Abstract
Novel classes of small and long non-coding RNAs (ncRNAs) are being characterized at a rapid pace, driven by recent paradigm shifts in our understanding of genomic architecture, regulation and transcriptional output, as well as by innovations in sequencing technologies and computational and systems biology. These ncRNAs can interact with DNA, RNA and protein molecules; engage in diverse structural, functional and regulatory activities; and have roles in nuclear organization and transcriptional, post-transcriptional and epigenetic processes. This expanding inventory of ncRNAs is implicated in mediating a broad spectrum of processes including brain evolution, development, synaptic plasticity and disease pathogenesis.
Our understanding of the architecture, activity and regulation of the eukaryotic genome has been revolutionized by the recent advent of advanced sequencing technologies and findings from large-scale consortia focused on characterizing functional genomic elements, such as ENCODE1 and FANTOM2,3. We now recognize the extraordinary complexity and flexibility of the genome. It encodes not only protein-coding genes with multiple transcription start sites, alternative promoter and enhancer elements, splicing initiation and donor sites, as well as variable 3′-untranslated regions (UTRs), but also unexpectedly large numbers of non-coding RNAs (ncRNAs), which have myriad regulatory and other functions and similarly serve as substrates for transcriptional and post-transcriptional diversification (BOX 1). Moreover, high-resolution transcriptomic studies have revealed, surprisingly, that the vast majority of the genome is transcribed in both sense and antisense orientations and expressed in a highly cell type-, subcellular compartment-, developmental stage- and environmental stimulus-specific manner. Each nucleotide can participate in context-dependent transcription that is mediated by specific RNA polymerases that are responsible for giving rise to multiple interlaced and overlapping transcripts4. These distinct RNAs can be coordinately or independently regulated, and they can act autonomously or be functionally interrelated, with one RNA modulating the expression and activity of other transcripts derived from the same genomic locus. It is also clear that the strict dichotomy between protein-coding and non-coding transcripts is false. Some ncRNAs contain open reading frames and can be translated and, in addition to encoding proteins, bifunctional RNA transcripts (such as the steroid receptor RNA activator and tumour protein 53 (TP53) mRNA) participate in meaningful cellular regulatory and functional processes, rather than serving simply as passé intermediates for translation5–7.
Box 1 | Life cycle of non-coding RNAs.
Non-coding RNAs (ncRNAs) are subject to diverse transcriptional and epigenetic regulatory mechanisms, biogenesis and maturational pathways, post-transcriptional processing, conformational changes and intra- and inter-cellular trafficking, imbuing the non-coding transcriptome with extraordinary functional diversity and environmental responsiveness. ncRNA expression is controlled by transcriptional and epigenetic factors, including those that coordinately regulate protein-coding genes (repressor element 1-silencing transcription factor (REST), cAMP response element-binding (CREB), tumour protein 53 (TP53) and zinc finger protein 143 (ZNF143))11,20,138,139. Specific RNA polymerase enzymes transcribe ncRNAs4. Various classes of ncRNAs can also target each other for post-transcriptional regulation31,140. Furthermore, a range of post-transcriptional processes (such as alternative splicing, polyadenylation, 5′ capping, non-templated modifications and RNA editing) can modify ncRNAs2,24,141. The roles of these modifications are mostly uncharacterized.
Nevertheless, they are likely to have important functional consequences, particularly in the nervous system. For example, adenosine-to-inosine RNA editing targets ncRNA transcripts, including microRNAs (miRNAs), leading to alterations in miRNA–target mRNA interactions, and targets ncRNAs derived from retrotransposons (Alu and LINE-1 (L1) sequences), which are the principal substrates for primate RNA editing141. Intriguingly, these sequences have undergone substantial evolutionary expansion in primates; the highest levels of RNA editing are present in human brain142, suggesting that RNA editing of retrotransposons mediated human brain evolution, has seminal roles in brain functioning and might even promote encoding of salient environmental information back into the neuronal genome. Moreover, ncRNAs can undergo nuclear–cytoplasmic, nuclear–mitochondrial and axodendritic trafficking via ribonucleoprotein complexes, which promote the coordinated spatial and temporal distribution and functioning of particular combinations of ncRNAs, mRNAs and RNA-binding proteins143. Additionally, ncRNAs can be involved in intercellular communication through transport to adjacent nerve cells, more distant somatic sites and the germline via exosomes released by multiple cell types and via other transport processes133,134. These observations imply that understanding ncRNAs and their roles in the nervous system will require interrogating, in greater detail, the life cycles of these molecules and the mechanisms responsible for coordinating these dynamic processes.
This renaissance in RNA biology is of prime importance for the CNS because neural cells are highly transcriptionally active, exhibiting robust expression of ncRNAs8,9, and also because ncRNAs have played a part in the evolution of human brain form and function. Indeed, the fastest evolving regions of the primate genome are non-coding sequences that can give rise to ncRNAs that are primarily implicated in modulating neural development genes10. In addition, because of their potential roles in regulating individual genes, as well as large gene networks (see below), ncRNAs confer neural cells with the capacity to exert very precise control over the spatiotemporal deployment of genes, which is crucial for executing complex neurobiological processes. For example, microRNAs (miRNAs), such as miR-124 and miR-9/9*, are highly integrated into the cAMP responsive element-binding protein (CREB), repressor element 1 (RE1)-silencing transcription factor (REST) and REST corepressor 1 (CoREST) transcriptional networks that mediate neural cell fate decisions11,12.
The evolving non-coding RNA landscape
tRNAs and ribosomal RNAs (rRNAs) are examples of well-known classes of ncRNAs. However, modern studies have demonstrated that these represent only the ‘tip of the iceberg’. The existence of many additional classes of ncRNAs has more recently been recognized (FIGS 1,2; TABLE 1). Most laboratory techniques for isolating RNA are based on size fractionation, which has led to the designation of recently identified classes of ncRNAs as long or small. There are also alternative approaches that can distinguish biologically relevant ncRNAs, such as capturing molecules associated with RNA-binding proteins (RBPs)13. Algorithms using sequence conservation, epigenetic marks, structural and other features can also predict ncRNAs14,15. Thus, it is likely that many novel classes remain to be discovered, including those expressed at low levels and in a highly context-specific manner16. The following sections discuss the major classes of ncRNAs, including the most recently identified, abundant and versatile class, long ncRNAs (lncRNAs).
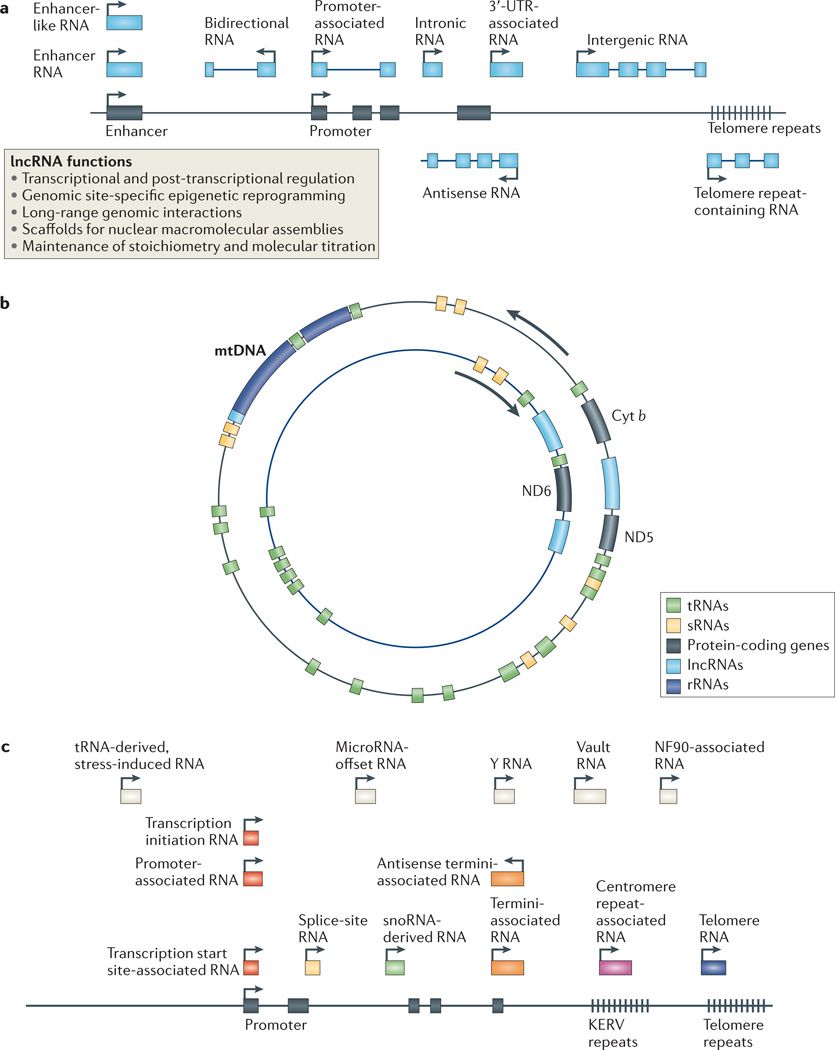
Figure 1. Emerging classes of non-coding RNAs.
a | Long non-coding RNAs (lncRNAs; shown in blue) mediate a broad array of genomic and cellular functions and are independently transcribed from: intergenic regions; in antisense, overlapping, intronic and bidirectional orientations to protein-coding genes (black); from gene-regulatory regions, including gene promoters, enhancers and untranslated regions (UTRs); and from specific chromosomal regions, including telomeres (arrows indicate direction of transcription). b | The mitochondrial genome contains small ncRNAs (such as ribosomal RNA (rRNA), tRNA and others) and lncRNAs transcribed from both heavy (outer) and light (inner) strands. Cytochrome b (Cyt b) and NADH dehydrogenase 5 (ND5) and ND6 are mitochondrial protein-coding genes that are associated with mitochondrial lncRNAs transcribed from the complementary mitochondrial DNA (mtDNA) strand. c | Small ncRNAs mediate RNAi and act as guides for RNA modifications (see FIG. 2), and additional small ncRNAs are derived from protein-coding gene (black) regulatory regions and gene boundaries, including 5′ regulatory regions (red), gene termini (orange), intron–exon junctions (yellow) and introns (green). They are also derived from structural components of chromosomes, including centromeres (purple) and telomeres (blue), and can be the cleavage products of other ncRNAs or originate from other sources (white). KERV, kangaroo endogenous retrovirus; NF90, nuclear factor 90; snoRNA, small nucleolar RNA; sRNAs, small RNAs.
