Abstract
Orosomucoid like 3 (ORMDL3) has been strongly linked with asthma in genetic association studies, but its function in asthma is unknown. We demonstrate that in mice ORMDL3 is an allergen and cytokine (IL-4 or IL-13) inducible endoplasmic reticulum (ER) gene expressed predominantly in airway epithelial cells. Allergen challenge induces a 127-fold increase in ORMDL3 mRNA in bronchial epithelium in WT mice, with lesser 15-fold increases in ORMDL-2 and no changes in ORMDL-1. Studies of STAT-6–deficient mice demonstrated that ORMDL3 mRNA induction highly depends on STAT-6. Transfection of ORMDL3 in human bronchial epithelial cells in vitro induced expression of metalloproteases (MMP-9, ADAM-8), CC chemokines (CCL-20), CXC chemokines (IL-8, CXCL-10, CXCL-11), oligoadenylate synthetases (OAS) genes, and selectively activated activating transcription factor 6 (ATF6), an unfolded protein response (UPR) pathway transcription factor. siRNA knockdown of ATF-6α in lung epithelial cells inhibited expression of SERCA2b, which has been implicated in airway remodeling in asthma. In addition, transfection of ORMDL3 in lung epithelial cells activated ATF6α and induced SERCA2b. These studies provide evidence of the inducible nature of ORMDL3 ER expression in particular in bronchial epithelial cells and suggest an ER UPR pathway through which ORMDL3 may be linked to asthma.
Keywords: macrophage, eosinophil
Moffatt et al. (1) were the first to report in 2007 a genome-wide association study (GWAS) of asthma, which demonstrated that multiple markers on chromosome 17q21 (linked to the gene orosomucoid like 3; ORMDL3) were strongly and reproducibly associated with asthma in three different asthma cohorts. The identification of ORMDL3 as an asthma-associated gene has been confirmed in additional GWAS studies and in genetic association studies in populations of diverse ethnic backgrounds (2–4). In addition to the association of ORMDL3 with asthma, it is also associated with childhood onset of asthma (1) and exposure of children with asthma to environmental tobacco smoke (2). At present the function of ORMDL3 in the lung and in asthma is unknown.
ORMDL3 is one of the three-member ORDML gene family (ORMDL-1, ORMDL-2, and ORMDL3), which encode transmembrane proteins located at the endoplasmic reticulum (ER) (5). ORMDL-1 (chromosome 2) (5) and ORMDL-2 (chromosome 12) (5) are on different chromosomes from ORMDL3 (chromosome 17) (5) and have not been linked to asthma. Both humans and mice express the same three ORMDL family members with ORMDL3 exhibiting 96% identity between mouse and man (5). ORMDL3 is a 153-aa protein with two predicted transmembrane domains (5). In limited studies of ORMDL3 expression in normal human fetal and adult tissue, mRNA transcripts of ORMDL3 have been detected in lung, and in several other organs (liver, pancreas, and kidney), but not skeletal muscle, heart, or brain as assessed by nonquantitative RT-PCR (1, 5). In yeast, double knockouts of two ORM genes (ORM-1 and ORM-2 proteins, the yeast equivalent of ORMDL-1 and ORMDL-2) results in slower growth of the ORM mutant yeast and sensitivity to toxic compounds that can be rescued by transfection of human ORMDL3 (5). At present there are no studies in lung cells to provide insight into the pathways regulated by ORMDL3 that could influence lung function, but recent studies in yeast (Saccharomyces cerevisiae) have identified that ORM1/2 proteins in the ER are negative regulators of sphingolipid synthesis (6).
At present no studies have reported whether lung cells express ORMDL3 and what pathways it might regulate in the lung after allergen exposure. We have made the unique observation that ORMDL3 is an allergen (and Th2 cytokine) inducible STAT-6–dependent gene expressed in the lung, in particular in airway epithelial cells. In addition, we demonstrate that transfection of ORMDL3 into normal human lung bronchial epithelial cells induces expression of genes with potential importance to the pathogenesis of asthma including metalloproteases (MMP-9 and ADAM-8) (7–11), CC chemokines (CCL-20 also known as MIP-3α) (12–15), CXC chemokines (IL-8 and CXCL-10) (16–21), and oligoadenylate synthetases (OAS) anti-viral regulated genes not previously implicated in asthma. Finally, we demonstrate that ORMDL3 transfection in lung epithelial cells activates the ATF6 pathway, one of three branches of the ER localized unfolded protein response (22, 23), whereas siRNA knockdown of ATF-6 in lung epithelial cells inhibited expression of SERCA2b (sarco/endoplasmic reticulum Ca2+ ATPase), which has been implicated in remodeling in asthma (24). The ORMDL3 activation of ATF6 (with induction of SERCA2b), and the ORMDL3 induction of metalloproteases and chemokines, provides a mechanism to link an ER localized protein such as ORMDL3 to the pathogenesis of asthma.
Results
ORMDL3 Is an Allergen Inducible Gene in Mouse Airways in Vivo.
Although ORMDL3 mRNA is constitutively expressed in the lungs of WT mice at baseline, there is a significant induction of ORMDL3 mRNA expression in the lung after OVA allergen challenge as assessed by quantitative PCR (qPCR) (Fig. 1A). The increased ORMDL3 mRNA expression we detected could be due to either increased numbers of cells expressing the same amount of ORMDL3 mRNA recruited to the allergen-challenged lung and/or up-regulation of ORMDL3 mRNA expression in individual cell types. To address this question, we performed in vitro studies with a lung epithelial cell line (A549) (Fig. 1B) and a mouse macrophage cell line (RAW 264.7) (Fig. 1C), which demonstrated that levels of ORMDL3 can be up-regulated in A549 epithelial cells (incubated with tobacco smoke containing media, because exposure to tobacco smoke has been linked to ORMDL3 and asthma) (2), and RAW 264.7 macrophages (stimulated with LPS) as assessed by qPCR.
