 | 3 | 3·009 ± 0·027 |
| S. eminentoides L.Houston | – | 4x§ | 4x | 5 | 1 | 3·002 ± 0·030 |
| S. cf. eminentoides | – | – | 4x | 5 | 1 | 3·017 ± 0·020 |
| S. hybrida L. | 68 | 4x | 4x | 2 | 1 | 2·946 ± 0·053 |
| S. intermedia (Ehrh.) Pers. | 68 | 4x | 4x | 4 | 3 | 3·018 ± 0·054 |
| S. lancastriensis E.F.Warb. | 68 | 4x | 4x | 10 | 4 | 3·000 ± 0·029 |
| S. latifolia (Lam.) Pers. | 34, 68 | 2x, 4x | 4x | 1 | 1 | 3·143 |
| S. leighensis T.C.G.Rich | – | 3x§ | 3x | 2 | 2 | 2·296 ± 0·000 |
| S. leptophylla E.F.Warb. | 68 | 4x | 4x | 5 | 1 | 3·007 ± 0·049 |
| S. leyana Wilmott | 51 | 3x | 3x | 4 | 1 | 2·265 ± 0·013 |
| S. margaretae M.Proctor | 68 | 4x | 4x | 6 | 4 | 3·014 ± 0·036 |
| S. minima (Ley) Hedl. | 51 | 3x | 3x | 4 | 1 | 2·275 ± 0·015 |
| S. mougeotii Soy.-Will. & Godr. | 68 | 4x | 4x | 5 | 3 | 3·018 ± 0·046 |
| S. ‘Observatory Hill taxon’ | – | – | 3x | 12 | 1 | 2·255 ± 0·018 |
| S. parviloba T.C.G.Rich | – | – | 4x | 1 | 1 | 3·011 |
| S. cf. parviloba | – | – | 3x | 2 | 1 | 2·248 ± 0·004 |
| S. porrigentiformis E.F.Warb. (s.s.) | 68 | 4x | 4x | 23 | 9 | 3·018 ± 0·030 |
| S. porrigentiformis (agg.) | – | – | 4x | 5 | 7 | 3·011 ± 0·015 |
| S. porrigentiformis ‘Symonds Yat clone’ | – | – | 3x | 11 | 3 | 2·269 ± 0·017 |
| S. cf. porrigentiformis | – | – | 5x | 1 | 1 | 3·807 |
| S. ‘Portishead taxon’ | – | – | 4x | 19 | 4 | 3·016 ± 0·029 |
| S. rupicola (Syme) Hedl. | 68 | 4x | 4x | 10 | 4 | 3·068 ± 0·024 |
| S. rupicoloides L.Houston | – | – | 4x | 3 | 1 | 3·013 ± 0·013 |
| S. saxicola T.C.G.Rich | – | – | 3x | 3 | 1 | 2·247 ± 0·004 |
| S. stenophylla M.Proctor | – | – | 5x | 5 | 2 | 3·696 ± 0·027 |
| S. subcuneata Wilmott | 51, approx. 68 | 3x, 4x | 3x | 12 | 7 | 2·346 ± 0·028 |
| S. torminalis (L.) Crantz | 34 | 2x | 2x | 14 | 7 | 1·612 ± 0·011 |
| S. vexans E.F.Warb. | 68 | 4x | 4x | 5 | 3 | 3·024 ± 0·030 |
| S. wilmottiana E.F.Warb. | 51 | 3x | 3x | 1 | 1 | 2·278 |
| S. cf. wilmottiana | – | – | 3x | 1 | 1 | 2·275 |
* Chromosome numbers extracted from Rich et al. (2010) and the Index to Plant Chromosome Numbers (IPCN electronic database; http://www.tropicos.org/Project/IPCN).
† Ploidy inferred by FCM.
‡ Fluorescence intensity ratio (target sample peak/internal standard peak) = relative DNA content.
§ Ploidy inferred from molecular data (Houston et al., 2009; Rich et al., 2010; Robertson et al., 2010).
The linear relationship between the relative DNA content and ploidy was confirmed (Supplementary Data Fig. S1; R2 = 0·99, P < 0·0001). Of the samples collected, tetraploids were most frequent (41 % of the samples analysed), followed by triploids (29 %), diploids (26 %) and pentaploids (4 %), but note that we focused on individuals which we expected to be polyploids. Spatial ploidy coexistence was frequent in Sorbus populations, and in approx. 70 % of the sites visited at least two cytotypes were reported (see Supplementary Data Table S1). Craig y Cilau National Nature Reserve in Wales harboured all four ploidies.