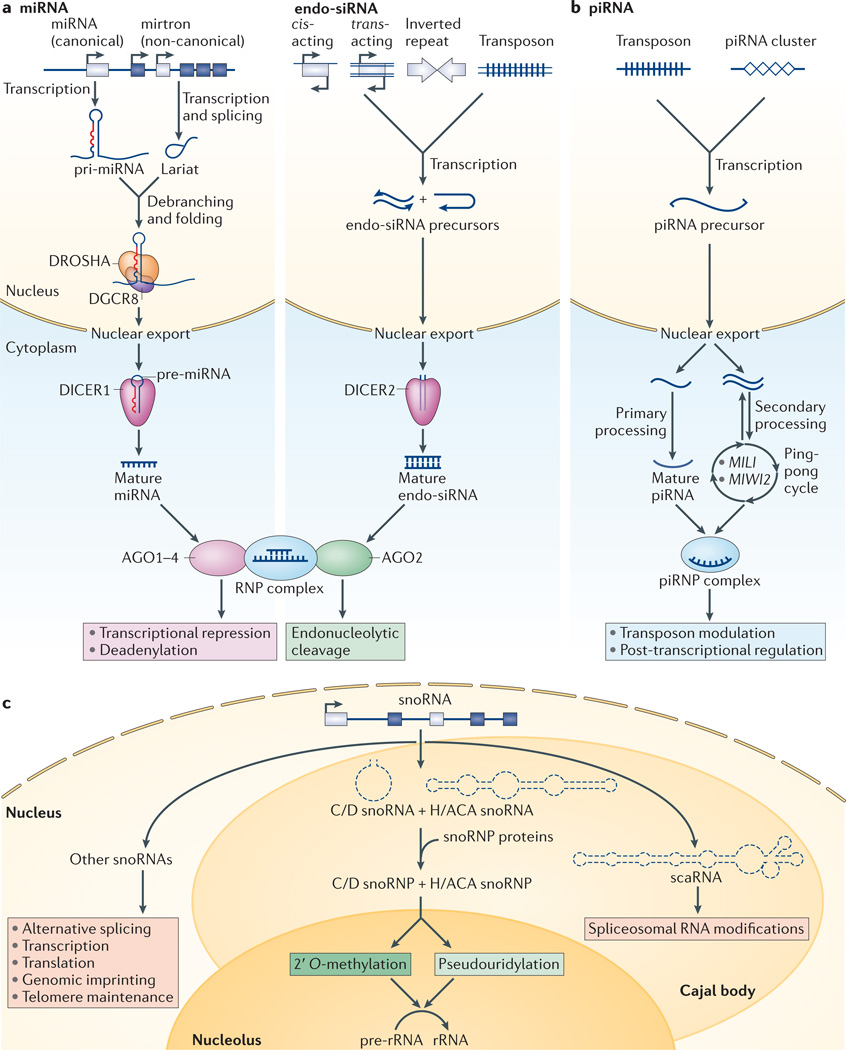
Figure 2. Non-coding RNAs mediating RNAi and RNA modifications.
Biogenesis and functions of non-coding RNAs mediating RNAi are shown. a | Left panel: microRNAs (miRNAs) are transcribed as primary miRNA (pri-miRNA; canonical pathway) or from introns (mirtrons; non-canonical pathway). After pri-miRNA and mirtron processing by the DROSHA–DiGeorge syndrome critical region 8 (DGCR8) complex or lariat debranching or folding enzymes, respectively, and nuclear export, precursor miRNAs (pre-miRNAs) are processed by DICER1 and undergo ribonucleoprotein (RNP) assembly with Argonaute proteins 1–4 (AGO1–4). Right panel: endogenous small interfering RNAs (endo-siRNAs) are generated from cis- or trans-acting sense–antisense pairs, inverted repeats and transposons. endo-siRNAs undergo nuclear processing and export, cytoplasmic processing by DICER2, and RNP assembly with AGO2. miRNAs promote deadenylation and translational repression and endo-siRNAs promote endonucleotytic cleavage, respectively. b | PIWI-interacting RNAs (piRNAs) are generated from transposons and piRNA clusters. piRNA precursors undergo nuclear processing and export, primary or cyclic secondary processing (by the PIWI proteins MILI and MIWI2) and piRNP complex assembly. piRNAs mediate transposon modulation and post-transcription regulation. c | Roles of small nucleolar RNAs (snoRNAs) are shown. snoRNAs are transcribed from introns, processed into C/D and H/ACA snoRNAs, undergo small nucleolar ribonucleoprotein (snoRNP) assembly in Cajal bodies and promote methylation and pseudouridylation of pre-ribosomal RNA (pre-rRNA) in the nucleolus. Small Cajal body-specific RNAs (scaRNAs) promote similar splicesosomal RNA modifications and other snoRNAs display additional functions.
Table 1.
Examples of the diversity and functionality of emerging classes of non-coding RNAs
| Class | Description | Refs |
|---|---|---|
| Antisense lncRNAs | lncRNAs derived from antisense transcription, present at ~70% of mammalian genomic loci and implicated in the regulation of sense protein-coding genes |
3 |
| Antisense termini-associated short RNAs |
Small ncRNAs with 5′ poly-U tails originating from 3′ termini of genes in antisense orientation, suggesting transcription by RNA-dependent RNA polymerase |
45 |
| Centromere repeat-associated small interacting RNAs |
Small ncRNAs (34–42 nucleotides) derived from centromeric repeats with putative roles in local epigenetic modifications and heterochromatin formation |
47 |
| Endogenous small interfering RNAs |
Small DICER-dependent ncRNAs (21–26 nucleotides) associated with Argonaute proteins (AGO2)and involved in post-transcriptional and epigenetic silencing of protein-coding genes and transposons |
33 |
| Enhancer RNAs | lncRNAs transcribed from enhancer domains of, and expressed coordinately with, activity-dependent neuronal genes |
109 |
| Enhancer-like long ncRNAs | lncRNAs exhibiting enhancer activity, particularly for genes regulating development and differentiation |
21 |
| Large intergenic RNAs | lncRNAs derived from intergenic regions that act as guides for recruiting PRC2, CoREST and other chromatin-modifying complexes |
17,18 |
| lncRNAs | Large family of ncRNAs (>200 nucleotides) with diverse functional roles | 24 |
| MicroRNA-offset RNAs | Small ncRNAs (~20 nucleotides) produced from microRNA precursors and exhibiting independent expression relative to associated microRNAs, implying distinct functional roles |
46 |
| MicroRNAs | Small DICER-dependent ncRNAs (20–23 nucleotides) associated with Argonaute proteins (AGO1-4) and involved in post-transcriptional silencing of protein-coding genes and ncRNAs |
28,29 |
| Mitochondrial ncRNAs | Small ncRNAs and lncRNAs generated from both strands of the mitochondrial genome, regulated by nuclear-encoded mitochondrial proteins and expressed in cell type- and tissue-specific patterns |
23,51 |
| PIWI-interacting RNAs | Small ncRNAs (26–30 nucleotides) associated with the PIWI subclass of Argonaute proteins and involved in silencing of mRNAs and transposons |
34 |
| Promoter-associated long RNAs |
lncRNAs transcribed from promoter domains of protein-coding genes, particularly cell cycle modulators, and capable of recruiting regulatory factors |
20,150 |
| Promoter-associated small RNAs | Small ncRNAs (20–200 nucleotides) possessing 5′ ends that coincide with the transcription start sites of protein-coding genes and ncRNAs |
43 |
| Small nucleolar RNAs | Small ncRNAs derived from intronic regions with roles in promoting RNA modifications, including pseudouridylation (H/ACA snoRNAs) and methylation (C/D snoRN As), as well as pre-mRNA processing |
36 |
| Small RNAs derived from small nucleolar RNAs |
Small ncRNAs derived from 3′ ends of H/ACA snoRNAs (20–24 nucleotides) and 5′ ends of C/D snoRNAs (17–19 nucleotides and >27 nucleotides) with microRNA-like functions |
41 |
| Small nuclear factor 90-associated RNAs |
Small ncRNAs (117 nucleotides) associated with the nuclear factor 90 RBP and exhibiting accelerated evolution and expansion in hominids and region-specific expression in human brain |
151 |
| Splice-site RNAs | Small ncRNAs (17–18 nucleotides) derived from splice sites of highly transcribed genes, and expressed in developmental stage- and region-specific patterns |
46 |
| Telomere small RNAs | Small DICER-independent telomere-specific ncRNAs (~24 nucleotides) | 48 |
| Telomere repeat-containing RNAs |
lncRNAs transcribed from telomeric repeats and dynamically regulated during the cell cycle with roles in heterochromatin formation and telomere functioning |
22 |
| Termini-associated short RNAs |
Small ncRNAs originating from 3′ termini of protein-coding genes and ncRNAs | 24 |
| Transcription initiation RNAs | Small ncRNAs (18 nucleotides) originating downstream of transcription start sites of protein-coding genes, implicated in modulating CTCF localization and nucleosome density |
44,52 |
| Transcription start site associated RNAs | Small ncRNAs (20–90 nucleotides) originating from the −250 to +50 position relative to the transcription start site of protein-coding genes | 152 |
| tRNA-derived RNA fragments | Small ncRNAs (19–40 nucleotides) derived from cleavage of mature tRNAs constitutively by DICER and in response to stress by angiogenin (in humans), promoting stress granule assembly and inhibiting mRNA translation |
49 |
| 3′-untranslated region-associated ncRNAs |
lncRNAs derived from 3′-untranslated regions of protein-coding transcripts and exhibiting independent developmental stage- and tissue-specific expression profiles |
19 |
| Vault RNAs | Small ncRNAs (88–100 nucleotides) integral to the vault RNP complex with putative roles inmultidrug resistance, apoptosis resistance and innate immunity |
153,154 |
| YRNAs | Small ncRNAs (~100 nucleotides) required for DNA replication, cleaved into microRNAs and implicated in Ro RBP localization and function |
155,156 |
CoREST, repressor element 1-silencing transcription factor corepressor 1; CTCF, CCCTC-binding factor; lncRNA, long non-coding RNA; ncRNA, non-coding RNA; PRC2, Polycomb repressive complex 2; RBP, RNA-binding protein.