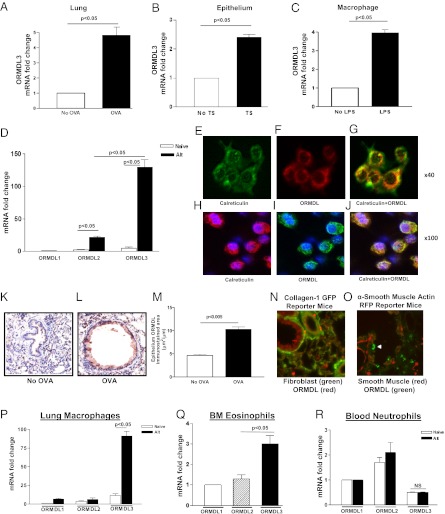
Fig. 1.
Allergen challenge induces ORMDL3 expression in epithelium in vitro and in vivo. (A) WT mice were challenged with OVA allergen by inhalation. RNA was extracted from lungs. qPCR for ORMDL3 was normalized to the housekeeping gene GAPDH. No OVA vs. OVA (P < 0.05). A549 lung epithelial cells incubated with tobacco smoke media (TS) (B) or mouse macrophage cell line RAW 264.7 (C) incubated with LPS for 3 h. RNA was extracted. qPCR for ORMDL3 was normalized to GAPDH. P < 0.05 for No TS vs. TS and No LPS vs. LPS. (D) WT mice were challenged with Alternaria allergen by inhalation. RNA was extracted from bronchial brushing derived epithelial cells. qPCR for ORMDL-1, ORMDL-2, and ORMDL3 was normalized to the housekeeping gene GAPDH. Alternaria allergen challenge induced a 127-fold increase in levels of expression of ORMDL3 mRNA (P < 0.01 Alt vs. naïve). Levels of ORMDL-1 did not change, whereas levels of ORMDL-2 increased (P < 0.05 Alt vs. naïve), but less than that noted with ORMDL3 (Alt ORMDL3 vs. Alt ORMDL-2; P < 0.05). Results are from three separate experiments with four mice per group. Double-label immunofluorescence microscopy (E–J) and double-label confocal immunofluorescence microscopy (H–J) was performed on human lung epithelial cells (A549) with an anti-ORMDL Ab and an anti-calreticulin Ab (detects ER). (K) Lung sections from non-OVA challenged WT mice have low levels of ORMDL immunostaining. In contrast, lung sections from OVA challenged WT mice (L) have increased ORMDL immunostaining particularly in airway epithelium and in peribronchial mononuclear cells as quantified (M) by light microscopy and image analysis (P < 0.005 vs. no OVA). Lungs from collagen 1 GFP reporter mice (N) have fibroblasts that fluoresce green but do not colocalize with the anti-ORMDL Ab (red). (O) Lungs from α-smooth muscle actin RFP reporter mice have smooth muscle that fluoresce red but does not colocalize with the anti-ORMDL Ab (green; white arrowheads points to green ORMDL+ cell, likely macrophage). RNA was extracted from either (P) BAL macrophages, (Q) bone marrow derived eosinophils, or (R) peripheral blood neutrophils. qPCR for ORMDL1, 2, 3 was normalized to the housekeeping gene GAPDH. ORMDL3 is significantly induced in BAL macrophages (P < 0.05, No Alternaria vs. Alternaria) (P), but not in neutrophils (R). Bone marrow eosinophils (Q) express higher levels of ORMDL3 compared to ORMDL2 or ORMDL1 (P < 0.05). qPCR results are from two separate experiments with four mice per group.
WT Mouse Bronchial Epithelial Cells Up-Regulate ORMDL3 mRNA Expression in Vivo After Allergen Challenge.
Because our in vitro studies demonstrated that ORMDL3 mRNA could be regulated in A549 lung epithelial cells in vitro, we examined whether in vivo primary bronchial epithelial cells derived from WT mice challenged with allergen-expressed ORMDL3 mRNA as assessed by qPCR. To obtain purified populations of bronchial epithelial cells, we used a bronchial brushing technique previously described in this laboratory that results in a >95% pure population of epithelial cells as assessed by expression of the epithelial specific gene E-cadherin (25). These studies demonstrated that there was a significant 127-fold induction of ORMDL3 mRNA in bronchial epithelial cells in vivo in WT mice challenged with Alternaria allergen (Fig. 1D). In contrast to the significant induction of ORMDL3 mRNA in bronchial epithelial cells by allergen challenge, ORMDL-1 mRNA was not induced in bronchial epithelial cells, and the induction of ORMDL-2 mRNA was ∼15% that of ORMDL3 mRNA (Fig. 1D). Thus, ORMDL3 mRNA is the predominant ORMDL family member induced in bronchial epithelium after allergen challenge.
WT Mouse Bronchial Epithelial Cells Up-Regulate ORMDL Protein Expression in Vivo After Allergen Challenge.
