Abstract
A group of candidate tumor suppressor genes (designated CACNA2D2, PL6, 101F6, NPRL2, BLU, RASSF1, FUS1, HYAL2, and HYAL1) has been identified in a 120-kb critical tumor homozygous deletion region (found in lung and breast cancers) of human chromosome 3p21.3. We studied the effects of six of these 3p21.3 genes (101F6, NPRL2, BLU, FUS1, HYAL2, and HYAL1) on tumor cell proliferation and apoptosis in human lung cancer cells by recombinant adenovirus-mediated gene transfer in vitro and in vivo. We found that forced expression of wild-type FUS1, 101F6, and NPRL2 genes significantly inhibited tumor cell growth by induction of apoptosis and alteration of cell cycle processes in 3p21.3 120-kb region-deficient (homozygous) H1299 and A549 cells but not in the 3p21.3 120-kb region-heterozygous H358 and the normal human bronchial epithelial cells. Intratumoral injection of Ad-101F6, Ad-FUS1, Ad-NPRL2, and Ad-HYAL2 vectors or systemic administration of protamine-complexed vectors significantly suppressed growth of H1299 and A549 tumor xenografts and inhibited A549 experimental lung metastases in nu/nu mice. Together, our results, coupled with other studies demonstrating a tumor suppressor role for the RASSSF1A isoform, suggest that multiple contiguous genes in the 3p21.3 120-kb chromosomal region may exhibit tumor suppressor activity in vitro and in vivo.
INTRODUCTION
Lung cancers develop after a multistage process involving a variety of genetic and epigenetic changes in dominant oncogenes and TSGs3 (1, 2). Several of these changes can be found in smoking damaged respiratory epithelium in preneoplastic lesions, normal appearing epithelium, and in persons even before lung cancer develops (3–6). In these and related studies, allelic loss of chromosome region 3p (particularly 3p21.3) was found to be a frequent and early event in the development of several cancers, including lung and breast cancers (4–9). Several 3p genes have been extensively studied and include FHIT at 3p14.2, RARB at 3p24, and VHL at 3p25 (summarize for lung cancer in Ref. 1). These results directed an intensive TSG search of the 3p21.3 region for one or more genes that could function as “gatekeepers” in the molecular pathogenesis of lung cancer, as well as several other human cancers.
Identification of nested 3p21.3 homozygous deletions in small cell lung cancers and a breast cancer line directed positional cloning efforts to a 630-kb region, which was narrowed subsequently to a 120-kb subregion by a breast cancer homozygous deletion (10, 11). This defined 3p21.3 region undergoes allele loss in ~80% of primary lung cancers and ~40% of preneoplastic or normal epithelial samples of smoking-damaged lung, marking it as one of the first sites to be involved (6). In addition, patients whose peripheral blood lymphocytes showed greater damage in this 3p21.3 region after in vitro treatment with the carcinogen benzo-a-pyrene diol epoxide had an increased risk of having lung cancer, suggesting the potential for genetic polymorphisms in this region predisposing to lung cancer development (12). The 630-kb region contains ≥25 genes, whereas 9 genes are located in or on the border of the 120-kb 3p21.3 region (10). This group of potential TSGs includes CACNA2D2 (GenBank no. AF040709), PL6 (U09584), 101F6 (AF040704), NPRL2 (AF040707), FUS1 (AF055479), BLU (U70880), RASSF1 (AF040703, RASSF1C and AF102770, RASSF1A), HYAL2 (U09577), and HYAL1 (U03056). The RASSF1A isoform of the RASSF1 gene has been studied extensively for promoter methylation in a variety of tumors, including lung and breast cancer, found to be frequently epigenetically inactivated in these tumors, and shows the ability to suppress lung cancer malignant growth (13, 14). The FUS1 gene was also found to suppress the growth of NSCLCs in vitro (15). However, there have been no detailed tests comparing the activity of several of the genes in this small region or in testing their effect on lung cancer xenografts (local tumors or metastases) in vivo. At the start of the search for a 3p21.3 TSG, everyone expected that one gene would be found that frequently suffered mutations. However, from detailed studies of the genes in the region, that was not the case (10). In addition, the possibility of haploinsufficiency needed to be considered. Thus, it was important to further define the tumor suppressing capability of these genes both in vitro and in vivo. Such identification, which is the focus of the current report, would target the gene(s) for development as new tools for the early detection, monitoring of prevention efforts, prognosis, and therapy of lung and other cancers.