Species cytotype identification
The results for the inferred DNA ploidies of the studied Sorbus taxa are summarized in Tables 2 and 3, and detailed information for each specimen assessed can be found in Supplementary Data Table S1.
Table 3.
Karyological information in known Sorbus hybrids
| Taxon | 2n* | Ploidy | Ploidy based on FCM | Ratio observed (mean ± s.d.) | Ratio expected | Difference (obs. – exp.) |
|---|---|---|---|---|---|---|
| S. × thuringiaca (S. aucuparia× S. aria) | 34 | 2x | 2x | 1·505 | 1·506 | –0·001 |
| S. × tomentella (S. aria × S. torminalis) | 34 | 2x | 2x | 1·552 ± 0·007 | 1·551 | –0·001 |
| S. × avonensis (S. aria × S. porrigentiformis) | – | 3x† | 3x | 2·252 ± 0·058 | 2·250 | 0·004 |
| S. × liljeforsii (S. aucuparia × S. intermedia) | 51 | 3x | 3x | 2·349 | 2·270 | 0·079 |
| S. × robertsonii (S. aria × S. eminens) | – | 3x† | 3x | 2·271 | 2·260 | 0·011 |
| S. × houstoniae (S. aria × S. bristolensis) | – | 4x† | 4x | 3·072 | 3·063 | 0·009 |
| S. × motleyi (S. aucuparia × S. leyana) | – | – | 4x | 3·039 ± 0·013 | 3·026 | 0·013 |
* Chromosome numbers extracted from Rich et al. (2010).
† Ploidy levels inferred from molecular data (Houston et al., 2009; Robertson et al., 2010).
Note: the ratios expected were calculated on the basis of the parental genomes. The calculations in diploid and triploid hybrids were made by summing the relative ratio of reduced genomes in both parents. In the tetraploids, the calculations resulted from the sum of an unreduced triploid and reduced diploid parents.
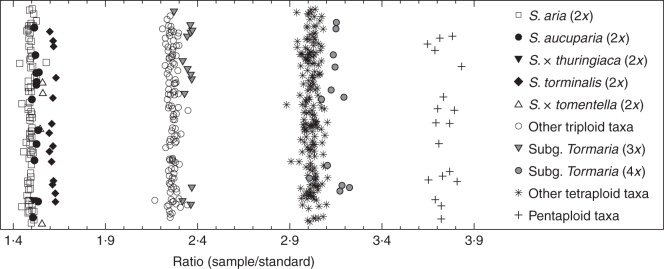
Of the diploids, S. aria (but see below), S. aucuparia and S. torminalis were found to be constant in the sampling (see Supplementary Data Table S1), with their FCM profiles discriminating between species (K = 57·732, P < 0·0001; Supplementary Data Fig. S2A), most clearly in the case of S. torminalis (Fig. 4). The expected results were confirmed in the homoploid hybrids S. × thuringiaca (S. aria × S. aucuparia) and S. × tomentella (S. aria × S. torminalis), the relative DNA contents of which fell between those of the hypothesized parents (Figs 1 and 4, Supplementary Data Fig. S2B). However, 14 specimens from seven sites collected as dubious S. aria were triploid. The taxonomic utility of the FCM records at higher levels was also tested in triploids and tetraploids (Supplementary Data Fig. S2C, D). Although polyploids of subgenus Tormaria (Fig. 1, subgenus Aria × subgenus Torminaria) were generally discriminated (Fig. 4; 3x, K = 13·133, P = 0·0013; 4x, K = 36·682, P < 0·0001), those belonging to subgenera Aria and Soraria (subgenus Aria × subgenus Sorbus) had overlapping genome sizes due to the similar nuclear DNA content of the parental taxa and could not be distinguished (Supplementary Data Fig. S2C, D).
Fig. 4.
Scatterplot of the flow cytometric data obtained illustrating the differences among ploidy levels in Sorbus.