To determine which lung cells express ORMDL proteins, we generated a rabbit polyclonal Ab that detects ORMDL3 of expected molecular weight on Western blot (Fig. S1). To determine whether this anti-ORMDL Ab detected all three closely related ORMDL family members (ORMDL-1, ORMDL-2, and ORMDL3), we used HEK 293 cells transfected with either ORMDL-1, -2, or -3 and performed Western blots with the respective cell lysates by using the polyclonal anti-ORMDL Ab we generated, and a second commercially obtained polyclonal anti-ORMDL3 Ab (Abgent). The anti-ORMDL Abs recognized all three ORMDL members equivalently (Fig. S1), and, therefore, cannot be used to distinguish expression of individual ORMDL family members that are >80% identical at the amino acid level (5). Preincubating the anti-ORMDL Ab with an ORMDL3 peptide before performing the Western blot inhibited detection of ORMDL3 and ORMDL-1, or ORMDL-2 (Fig. S1). We therefore refer to these Abs as anti-ORMDL Abs in studies using these Abs.
We performed double-label immunofluorescence microscopy with an anti-calreticulin Ab (an ER marker), which demonstrated that ORMDL was colocalized with calreticulin in the ER in lung A549 epithelial cells (Fig. 1 E–G). Reconstruction of 3D images of confocal microscopy confirmed colocalization of ORMDL and calreticulin in the ER (Fig. 1 H–J).
Although the anti-ORMDL Ab was not specific for individual ORMDL family members, when used in immunohistochemistry experiments, it demonstrated that not all lung cells expressed equivalent levels of ORMDL proteins. For example, airway epithelial cells were the predominant cell type in the lung that expressed ORMDL after allergen challenge as assessed by IHC (Fig. 1 K–M). Non-OVA challenged WT mice expressed low levels of ORMDL in the lung as assessed by IHC (Fig. 1K). However, ORMDL was significantly up-regulated in airway epithelium in OVA-challenged WT mice (Fig. 1L). Levels of ORMDL immunostaining in airway epithelium quantitated by image analysis demonstrated significantly higher levels in OVA-challenged WT mice (P < 0.05 vs. non-OVA) (Fig. 1M). ORMDL was not detected in structural cells such as lung fibroblasts (collagen-1 GFP reporter mice) (Fig. 1N) or lung smooth muscle (alpha smooth muscle actin RFP reporter mice) (Fig. 1O).
To determine whether ORMDL expression was induced in lung cell types important to allergic inflammation, we performed FACS analysis on single-cell suspensions of lung cells (gated to detect either epithelium, macrophages, eosinophils, or neutrophils) before and after Alternaria challenge. These FACS studies demonstrated that all these cell types expressed baseline ORMDL (Fig. S2). However, inducible ORMDL was detected by FACS in lung epithelium (Fig. S2A), lung macrophages (Fig. S2B), and lung eosinophils (Fig. S2C), but not lung neutrophils (Fig. S2D). Because inducible ORMDL was detected in epithelium, macrophages, and eosinophils, but not neutrophils, we examined levels of ORMDL family member mRNA expression in purified populations of these cell types. ORMDL3 mRNA was the predominant ORMDL family member induced in bronchial epithelium (Fig. 1D) and BAL macrophages (Fig. 1P). In contrast, the predominant ORMDL family member expressed by peripheral blood neutrophils was ORMDL-2 (Fig. 1R), whereas in the case of bone marrow-derived eosinophils, ORMDL3 was predominant (Fig. 1Q).
Th2 Cytokines IL-4 and IL-13 Induce Expression of ORMDL3 mRNA in Bronchial Epithelium in Vivo.
Because allergen challenge induced expression of ORMDL3 mRNA in bronchial epithelium (Fig. 1D), we examined whether administration of individual cytokines known to be expressed after allergen challenge (IL-4, IL-13, TNF-α), and known to have receptors on airway epithelium, could induce expression of ORMDL3. These studies demonstrated that in vivo administration of either IL-4 or IL-13 intranasally could induce significant expression of ORMDL3 mRNA in WT bronchial epithelium as assessed by qPCR (Fig. S3A). In contrast, administration of TNF-α did not induce expression of ORMDL3 (Fig. S3A). Studies using IHC demonstrated that IL-4, and IL-13, but not TNF-α, induced significant epithelial ORMDL expression in WT mice (Fig. S3B). IL-4, IL-13, and TNF-α did not induce ORMDL-1 or ORMDL-2 mRNA (Fig. S3A).
Allergen Induction of ORMDL3 in Airway Epithelium Depends on STAT-6 and Is Independent of NF-κB.
Because IL-4 and IL-13 signal through the transcription factor STAT-6 (25), whereas TNF signals through NF-κB (26), we examined whether allergen induction of ORMDL3 in airway epithelium depended on STAT6 rather than NF-κB. In studies of bronchial epithelial cells derived by bronchial brushing, levels of ORMDL3 mRNA expression were significantly reduced in STAT6-deficient mice compared with WT mice challenged with Alternaria (Fig. S3C). Levels of ORMDL-1 were not increased in WT or STAT6-deficient mice, whereas the low levels of ORMDL-2 induced in WT mice depended on STAT6 for induction (Fig. S3C).