In this study, we used recombinant adenoviruses to introduce WT 3p21.3 genes into NSCLC tumor cell lines or tumor xenografts, where 3p21.3 120-kb region genes are either heterozygous or homozygous to characterize their potential tumor suppressing function in vitro and in vivo. We demonstrate that introduction of individual WT 3p21.3 genes by recombinant adenoviral vector-mediated transfer into lung cancer cells with loss of heterozygosity at the3p21.3 120-kb region inhibited tumor cell growth and induced apoptosis in vitro. Moreover, intratumoral injection of recombinant adenoviral vectors containing WT 3p21.3 genes significantly suppressed growth of human NSCLC xenografts, whereas systemic administration of protamine-complexed adenoviral vectors of 3p21.3 genes efficiently inhibited development of experimental metastases of lung cancer cells in xenograft mouse models. Together, our results strongly suggest that multiple contiguous genes in the 3p21.3 chromosomal region may function as TSGs in vitro and in vivo.
MATERIALS AND METHODS
Cell Lines and Cell Culture
Four human NSCLC cell lines with varied 3p21.3 and p53 gene status, A549 (homozygous for multiple 3p21.3 region markers and WT p53), NCI-H1299 (homozygous for multiple 3p21.3 region markers and homozygous deletion of p53), NCI-H358 (retained heterozygosity of multiple 3p21.3 region markers and homozygous deletion of p53), and NCI-H460 (homozygous for multiple 3p21.3 region markers and WT p53), and a normal HBEC line, were used for in vitro and in vivo experiments. The multiple 3p21.3 polymorphic markers that were used for typing the lung cancer lines are located in the 630-kb homozygous deletion region in which the 120-kb region containing the six genes studied in this report reside and have been described previously (16). The homozygosity of multiple markers is consistent with loss of heterozygosity in this region. In this report, lung cancer cell lines with such homozygosity are referred to as “3p21.3-deficient” cells. The A549 line was maintained in Ham’s F12 medium supplemented with 10% FCS. The H1299, H358, and H460 lines were maintained in RPMI 1640 supplemented with 10% FCS and 5% glutamine. Normal HBECs were obtained from Clonetics, Inc. (Walkersville, MD) and cultured in the medium supplied by the manufacturer according to the instructions provided.
Construction of Recombinant Ad-3p21.3 Gene Vectors
The recombinant Ad-3ps were constructed using our recently developed ligation-mediated plasmid adenovirus vector construction system, named herein pAd-RAP and pAd-RAP-Shuttle (detailed structures of plasmids will be provided on request). The 3p21.3 genes were assembled as a mammalian expression cassette that is driven by a cytomegalovirus promoter and tailed with Bovine Growth Hormone poly(A) signal sequence. The resulting Ad-3p vectors were named Ad-101F6, Ad-NPRL2, Ad-BLU, Ad-RASSF1C, Ad-FUS1, Ad-HYAL1, and Ad-HYAL2. Sequences of 3p21.3 genes in the viral vectors were confirmed by automated DNA sequencing. A vector expressing GFP gene (Ad-GFP), and a vector carrying the β-galactosidase gene LacZ (Ad-LacZ), were used to monitor efficiency of transduction by the viral vectors and as nonspecific transgene expression controls. Ad-EV, an E1-deleted empty vector, was used as a negative control. Ad-p53, a vector containing the WT p53 gene, was used as a positive tumor suppressor control. Viral titers were determined by both absorbance measurement (i.e., vp/ml) and plaque assay (i.e., pfu/ml).
Cell Viability Assay
Inhibition of tumor cell growth by treatment with various Ad-3p vectors was analyzed by quantitatively determining cell viability using an improved 2,3-bis[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazoli-um-5-carboxanilide inner salt (XTT) assay (Roche Molecular Biochemicals, Indianapolis, IN; Ref. 17) as described previously (18). Percentage of cell viability was calculated in terms of the absorbency of treated cells relative to the absorbency of untreated control cells. Experiments were repeated at least three times with quadruplicate samples for each treatment in each individual experiment.
