Abstract
CEP192 is a centrosome protein that plays a critical role in centrosome biogenesis and function in mammals, Drosophila and C. elegans.1-6 Moreover, CEP192-depleted cells arrest in mitosis with disorganized microtubules, suggesting that CEP192’s function in spindle assembly goes beyond its role in centrosome activity and pointing to a potentially more direct role in the regulation of the mitotic microtubule landscape.7 To better understand CEP192 function in mitosis, we used mass spectrometry to identify CEP192-interacting proteins. We previously reported that CEP192 interacts with NEDD1, a protein that associates with the γ-tubulin ring complex (γ-TuRC) and regulates its phosphorylation status during mitosis.8 Additionally, within the array of proteins that interact with CEP192, we identified the microtubule binding K63-deubiquitinase CYLD. Further analyses show that co-depletion of CYLD alleviates the bipolar spindle assembly defects observed in CEP192-depleted cells. This functional relationship exposes an intriguing role for CYLD in spindle formation and raises the tantalizing possibility that CEP192 promotes robust mitotic spindle assembly by regulating K63-polyubiquitin-mediated signaling through CYLD.
Keywords: CEP192, CYLD, mitosis, spindle, microtubules, ubiquitination, K63, centrosome, proteomics
Introduction
CEP192 is required for centrosome maturation at the onset of mitosis. Mitotic centrosomes in CEP192-depleted cells do not recruit pericentriolar material (PCM) and are unable to nucleate microtubules. Moreover, although microtubules are generated in the vicinity of chromosomes in these cells, contrarily to what is observed in cells lacking centrosomes by centriole defects or physical ablation,9-11 they fail to self-organize into bipolar spindles.5,6 This observation suggests that CEP192, in addition to regulating centrosome maturation, has a more specific role in the organization of the mitotic microtubule landscape.7
Although significant advances have been made concerning our understanding of CEP192 importance in spindle assembly, the molecular mechanisms underlying the critical role of CEP192 in this process are only now starting to be unraveled. We have recently found that CEP192 interacts with the microtubule binding protein NEDD1 and regulates its mitotic phosphorylation.8 Additionally, CEP192 interacts with the mitotic kinase AURKA and controls its activation.12 Nevertheless, a precise molecular mechanism that explains why and how CEP192 affects spindle assembly, and particularly microtubule organization, remains elusive.
CEP192 physically interacts with the K63-deubiquitinase CYLD
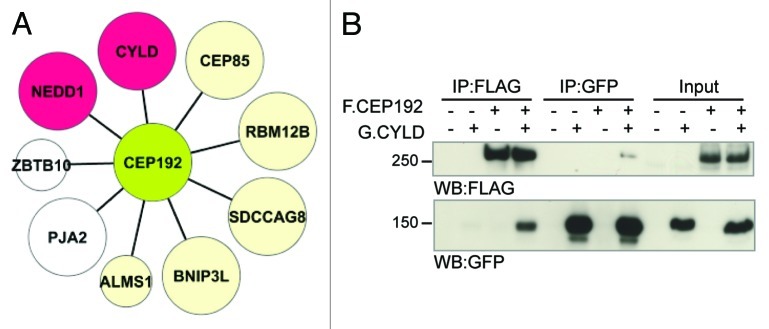
To gain insights on CEP192 function, we used mass spectrometry to identify CEP192-interacting proteins. Our analysis revealed that, in addition to NEDD1,8 CEP192 associates with an array of proteins of diverse activity (Fig. 1A; Fig. S1). Indeed, CEP192 associates with the centrosome proteins CEP85/CCDC21, ALMS1 and SDCCAG8. Interestingly, both ALMS1 and SDCCAG8 are implicated in cilia function.13,14 Critically, we found that CEP192 associates with the K63-deubiquitinating enzyme (DUB) CYLD, in line with the recently described DUB interactome15,16 (Fig. 1A). This interaction was confirmed by immunoprecipitation followed by western blot (Fig. 1B).