We also used IHC to examine ORMDL expression in STAT-6 bone marrow chimeric mice (25) (Fig. S3 D–G). WT (Fig. S3D) and STAT-6 chimeric mice (with STAT-6 expressed in airway epithelium but not in bone marrow cells) (Fig. S3E) had significant increased levels of ORMDL expression in airway epithelium after allergen challenge. In contrast, STAT-6 chimeric mice (with STAT-6 expressed in bone marrow cells, but not expressed in airway epithelium) had significantly reduced ORMDL expression in airway epithelium after allergen challenge (Fig. S3 F and G). Studies in CC10-Cretg /IkkβΔ/Δ mice in which NF-κB signaling through IκB-kinase-β (IKK-β) is selective ablated in airway epithelial cells (26) demonstrated that CC10-Cretg/IkkβΔ/Δ and WT mice expressed similar levels of ORMDL as assessed by IHC after allergen challenge (Fig. S3 H–J), indicating that inducible ORMDL in airway epithelium was not regulated by NF-κB.
To determine whether STAT-6 directly or indirectly regulates ORMDL3 expression, we analyzed the predicted promoter region of ORMDL3 to identify potential STAT6 transcription factor binding sites immediately 5′ of the transcriptional start site by using the published ORMDL3 sequence (27) in conjunction with TFSEARCH (www.cbrc.jp/research/db/TFSEARCH.html) and Genomatix (www.genomatix.de/en/index.html), but we did not identify a STAT6 binding site. We extended this bioinformatic analyses on up to 4,000 bp 5′ of the transcriptional start site by using these programs as well as TESS (www.cbil.upenn.edu/cgi-bin/tess/tess) and the University of California, Santa Cruz genome browser. Although a large number of binding sites were identified consistent with potential regulatory sequence, we did not identify any STAT6 binding sites in this sequence. These studies suggest that STAT6 is not directly regulating ORMDL3 expression, but rather influencing ORMDL3 expression indirectly. STAT6 is known to regulate the expression of at least 452 known genes (28), one or more of which could potentially be regulating ORMDL3 expression.
Transfection of ORMDL3 in Primary Normal Human Bronchial Epithelial Cells in Vitro Induces Metalloproteases, Chemokines, and OAS Genes.
Because our in vivo studies demonstrated that ORMDL3 was an inducible gene in airway epithelium after allergen or Th2 cytokine challenge, we examined whether ORMDL3 regulated expression of metalloproteases, chemokines, and OAS antiviral genes important to allergic inflammation, remodeling, and antiviral responses expressed by primary normal human bronchial epithelial cells. Primary normal human bronchial epithelial cells were efficiently transfected with either a full-length ORMDL3 cDNA containing a terminal GFP tag, or as a negative control the empty vector containing the GFP tag. The transfection efficiency was ∼72% (Fig. 2A).
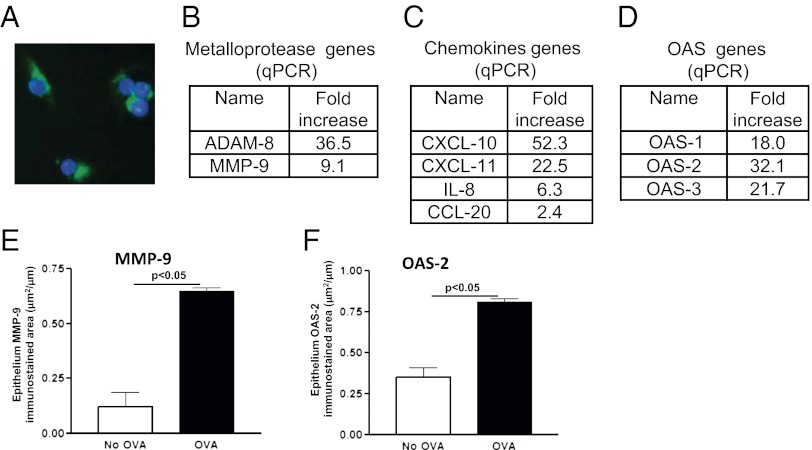
Fig. 2.
ORMDL3 transfection into lung epithelium regulates expression of metalloproteases, chemokines, and OAS genes. (A) Lung epithelial cells (A549) transfected with GFP tagged full-length ORMDL3 (green color; blue nuclei). (B–D) qPCR was performed on primary human bronchial epithelial cells transfected with either full-length ORMDL3 or with the empty vector control. The genes that were highly induced by ORMDL3 transfection include metalloproteases (B), chemokines (C), and OAS (D) genes. Immunohistochemistry was used to demonstrate increased MMP-9 (E) and OAS-2 (F) expression in airway epithelium in vivo in WT mice challenged with OVA.
Using qPCR, we observed that the primary normal human bronchial epithelial cells transfected with ORMDL3 but not the empty vector control expressed a gene signature characterized by high levels of expression of metalloproteases (MMP-9, ADAM-8), CC chemokines (CCL-20), CXC chemokines (IL-8, CXCL-10, CXCL-11), and OAS regulated genes (Fig. 2 B–D). In addition, we demonstrated by IHC that several of these ORMDL3 regulated genes were expressed in WT mice after allergen challenge (Fig. 2 E and F).
Transfection of ORMDL3 in A549 Epithelial Cells in Vitro Activates ATF6, a Signaling Branch of the ER Localized Unfolded Protein Response.
Because ORMDL3 is an ER protein, we examined whether transfection of ORMDL3 into human airway epithelial cells (A549 cells) activated ATF6, a signaling branch of the ER localized unfolded protein response, whose target genes in mice include SERCA-2 (23), a gene implicated in airway remodeling in asthma (24). In addition, ATF6 regulates expression of IL-6 (23), a cytokine we detected at increased levels in human subjects with asthma (29) and which has also been implicated in mouse models of asthma (30).