Analysis of Apoptosis and Cell Cycle Kinetics
Induction of apoptosis in tumor cells treated by various Ad-3p vectors was analyzed by flow cytometry (FACS) using TUNEL reaction with FITC-labeled dUTP (Roche Molecular Biochemicals, Mannheim, Germany). Cells were processed for FACS analysis for apoptosis and cell cycle kinetics as described previously (19).
Animal Studies
All animals were maintained, and animal experiments were performed under NIH and institutional guidelines established for the Animal Core Facility at the University of Texas M. D. Anderson Cancer Center. Procedures for A549 and H1299 s.c. tumor inoculations in nu/nu mice were described previously (19). When average tumor size reaches ~0.5 cm in diameter, mice were injected intratumorally three times within a week with various Ad-3p and control vectors at a dose of 3 × 1010 pfu (3 × 1012 vp)/tumor in a volume of 0.2 ml. Differences in tumor volumes between treatment groups were analyzed with a mixed model ANOVA using the Statistica software (StatSoft, Inc., Tulsa, OK). A difference was considered to be statistically significant when P < 0.05.
An experimental A549 lung metastasis model was used to study the effects of 3p21.3 genes on development of metastases. Briefly, nu/nu mice were inoculated with A549 cells (1–2 × 106) in 200 μl of PBS via tail vein injection. Pulmonary experimental metastatic tumor colonies were formed 7–10 days after inoculation. Then, protamine-complexed Ad-3p (P-Ad3p) vectors or control complexes were administered systemically to animals by i.v. injection for three times within a week at each a dose of 2–5 × 1010 vp/200–500 μg of protamine in a total volume of 200 μl/animal. The P-Ad complexes are prepared by mixing an equal volume of the adenoviral vector (1 × 1010 vp) and the protamine sulfate (100 μg; Fujisawa USA, Inc., Deerfield, IL) in room temperature for 15 min and then bringing it to a total volume of 200 μl with PBS. Two weeks after the last injection, the animals were euthanized, and their lung metastatic tumors were stained with India ink. Tumor colonies on lung surfaces were counted under a dissecting microscope without knowledge of the treatment groups, and the lung tissues were sectioned for further pathologic and immunohistochemical analysis.
RESULTS
Effects of Forced Expression of 3p Genes on Tumor Cell Growth
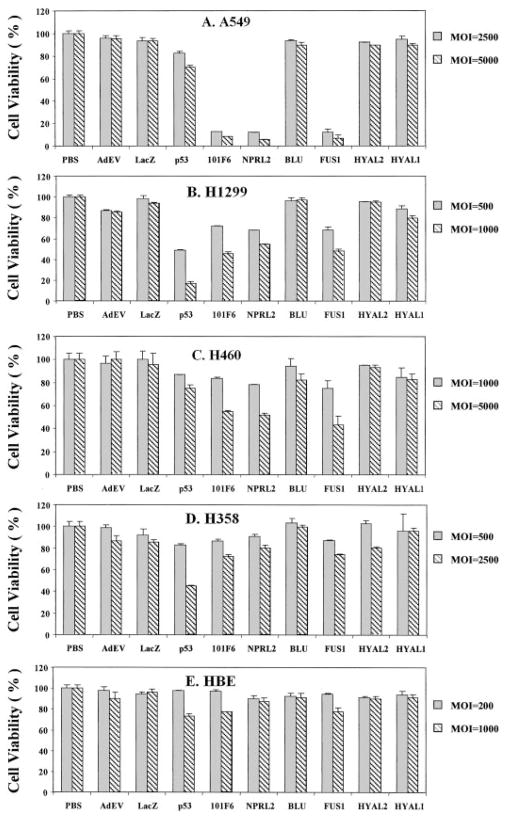
To test the hypothesis that one or more of the 3p genes function as tumor suppressors in vitro, we performed a series of experiments to study the effects of expression of the 3p21.3 genes on cell proliferation in several types of Ad-3p-transduced human NSCLC and normal HBEC cells. Cells from each line were transduced in vitro by Ad-101F6, Ad-FUS1, Ad-NPRL2, Ad-BLU, Ad-RASSF1C, Ad-HYAL2, and Ad-HYAL1 vectors at various MOIs in units of vp/c; cells were also treated with PBS, Ad-EV, Ad-LacZ, or Ad-p53 as mock, negative, nonspecific, or positive controls, respectively. The ratio of vp/ml:pfu/ml in our adenoviral preparations is ~100:1. The transduction efficiency was determined by examining GFP-expressing cells in the Ad-GFP-transduced cell population under a fluorescence microscope and was found to be >80% at the highest MOI applied for each cell line.