Of the polyploid apomictic species in subgenus Aria, three were inferred as triploid and ten as tetraploid cytotypes (Table 2). The hybrids S. × robertsonii (S. aria × S. eminens) and S. × avonensis (S. aria × S. porrigentiformis) were triploid as expected (Table 3), and S. cambrensis and S. stenophylla were shown to be fixed pentaploids. Different cytotypes were found in S. parviloba, one tetraploid (the type tree) and two triploids. Specimens collected as members of the S. porrigentiformis group included both triploid and tetraploid cytotypes.
In subgenus Soraria (members of which generally originated as subgenus Aria × subgenus Sorbus), S. minima and S. leyana were triploid. The backcross between S. leyana and S. aucuparia (= S. × motleyi) was tetraploid, as were S. cuneifolia and S. mougeotii. Sorbus hybrida was also tetraploid, but the sample collected as a backcross with S. aucuparia (of cultivated origin) was unexpectedly diploid. Sorbus intermedia, generally included in this subgenus [although it also has S. torminalis in its genome (Fig. 1; Rich et al., 2010)], was tetraploid, and its backcross with S. aucuparia (= S. × liljeforsii) was triploid as expected. Sorbus anglica, previously reported as both triploid and tetraploid, was found to be consistently tetraploid. One possible S. anglica × S. aria hybrid from Cheddar was found to be triploid and will be investigated in more detail.
Results for subgenus Tormaria (members of which generally originated as subgenus Aria × subgenus Torminaria) were as follows: S. bristoliensis was confirmed to be triploid and its backcross with S. aria (= S. × houstoniae) tetraploid. The latter cytotype was also found in S. admonitor, S. devoniensis and S. latifolia. Although S. subcuneata had previously been reported at different ploidies (3x, 4x), the samples studied here were consistently triploid.
DISCUSSION
Efficacy of FCM for ploidy estimation in Sorbus
Regarding the utility of FCM, unlike in Crataegus (also Rosaceae; Talent and Dickinson, 2005), in which assessment of ploidy was sometimes difficult due to overlapping ranges of genome size, DNA ploidy inferred in Sorbus was highly reliable and clearly differentiated between cytotypes (Figs 3 and 4). This situation has been mainly reported in surveys restricted at either the species (Balao et al., 2009; Cosendai and Hörandl, 2010) or small aggregate (Marhold et al., 2010; Trávníček et al., 2011) levels, but in large complexes such in Sorbus, where multiple hybridization episodes are frequent, it would not be surprising to have found more complex FCM profiles as a result of different evolutionary histories. Furthermore, the similar genome sizes found in the diploid sexual species (S. aria, S. aucuparia, S. torminalis), which are in agreement with the records of Siljak-Yakovlev et al. (2010) from the Balkan region, should be regarded as the main reason why the relative DNA contents fitted almost perfectly to ploidy allocations.
Ploidy diversity in Sorbus
The diploid chromosome number in Sorbus is 2n = 34, in agreement with the currently accepted hypothesis that chromosome number in subfamily Maloideae arose via aneuploidy from x = 18 (Evans and Campbell, 2002). Chromosome counts in several species of the genus revealed the existence of di-, tri- and tetraploid taxa and one ‘pentaploid’ seedling (Bailey et al., 2008; Rich et al., 2010). The incidence of polyploid taxa in the investigated Sorbus is comparable with that reported in other related genera such as the above mentioned Crataegus (Talent and Dickinson, 2005) and other Rosaceae (Dickson et al., 1992; Dickinson et al., 2007). The present data confirm that diploidy is restricted to the parental taxa and their homoploid hybrids. Although Siljak-Yakovlev et al. (2010) recorded a triploid S. torminalis from Serbia, we have not found any polyploid specimen for the species in our sampling. In fact, and given the stability in ploidy of the British S. torminalis samples investigated, a hybrid origin for this isolated triploid should be considered. Similar taxonomic issues have arisen when considering polyploid specimens of S. aria. Some authors still include the apomictic polyploids within S. aria sensu stricto (Aldasoro et al., 2004), but with our species concept, S. aria is restricted to sexual diploid individuals.