ORMDL3 transfection into A549 airway epithelial cells activated ATF6 as assessed by immunoflurescence microscopy detection of increased nuclear localized ATF6 (Fig. 3A). Furthermore, activity of the nuclear localized ATF6 was detected by a unfolded protein response (UPR) reporter assay containing the ER stress element (ERSE) derived from the GRP78/BiP promoter fused to luciferase (Fig. 3B). Transfection of ORMDL3 selectively activated ATF6 but not the PERK or Ire1 pathways of the UPR (Fig. 3C). Neither transfection of control empty vector (Fig. 3 A–C), nor transfection of another ER resident gene Sec61α-GFP into A549 lung epithelial cells (Fig. S4), activated any of the three UPR branches.
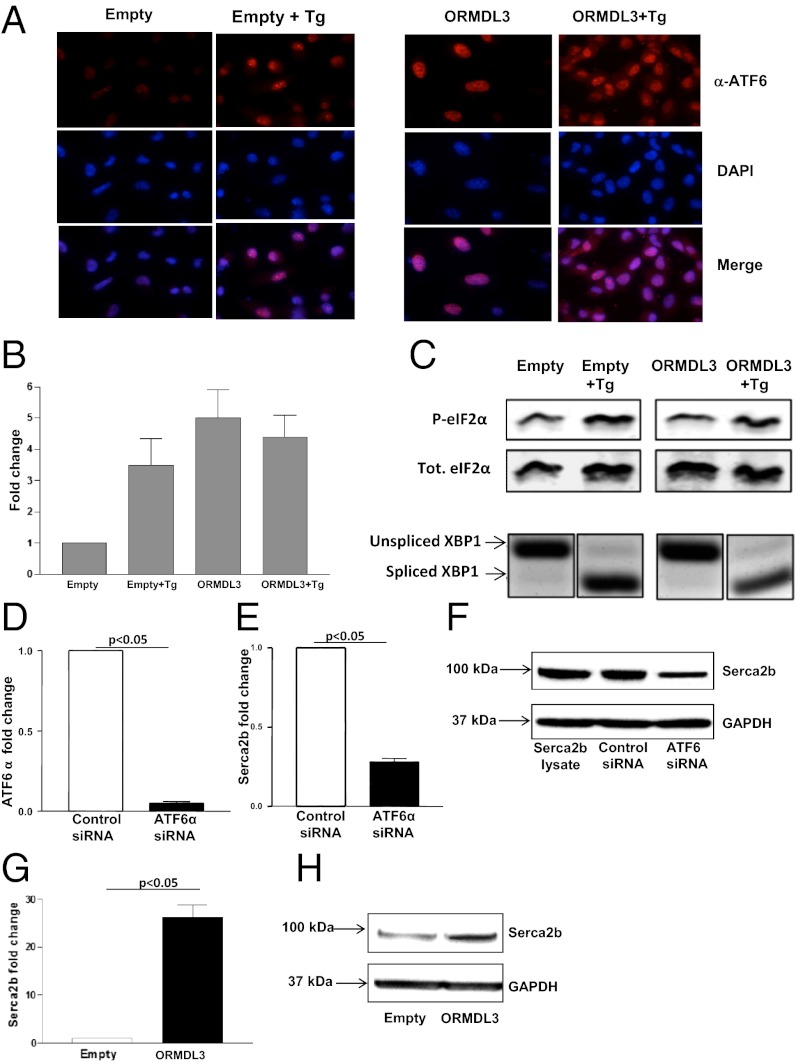
Fig. 3.
Activation of ATF6 branch of UPR in lung epithelial cells transfected with ORMDL3. Lung epithelial cells (A549) were transfected with either full-length ORMDL3 or empty vector and treated with 200 nM thapsigargin (Tg), a known activator of the UPR, for 1 h. (A) Immunofluorescence against ATF6 (red) was performed. Active ATF6 is shown by nuclear localization as depicted by colocalization with DAPI (blue). (B) Lung epithelial cells (A549) were cotransfected with a reporter containing an ATF6 binding site, ERSE fused to a luciferase gene, and either empty vector or full-length ORMDL3 and treated with Tg for 12 h. Luciferase activity was then assayed, indicating ATF6 activity (n = 2 experiments). (C) Activation of PERK is assessed by levels of phosphorylated eIF2α on SDS/PAGE by Western blot. Blot with total elF2α showed overall protein level was not changed. Activation of Ire1 was detected by PCR because it removes the UPR intron from the unspliced form of XBP1 (XBP1u) to generate the spliced form of XBP1 (XBP1s) mRNA. (D–F) A549 lung epithelial cells were transfected with ATF6α or control (scrambeled) siRNA. ATF6α siRNA inhibited ATF6α mRNA expression (D) and SERCA2b expression as assessed by qPCR (E), and Western blot (F). (G and H) A549 lung epithelial cells were transfected with ORMDL3 or control empty vector. ORMDL3 transfection induced SERCA2b expression as assessed by qPCR (G) and Western blot (H).
siRNA Knockdown of ATF-6α in A549 Human Lung Epithelial Cells Inhibits Expression of SERCA2b.
Because our in vitro studies demonstrated that ORMDL3 activated the ATF6α pathway, which is known to activate SERCA-2 in mice (23), we examined whether siRNA knockdown of ATF-6α in human lung epithelial cells (A549) inhibited expression of SERCA2b. These studies demonstrated that siRNA knockdown of ATF-6α was efficient in A549 lung cells (Fig. 3D), and that this knockdown resulted in significant inhibition of SERCA2b expression as assessed by qPCR (Fig. 3E) and Western blot (Fig. 3F).