Long non-coding RNAs
lncRNAs are transcripts of at least 200 nucleotides in length, although they can be orders of magnitude longer. lncRNAs are transcribed from intergenic regions (large intergenic ncRNAs (lincRNAs)) 17,18; in antisense, overlapping, intronic and bidirectional orientations relative to protein-coding genes; from gene regulatory regions (UTRs19, promoters20 and enhancers21); and from specific chromosomal regions (telomeres22) (FIG. 1a). Intriguingly, lncRNAs are also derived from the mitochondrial genome23 (FIG. 1b). lncRNAs are subject to post-transcriptional processing, potentially undergoing 5′ capping, polyadenylation, alternative splicing, RNA editing and trafficking2,24. Many lncRNAs exert regulatory effects on the genomic loci they are derived from or on neighbouring loci. For example, lincRNAs can have enhancer-like activity21. By contrast, other lncRNAs, such as HOX transcript antisense RNA (HOTAIR)25 and X (inactive)-specific transcript (XIST)26, promote the establishment of repressive chromatin environments across large genomic regions and even entire chromosomes, respectively. Indeed, lncRNAs have a broad range of functions, including roles in transcriptional and epigenetic mechanisms via the recruitment of transcription factors and chromatin-modifying complexes to specific nuclear and genomic sites; alternative splicing and other post-transcriptional RNA modifications through the assembly of nuclear domains containing RNA-processing factors; nuclear–cytoplasmic shuttling; and translational control27. lncRNAs can also act as precursors for small ncRNAs, such as small nucleolar RNAs (snoRNAs) and miRNAs.
An elegant framework for categorizing the emerging functions of lncRNAs was recently proposed. It describes lncRNAs as: signals for integrating temporal, spatial, developmental and stimulus-specific cellular information; decoys with the ability to sequester a range of RNA and protein molecules, thereby inhibiting their functions; guides for genomic site-specific and more widespread recruitment of transcriptional and epigenetic regulatory factors; and scaffolds for macromolecular assemblies with varied functions27.
Small non-coding RNAs with roles in RNAi pathways
The most important small ncRNAs are those involved in post-transcriptional regulation of target RNAs via RNAi. These include miRNAs, endogenous small interfering RNAs (endo-siRNAs) and PIWI-interacting RNAs (piRNAs) (FIG. 2a,b). Mature miRNAs are 20–23 nucleotide single-stranded RNAs (ssRNAs), canonically derived from longer primary transcripts (pri-miRNA)28,29. primiRNAs are processed by the DROSHA (also known as ribonuclease 3)-containing microprocessor complex, exported into the cytoplasm by exportin 5, and cleaved by the DICER1 ribonuclease. Mature miRNAs are loaded onto Argonaute proteins and integrated into diverse multicomponent RNA-induced silencing complexes (RISCs). miRNAs bind to imperfectly complementary sequences, predominantly in 3′-UTRs of target mRNAs, leading to repression or degradation of these transcripts. Not only can an individual miRNA regulate hundreds of different mRNAs but multiple miRNAs can also target a single mRNA.
This model for understanding miRNA processing and mechanism of action is still evolving. In fact, DROSHA- and DICER1-independent biogenesis pathways have been described. miRNA precursors are subject to post-transcriptional modification via RNA editing, which can affect their maturation and target gene interactions30. miRNAs can also target and repress other ncRNAs. For example, miR-671 silences the expression of an antisense lncRNA transcribed from the cerebellar degeneration-related protein 1 locus31. Moreover, high-resolution RNA-sequencing analyses, including those carried out using human brain tissues, have revealed most miRNAs have length and sequence heterogeneity32. These miRNA variants (isomiRs) are present in developmental stage- and tissue-specific profiles and selectively associate with particular Argonaute RBPs, implying nuanced and context-dependent functional roles.
endo-siRNAs are 21–26 nucleotide double-stranded RNAs (dsRNAs) that are cleaved from longer dsRNA intermediates in a DICER2-dependent manner and incorporated into RISC, in which they function as single-stranded entities33. endo-siRNAs are derived from various genomic locations and are implicated in gene regulation and genome defence via silencing of mRNAs and transposon-derived ncRNAs, respectively, as well as other emerging functions. In contrast to miRNAs, endo-siRNAs act on RNA molecules containing perfectly complementary sequences. piRNAs are 26–30 nucleotide ssRNAs that are DICER-independent and generated via biogenesis pathways, including a series of amplification steps, termed the ‘ping-pong’ cycle34. piRNAs derived from transposon elements are implicated in regulating transposon activity, whereas piRNAs derived from piRNA clusters are implicated in modulating gene expression. Although piRNAs were initially found in germ cells, recent studies have established that piRNAs are expressed in somatic cells, including neurons35.
Small non-coding RNAs as guides for modifying other RNAs
snoRNAs guide RNA-modifying enzyme complexes to other RNA molecules (such as rRNAs) in the nucleolus36,37 (FIG. 2c). These functions are mediated by the formation of duplexes between snoRNAs and RNAs with complementary sequences. The H/ACA and C/D box snoRNAs promote pseudouridinylation and methylation, respectively. Additionally, snoRNAs have other functions. For example, snoRNAs from the brain-specific snoRNA C/D box 115 cluster promote alternative splicing through interactions with non-canonical protein partners38,39. Small Cajal body RNAs are variants of snoRNAs that mediate alternative splicing in nuclear Cajal bodies40. Interestingly, the majority of snoRNA loci can also give rise to miRNA-like small RNAs, suggesting the existence of complex crosstalk between snoRNA-mediated RNA processing and RNAi pathways41,42.
Additional classes of small non-coding RNAs
Additional classes of poorly characterized small ncRNAs (FIG. 1b,c) include those derived from gene-regulatory regions and gene boundaries (subclasses of promoter-associated small RNAs43, such as transcription initiation RNAs (tiRNAs)44, termini-associated short RNAs, antisense termini-associated short RNAs45 and splice-site RNA (spliRNA)46); structural components of chromosomes (centromere-associated RNAs and telomere small RNAs47,48); cleavage of other ncRNAs (tRNA fragments49); and other sources (mitochondrial ncRNAs50,51 and miRNA-offset RNAs46). Functional studies have begun to interrogate these factors. For example, deleting tiRNAs that are associated with binding sites for RNA polymerase II and CCCTC-binding factor (CTCF; also known as transcriptional repressor CTCF) alters CTCF binding and nucleosome density at genomic loci proximal to sites of tiRNA biogenesis52. In addition, the biological processes associated with ncRNAs may be inferred by studying their genomic contexts. For example, spliRNAs, which are associated with splice sites of alternatively spliced genes, may have a role in modulating developmental stage- and tissue-specific alternative splicing. Ongoing studies are focused on understanding the evolution, biogenesis and functions of these diverse classes of ncRNAs and their roles in the CNS and other organ systems.
Evolutionary aspects of brain organization
Increasingly large proportions of non-coding sequences constitute the genomes of organisms with correspondingly greater degrees of developmental complexity53. It has further been suggested that an expansion in the inventory of ncRNAs — particularly miRNAs, which have been continuously acquired throughout vertebrate evolution and remain fixed within most lineages — was responsible for the emergence of vertebrate complexity54.
Comparative genomic and transcriptomic analyses suggest that ncRNAs are partly responsible for evolutionary innovations in brain and the cognitive and behavioural repertoires of higher organisms. For example, non-coding sequences associated with human neural genes exhibit prominent signatures of positive selection and accelerated evolution10. The highly accelerated region 1A (HAR1A) lncRNA is derived from one of 563 genomic regions designated human accelerated regions, because they evolved rapidly since divergence from the great apes. HAR1A expression correlates with that of reelin, suggesting that it similarly coordinates the establishment of regional forebrain organization. By contrast, evolutionarily constrained genomic elements also give rise to ncRNAs. For example, Evf2 (also known as DLX6 antisense RNA 1) is an lncRNA transcribed from an ultra-conserved enhancer that affects forebrain development by modulating expression of the distalless homeobox 5/6 transcription factors55. Loss of function of Evf2 in mice leads to a transient decrease in the population of GABAergic interneurons within the early postnatal hippocampus and dentate gyrus. Although the numbers of these interneurons later normalize, defects in synaptic inhibition are present throughout life, high-lighting the role of Evf2 in neuronal functioning. Analysis of other highly conserved lncRNA transcripts in birds, marsupials and eutherian mammals reveals remarkable similarities in the spatiotemporal expression profiles of orthologous lncRNAs and indicates ancient roles during brain development56.
Recent studies also implicate miRNAs in mediating neural gene expression changes during evolution. Indeed, a range of miRNAs are conserved only in primates, exhibit lineage-specific expansion or are human-specific and are preferentially expressed in brain57–60. One interesting study revealed that a substantial portion of intergenic transcripts conserved between human, chimpanzee and rhesus macaque brains are alternative or extended 3′-UTRs of known genes and represent putative sites for differential miRNA regulation59. A corresponding analysis of the prefrontal cortex and cerebellum of adult humans, chimpanzees and rhesus macaques identified substantial and parallel degrees of divergence in the expression of miRNAs57. This divergence in miRNA expression seems, in part, to explain differences in brain mRNA and protein expression levels between species. Indeed, a recent comparative genomic analysis that reported the existence of 220 candidate RNA structural families, 280,000 non-coding regions from transposable elements and >1,000 primate- and human-accelerated regions61 suggests that ncRNAs served as central effectors of brain evolution. Moreover, miRNAs that exhibit human-specific expression profiles preferentially target genes involved in neural functions. These range from neural stem cell (NSC) proliferation and differentiation to axon guidance and long-term potentiation (LTP) (discussed below).
Neural development, maintenance and plasticity
ncRNAs are expressed in precise regional, cellular, subcellular and temporal patterns in developing and adult brains, reflecting their widespread and diverse influence. Whereas most studies have focused on defining neurobiological roles for miRNA62, recent studies have also begun to characterize the expression and function of lncRNAs63–65 and other classes of ncRNAs66. lncRNAs that are found in developing and adult brains, including those found in specific cortical laminae, have expression profiles that correlate with factors involved in promoting brain development and organization and in transcriptional regulation of neural genes. For example, SOX2OT (sex-determining region Y-box 2 overlapping transcript) and SOX2DOT (sex-determining region Y-box 2 distal overlapping transcript) are lncRNAs transcribed from the same genomic locus as SOX2 and are implicated in its regulation67.