Tetraploids and (to a lesser extent) triploids are the prevalent cytotypes of the apomictic taxa, accounting for most of the species richness in Britain. Additionally, although they are at low frequency, this extensive survey has confirmed the existence of pentaploid cytotypes in adult individuals at different sites (Supplementary Data Table S1), a cytotype previously only reported in a single seedling of the S. porrigentiformis group grown from Craig y Cilau (Wales). This illustrates the convenience of FCM for rapid ploidy screening from field conditions. As most apomictic Sorbus taxa use pollen for endosperm fertilization (pseudogamy), crosses involving different ploidies might prove problematic for endosperm balancing and sometimes result in seed abortion (Cosendai and Hörandl, 2010). This limitation could be the reason why pentaploids are poorly represented in Sorbus. However, given the high incidence of triploids in the data set and that some other pseudogamous Rosaceae (e.g. Crataegus; Talent and Dickinson, 2007) are not as sensitive to endosperm imbalance, other limiting factors cannot be discarded. Further investigation into the mechanism of apomixis in hybrid Sorbus would be rewarding in leading to improved understanding of how some cytotypes might occur, despite the disadvantages relating to endosperm formation.
Expected and unexpected results: new taxa or simple misidentifications?
Subgenus Aria is perhaps the most complex of the sections in Britain and it is not always possible to be certain of the identity from some of the immature or shaded plants that we sampled. This could explain why 14 specimens from seven sites collected as S. aria were 3x and not 2x, indicating that they are probably hybrids and only future molecular approaches will help to unravel their position within the Aria aggregate (Fig. 1). In addition, some potential new taxa were found: a triploid clone by the Observatory in the Avon Gorge, a triploid S. porrigentiformis relative from the Symonds Yat area and a stable tetraploid taxon from Portishead. This last named was suggested to be S. × robertsonii by Rich et al. (2010), but as their ploidy differs they must have different origins. Another interesting group of taxa is the S. porrigentiformis aggregate, in which we confirmed the existence of triploid and tetraploid cytotypes. The previously existing pentaploid count (2n = approx. 87) is more likely to refer to S. cambrensis, which we have confirmed to be consistently pentaploid. As several species have already been segregated from the complex (e.g. S. × cheddarensis, S. leighensis, S. saxicola; Rich et al., 2010), further molecular investigation would be useful in unravelling the aggregate and shedding light on the need (if any) for additional taxonomic rearrangements.
Another taxon in subgenus Aria in which conflicting cytotypes were found was S. parviloba. Its chromosome number is unknown but ploidy assessments revealed both triploid and tetraploid cytotypes. Given (1) the small population size of this endemic to Coldwell Rocks in Gloucestershire, provisionally categorized as ‘critically endangered’ sensu IUCN, and (2) our doubts in identifying two of the sampled trees, additional investigation will provide stronger evidence to confirm whether these are true established cytotypes or illustrate potential hybridization with neighbouring species. In addition, some trees in the Wye Valley previously regarded as backcrosses between S. × tomentella and S. aria (Price and Rich, 2007) were found to be triploid, in agreement with some limited nuclear microsatellite data (Bentham-Green, 2006).
Within subgenus Soraria there were also cases which are worthy of comment. Sorbus × thuringiaca nm. pinnatifida (=S. × pinnatifida) was originally believed to have arisen from a cross between S. aucuparia and S. intermedia (Fig. 1; Richards, 1975), but Nelson-Jones et al. (2002) rejected this assumption based on nuclear microsatellite results. The cytotype we inferred for the taxon evaluated (2x) correlates with its presumed origin (S. aria × S. aucuparia, cf. Rich et al., 2010); however, further investigation is required to discard hypothetical multiple origins for this cultivated hybrid. Sorbus anglica, previously reported to occur at both triploid and tetraploid levels, was found to be consistently tetraploid despite collection of material from two of the three sites at which triploids had previously been reported (Bailey et al., 2008). Robertson et al. (2010), based on molecular data, postulated that this species arose from a cross between S. aucuparia and a representative of the S. porrigentiformis aggregate (3x, 4x). Although our results are not conclusive, the sole cytotype found suggests that a triploid S. porrigentiformis cytotype would be involved in its origin, and that triploid chromosome counts reported might reflect seedlings having different ploidies to the parent from which the seeds were collected.
Hybrid taxa: how does FCM information help?