Transfection of ORMDL3 in A549 Epithelial Cells Induces Expression of SERCA2b.
Transfection of ORMDL3 in A549 epithelial cells induced expression of SERCA2b as assessed by qPCR (Fig. 3G) and Western blot (Fig. 3H).
Discussion
In this study we demonstrate that ORMDL3 is an inducible bronchial epithelial gene regulating pathways of potential importance to asthma including expression of metalloproteases (MMP-9, ADAM-8), CC chemokines (CCL-20), CXC chemokines (IL-8, CXCL-10, CXCL-11), OAS genes, and SERCA2b a gene implicated in airway remodeling in asthma (24). Induction of ORMDL3 in lung epithelium depends on Th2 cytokines, STAT-6, and selectively activates ATF6, an UPR pathway transcription factor. The link between lung epithelial ORMDL3, ATF-6, and SERCA2b expression was demonstrated in studies in which transfection of ORMDL3–induced activation of ATF6 and expression of SERCA2b, whereas knockdown of ATF6α inhibited SERCA2b expression. These studies provide an ER localized mechanism for ORMDL3 to interact with ATF6 to modulate levels of SERCA2b in lung epithelium. These studies provide evidence of the Th2 cytokine inducible nature of ORMDL3 ER expression in bronchial epithelial cells in the lung and suggest ATF-6α–dependent (Serca 2b) and ATF-6α–independent pathways (metalloproteases, chemokines, OAS) through which ER localized ORMDL3 may be linked to asthma.
Transfection of ORMDL3 into primary human lung bronchial epithelial cells induced increased expression of genes with potential importance to asthma including metalloproteases (MMP-9, ADAM-8) (7–11), CC chemokines (CCL-20) (12–15), and CXC chemokines (IL-8, CXC-10) (16–21), suggesting potential downstream pathways from ORMDL3 pertinent to the pathogenesis of asthma. The importance of these ORMDL3–regulated genes to asthma is suggested from studies demonstrating increased expression of many of these mediators in the airways of human asthmatics (MMP-9, ADAM-8, CCL-20, CXCL-10, IL-8) (7–17), induction of these mediators by allergen inhalation in asthmatics (MMP-9, CCL-20, CXCL-10) (7, 12, 19), and inhibition of asthma outcomes in mice deficient in these genes (MMP-9, ADAM-8, CXCL-10) (8, 9, 11, 20, 21). In addition to the important role of CCL-20 on T-cell recruitment (12), and epithelial mucus production (14), in mouse models of asthma, there is a critical link between CCL-20 and TRAIL for Th2 cell activation and allergic airway inflammation (15). Although asthma has predominantly been associated with expression of CC chemokines, both CC and CXC chemokines have been linked to asthma in studies in humans with asthma (12, 13, 16–19), and in studies in animal models (13, 20, 21) (i.e., CXCL-10 KO have significant reduction in Th2-type allergic airway inflammation and AHR) (20).
In addition to identifying genes linked to asthma, the transfection of ORMDL3 into human bronchial epithelial cells also induced expression of OAS genes (31), a pathway not previously linked to ORMDL3. OAS proteins are pattern recognition receptors for the viral pattern associated molecular pattern, double-stranded RNA. In humans, the OAS gene family has three enzymatically active copies, OAS-1, OAS-2, and OAS-3 (31), all of which were significantly up-regulated in human lung epithelial cell transfected with ORMDL3. OAS catalyze the polymerization of ATP into 2′-5′-linked oligoadenylates, which activate a constitutively expressed latent endonuclease, RNaseL, to block viral replication at the level of mRNA degradation. OAS-1 is up-regulated by a wide range of viruses and has been shown to play an antiviral role during viral infections including in studies with the asthma associated virus RSV (32). At present it is not known how up-regulation of the OAS pathway by ORMDL3 may influence asthma outcomes. One possibility is that the induction of the OAS pathway may be a protective antiviral pathway in asthma inhibiting viral triggered exacerbations. Alternatively, OAS through up-regulation of IFN pathways may bias immune responses against the development of Th2 responses.
Because we demonstrated that ORMDL3 is an ER-inducible protein, we also examined whether ORMDL3 influenced the function of the ER. In addition to influencing protein folding, the ER responds to the accumulation of unfolded proteins in its lumen or disruption of ER functional capacity (collectively termed ER stress) by activating intracellular signal transduction pathways called the UPR (22). Together at least three mechanistically distinct arms of the UPR (IRE1, PERK, ATF6) regulate the expression of numerous genes that function within the ER secretory pathway but also affect broad aspects of cell fate and metabolism of proteins, amino acids, and lipids (22). In this study, we have made the observation that the ATF6 pathway of the UPR is selectively activated by ORMDL3. ATF6 (consisting of the closely related ATF6α and ATF6β in mammals) (23) is a transcription factor known to regulate genes involved in protein folding (22). Interestingly, our studies demonstrate that ATF6α regulates expression of SERCA2b, which has been implicated in airway remodeling in asthma (24). In addition, in mice ATF6 regulates expression of IL-6, a cytokine we detected at increased levels in human subjects with asthma (29) and which has also been implicated in mouse models of asthma (30). Prior studies of ORMDL3 in ER signaling using HEK 293 cells (human embryonic kidney cells transformed with adenovirus) have investigated the role of ER pathways (IRE1, PERK) other than ATF6 and suggested that the main impact of ORMDL3 is on the PERK/eIF2α pathway (33). These studies also demonstrated that ORMDL3 interacted with SERCA-2 and inhibited its function (33). Our results may differ from these studies because we examined the ER ATF6 pathway (not examined in prior study) (33) and used cell types pertinent to ORMDL3 expression in asthma (airway epithelium rather than transformed embryonic kidney fibroblasts used in prior study).