Figure 1. CYLD interacts physically with CEP192. (A) CEP192 interaction partners identified by immunoprecipitation and mass spectrometry analysis (see also, Fig. S1 and Table S1). Large nodes represent hits found in three out of three experiments, while the small nodes indicate hits found in two out of three experiments. Depletion of the proteins colored in beige using RNAi did not affect spindle assembly (data not shown). Nodes in red represent hits with a known (NEDD1) or novel (CYLD) role in spindle formation. (B) FLAG-CEP192 and GFP-CYLD were expressed in HEK293 cells. Immunoprecipitation of either protein followed by western blot confirm their physical interaction.
CYLD co-depletion restores spindle assembly defects in CEP192-depleted cells
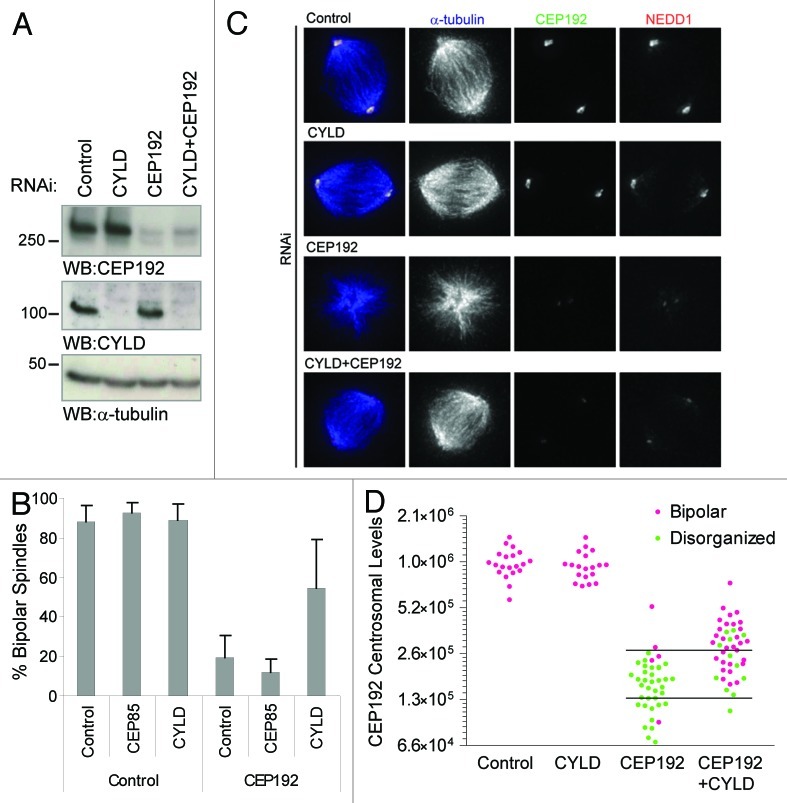
With the exception of NEDD1, depletion of the novel CEP192 interactors analyzed did not perturb spindle assembly (Fig. 1A and data not shown). Excitingly, however, co-depletion of CYLD alleviated the spindle assembly defects observed in CEP192 RNAi-treated cells (Fig. 2A–C). These results were confirmed using two different CYLD-specific RNAi reagents (Fig. S2A). Furthermore, we measured the levels of CEP192 at mitotic centrosomes under these conditions and found that 60% of cells with comparably low levels of CEP192 can assemble bipolar spindles upon CYLD co-depletion (Fig. 2D; Fig. S2B and C). Taken together, these results suggest that CYLD function perturbs bipolar spindle assembly in CEP192-depleted cells, raising the interesting possibility that CEP192 activity is required to antagonize CYLD function.
Figure 2. CYLD co-depletion restores spindle assembly defects in CEP192-depleted cells. (A) western blot showing depletion of CEP192 and CYLD in HeLa cells transfected under the indicated RNAi conditions. For single depletion of CEP192 or CYLD we added non-targeting (control) RNAi to make the total RNAi quantity equal to the condition used for double CYLD+CEP192 depletion. (B) Quantification of the spindle phenotypes in HeLa cells transfected with the indicated esiRNA. Co-depletion of CEP85 with CEP192 was used as a negative control. We show the average and standard deviation (s.d.) of at least three independent experiments. (C) HeLa cells transfected with the corresponding RNAi and stained with antibodies anti-α-tubulin, CEP192 and NEDD1. (D) Quantification of CEP192 centrosomal levels in cells treated as in (C). Each dot represents a cell where centrosomal CEP192 intensity and spindle morphology were scored. Horizontal lines in the graph mark the range of CEP192-depleted levels that we consider for the quantifications in Figure S2C. We show one representative experiment out of five.