The hybrid origin for many Sorbus species has been frequently proposed (see Fig. 1 for illustration). However, few taxa have been taxonomically recognized as hybrids in Britain (Table 3; Rich et al., 2010) due to the involvement of apomixis. One of the useful aspects of FCM is that it allows discrimination between taxa and their hybrids. In fact, the diploid hybrids studied yielded relative DNA contents similar to those of the non-hybrid assumed parents. These values were almost exactly intermediate (Supplementary Data Fig. S2B) and were in agreement with the ratios we calculated using the putative parents (Table 3). A similar situation was observed in the known triploid and tetraploid hybrids investigated. The high concordance between the values observed and predicted not only confirms their homoploid and polyploid hybrid origin, respectively, but also supports their parentage as postulated by morphological traits (Rich et al., 2010) and molecular approaches (Nelson-Jones et al., 2002; Chester et al., 2007). However, the slight deviation observed in S. × liljeforsii, rather than being the result of genomic reorganization, could be influenced by the lack of complete agreement in the multiple origins suggested for the parental S. intermedia (Challice and Kovanda, 1978; Lemche, 1999). The ploidy survey we carried out also brought to light several specimens displaying unexpected ploidy, which were considered as potential interspecific crosses (Supplementary Data Table S1). In these cases, any inference about their origin based on the observed ploidy would be speculative, but provides new insight into the ability to hybridize in Sorbus and will be of great help in improving our understanding and interpretation of further research using molecular markers such as nuclear and plastid microsatellites.
Concluding remarks and future research
British Sorbus is revealed as an interesting example of a polyploid complex enhanced by recurrent hybridization episodes. A noteworthy coexistence of cytotypes (2x, 3x, 4x and 5x) has been found, probably facilitated by the overlapping distribution of many of the species studied, and resulting from the potential hybrid origin of many apomictic polyploids. As in former investigations, FCM has been found to be highly effective in estimating the relative DNA content of the species under study to infer ploidy. The data presented here provide a solid baseline for forthcoming molecular research aimed at gaining a better understanding of the genetic pathways controlling the formation and establishment of polyploids.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank M. W. Chase, M. Christenhusz, S. Ludwig, N. W. Poulton, I. Trotman, S. Whild and M. Wilcox for help in collecting material, and the National Botanic Garden of Wales and the Finnish Museum of Natural History (M. Christenhusz) for access to their living collections. We also acknowledge the Royal Botanic Gardens, Kew, Natural England and National Museum Wales for financial support. Two anonymous referees are also acknowledged for their helpful comments to an earlier version of the manuscript.
LITERATURE CITED
- Aldasoro JJ, Aedo C, Garmendia FM, Hoz FPdl, Navarro C. Revision of Sorbus subgenera Aria and Torminaria (Rosaceae–Maloideae) Systematic Botany Monographs. 2004;69:1–148. [Google Scholar]
- Bailey JP, Kay QON, McAllister H, Rich TCG. Chromosome numbers in Sorbus L. (Rosaceae) in the British Isles. Watsonia. 2008;27:69–72. [Google Scholar]
- Balao F, Casimiro-Soriguer R, Talavera M, Herrera J, Talavera S. Distribution and diversity of cytotypes in Dianthus broteri as evidenced by genome size variations. Annals of Botany. 2009;104:965–973. doi: 10.1093/aob/mcp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennert HW, Horn K, Kauth M, et al. Flow cytometry confirms reticulate evolution and reveals triploidy in Central European Diphasiastrum taxa (Lycopodiaceae, Lycophyta) Annals of Botany. 2011;108:867–876. doi: 10.1093/aob/mcr208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentham-Green S. Bristol, UK: University of; 2006. Introgressive hybridisation between Sorbus aria and S. torminalis in Wye Valley. Undergraduate thesis. [Google Scholar]
- Challice JS, Kovanda M. Chemotaxonomic survey of the genus Sorbus in Europe. Naturwissenschaften. 1978;65:11. [Google Scholar]