Other classes of ncRNAs are also likely to have important roles in shaping brain development. For example, one remarkable study recently identified thousands of retrotransposon reintegration sites for LINE-1 (L1), Alu and composite SVA (SINE, VNTR and Alu) elements in the human caudate and hippocampus68. These findings are consistent with previous reports suggesting that retrotransposon activity is important for neural development and plasticity69–71. In addition, they also indicate that retrotransposon-derived ncRNAs and ncRNAs that are responsible for regulating retrotransposition events (via endo-siRNAs and piRNAs) mediate somatic mosaicism in neurons and act as regulatory control systems for the establishment, maintenance and plasticity of the transcriptional architecture of the brain.
Neural stem cell maintenance and differentiation
ncRNAs have important roles in regulating stem cell maintenance and differentiation programmes, including neural lineage restriction, cell fate specification and terminal differentiation. Numerous studies have focused on defining how miRNA factors are incorporated into the complex transcriptional and epigenetic circuitry regulating neural cell fate decisions62. A key experimental strategy involves targeting the miRNA biogenesis factor DICER1. Dicer1-knockout animals are associated with a range of neural developmental defects, including abnormalities in brain size and cytoarchitecture and extensive apoptosis72–76. Complementary approaches have elucidated the seminal neurobiological roles of individual miRNAs (let-7, miR-9, miR-10, miR-17, miR-34, miR-124, miR-125, miR-132, miR-324-5p and miR-326)62. For example, in zebrafish, miR-9 modulates midbrain–hindbrain boundary (MHB) patterning during development, the balance between maintenance and differentiation of neural stem and progenitor cells, and their migration77. miR-9 promotes the progression of neurogenesis in the MHB progenitor pool by targeting the antineurogenic genes (her5 and her9)78. In the mouse, miR-9 has a bidirectional negative feedback relationship with the orphan nuclear receptor TLX, which mediates NSC self-renewal and proliferation, thereby promoting neural differentiation79. Moreover, introducing miR-9 into the embryonic brain leads NSCs to differentiate prematurely and migrate into the cortical plate. In addition to TLX, miR-9 targets forkhead box protein G1 (FOXG1) and promotes differentiation of Cajal–Retzius cells in the medial pallium80. Mice with ablation of miR-9 exhibit impairments in the production of Cajal–Retzius cells and early born cortical neurons, suppression of progenitor proliferation in ventricular and subventricular zones, defects in neocortical lamination, enhancement of neural progenitor cell (NPC) proliferation in the subpallium, shifting of the pallial–subpallial boundary and misrouting of thalamocortical and corticofugal axons81.
The extent to which miRNAs serve as key nodes in networks responsible for establishing cell identity is highlighted by reports showing that the expression of a small number of miRNAs can promote reprogramming of somatic cells into induced pluripotent stem cells (iPSCs)82,83 and, conversely, how other miRNAs can act as barriers for somatic cell reprogramming84. Two seminal studies demonstrated that the introduction of miR-124 together with either a particular combination of transcription factors or miR-9/9* in human fibroblasts can reprogramme them into neuronal cells85,86. The mechanisms of action for these two miRNAs may involve modulating the composition of ATP-dependent Brg1/ Brm-associated factor (BAF) chromatin-remodelling complexes, which are comprised of specific combinations of subunits and are essential for particular stages of neuronal lineage maturation87. miR-124 and miR-9/9* are also crucial for modulating neural gene expression because they are integrated with the circuitry of CREB, REST and CoREST, which are master transcriptional and epigenetic regulators of neural genes and neural cell fate decisions11,12 (FIG. 3). miRNAs also regulate glial lineage elaboration. For example, Dicer1 depletion leads to disruptions in the maturation of oligodendroglial (OL) and astroglial lineage species, which can be rescued by reintroducing particular miRNAs73–75. Given the emerging roles of REST and CoREST in NSC maintenance, lineage restriction, neuronal and glial subtype specification and differentiation88, these observations suggest the existence of intricate regulatory relationships between these multifunctional factors and ncRNAs in promoting glial lineage specification and maturation analogous to developmental processes in neuronal cells.
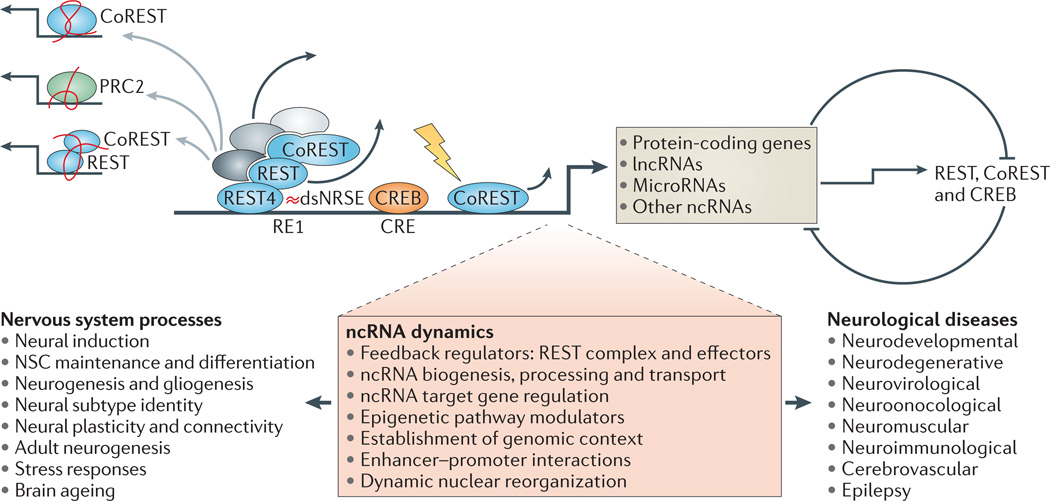
Figure 3. Non-coding RNA dynamics mediate diverse nervous system processes and neurological disease states.
Non-coding RNAs (ncRNAs) engage in a complex range of molecular and cellular functions (shown in the pink shaded box). They are also highly integrated, at multiple levels, into the mechanisms and circuitry that underlie neurobiological processes in health and disease. The expression of microRNAs, long ncRNAs (lncRNAs) and possibly other classes of ncRNAs, is similar to the expression of protein-coding genes: it is subject to developmental, constitutive and activity-dependent regulation (yellow lightning bolt) by key modulators of neural gene transcription (including repressor element 1 (RE1)-silencing transcription factor (REST), REST co-repressor 1 (CoREST)) and cAMP response element-binding (CREB)). In turn, ncRNAs influence the expression of these and other seminal neural factors via gene silencing, forming bidirectional regulatory relationships (curved lines, figure right), and by affecting their post-transcriptional RNA processing (alternative splicing (REST4)). ncRNAs can also influence the activity and deployment of neural factors. For example, the small modulatory ncRNA double-stranded neuron-restrictive silencer element (dsNRSE) (red waves) associates with and regulates the REST complex and its modular cofactors (grey ovals), and lncRNAs (red strings) promote the genome-wide deployment of chromatin-modifying complexes (including REST, PRC2 (Polycomb repressor complex 2) and CoREST–REST) (grey arrows). CRE, cAMP response element; NSC, neural stem cell.
Complementary efforts have been directed towards characterizing the parts played by other classes of ncRNAs in the establishment of neural cell identity. For example, isolating small ncRNAs from hippocampal NSCs identified a small double-stranded ncRNA derived from the RE1 (also known as NRSE) REST-binding sequence, which is implicated in modulating REST activity and promoting hippocampal neurogenesis89. By contrast, genomic approaches first identified dynamic expression profiles for substantial numbers of lncRNAs in developing neural cell types90. Subsequent studies focused on determining the functions of a few of these lncRNAs in neural cell fate specification and maturation91,92. We used a custom designed microarray platform to identify hundreds of lncRNAs that are dynamically regulated during GABAergic neurogenesis and OL lineage elaboration93. Many of these lncRNAs are transcribed from genomic loci associated with, and exhibit expression patterns that correlate with, key neural developmental genes. Another functional study analysed chromatin-state maps and reported the presence of more than 1,500 lincRNAs in NPC cells (and other cell types), including those associated with hippocampal development, OL maturation and GABAergic neuronal function17. An RNA-seq analysis carried out using a human iPSC paradigm showed that more than 1,500 lncRNAs are dynamically regulated during differentiation of iPSCs into ‘glutamatergic neurons’ (REF. 94). A large proportion of these lncRNAs are implicated in recruiting chromatin-remodelling complexes (including REST, CoREST and Polycomb repressive complex 2) to their genomic sites of action18,95 (FIG. 3). These observations provide insight into the complex interrelationships between ncRNAs and multifunctional epigenetic and transcriptional regulatory factor complexes coordinating neuronal and glial lineage elaboration.
Synaptic development, maintenance and plasticity
Numerous aspects of synaptic function are influenced by ncRNAs. Indeed, there is a bidirectional relationship between the two: ncRNAs mediate synaptic processes, and synaptic activity influences ncRNA expression, intracellular transport and function62,96,100 (FIG. 4). For example, various miRNAs target CREB, which is vital for LTP and memory11. In turn, miRNAs can also be regulated by CREB activity. Specific miRNAs are enriched in distinct subcellular compartments and can participate in the spatiotemporal control of protein synthesis underlying axon guidance, dendritic elaboration and synaptic plasticity97. Reciprocally, various miRNAs are subject to regulation by LTP and long-term depression98,99. Examining individual miRNAs reveals diverse roles in modulating dendritic growth and arborization (miR-124, miR-125b, miR-132, miR-134, miR-137 and miR-485), synapse formation (let-7, miR-125b, miR-132, miR-137, miR-138 and miR-485) and synaptic function and plasticity (miR-1, miR-132, miR-134, miR-181a, miR-219, miR-284 and miR-485)62,96,100. Manipulating miRNA biogenesis and maturation pathways similarly highlights the importance of miRNAs in synaptic structure and physiology. For example, in Drosophila melanogaster, hypomorphic mutations of miRNA pathway components drosha, dgcr8 (DiGeorge syndrome critical region 8; also known as pasha) and Dicer1 lead to reduced levels of miRNAs and defects in synaptic transmission in photoreceptor neurons, but no neural developmental effects101. In mice, using conditional and inducible strategies to inactivate Dicer1 in forebrain neurons yields alterations in dendritic spine morphology72,102. Mice with monoallelic deletions of Dgcr8 have decreased levels of expression for a subset of miRNAs involved in synaptic development and functioning103. Accordingly, pyramidal neurons in the cortex of these mice exhibit impairments in dendritic morphology and excitatory synaptic transmission. These observations suggest that other short ncRNAs, which are dependent on these biogenesis and maturation pathways, also have roles in establishing these abnormal synaptic phenotypes.