Cell proliferation was analyzed by using the XTT assay to determine the number of viable cells at 1, 2, 3, and 5 days after transduction (only data for day 5 at various MOIs are shown; Fig. 1). In all cases, the viability of transduced cells was compared with that of untransduced (PBS treated) control cells (whose viability was set at 100%). As can be seen in Fig. 1, cell viability was reduced significantly in Ad-101F6-, Ad-Fus1-, and Ad-NPRL2-transduced A549 and H460 cells, which show homozygosity for multiple 3p21.3 markers and contain WT p53, and H1299 cells, which exhibit 3p21.3 homozygosity but also have a homozygous deletion of p53. A modest reduction of cell viability was shown in Ad-RASSF1C-transduced H1299 cells (data not shown). However, no significant effect on growth was observed in any of these cells transduced with Ad-HYAL1, Ad-HYAL2, Ad-BLU, Ad-EV, or Ad-LacZ. These results suggest that exogenous expression of some but not all WT 3p21.3 genes can inhibit 3p-deficient tumor cell growth in vitro.
Fig. 1.
Effects of exogenous expression of 3p21.3 genes on tumor cell growth in Ad-3p-transduced human lung cancer cells and normal bronchial epithelial cells. Cells were transduced with adenoviral vectors of 3p21.3 genes 101F6, NPRL2, BLU, FUS1, HAYL2, and HYAL1, control genes LacZ and p53, and empty vector, Ad-EV, at various MOIs (vp/c) as indicated, and PBS alone was used as a mock control. The cell viability was expressed as the percentage of viable adenoviral vector-transduced cells in relation to PBS-treated control cells (100%). Bars, SDs of the mean in at least three individual experiments. Treatments were given in quadruplicate for each experiment. The significance of the difference in cell viability between vector-treated cells and the Ad-EV-, Ad-LacZ-, or PBS-treated controls was analyzed by two-sided Student’s t test. P < 0.05 was taken as significant. The differences between the cell viability of the Ad-EV- and Ad-LacZ-transduced cells versus PBS-treated controls were not significant (P = 0.25–0.95 from different time points and cell lines). The differences between the cell viability of the Ad-101F6-, Ad-Fus1-, and Ad-NPRL2-transduced cells versus the Ad-EV- and Ad-LacZ-transduced or PBS-treated controls at the same MOI were significant in A549, H1299, and in H460 at both 3 and 5 days post-transduction (P ≤ 0.0001–0.005) but not significant in H358 and HBEC cell lines at both 3 and 5 days post-transduction (P ≥ 0.10–0.95, from different time points and cell lines), respectively. The effects of Ad-BLU, Ad-HYAL2, and Ad-HYAL1 on cell viability were not significant in all cell lines (P > 0.45), compared with those of Ad-EV and Ad-LacZ.
To clarify the specificity of the observed inhibitory effects on tumor cell growth and examine the potential cytotoxicity of the exogenously expressed 3p21.3 genes on normal cells, we analyzed the effects of these 3p21.3 genes on cell proliferation in 3p21.3 heterozygous H358 cells and normal HBECs (Fig. 1). As shown in Fig. 1, HBECs transduced with all Ad-3p genes at MOIs that generated >80% transduction efficiency had reductions in cell number after 5 days of transduction of <10%, whereas H358 cells transduced with the same vectors had losses of <20% when compared with the untransduced control cells. Similar levels of losses of cell numbers were observed in H358 and HBEC cells transduced with control vectors Ad-EV and Ad-LacZ. As a positive control, H358 cells, which are homozygously deleted for p53, showed reduced cell numbers when transduced with the Ad-p53 vector. These results, coupled with the lack of effect with Ad-LacZ, Ad-HYAL2, Ad-HYAL1, Ad-RASSF1C, and Ad-BLU, demonstrate the specificity of the tumor suppressing function of 3p21.3 genes FUS1, NPRL2, and 101F6 in 3p-deficient tumor cells and indicate that no generalized cytotoxicity was associated with exogenous expression of these WT 3p21.3 120-kb region genes.