By processing K63-polyubiquitin chains, CYLD regulates NFκB, JNK, p38MAPK and Akt signaling pathways, thereby controlling multiple cellular processes including cell proliferation and inflammation.16,17 CYLD also regulates the timely entry of the cell in mitosis, possibly via PLK1.18 On the other hand, CYLD directly binds to interphase microtubules through its Cap-Gly domains and promotes tubulin polymerization, microtubule stabilization and cell migration.19,20 However, CYLD does not appear to localize to the mitotic spindle and had not been implicated, until now, in the process of spindle formation. Although further mechanistic studies will be required to fully comprehend the relationship between CYLD and CEP192, with the available data, we propose a model by which CEP192 antagonizes CYLD function at the onset of mitosis to either promote de-polymerization of interphase microtubules (stabilized otherwise by CYLD,19,20 model in Fig. S2D) and/or preserve the ubiquitination levels of CYLD substrates implicated in spindle assembly.
Final remarks
Our proteomic analysis identified an array of centrosomal and microtubule binding proteins that physically interact with CEP192. Although additional analyses are required to determine the functional relevance of CEP192 binding to CEP85/CCDC21, ALMS1 or SDCCAG8, we believe they could determine particular aspects of centrosome biogenesis. Moreover, our discoveries unravel a pleiotropic and complex role of CEP192 in microtubule dynamics, as CEP192 may not only induce microtubule nucleation by regulating NEDD1 and AURKA,8,12 but also control microtubule polymerization/stability by antagonizing CYLD.
Noticeably, the arising possibility that CEP192 controls spindle formation by regulating CYLD DUB activity suggests that novel CYLD substrates modified by K63-ubiquitination could have a direct role in bipolar spindle assembly. This observation highlights the need to investigate two complex and unexplored aspects of mitotic regulation: how ubiquitination-based signaling contributes to spindle assembly and how centrosomes control these posttranslational modifications.
Materials and Methods
cDNA clones and transfections
CEP192 cDNA (ENSP00000317156, 1941 a.a) was cloned into pcDNA3 fused with FLAG. CYLD cDNA (ENSP00000392025, 956 a.a.) was cloned into pcDNA3 fused to GFP. Plasmid transfections were performed in HEK293 cells using Lipofectamine 2000 (Invitrogen) for 36 h.
Affinity purification and mass spectrometric analysis
HEK293 cells growing in p150 plates were transfected with 12 μg of FLAG-CEP192. We used 6–8 p150 plates per experiment and allowed protein expression for 24 h. Lysis buffer contained 50 mM Hepes-KOH pH 8, 100 mM KCl, 2 mM EDTA, 0.5% NP40, 10% glycerol, 10 mM NaF, 50 mM β-glycerophosphate, 5 nM okadaic acid, 5 nM calyculinA, 1 mM DTT and protease inhibitors (Roche; 04693132001). FLAG-CEP192 was immunoprecipitated using anti-FLAG M2 agarose beads (Sigma; A2220) for 4 h at 4°C. Immunoprecipitated complexes were eluted in NH4OH and digested with 1 μg of trypsin (Sigma, T7575) at 37°C overnight.21 Samples were loaded onto capillary columns packed with Magic C18AQ 100 A 5 μm (Michrom Bioresource; 00015922). MS/MS data were acquired in data-dependent mode (over a 2 h acetonitrile 2–40% gradient) on a ThermoFinnigan LTQ, equipped with a Proxeon Nano Source and an Agilent 1100 capillary pump. Spectra were matched against the human and adenovirus complement of the RefSeqV53 database using the Mascot search engine (Matrix Science). Search parameters allowed for two missed tryptic cleavages as well as for asparagine deamidation and methionine oxidation. The fragment mass tolerance was 0.6 Da (monoisotopic mass), and the mass window for the precursor was set to 3 Da. All results were transferred into the Samuel Lunenfeld Research Institute relational database for interaction proteomics ProHits22 for additional filtering as defined in Supplementary Material.