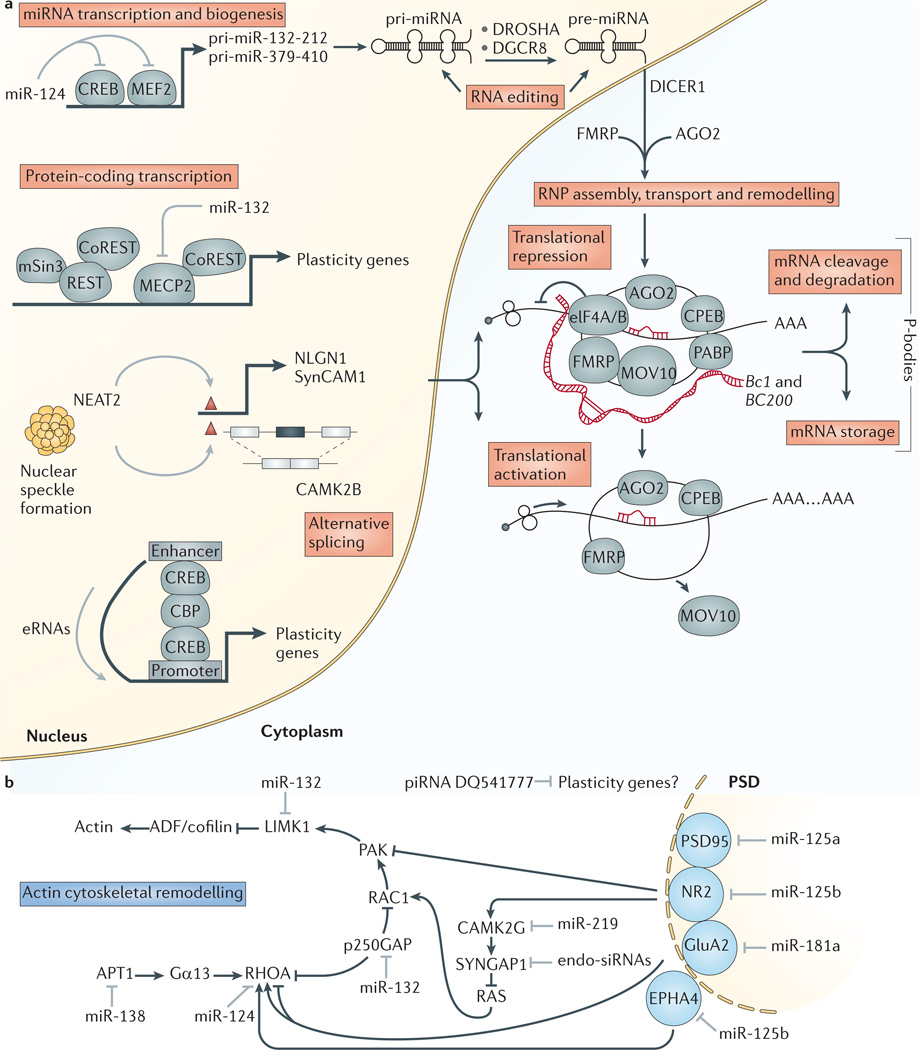
Figure 4. Role of non-coding RNAs in synaptic plasticity.
a | Diverse non-coding RNA (ncRNA) mechanisms are involved in activity-dependent processes underlying synaptic plasticity (shown in pink shaded boxes). MicroRNAs (miRNAs) contribute to synaptic plasticity through transcription of primary miRNA (pri-miRNA) clusters; processing of pri-miRNAs (by DROSHA and DiGeorge syndrome critical region 8 (DGCR8)) and pre-miRNAs (by DICER1); RNA editing; ribonucleoprotein (RNP) assembly, transport and remodelling; translational repression and activation; and associated mRNA storage, cleavage and degradation in P-bodies. miRNAs also modulate transcription of pri-miRNA clusters and protein-coding plasticity genes through regulation of activity-dependent transcription factors (including cAMP response element-binding (CREB) and myocyte enhancer factor 2 (MEF2)) and methyl-CpG binding proteins (including methyl-CpG-binding protein 2 (MECP2)). The long ncRNAs, brain cytoplasmic RNA 1 (Bc1) and its primate analogue BC200 (red), promote translational repression by binding to RNP components (including eukaryotic translation initiation factor 4A/B (elF4A/B), fragile X mental retardation 1 protein (FMRP) and polyadenylate binding protein (PABP)). Nuclear-enriched abundant transcript 2 (NEAT2) forms nuclear speckles, which are nuclear domains that regulate transcription of plasticity genes (such as, neuroligin 1 (NLGN1) and synaptic cell adhesion molecule 1 (SynCAM1)) and alternative splicing of plasticity genes (such as calcium/calmodulin-dependent protein kinase type II-β (CAMK2B)). Activity-dependent enhancer RNAs (eRNAs) promote transcription of plasticity genes through enhancer–promoter interactions. b | Within the spine head and shaft, modulation of local protein synthesis of neurotransmitter receptors, scaffolding proteins, calcium-dependent signalling molecules and actin cytoskeletal remodelling factors by miRNAs, endogenous small interfering RNAs (endo-siRNAs) and PIWI-interacting RNAs (piRNAs) contributes to synaptic plasticity by regulating spine morphology and synaptic transmission. ADF, actin-depolymerizing factor (also known as gelosin); AGO2, Argonaute 2; APT1, acyl-protein thioesterase 1; CBP, CREB-binding protein; CoREST, REST co-repressor 1; CPEB, cytoplasmic polyadenylation element-binding protein; EPHA4, Eph receptor A4; GluA2, AMPA-type glutamate receptor subunit 2 (also known as GluR2); Gα13, guanine nucleotide-binding protein-α 13; LIMK1, LIM domain kinase 1; MOV10, Moloney leukaemia virus 10 (also known as putative helicase MOV-10); mSin3, paired amphipathic helix family proteins; NR2, glutamate receptor, ionotropic, N-methyl d-aspartate 2; PAK, p21 protein (CDC42/RAC)-activated kinase; PSD, postsynaptic density; RAC1, Ras-related C3 botulinum toxin substrate 1; REST, repressor element 1-silencing transcription factor; RHOA, RAS homologue family member A; RISC, RNA-induced silencing complex; SYNGAP1, synaptic RAS GTPase-activating protein 1.
miRNAs and associated factors are often packaged into cytoplasmic RNA–protein complexes (neuronal RNA granules) that mediate subcellular targeting, metabolism and effector functions96. Studies of RBPs — many of which are linked to neurological diseases — have therefore been used to elucidate how miRNAs promote synaptic development and plasticity. The fragile X mental retardation 1 protein (FMRP) and related family members (FXR1P (also known as FMR1) and FXR2P (also known as FMR2)) provide salient examples. This family of RBPs is integrated with miRNA pathways at multiple levels and promotes the stabilization, dendritic targeting and local synaptic translation of neuronal mRNAs104,105. These RBPs interact with miRNA biogenesis (through DICER1) and effector (RISC) mechanisms, directly associate with particular miRNAs and are even subject to regulation by miRNAs. In fact, FXR1P is involved in the biogenesis of miR-9 and miR-124 (REF. 106) and, in turn, its expression is modulated by various miRNAs107. Ataxin 2 (ATX2) is another example of a protein involved in neurological disease that links RNA granules and miRNA pathways with synaptic functions. ATX2 and its homologues are RBPs that interact with a range of RNA processing factors (including polyadenylate binding protein (PABP) and maternal expression at 31B (ME31B)) and have established roles in RNA granule assembly and translational regulation. A recent study further demonstrated that D. melanogaster ATX2 is required for synaptic plasticity in a long-term olfactory habituation paradigm108. It suggested that ATX2 specifically regulates the Argonaute 1 (AGO1)–RISC pathway of miRNA-mediated translational repression through effects mediated by ME31B and PABP, which interact with the core RISC proteins, GW182 (also known as TNRC6A) and AGO1.
Other ncRNAs are also implicated in modulating synaptic development, maintenance and plasticity. One intriguing report identified thousands of ncRNAs in neuronal cells derived from enhancer elements, called enhancer RNAs (eRNAs)109. The transcription of eRNAs is activity-dependent and correlates with the expression of proximally located protein-coding genes, suggesting that the process of eRNA transcription and/or the eRNA transcripts are responsible for promoting activity-dependent gene transcription. Additionally, specific lncRNAs located in the nucleus or those that are trafficked to dendritic compartments can mediate essential synaptic processes. For example, nuclear-enriched abundant transcript 2 (NEAT2; also known as MALAT1) is a nuclear lncRNA that modulates synaptogenesis by regulating synaptic gene expression (of neuroligin 1 and synaptic cell adhesion molecule 1 (SynCAM1); also known as CADM1) and alternative splicing (of calcium/calmodulin-dependent protein kinase II-β) via sarcoplasmic reticulum-splicing factors in nuclear speckle domains110. By contrast, rodent brain cytoplasmic RNA 1 (Bc1) and its primate analogue BC200 (also known as BCYRN1) are actively transported to the somatodendritic domains of neurons, where they repress local protein synthesis. They disrupt translational initiation complex assembly by inhibiting the catalytic activity of eukaryotic initiation factor 4A and binding to eukaryotic initiation factor 4B, thus preventing its interaction with the 18S rRNA of the ribosome111. Moreover, mice with deletion of Tsx, a lncRNA transcribed from the X-inactivation locus, exhibit changes in associative learning and memory for conditioned fear, suggesting that Tsx modulates short-term hippocampal memory112. A number of atypical piRNAs113 are expressed in mouse brain in specific regional, cellular and subcellular distributions, and suppression of the most highly expressed hippocampal dendritic piRNA, DQ541777, reduced dendritic spine size, suggesting a role for this factor in activity-dependent translational regulation66. Moreover, high-throughput sequencing of short ncRNAs from hippocampi of mice undergoing olfactory discrimination training, a hippocampal-dependent task, found that a number of short ncRNAs increase more than 100-fold with training, including various endo-siRNAs, snoRNA-derived ncRNAs and other small RNAs114. Conspicuously, a considerable number of these endo-siRNAs are derived from genomic loci encompassing genes that regulate synaptic plasticity, and the most abundant siRNAs identified in this study are associated with a gene involved in synapse development and plasticity, synaptic RAS GTPase-activating protein 1 (SYNGAP1), which encodes a RAS GTPase-activating protein that is highly expressed at the postsynaptic density and implicated in modulating α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor trafficking and excitatory synaptic transmission. The SYNGAP1 gene locus is also linked to mental retardation and autism, highlighting the potential for ncRNAs to be derived from disease-associated loci and to affect disease pathways.