Expression of 3p21.3 genes in Ad-3p-transduced H1299 and normal HBEC cells was verified by quantitative real-time reverse transcription-PCR. Known concentrations of human Raji total RNA and a TaqMan probe for glyceraldehyde-3-phosphate dehydrogenase cDNA were used to make a standard curve as an internal control. The increase in levels of transcripts of exogenously expressed 3p21.3 genes relative to those of endogenously expressed mRNAs in HBEC is ~10 –15-, 30 –50-, and 50 – 80-fold at an MOI of 100, 500, and 1000 vp/cell, respectively (data not shown). The levels of expression in transduced H1299 cells were similar to those in HBEC (data not shown). In addition, there was an association between increased levels of expression of these 3p21.3 genes and increased MOIs of the corresponding Ad-3p vectors transduced in both H1299 and HBEC cells. The expression of FUS1 and 101F6 proteins after transfection was detected by Western blot analysis using available polyclonal antibodies raised against the oligopeptides derived from their deduced amino acid sequences (data not shown).
Induction of Apoptosis by 3p Genes in Ad-3p-transduced Tumor Cells
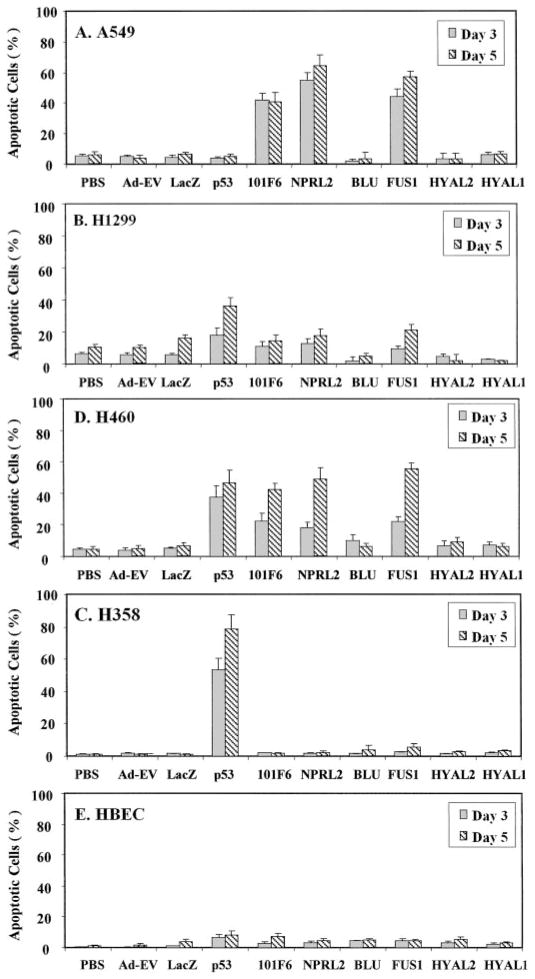
The ability of exogenously expressed 3p21.3 genes to induce apoptosis in the Ad-3p-transduced H1299, A549, H460, H358, and HBEC cells was analyzed by FACS using the TUNEL reaction (Fig. 2). Induction of apoptosis was detected in Ad-101F6-, Ad-FUS1-, and Ad-NPRL2-transduced A549 (Fig. 2A), H1299 (Fig. 2B), and H460 (Fig. 2C) cells but not in H358 (Fig. 2D) and HBEC (Fig. 2E) cells. The apoptotic cell populations increased with increased duration of transduction; >15–20, 40–65, and 75% of cells were apoptotic 5 days after transduction with Ad-101F6, Ad-FUS1, and Ad-NPRL2 in the transduced H1299, A549, and H460 cells, respectively, whereas only ~7 and 10% of cells treated with PBS alone and transduced with Ad-EV vector, respectively, were TUNEL positive after the same time interval. The levels of apoptosis induction by Ad-101F6, Ad-FUS1, and Ad-NPRL2 were quantitatively greater in A549 and H460 cell lines with WT p53 genes (Fig. 2, A and C) than that in H1299 cell line deleted for p53 gene (Fig. 2B). Levels of apoptosis in A549 and H460 cells were comparable with those induced by Ad-p53 in p53-deficient H1299 and H358 cells (Fig. 2, B and D). However, no significant induction of apoptosis was observed in any tumor cell line transduced by Ad-BLU, Ad-RASSF1, Ad-HYAL2, and Ad-HYAL1 (Fig. 2). The levels and time of induction of apoptosis in cells transduced by these Ad-3p vectors correlated with those of reduction of cell numbers in cells treated with the same vectors (Fig. 1), suggesting that suppression of tumor cell growth by these 3p21.3 genes is mediated directly or indirectly through a mechanism of apoptosis.