One of the limitations of this study is the inability of the two anti-ORMDL Abs used for FACS and IHC to distinguish between ORMDL3 and the closely related ORMDL-1 and ORMDL-2, which share significant amino acid identity (5). Nevertheless, the FACS and IHC studies using these Abs demonstrate that ORMDL is induced in WT mice bronchial epithelium after allergen challenge. When the results of the FACS and IHC studies of ORMDL expression are coupled with results examining ORMDL-1, ORMDL-2, and ORMDL3 mRNA expression in these cell types, it strongly suggests that allergen challenge is predominantly inducing ORMDL3. For example, allergen challenge in WT mice resulted in a 127-fold induction of ORMDL3 mRNA by bronchial epithelial cells in vivo, with no induction of ORMDL-1, and only minimal induction of ORMDL-2 mRNA. In addition to lung epithelial cells, lung macrophages and lung eosinophils expressed both constitutive and inducible ORMDL. In contrast, lung neutrophils expressed constitutive ORMDL, which was not inducible with allergen challenge. Based on qPCR studies, ORMDL-2 was the predominant family member expressed by neutrophils.
Studies with STAT6-deficient mice demonstrated that the induction of ORMDL3 mRNA in airway epithelial cells in vivo depended on STAT6. Interpreting our STAT6 chimera studies in conjunction with the available ORMDL3 promoter bioinformatic database suggest that STAT6 is not directly regulating ORMDL3 expression (no STAT6-binding sites identified in promoter region of ORMDL3) but rather that a resident lung cell expresses a STAT6-dependent mediator that is regulating ORMDL3 expression. Potential candidate lung cells that express STAT6 and may generate a mediator regulating ORMDL3 include epithelial and/or non-epithelial cells (T cells, B cells, mast cells, macrophages, and natural helper cells). STAT6 is known to regulate the expression of at least 452 known genes (28), one or more of which could potentially be regulating ORMDL3 expression and will require further study.
In summary, these studies provide evidence of the allergen, Th2 cytokine, and STAT6-dependent inducible nature of ORMDL3 expression in airway epithelium in the lung. In addition, we demonstrated that ORMDL3 selectively regulates activation of the ER localized transcription factor ATF6α signaling branch, a pathway we have demonstrated in human lung epithelial cells is linked to SERCA-2b, which is implicated in airway remodeling in asthma (24). Finally, our studies demonstrate that ORMDL3 activates several metalloprotease, chemokine, and OAS-regulated genes, several of whom have also been implicated in the pathogenesis of asthma (7–24). Overall, these studies provide evidence for a mechanism to link an ER localized protein such as ORMDL3 to the pathogenesis of asthma.
Materials and Methods
Mouse Model of Allergen-Induced Asthma.
The mouse models of acute OVA, Alternaria, and Th2 cytokine administration induced eosinophilic airway inflammation in WT and mutant mice have been described and are detailed in the SI Materials and Methods.
Detection of ORMDL3 mRNA and ORMDL Protein.
The methods for qPCR, IHC, immunofluorescence microscopy, confocal microscopy, FACS, transfection, and siRNA knockdown have been described and are detailed in SI Materials and Methods.
UPR.
The methods for investigation of activation of ATF6, Ire1, or PERK UPR pathways have been described and are detailed in SI Materials and Methods.
Statistical Analysis.
All results are presented as mean ± SEM. A statistical software package (GraphPad Prism) was used for the analysis. P values of < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
Support was provided by National Institutes of Health Grants AI 38425, AI 70535, and AI 72115 (to D.H.B); GM087415; and American Cancer Society Grants RSG10-027-01 (to M.N.) and 1K08 AI080938 (to T.D.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204151109/-/DCSupplemental.