Neurological disease pathogenesis
It is well known that defects in ncRNAs can lead to disease115. For example, mitochondrial tRNA mutations are the primary causes of mitochondrial encephalomyopathies116. However, our appreciation for the evolving ncRNA landscape and the central roles ncRNAs have in neurobiological processes has had a major impact on the study of CNS disease mechanisms and the development of novel clinical applications. Indeed, a rapidly expanding body of evidence suggests that miRNAs and other classes of ncRNAs, along with associated factors, are involved in the pathophysiology of every major class of neurological and psychiatric disorder (TABLE 2; see Supplementary information S1 (table)).
Table 2.
Roles of non-coding RNAs in nervous system disorders
| Paradigms for linking non-coding RNAs to disease | Refs |
|---|---|
| Genetic variation in non-coding RNA genes | 116-121 |
| Genetic variation in disease-associated protein-coding genes affects interactions with non-coding RNAs |
122-124 |
| Epigenetic deregulation of non-coding RNA genes | 125 |
| Perturbations in factors that modulate non-coding RNA biogenesis, maturation and function |
126 |
| Genomic context links non-coding RNA genes to disease-causing genes and susceptibility loci |
127 |
| Differential expression of non-coding RNAs in disease-related central and peripheral tissues |
130-134 |
Genetic variation in ncRNA genes can promote CNS disease onset. For example, eliminating the Bc1 gene in mice leads to neuronal hyperexcitability, increased cortical gamma frequency oscillations and susceptibility to epileptogenesis117. Expansion repeats in the ATXN8OS lncRNA gene are partly responsible for the pathology of spinocerebellar ataxia type 8 through a toxic RNA gain-of-function mechanism that includes formation of ribonuclear inclusions in the cerebellum and deregulation of muscleblind-like splicing regulator 1-mediated alternative splicing118. Moreover, genome-wide association studies have demonstrated that the 9p21 genomic locus is a hotspot for the risk of stroke, glioma, plexiform neurofibroma and other disorders119–121. This locus encodes the cyclin-dependent kinase inhibitors CDKN2A and CDKN2B and the lncRNA ANRIL (also known as CDKN2B-AS1), but analysing single-nucleotide polymorphisms (SNPs) and transcript levels suggests that, whereas multiple sites within the 9p21 locus may influence disease vulnerability, variations in ANRIL and levels of ANRIL expression correlate with disease most significantly. These observations underscore the concept that genetic variation at a single locus can affect the expression and function of many distinct protein-coding and non-coding transcripts, and studies of genetic epidemiology must be interpreted in this context.
Correspondingly, genetic variation present in either miRNAs or their targets can change regulatory interactions between these molecules and, thereby, affect disease processes122–124. For example, a risk of Parkinson’s disease conferring an SNP in the fibroblast growth factor 20 (FGF20) gene disrupts the binding of miR-433 to the FGF20 mRNA 3′-UTR. Deregulated expression of FGF20 is correlated with increased levels of α-synuclein, which can lead to Parkinson’s disease. Similarly, a Tourette’s syndrome-linked variant within the miR-189 binding site of the 3′-UTR of SLIT and NTRK-like (SLITRK1) leads to increased repression of SLITRK1 (REF. 124). These findings have important implications for characterizing disease-linked non-coding DNA regions because an individual miRNA can target multiple protein-coding transcripts and because a single protein-coding transcript can be modulated by multiple miRNAs. Furthermore, these observations support the more general hypothesis that genetic variation can alter biophysical relationships between ncRNA transcripts and their DNA, RNA and protein partners, considerably or more subtly influencing regulatory and functional interactions and modifying disease susceptibility.
Additionally, epigenetic deregulation of ncRNA genes can be associated with CNS diseases115. For example, it has been suggested that DNA methylation modulates the expression of the tumour suppressor miRNAs miR-124 and miR-137 in anaplastic astrocytoma and glioblastoma125. ncRNAs are also subject to epigenetic regulation via mechanisms other than DNA methylation, such as histone and chromatin modification, and it is likely that future studies will reveal that disease-linked abnormalities in ncRNA activity are associated with deregulation of these processes. Perturbations in factors responsible for modulating ncRNA metabolism and function can also lead to pathology. For example, age-related macular degeneration is characterized by decreased levels of DICER1 in the eye that result in impaired degradation of Alu retrotransposon-derived ncRNAs, which are directly toxic to the retinal pigmented epithelium126.
Genomic context can also link ncRNA genes to CNS disease-causing genes and susceptibility loci. In some cases, mechanistic relationships between overlapping or proximally located ncRNA and protein-coding genes have been identified. For example, SCA7/ATXN7 antisense RNA 1 (SCAANT1) is an lncRNA transcribed antisense to the gene mutated in spinocerebellar ataxia type 7 (ATXN7), and it functions as a repressor of ATXN7 transcription127. Convergent bidirectional transcription has similarly been observed at a number of loci associated with neurological diseases and is, notably, implicated in promoting instability of expansion repeats128.
Last, ncRNAs can be differentially expressed in disease-related central and peripheral tissues associated with a range of CNS disorders, including neurovascular, neurodevelopmental, neurodegenerative, neuropsychiatric and neuroimmunological disorders and brain tumours62,129. It is debatable to what extent these correlations represent causal relationships. Nevertheless, target genes of deregulated ncRNAs are often involved in cellular pathways linked to the pathophysiology of corresponding disease states. For example, a recent study demonstrated that in peripheral blood mononuclear cells from patients with Parkinson’s disease, miRNAs were differentially expressed, with higher levels of miRNAs for predicted target genes that are involved in pathways associated with the disease, including those in the protein ubiquitylation and glycosphingolipid biosynthesis pathways130. This example suggests that ncRNA expression profiles in peripheral tissues might reflect CNS disease states. Evidence from other disorders, such as stroke, has in fact demonstrated that miRNA expression profiles present in the brain after cerebral ischaemia correspond with those in the blood (in animal models131 and in stroke patients132). Moreover, ncRNAs derived from neural cells can circulate in the blood and cerebrospinal fluid in membrane-bound exosomes133,134. These overall observations suggest that ncRNAs might represent peripheral biomarkers for CNS diseases.
However, whether safe and effective diagnostic and treatment strategies for neurological and psychiatric diseases can be developed by targeting ncRNAs (and their biogenesis and effector pathways) is yet to be determined. miRNA-profiling assays are already commercially available for cancer diagnosis and prognostication. They are also being developed for other ncRNAs and other systemic disorders. Therapeutic approaches targeting ncRNAs are also being explored, including those focused on designing and delivering oligonucleotide molecules with the ability to repress or replace ncRNAs115 and on identifying small molecules that can influence ncRNA biogenesis and effector pathways135. Although these strategies are promising, selective delivery into the CNS remains the most considerable challenge, and it has not been addressed effectively although methods using viral delivery systems, chemical modification and conjugation strategies, aptamers, nanotechnologies and barrier modulation are currently being investigated.
Perspectives
The CNS is defined by extraordinary degrees of cellular diversification, synaptic and neural network connectivity and plasticity, and responsiveness to interoceptive and environmental signals. It is now clear that these features are mediated not only by cell type-, developmental stage- and stimulus-specific profiles of protein-coding mRNA expression, post-transcriptional modifications, localization and translation but also by ncRNAs that have similarly complex life cycles and a diverse and interrelated range of regulatory and functional roles in transcriptional, post-transcriptional, epigenetic and nuclear processes136,137 (FIG. 3). In fact, because the proportion of non-coding DNA sequence present in the genome correlates with organismal complexity53 (in contrast to protein-coding genes), it is intriguing to speculate that ncRNAs preferentially mediated the accelerated and asymmetric evolution of the human CNS and underlie the unique functional repertoire of the brain. Further elucidating how ncRNAs operate at molecular, cellular and more hierarchical (neural network activity and topology) levels may therefore provide novel insights for answering seminal neurobiological questions (BOX 2). Chief among these may be uncovering the genetic and evolutionary bases of complex CNS diseases, which have remained elusive. Indeed, the concept that each nucleotide — in addition to being involved in diverse DNA–protein, DNA–RNA and DNA–DNA interactions in local and more global chromatin environments within the nucleus — can give rise to multiple ncRNAs with divergent functions suggests a paradigm in which genetic variation in even a single base pair can both subtly and more dramatically affect the execution of myriad cellular programmes, potentially accounting for how complex CNS disease states unfold and co-morbidities arise.
Box 2 | Live cellular imaging of non-coding RNAs.
The development of techniques for live cellular imaging and high-resolution analysis of single RNA molecules is crucial for better understanding the in vivo spatiotemporal dynamics of non-coding RNAs (ncRNAs). Strategies for RNA imaging in live cells include using light microscopy (wide-field, confocal or total internal reflection microscopy) to detect fluorescence of reporter probes (molecular beacons or MS2 labels). Oligonucleotide-based molecular beacons fluoresce upon hybridization with target RNAs. MS2 labelling exploits the high-affinity interaction between the MS2 bacteriophage coat protein and its cognate MS2 RNA-binding sequence. The MS2 protein can be fused with a fluorescent protein and used to label RNAs engineered with MS2-binding sequences. One interesting study investigated the behaviour of the human telomere repeat-containing RNA (TERRA) ncRNA, using a molecular beacon-based approach144. It was observed that TERRA localizes to telomeric DNA and forms a parallel G-quadruplex structure, which has roles in modulating telomerase activity and gene expression. Other studies have uncovered how the X (inactive)-specific transcript (XIST) long ncRNA (lncRNA), mediates X chromosome inactivation with MS2 labelling145 and how the nuclear paraspeckle assembly transcript 1 (NEAT1) lncRNA establishes paraspeckle nuclear domains using direct visualization of paraspeckle proteins and fluorescence recovery after photobleaching146. Various studies have also reported imaging microRNAs (miRNAs) with fluorescent proteins, luciferase reporters and molecular beacons147. One described imaging miR-124 during neuronal differentiation using a molecular beacon with an miR-124-binding sequence148. Intriguingly, this molecular beacon was linked to a magnetic nanoparticle, enabling the imaging of miRNAs in live animals by magnetic resonance imaging. The same authors also reported how a molecular beacon targeting the glioma-promoting miR-221 can be conjugated to a magnetic nanoparticle and to an aptamer that specifically binds to nucleolin, which is expressed on glioma cells149. This ‘theragnostic’ device can potentially be used to target glioma cells selectively and to image and even inhibit miR-221 activity. The widespread adoption of these and other innovative quantitative real-time in vitro and in vivo imaging technologies — those exploiting different probe designs (nanoparticles, quantum dots, organic dyes and chemical tags) and advanced optical methods — will help to characterize specific patterns and kinetics for dynamic ncRNA transcription, processing, transport and molecular interactions in health and disease and to develop novel diagnostics and therapeutics.