Fig. 2.
Induction of apoptosis by exogenous expression of 3p21.3 genes in Ad-3p-transduced human NSCLC cells and normal HBECs. Apoptosis was analyzed by FACS, using TUNEL reaction with FITC-labeled dUTP. Cells were transduced with adenoviral vectors of 3p21.3 genes at an MOI (vp/c) of 5000 for A549 (A), 1000 for H1299 (B), 5000 for H460 (C), 2500 for H358 (D), and 1000 for HBEC (E), respectively, and PBS, Ad-EV, and p53 were used as controls. Cells were harvested and analyzed for apoptosis at the indicated days post-transduction. The rate of apoptosis is expressed as the percentage of FITC-labeled cells in the total cell population. Bars, SDs of the mean in two or three repeated experiments with triplicate treatments and TUNEL reactions for each experiment. The significance of the difference in apoptosis between vector-treated cells and the Ad-EV-, Ad-LacZ-, or PBS-treated controls was analyzed by two-sided Student’s t test. P < 0.05 was considered significant. The differences between the apoptosis induced by the Ad-EV- and Ad-LacZ-transduced cells versus PBS-treated controls were not significant (P = 0.925–0.675 from different time points and cell lines). The differences between the apoptosis induced in the Ad-101F6-, Ad-FUS1-, and Ad-NPRL2-transduced cells versus the Ad-EV-, Ad-LacZ-, or PBS-treated controls were significant in A549 and H460 cells at both 3 and 5 days post-transduction (P ≤ 0.0001–0.005) and significant versus the Ad-EV- and PBS-treated cells in H1299 at 5 days post-transduction (P ≤ 0.02) but not significant in H358 and HBEC cell lines at both 3 and 5 days post-transduction at all time points (P ≥ 0.85–0.95), respectively. Induction of apoptosis in Ad-p53-transduced H358 cells was significant at all time points compared with all other treatments (P < 0.0001). Induction of apoptosis in cells treated with Ad-BLU, Ad-HYAL2, and Ad-HYAL1 was not significant compared with those treated with PBS, Ad-EV, or Ad-LacZ, in all cell lines at all time points (P > 0.85).
Suppression of Tumor Growth by Intratumoral Injection of Ad-3p Vectors
To determine whether the observed inhibitory effects of these 3p21.3 genes on tumor cell proliferation in vitro could be demonstrated on tumor growth in vivo, we evaluated the efficacy of 3p21.3 genes in suppressing tumor growth by direct intratumoral injection of Ad-3p21.3 gene vectors, along with PBS and Ad-EV, Ad-LacZ, and Ad-p53 vectors as controls, into A549 or H1299 tumor xenografts in nu/nu mice (Fig. 3). The growth of tumors was recorded from the first injection until 23 days after the last injection. Tumor volumes were normalized by calculating the percentage increase in tumor volume after treatment relative to volume at the beginning of treatment in each group. In both A549 (Fig. 3A) and H1299 (Fig. 3B) tumor models, all of the tumors treated with Ad-101F6, Ad-FUS1, or Ad-NPRL2 showed significantly suppressed growth (P < 0.001), compared with mouse groups treated with Ad-LacZ or Ad-EV controls, whereas no significant effect was observed in Ad-HYAL1-, Ad-BLU (data not shown)-, and Ad-HYAL2 (data not shown)-treated tumors. Moreover, a significantly stronger inhibition of tumor growth was shown in A549 tumors treated with Ad-101F6 and Ad-NPRL2 vectors than in tumors treated with the Ad-p53 vector (Fig. 3A).
Fig. 3.