- Chester M, Cowan RS, Fay MF, Rich TCG. Parentage of endemic Sorbus L. (Rosaceae) species in the British Isles: evidence from plastid DNA. Botanical Journal of the Linnean Society. 2007;154:291–304. [Google Scholar]
- Cosendai AC, Hörandl E. Cytotype stability, facultative apomixis and geographical parthenogenesis in Ranunculus kuepferi (Ranunculaceae) Annals of Botany. 2010;105:457–470. doi: 10.1093/aob/mcp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson TA, Lo E, Talent N. Polyploidy, reproductive biology, and Rosaceae: understanding evolution and making classifications. Plant Systematics and Evolution. 2007;266:59–78. [Google Scholar]
- Dickson EE, Arumuganathan K, Kresovich S, Doyle JJ. Nuclear DNA content variation within the Rosaceae. American Journal of Botany. 1992;79:1081–1086. [Google Scholar]
- Doležel J, Bartoš J, Voglmayr H, Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytometry. 2003;51A:127–128. doi: 10.1002/cyto.a.10013. [DOI] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J, Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols. 2007;2:2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- Ebihara A, Ishikawa H, Matsumoto S, et al. Nuclear DNA, chloroplast DNA, and ploidy analysis clarified biological complexity of the Vandenboschia radicans complex (Hymenophyllaceae) in Japan and adjacent areas. American Journal of Botany. 2005;92:1535–1547. doi: 10.3732/ajb.92.9.1535. [DOI] [PubMed] [Google Scholar]
- Evans RC, Campbell CS. The origin of the apple subfamily (Maloideae; Rosaceae) is clarified by DNA sequence data from duplicated GBSSI genes. American Journal of Botany. 2002;89:1478–1484. doi: 10.3732/ajb.89.9.1478. [DOI] [PubMed] [Google Scholar]
- Hörandl E, Cosendai AC, Temsch EM. Understanding the geographic distributions of apomictic plants: a case for pluralistic approach. Plant Ecology & Diversity. 2008;1:309–320. doi: 10.1080/17550870802351175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston L, Robertson A, Jones K, Smith SCC, Hiscock SJ, Rich TCG. An account of the Whitebeams (Sorbus L., Rosaceae) of Cheddar Gorge, England, with description of three new species. Watsonia. 2009;27:283–300. [Google Scholar]
- Kron P, Suda J, Husband BC. Applications of flow cytometry to evolutionary and population biology. Annual Review of Ecology Evolution and Systematics. 2007;38:847–876. [Google Scholar]
- Leitch IJ, Hanson L, Lim KY, et al. The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae) Annals of Botany. 2008;101:805–814. doi: 10.1093/aob/mcm326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemche EB. The origins and interactions of British Sorbus species. Cambridge, UK: Darwin College; 1999. PhD thesis. [Google Scholar]
- Liljefors A. Cytological studies in Sorbus. Acta Horti Bergiani. 1955;17:47–113. [Google Scholar]
- Loureiro J, Rodriguez E, Doležel J, Santos C. Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Annals of Botany. 2007;100:875–888. doi: 10.1093/aob/mcm152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro J, Trávníček P, Rauchová J, et al. The use of flow cytometry in the biosystematics, ecology and population biology of homoploid plants. Preslia. 2010;82:3–21. [Google Scholar]
- Marhold K, Jongepierova I, Krahulcova A, Kucera J. Morphological and karyological differentiation of Gymnadenia desnsiflora and G. conopsea in the Czech Republic and Solvakia. Preslia. 2005;77:159–176. [Google Scholar]
- Marhold K, Kudoh H, Pak J-H, Watanabe K, Spaniel S, Lihova J. Cytotype diversity and genome size variation in eastern Asian polyploid Cardamine (Brassicaceae) species. Annals of Botany. 2010;105:249–264. doi: 10.1093/aob/mcp282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques I, Nieto-Feliner G, Martins-Loução MA, Fuertes Aguilar J. Genome size and base composition variation in natural and experimental Narcissus (Amaryllidaceae) hybrids. Annals of Botany. 2012;109:257–264. doi: 10.1093/aob/mcr282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson-Jones EN-J, Briggs DB, Smith AS. The origin of intermediate species of the genus Sorbus. Theoretical and Applied Genetics. 2002;105:953–963. doi: 10.1007/s00122-002-0957-6. [DOI] [PubMed] [Google Scholar]