Supplementary Material
Acknowledgements
The authors regret that space constraints have prevented the citation of many relevant and important references. M.F.M. is supported by grants from the US National Institutes of Health (NS071571, HD071593 and MH66290), as well as by the F.M. Kirby, Alpern Family, Mildred and Bernard H. Kayden and Roslyn and Leslie Goldstein Foundations.
Glossary
- Non-coding RNAs(ncRNAs)
RNA molecules belonging to an increasing number of different classes that function explicitly as RNAs, rather than as proteins, in a wide variety of regulatory, structural and functional processes.
- Open reading frames
DNA sequences that are present between start, or initiation, codons and stop, or termination, codons. It is implied that these sequences are translated into proteins.
- Chromatin
The structure of the genome in the nucleus formed by DNA, histones, non-histone proteins and associated factors that can undergo dynamic changes locally and more globally into open and closed conformations, which promote the execution of specific cellular programmes, such as transcription and DNA replication and repair.
- Epigenetic mechanisms
Multilayered cellular processes that modulate gene expression and function in response to interoceptive and environmental stimuli during development, adult life and ageing, including DNA methylation, post-translational histone modifications, ATP-dependent nucleosome and higher-order chromatin remodelling, non-coding RNA deployment and nuclear reorganization.
- Nuclear domains
A range of dynamically forming macromolecular nuclear assemblies found prominently in neural cells and consisting of chromatin and diverse factors involved in transcriptional and epigenetic regulation and post-transcriptional RNA processing, such as transcription factories, Cajal bodies, promyelocytic leukaemia nuclear bodies, nuclear stress bodies, nuclear speckles and paraspeckles.
- Argonaute proteins
Proteins of the AGO and PIWI subfamilies that interact with microRNAs and endogenous small interfering RNAs and with PIWI-interacting RNAs, respectively, and have key roles in mediating post-transcriptional RNA silencing.
- Retrotransposon
Mobile genetic element comprising a substantial proportion of the human genome transcribed into RNA intermediates and subsequently reintegrated into the genome, including LINE and SINE subclasses, such as Alu elements.
- Somatic mosaicism
The presence of distinct genotypes in different somatic cells and tissues of an individual organism. Mosaicism arises from somatic mutations that can be generated by endogenous factors, such as mobile genetic elements and exogenous factors.
- Bidirectional transcription
Transcription that occurs on both the positive and negative strands of DNA simultaneously, where the direction of RNA polymerase progression along each strand is either is convergent or divergent.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Mark F. Mehler’s homepage: http://www.einstein.yu.edu/faculty/profile.asp?id=1583
SUPPLEMENTARY INFORMATION
See online article: S1 (table)
References
- 1. Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. This paper reported the results of the pilot phase of the ENCODE project, which aims to define the regulatory and functional landscape of the human genome, and provided evidence supporting the conclusion that the human genome is pervasively transcribed.
- 2.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 3. Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. References 2 and 3 reported data from the FANTOM3 project, which focused on characterizing the mammalian transcriptome and the organization of transcriptional and regulatory units in the genome, and identified the diversity of protein-coding and ncRNA transcripts that arise from overlapping transcription on both strands.
- 4.White RJ. Transcription by RNA polymerase III: more complex than we thought. Nature Rev. Genet. 2011;12:459–463. doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- 5.Candeias MM. The can and can’t dos of p53. RNA. Biochimie. 2011;93:1962–1965. doi: 10.1016/j.biochi.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Cooper C, et al. Steroid receptor RNA activator bi-faceted genetic system: heads or tails? Biochimie. 2011;93:1973–1980. doi: 10.1016/j.biochi.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Kageyama Y, Kondo T, Hashimoto Y. Coding versus non-coding: translatability of short ORFs found in putative non-coding transcripts. Biochimie. 2011;93:1981–1986. doi: 10.1016/j.biochi.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 8. Kapranov P, et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol. 2010;8:149. doi: 10.1186/1741-7007-8-149. This single-molecule sequencing study determined that, by mass, most of the non-ribosomal RNA found in a range of human cell types is unannotated or poorly functionally characterized ‘dark matter RNA’.
- 9.Gustincich S, et al. The complexity of the mammalian transcriptome. J. Physiol. 2006;575:321–332. doi: 10.1113/jphysiol.2006.115568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollard KS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 11.Wu J, Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7:R85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/ miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burroughs AM, et al. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8:158–177. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Washietl S. Sequence and structure analysis of noncoding RNAs. Methods Mol. Biol. 2010;609:285–306. doi: 10.1007/978-1-60327-241-4_17. [DOI] [PubMed] [Google Scholar]
- 15.Parker BJ, et al. New families of human regulatory RNA structures identified by comparative analysis of vertebrate genomes. Genome Res. 2011;21:1929–1943. doi: 10.1101/gr.112516.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mercer TR, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nature Biotech. 2011;30:99–104. doi: 10.1038/nbt.2024. This study used a targeted RNA capture and sequencing strategy, termed ‘RNA CaptureSeq’, to analyse the human transcriptome at extremely high resolution and revealed the presence of extraordinarily complex and previously unannotated profiles of ncRNA transcription, even from relatively well-studied genomic loci encompassing the p53 and HOX genes.
- 17.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. This paper showed that a significant number of lincRNAs bind to PRC2- and CoREST-containing chromatin modifying complexes and act as guides for recruiting them to specific genomic loci.
- 19.Mercer TR, et al. Expression of distinct RNAs from 3' untranslated regions. Nucleic Acids Res. 2011;39:2393–2403. doi: 10.1093/nar/gkq1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung T, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 23.Rackham O, et al. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA. 2011;17:2085–2093. doi: 10.1261/rna.029405.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 25.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nature Rev. Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 27.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 29.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 30.Peng Z, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nature Biotech. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 31.Hansen TB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marti E, et al. A myriad of miRNA variants in control and Huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 2010;38:7219–7235. doi: 10.1093/nar/gkq575. This paper showed that, in the human brain, a significant number of miRNAs exhibit sequence and length variability. These ‘isomiRs’ are highly deregulated in cortical and striatal tissues from patients with Huntington’s disease.
- 33.Okamura K, et al. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim VN. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 2006;20:1993–1997. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- 35. Yan Z, et al. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 2011;39:6596–6607. doi: 10.1093/nar/gkr298. This study provides evidence supporting the expression of piRNAs in the brain.
- 36.Kiss T. Small nucleolar RNA-guided posttranscriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 38.Soeno Y, et al. Identification of novel ribonucleoprotein complexes from the brain-specific snoRNA MBII-52. RNA. 2010;16:1293–1300. doi: 10.1261/rna.2109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 40.Richard P, et al. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 2003;22:4283–4293. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taft RJ, et al. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ender C, et al. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 43.Taft RJ, Kaplan CD, Simons C, Mattick JS. Evolution, biogenesis and function of promoterassociated RNAs. Cell Cycle. 2009;8:2332–2338. doi: 10.4161/cc.8.15.9154. [DOI] [PubMed] [Google Scholar]
- 44.Taft RJ, et al. Tiny RNAs associated with transcription start sites in animals. Nature Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 45.Kapranov P, et al. New class of gene-termini-associated human RNAs suggests a novel RNA copying mechanism. Nature. 2010;466:642–646. doi: 10.1038/nature09190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taft RJ, et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nature Struct. Mol. Biol. 2010;17:1030–1034. doi: 10.1038/nsmb.1841. [DOI] [PubMed] [Google Scholar]
- 47.Carone DM, et al. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma. 2009;118:113–125. doi: 10.1007/s00412-008-0181-5. [DOI] [PubMed] [Google Scholar]
- 48.Cao F, et al. Dicer independent small RNAs associate with telomeric heterochromatin. RNA. 2009;15:1274–1281. doi: 10.1261/rna.1423309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sobala A, Hutvagner G. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip. Rev. RNA. 2011;2:853–862. doi: 10.1002/wrna.96. [DOI] [PubMed] [Google Scholar]
- 50.Landerer E, et al. Nuclear localization of the mitochondrial ncRNAs in normal and cancer cells. Cell Oncol. (Dordr.) 2011;34:297–305. doi: 10.1007/s13402-011-0018-8. [DOI] [PubMed] [Google Scholar]
- 51.Lung B, et al. Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 2006;34:3842–3852. doi: 10.1093/nar/gkl448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taft RJ, Hawkins PG, Mattick JS, Morris KV. The relationship between transcription initiation RNAs and CCCTC-binding factor (CTCF) localization. Epigenetics Chromatin. 2011;4:13. doi: 10.1186/1756-8935-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 54.Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc. Natl Acad. Sci. USA. 2008;105:2946–2950. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bond AM, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nature Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chodroff RA, et al. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11:R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu HY, et al. MicroRNA expression and regulation in human, chimpanzee, and macaque brains. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002327. e1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Somel M, et al. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 2010;20:1207–1218. doi: 10.1101/gr.106849.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu AG, et al. Intergenic and repeat transcription in human, chimpanzee and macaque brains measured by RNA-Seq. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000843. e1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Somel M, et al. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001214. e1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lindblad-Toh K, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fiore R, Khudayberdiev S, Saba R, Schratt G. MicroRNA function in the nervous system. Prog. Mol. Biol. Transl. Sci. 2011;102:47–100. doi: 10.1016/B978-0-12-415795-8.00004-0. [DOI] [PubMed] [Google Scholar]
- 63.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000617. e1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Belgard TG, et al. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee EJ, et al. Identification of piRNAs in the central nervous system. RNA. 2011;17:1090–1099. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amaral PP, et al. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA. 2009;15:2013–2027. doi: 10.1261/rna.1705309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. This study established the presence of significant degrees of genetic mosaicism in the human brain by identifying thousands of genomic reintegration sites for L1, Alu and SVA retrotransposons in the hippocampus and caudate nucleus of three individuals, using a ‘retrotransposon capture sequencing’ strategy.