Immunoprecipitation and western blot
For immunoprecipitation, we used FLAG-M2 agarose beads and goat anti-GFP antibody (kindly provided by D. Drechsel). For blotting we used the following antibodies: rabbit anti-FLAG (Sigma, F7425), mouse anti-GFP (Roche, 11814460001), rabbit anti-CEP192,5 rabbit anti-CYLD (Cell Signal, 4495) and mouse anti-α-tubulin (Sigma, T9026).
RNA silencing
EsiRNA (endoribonuclease-prepared siRNA) was generated as described.23 Targeted regions are encompassed by the primers: CEP192 (NM_032142): TTTTCAAGGGCTAGTATGTCTGA/GGATGTTATTCTGGGGTTCCT; CYLD (NM_015247.1): TGATGAAGATTGTGGCGTGT/ATGAACCTTTGTCCCCAACA; CEP85 (NM_022778): GGAGCTTTCAGTGCAAAACC/AATCCCCCAGAATTCC TCAC. Non-targeting esiRNA (AY_015988, luciferase): TGGTTTGGTTGTT GATGGAA/GTGCCTGGTGAAACTTGGTT. To confirm the specificity of the phenotype, siRNA SMARTpool targeting CYLD (LQ-004609, GAAGGTTGGAGAAACAATA, GGACATGGATAACCCTATT, AGAGATATCTACAGACTTT, GGAGAGTACTTGAAGATGT) and CEP85 (LQ-014214, GGGAGAAGCAGCAGCGUAU, CAGGAAUUGCAGCGAGAAA, GAGCAGAAAGUGCGAGAGA, UGGCAGAAGCGAUACGAUU) were purchased from Dharmacon. HeLa cells were transfected in 6-well plates with 0.4 μg of esiRNA, 80 pmol of siRNA and 4 μL of Lipofectamine RNAiMax (Invitrogen) for 72 h. In all experiments (Fig. 2; Fig. S2A–C), for single depletion of CEP192 or CYLD, we added non-targeting RNAi to make the total quantity equal to the conditions used for double CYLD+CEP192 depletion.
Microscopy and automatic quantification of signal intensities
Cells were fixed in cold methanol and stained using antibodies against α-tubulin (Serotec MCA77G), CEP1925 and NEDD1 (Abcam, ab57336). Three-dimensional images were acquired on a DeltaVision Core System (Applied Precision) equipped with an IX71 microscope (Olympus), a CCD camera (Roper Scientific; CoolSNAP HQ2 1024x1024) and 60x/1.42 NA objective (Olympus). Z stacks (0.4 μm apart) were collected, deconvolved using SoftWorx v4.0 (Applied Precision) and shown as maximum intensity projections. Automated analysis of fluorescence intensities was performed on 12-bit TIFF images using Acapella v2.18 (Perkin Elmer). Cellular and centrosomal masks were detected in the CEP192 channel using an adaptive threshold to outline cytosolic and centrosomal regions. Fluorescence intensity of CEP192 was analyzed using the detected masks.
Supplementary Material
Acknowledgments
We thank Christine Holley for esiRNA production. This work was funded by the Canadian Cancer Society (019562). M.G. was supported by Fundacion Caja Madrid (Spain).