- Pellicer J, Fay MF, Leitch IJ. The largest eukaryotic genome of them all? Botanical Journal of the Linnean Society. 2010;164:10–15. [Google Scholar]
- Price DT, Rich TCG. One-way introgressive hybridisation between Sorbus aria and S. torminalis (Rosaceae) in southern Britain. Watsonia. 2007;26:419–432. [Google Scholar]
- Proctor MCF. In: Sorbus L. (Rosaceae). British red data books. 1. Vascular plants. Wigginton MJ, editor. Peterborough: Joint Nature Conservation Commitee; 1999. p. 347. [Google Scholar]
- Proctor MCF, Proctor ME, Groenhof AC. Evidence from peroxidase polymorphism on the taxonomy and reproduction of some Sorbus populations in south-west England. New Phytologist. 1989;112:569–575. doi: 10.1111/j.1469-8137.1989.tb00352.x. [DOI] [PubMed] [Google Scholar]
- Rich TCG, Proctor MCF. Some new British and Irish Sorbus L. taxa (Rosaceae) Watsonia. 2009;27:207–216. [Google Scholar]
- Rich TCG, Houston L, Robertson A, Proctor MCF. Whitebeams, rowans and service trees of Britain and Ireland. A monograph of British and Irish Sorbus L. 1st edn. London: Botanical Society of the British Isles; 2010. [Google Scholar]
- Richards AJ. In: Sorbus L. In Hybridisation and the flora of the British Isles. Stace CA, editor. London: Academic Press; 1975. pp. 233–238. [Google Scholar]
- Robertson A, Rich TCG, Allen AM, et al. Hybridization and polyploidy as drivers of continuing evolution and speciation in Sorbus L. (Rosaceae) Molecular Ecology. 2010;19:1675–1690. doi: 10.1111/j.1365-294X.2010.04585.x. [DOI] [PubMed] [Google Scholar]
- Šafářová L, Duchoslav M. Cytotype distribution in mixed populations of polyploid Allium oleraceum measured at a microgeographic scale. Preslia. 2010;82:107–126. [Google Scholar]
- Siljak-Yakovlev S, Stevanovic V, Tomasevic M, Brownc SC, Stevanovic B. Genome size variation and polyploidy in the resurrection plant genus Ramonda: cytogeography of living fossils. Environmental and Experimental Botany. 2008;62:101–102. [Google Scholar]
- Siljak-Yakovlev S, Pustahija F, Šolic EM, et al. Towards a genome size and chromosome number database of Balkan flora: C-values in 343 taxa with novel values for 242. Advanced Science Letters. 2010;3:190–213. [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, et al. Polyploidy and angiosperm diversification. American Journal of Botany. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Stark C, Michalski SG, Babik W, Winterfeld G, Durka W. Strong genetic differenctiation between Gymnadenia conopsea and G. densiflora despite morphological similarity. Plant Systematics and Evolution. 2011;293:213–226. [Google Scholar]
- Suda J, Krahulcova A, Travnicek P, Krahulec F. Ploidy level versus DNA ploidy level: an appeal for consistent terminology. Taxon. 2006;55:447–450. [Google Scholar]
- Suda J, Weiss-Schneeweiss H, Tribsch A, Schneeweiss GM, Travnicek P, Schonswetter P. Complex distribution patterns of di-, tetra-, and hexaploid cytotypes in the European high mountain plant Senecio carniolicus (Asteraceae) American Journal of Botany. 2007;94:1391–1401. doi: 10.3732/ajb.94.8.1391. [DOI] [PubMed] [Google Scholar]
- Talent N, Dickinson TA. Polyploidy in Crataegus and Mespilus (Rosaceae, Maloideae): evolutionary inferences from flow cytometry of nuclear DNA amounts. Canadian Journal of Botany. 2005;83:1268–1304. [Google Scholar]
- Talent N, Dickinson TA. Endosperm formation in aposporous Crataegus (Rosaceae, Spiraeoideae, tribe Pyreae): parallels to Ranunculaceae and Poaceae. New Phytologist. 2007;173:231–249. doi: 10.1111/j.1469-8137.2006.01918.x. [DOI] [PubMed] [Google Scholar]
- Trávníček P, Eliášová A, Suda J. The distribution of cytotypes of Vicia cracca in Central Europe: the changes that have occurred over the last four decades. Preslia. 2010;82:149–163. [Google Scholar]
- Trávníček P, Kubátová B, Čurn V, et al. Remarkable coexistence of multiple cytotypes of the Gymnadenia conopsea aggregate (the fragrant orchid): evidence from flow cytometry. Annals of Botany. 2011;107:77–87. doi: 10.1093/aob/mcq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg EF, Kárpáti ZE. In: Sorbus L. Flora Europaea. Tutin TG, Heywood VH, Burges NA, Moore DM, Valentine DH, et al., editors. Vol. 2. Cambridge: Cambridge University Press; 1968. pp. 67–71. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.