- 69.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muotri AR, Zhao C, Marchetto MC, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. 2009;19:1002–1007. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Faunes F, et al. Expression of transposable elements in neural tissues during Xenopus development. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0022569. e22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davis TH, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tao J, et al. Deletion of astroglial Dicer causes non-cellautonomous neuronal dysfunction and degeneration. J. Neurosci. 2011;31:8306–8319. doi: 10.1523/JNEUROSCI.0567-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dugas JC, et al. Dicer1 and miR-219 are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao X, et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65:612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Giraldez AJ, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. This study was the first to establish the crucial part played by miRNAs in brain development.
- 77.Delaloy C, et al. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6:323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leucht C, et al. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nature Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- 79.Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nature Struct. Mol. Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shibata M, Kurokawa D, Nakao H, Ohmura T, Aizawa S. MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J. Neurosci. 2008;28:10415–10421. doi: 10.1523/JNEUROSCI.3219-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miyoshi N, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 83.Anokye-Danso F, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Choi YJ, et al. miR34 miRNAs provide a barrier for somatic cell reprogramming. Nature Cell Biol. 2011;13:1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoo AS, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ambasudhan R, et al. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–118. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatinremodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qureshi IA, Gokhan S, Mehler MF. REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle. 2010;9:4477–4486. doi: 10.4161/cc.9.22.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- 90.Blackshaw S, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rapicavoli NA, Poth EM, Blackshaw S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev. Biol. 2010;10:49. doi: 10.1186/1471-213X-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 2011;6:32. doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mercer TR, et al. Long noncoding RNAs in neuronalglial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lin M, et al. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0023356. e23356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Konecna A, Heraud JE, Schoderboeck L, Raposo AA, Kiebler MA. What are the roles of microRNAs at the mammalian synapse? Neurosci. Lett. 2009;466:63–68. doi: 10.1016/j.neulet.2009.06.050. [DOI] [PubMed] [Google Scholar]
- 97.Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA. 2010;16:1516–1529. doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Park CS, Tang SJ. Regulation of microRNA expression by induction of bidirectional synaptic plasticity. J. Mol. Neurosci. 2009;38:50–56. doi: 10.1007/s12031-008-9158-3. [DOI] [PubMed] [Google Scholar]
- 99.Wibrand K, et al. Differential regulation of mature and precursor microRNA expression by NMDA and metabotropic glutamate receptor activation during LTP in the adult dentate gyrus in vivo. Eur. J. Neurosci. 2010;31:636–645. doi: 10.1111/j.1460-9568.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cohen JE, Lee PR, Chen S, Li W, Fields RD. MicroRNA regulation of homeostatic synaptic plasticity. Proc. Natl Acad. Sci. USA. 2011;108:11650–11655. doi: 10.1073/pnas.1017576108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smibert P, et al. A Drosophila genetic screen yields allelic series of core microRNA biogenesis factors and reveals post-developmental roles for microRNAs. RNA. 2011;17:1997–2010. doi: 10.1261/rna.2983511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Konopka W, et al. MicroRNA loss enhances learning and memory in mice. J. Neurosci. 2010;30:14835–14842. doi: 10.1523/JNEUROSCI.3030-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schofield CM, et al. Monoallelic deletion of the microRNA biogenesis gene Dgcr8 produces deficits in the development of excitatory synaptic transmission in the prefrontal cortex. Neural Dev. 2011;6:11. doi: 10.1186/1749-8104-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cheever A, Ceman S. Translation regulation of mRNAs by the fragile X family of proteins through the microRNA pathway. RNA Biol. 2009;6:175–178. doi: 10.4161/rna.6.2.8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Muddashetty RS, et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu XL, et al. FXR1P but not FMRP regulates the levels of mammalian brain-specific microRNA-9 and microRNA-124. J. Neurosci. 2011;31:13705–13709. doi: 10.1523/JNEUROSCI.2827-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cheever A, Blackwell E, Ceman S. Fragile X protein family member FXR1P is regulated by microRNAs. RNA. 2010;16:1530–1539. doi: 10.1261/rna.2022210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McCann C, et al. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc. Natl Acad. Sci. USA. 2011;108:E655–E662. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. This paper identified thousands of eRNAs that are transcribed from the enhancer regions of activity-dependent neuronal genes.
- 110.Bernard D, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eom T, Berardi V, Zhong J, Risuleo G, Tiedge H. Dual nature of translational control by regulatory BC RNAs. Mol. Cell. Biol. 2011;31:4538–4549. doi: 10.1128/MCB.05885-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Anguera MC, et al. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002248. e1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smalheiser NR. The search for endogenous siRNAs in the mammalian brain. Exp. Neurol. 2012;235:455–463. doi: 10.1016/j.expneurol.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Smalheiser NR, Lugli G, Thimmapuram J, Cook EH, Larson J. Endogenous siRNAs and noncoding RNAderived small RNAs are expressed in adult mouse hippocampus and are up-regulated in olfactory discrimination training. RNA. 2011;17:166–181. doi: 10.1261/rna.2123811. This study provides evidence for the expression of endo-siRNAs in the brain and for their roles in mediating learning and memory.
- 115.Esteller M. Non-coding RNAs in human disease. Nature Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 116.Yarham JW, Elson JL, Blakely EL, McFarland R, Taylor RW. Mitochondrial tRNA mutations and disease. Wiley Interdiscip. Rev. RNA. 2010;1:304–324. doi: 10.1002/wrna.27. [DOI] [PubMed] [Google Scholar]
- 117.Zhong J, et al. BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J. Neurosci. 2009;29:9977–9986. doi: 10.1523/JNEUROSCI.3893-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Daughters RS, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000600. e1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 120.Zhang W, et al. Variants on chromosome 9p21.3 correlated with ANRIL expression contribute to stroke risk and recurrence in a large prospective stroke population. Stroke. 2012;43:14–21. doi: 10.1161/STROKEAHA.111.625442. [DOI] [PubMed] [Google Scholar]
- 121.Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Keavney B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000899. e1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang G, et al. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of α-synuclein. Am. J. Hum. Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Richardson K, Lai CQ, Parnell LD, Lee YC, Ordovas JM. A genome-wide survey for SNPs altering microRNA seed sites identifies functional candidates in GWAS. BMC Genomics. 2011;12:504. doi: 10.1186/1471-2164-12-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Abelson JF, et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 125.Silber J, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kaneko H, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sopher BL, et al. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding. RNA. Neuron. 2011;70:1071–1084. doi: 10.1016/j.neuron.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nakamori M, Pearson CE, Thornton CA. Bidirectional transcription stimulates expansion and contraction of expanded (CTG)*(CAG) repeats. Hum. Mol. Genet. 2011;20:580–588. doi: 10.1093/hmg/ddq501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Qureshi IA, Mattick JS, Mehler MF. Long noncoding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Martins M, et al. Convergence of miRNA expression profiling, α-synuclein interacton and GWAS in Parkinson’s disease. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0025443. e25443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 132.Tan KS, et al. Expression profile of microRNAs in young stroke patients. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0007689. e7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. This paper showed that glioblastoma cells release exosomes containing miRNAs, including those overexpressed in the tumour, in order to signal to local and distal sites and promote tumour progression.
- 134.Balaj L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nature Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li Y, He C, Jin P. Emergence of chemical biology approaches to the RNAi/miRNA pathway. Chem. Biol. 2010;17:584–589. doi: 10.1016/j.chembiol.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.St Laurent G, Wahlestedt C. Noncoding RNAs: couplers of analog and digital information in nervous system function? Trends Neurosci. 2007;30:612–621. doi: 10.1016/j.tins.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 137.Mattick JS. A new paradigm for developmental biology. J. Exp. Biol. 2007;210:1526–1547. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- 138.Anno YN, et al. Genome-wide evidence for an essential role of the human Staf/ZNF143 transcription factor in bidirectional transcription. Nucleic Acids Res. 2011;39:3116–3127. doi: 10.1093/nar/gkq1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Faghihi MA, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Paz-Yaacov N, et al. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc. Natl Acad. Sci. USA. 2010;107:12174–12179. doi: 10.1073/pnas.1006183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mansfield KD, Keene JD. The ribonome: a dominant force in co-ordinating gene expression. Biol. Cell. 2009;101:169–181. doi: 10.1042/BC20080055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xu Y, Suzuki Y, Ito K, Komiyama M. Telomeric repeat-containing RNA structure in living cells. Proc. Natl Acad. Sci. USA. 2010;107:14579–14584. doi: 10.1073/pnas.1001177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ng K, et al. A system for imaging the regulatory noncoding Xist RNA in living mouse embryonic stem cells. Mol. Biol. Cell. 2011;22:2634–2645. doi: 10.1091/mbc.E11-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nature Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang F, Niu G, Chen X, Cao F. Molecular imaging of microRNAs. Eur. J. Nuclear Med. Mol. Imag. 2011;38:1572–1579. doi: 10.1007/s00259-011-1786-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hwang do W, Song IC, Lee DS, Kim S. Smart magnetic fluorescent nanoparticle imaging probes to monitor microRNAs. Small. 2010;6:81–88. doi: 10.1002/smll.200901262. [DOI] [PubMed] [Google Scholar]
- 149.Kim JK, Choi KJ, Lee M, Jo MH, Kim S. Molecular imaging of a cancer-targeting theragnostics probe using a nucleolin aptamer- and microRNA-221 molecular beacon-conjugated nanoparticle. Biomaterials. 2012;33:207–217. doi: 10.1016/j.biomaterials.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 150.Wang X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Parrott AM, et al. The evolution and expression of the snaR family of small non-coding RNAs. Nucleic Acids Res. 2011;39:1485–1500. doi: 10.1093/nar/gkq856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kickhoefer VA, et al. Vault ribonucleoprotein particles from rat and bullfrog contain a related small RNA that is transcribed by RNA polymerase III. J. Biol. Chem. 1993;268:7868–7873. [PubMed] [Google Scholar]
- 154.Nandy C, et al. Epstein-Barr virus-induced expression of a novel human vault. RNA. J. Mol. Biol. 2009;388:776–784. doi: 10.1016/j.jmb.2009.03.031. [DOI] [PubMed] [Google Scholar]
- 155.Zhang AT, et al. Dynamic interaction of Y RNAs with chromatin and initiation proteins during human DNA replication. J. Cell Sci. 2011;124:2058–2069. doi: 10.1242/jcs.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sim S, et al. The zipcode-binding protein ZBP1 influences the subcellular location of the Ro 60-kDa autoantigen and the noncoding Y3 RNA. RNA. 2012;18:100–110. doi: 10.1261/rna.029207.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.