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21574
References
- 1.Dix CI, Raff JW. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr Biol. 2007;17:1759–64. doi: 10.1016/j.cub.2007.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giansanti MG, Bucciarelli E, Bonaccorsi S, Gatti M. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr Biol. 2008;18:303–9. doi: 10.1016/j.cub.2008.01.058. [DOI] [PubMed] [Google Scholar]
- 3.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O’Connell KF. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell. 2004;6:511–23. doi: 10.1016/S1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- 4.Pelletier L, Ozlü N, Hannak E, Cowan C, Habermann B, Ruer M, et al. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr Biol. 2004;14:863–73. doi: 10.1016/j.cub.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Zhu F, Lawo S, Bird A, Pinchev D, Ralph A, Richter C, et al. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr Biol. 2008;18:136–41. doi: 10.1016/j.cub.2007.12.055. [DOI] [PubMed] [Google Scholar]
- 6.Gomez-Ferreria MA, Rath U, Buster DW, Chanda SK, Caldwell JS, Rines DR, et al. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr Biol. 2007;17:1960–6. doi: 10.1016/j.cub.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 7.Gomez-Ferreria MA, Sharp DJ. Cep192 and the generation of the mitotic spindle. Cell Cycle. 2008;7:1507–10. doi: 10.4161/cc.7.11.5957. [DOI] [PubMed] [Google Scholar]
- 8.Gomez-Ferreria M, Bashkurov M, Helbig A, Larsen B, Pawson T, Gingras AC, et al. Novel NEDD1 phosphorylation sites regulate γ-tubulin bindingand mitotic spindle assembly. J Cell Sci. 2012 doi: 10.1242/jcs.105130. [DOI] [PubMed] [Google Scholar]
- 9.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, et al. Flies without centrioles. Cell. 2006;125:1375–86. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 10.Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/S0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- 11.Mahoney NM, Goshima G, Douglass AD, Vale RD. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr Biol. 2006;16:564–9. doi: 10.1016/j.cub.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 12.Joukov V, De Nicolo A, Rodriguez A, Walter JC, Livingston DM. Centrosomal protein of 192 kDa (Cep192) promotes centrosome-driven spindle assembly by engaging in organelle-specific Aurora A activation. Proc Natl Acad Sci USA. 2010;107:21022–7. doi: 10.1073/pnas.1014664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jagger D, Collin G, Kelly J, Towers E, Nevill G, Longo-Guess C, et al. Alström Syndrome protein ALMS1 localizes to basal bodies of cochlear hair cells and regulates cilium-dependent planar cell polarity. Hum Mol Genet. 2011;20:466–81. doi: 10.1093/hmg/ddq493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otto EA, Hurd TW, Airik R, Chaki M, Zhou W, Stoetzel C, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010;42:840–50. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massoumi R. Ubiquitin chain cleavage: CYLD at work. Trends Biochem Sci. 2010;35:392–9. doi: 10.1016/j.tibs.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Lim JH, Jono H, Komatsu K, Woo CH, Lee J, Miyata M, et al. CYLD negatively regulates transforming growth factor-β-signalling via deubiquitinating Akt. Nat Commun. 2012;3:771. doi: 10.1038/ncomms1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stegmeier F, Sowa ME, Nalepa G, Gygi SP, Harper JW, Elledge SJ. The tumor suppressor CYLD regulates entry into mitosis. Proc Natl Acad Sci USA. 2007;104:8869–74. doi: 10.1073/pnas.0703268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J, Huo L, Sun X, Liu M, Li D, Dong JT, et al. The tumor suppressor CYLD regulates microtubule dynamics and plays a role in cell migration. J Biol Chem. 2008;283:8802–9. doi: 10.1074/jbc.M708470200. [DOI] [PubMed] [Google Scholar]
- 20.Wickström SA, Masoumi KC, Khochbin S, Fässler R, Massoumi R. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. EMBO J. 2010;29:131–44. doi: 10.1038/emboj.2009.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Hakim AK, Bashkurov M, Gingras AC, Durocher D, Pelletier L. Interaction Proteomics Identify NEURL4 and the HECT E3 Ligase HERC2 as Novel Modulators of Centrosome Architecture. Mol Cell Proteomics. 2012;11:M111–, 014233. doi: 10.1074/mcp.M111.014233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu G, Zhang J, Larsen B, Stark C, Breitkreutz A, Lin ZY, et al. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat Biotechnol. 2010;28:1015–7. doi: 10.1038/nbt1010-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kittler R, Pelletier L, Ma C, Poser I, Fischer S, Hyman AA, et al. RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells. Proc Natl Acad Sci USA. 2005;102:2396–401. doi: 10.1073/pnas.0409861102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.