References
- 1.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 2.Bouzigon E, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–1994. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 3.Moffatt MF, et al. GABRIEL Consortium A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galanter J, et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177:1194–1200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hjelmqvist L, et al. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-6-research0027. RESEARCH0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breslow DK, et al. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly EA, Busse WW, Jarjour NN. Increased matrix metalloproteinase-9 in the airway after allergen challenge. Am J Respir Crit Care Med. 2000;162:1157–1161. doi: 10.1164/ajrccm.162.3.9908016. [DOI] [PubMed] [Google Scholar]
- 8.Lim DH, et al. Reduced peribronchial fibrosis in allergen-challenged MMP-9-deficient mice. Am J Physiol Lung Cell Mol Physiol. 2006;291:L265–L271. doi: 10.1152/ajplung.00305.2005. [DOI] [PubMed] [Google Scholar]
- 9.King NE, et al. Expression and regulation of a disintegrin and metalloproteinase (ADAM) 8 in experimental asthma. Am J Respir Cell Mol Biol. 2004;31:257–265. doi: 10.1165/rcmb.2004-0026OC. [DOI] [PubMed] [Google Scholar]
- 10.Foley SC, et al. Increased expression of ADAM33 and ADAM8 with disease progression in asthma. J Allergy Clin Immunol. 2007;119:863–871. doi: 10.1016/j.jaci.2006.12.665. [DOI] [PubMed] [Google Scholar]
- 11.Naus S, et al. The metalloprotease-disintegrin ADAM8 is essential for the development of experimental asthma. Am J Respir Crit Care Med. 2010;181:1318–1328. doi: 10.1164/rccm.200909-1396OC. [DOI] [PubMed] [Google Scholar]
- 12.Francis JN, Sabroe I, Lloyd CM, Durham SR, Till SJ. Elevated CCR6+ CD4+ T lymphocytes in tissue compared with blood and induction of CCL20 during the asthmatic late response. Clin Exp Immunol. 2008;152:440–447. doi: 10.1111/j.1365-2249.2008.03657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hastie AT, et al. National Heart, Lung, and Blood Institute Severe Asthma Research Program Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. J Allergy Clin Immunol. 2010;125:1028–1036, e13. doi: 10.1016/j.jaci.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S, Lewis C, Nadel JA. CCL20/CCR6 feedback exaggerates epidermal growth factor receptor-dependent MUC5AC mucin production in human airway epithelial (NCI-H292) cells. J Immunol. 2011;186:3392–3400. doi: 10.4049/jimmunol.1003377. [DOI] [PubMed] [Google Scholar]
- 15.Weckmann M, et al. Critical link between TRAIL and CCL20 for the activation of TH2 cells and the expression of allergic airway disease. Nat Med. 2007;13:1308–1315. doi: 10.1038/nm1660. [DOI] [PubMed] [Google Scholar]
- 16.Jatakanon A, et al. Neutrophilic inflammation in severe persistent asthma. Am J Respir Crit Care Med. 1999;160:1532–1539. doi: 10.1164/ajrccm.160.5.9806170. [DOI] [PubMed] [Google Scholar]
- 17.Norzila MZ, Fakes K, Henry RL, Simpson J, Gibson PG. Interleukin-8 secretion and neutrophil recruitment accompanies induced sputum eosinophil activation in children with acute asthma. Am J Respir Crit Care Med. 2000;161:769–774. doi: 10.1164/ajrccm.161.3.9809071. [DOI] [PubMed] [Google Scholar]
- 18.Ying S, et al. Expression and cellular provenance of thymic stromal lymphopoietin and chemokines in patients with severe asthma and chronic obstructive pulmonary disease. J Immunol. 2008;181:2790–2798. doi: 10.4049/jimmunol.181.4.2790. [DOI] [PubMed] [Google Scholar]
- 19.Bochner BS, Hudson SA, Xiao HQ, Liu MC. Release of both CCR4-active and CXCR3-active chemokines during human allergic pulmonary late-phase reactions. J Allergy Clin Immunol. 2003;112:930–934. doi: 10.1016/j.jaci.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Medoff BD, et al. IFN-gamma-inducible protein 10 (CXCL10) contributes to airway hyperreactivity and airway inflammation in a mouse model of asthma. J Immunol. 2002;168:5278–5286. doi: 10.4049/jimmunol.168.10.5278. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y, et al. Attenuation of antigen-induced airway hyperresponsiveness and inflammation in CXCR3 knockout mice. Respir Res. 2011;12:123. doi: 10.1186/1465-9921-12-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walter P, Ron D. The unfolded protein response: From stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 23.Adachi Y, et al. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- 24.Mahn K, et al. Diminished sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) expression contributes to airway remodelling in bronchial asthma. Proc Natl Acad Sci USA. 2009;106:10775–10780. doi: 10.1073/pnas.0902295106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doherty TA, et al. Alternaria induces STAT6-dependent acute airway eosinophilia and epithelial FIZZ1 expression that promotes airway fibrosis and epithelial thickness. J Immunol. 2012;188:2622–2629. doi: 10.4049/jimmunol.1101632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broide DH, et al. Allergen-induced peribronchial fibrosis and mucus production mediated by IkappaB kinase beta-dependent genes in airway epithelium. Proc Natl Acad Sci USA. 2005;102:17723–17728. doi: 10.1073/pnas.0509235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin R, et al. Mechanisms elevating ORMDL3 expression in recurrent wheeze patients: Role of Ets-1, p300 and CREB. Int J Biochem Cell Biol. 2012;44:1174–1183. doi: 10.1016/j.biocel.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Elo LL, et al. Genome-wide profiling of interleukin-4 and STAT6 transcription factor regulation of human Th2 cell programming. Immunity. 2010;32:852–862. doi: 10.1016/j.immuni.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Broide DH, et al. Cytokines in symptomatic asthma airways. J Allergy Clin Immunol. 1992;89:958–967. doi: 10.1016/0091-6749(92)90218-q. [DOI] [PubMed] [Google Scholar]
- 30.Kuhn C, 3rd, et al. Airway hyperresponsiveness and airway obstruction in transgenic mice. Morphologic correlates in mice overexpressing interleukin (IL)-11 and IL-6 in the lung. Am J Respir Cell Mol Biol. 2000;22:289–295. doi: 10.1165/ajrcmb.22.3.3690. [DOI] [PubMed] [Google Scholar]
- 31.Kristiansen H, Gad HH, Eskildsen-Larsen S, Despres P, Hartmann R. The oligoadenylate synthetase family: An ancient protein family with multiple antiviral activities. J Interferon Cytokine Res. 2011;31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- 32.Behera AK, Kumar M, Lockey RF, Mohapatra SS. 2′-5′ Oligoadenylate synthetase plays a critical role in interferon-gamma inhibition of respiratory syncytial virus infection of human epithelial cells. J Biol Chem. 2002;277:25601–25608. doi: 10.1074/jbc.M200211200. [DOI] [PubMed] [Google Scholar]
- 33.Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19:111–121. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.