Effects of intratumoral administration of adenoviral vectors of 3p21.3 genes on growth of human lung cancer A549 (A) and H1299 (B) s.c. tumors in nu/nu mice. When the tumor reached 5–10 mm in diameter at ~2 weeks after tumor inoculation, the tumor was injected with individual adenoviral vectors of 3p21.3 genes 101F6, NPRL2, FUS1, and HYAL1 or control vectors Ad-EV and p53, at a dose of 5 × 1010 vp/tumor each in 200 μl of PBS for three times within a week, respectively, and PBS alone was used as a mock control. Results were reported as the mean ± SD in 5–10 mice for each treatment group. Tumor volumes were normalized by the percentage increase of tumor sizes after treatment relative to those at the beginning of the treatment in each group. Mean tumor volumes ± SE from these experiments are shown. ANOVA was performed to determine statistical significance between each treatment group using Statistica software (Stat-Soft, Inc.), and P ≤ 0.05 was considered significant. The differences of the tumor volumes in the Ad-101F6-, Ad-FUS1-, and Ad-NPRL2-treated mice versus in the Ad-EV-treated mouse controls were statistically significant in both A549 and H1299 tumor models (P < 0.0001) after 5 days from the last injection but not significant in Ad-HYAL1 (P > 0.05 in both A549 and H1299 tumor models).
Inhibition of Development of Experimental Lung Metastases by Protamine-Adenovirus Complex-mediated 3p21.3 Gene Transfer
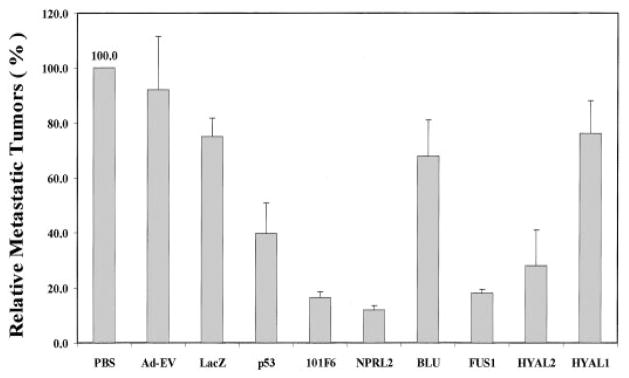
A novel formulation using protamine/adenovirus complexes (designated P-Ad) for enhanced systemic delivery of recombinant adenovirus in vivo was developed to further explore the potential of 3p21.3 genes in suppressing systemic metastases. An experimental A549 metastatic human lung cancer model (established by i.v. injection of tumor cells) was used to study the effects of 3p21.3 gene transfer on the development of lung metastases in nu/nu mice (Fig. 4). The adenoviral 3p21.3 gene vectors were complexed to protamine and delivered via i.v. injection. The development of A549 pulmonary metastases was inhibited significantly, and the numbers of metastatic tumor colonies found on the surfaces of lungs from mice inoculated with A549 cells were reduced by >80% in animals treated with P-Ad-101F6, P-Ad-FUS1, P-Ad-NPRL2, or P-Ad-HYAL2 (70% reduction), compared with those in control treatment groups (Fig. 4). However, no significant reduction of metastatic colony formation was observed in animals treated with P-Ad-HYAL1 and P-Ad-BLU. These data are consistent with results obtained from Ad-3p-treated s.c. tumor xenografts, further supporting the roles of these 3p21.3 genes in suppression of tumor growth and inhibition of tumor progression in vivo. Finally, we noted no systemic toxicity to the mice given the systemic injection of PAd-3p complexes.
Fig. 4.
Effect of systemic administration of protamine-Ad-3p complexes on development of A549 experimental lung metastases in nu/nu mice. All animals were i.v. injected with various protamine-adenoviral vector complexes every other 2 days for three times each at a dose of 3 × 1010 vp plus 300 μg of protamine in a total volume of 200 μl/animal, and PBS alone was used as a mock control. Each treatment group consisted of 5–10 animals. Lungs were harvested 2 weeks after the last injection, and metastastic colonies on the surfaces of lung were counted without knowledge of the treatment groups. Development of metastases was represented as the percentages of metastatic colonies formed in protamine-adenovirus complex-treated groups in relation to those in the PBS-treated group (as 100%). Bars, SE. Nonparametric t test (Wald-Wolfowitz Runs Test) was performed to determine statistical significance between each treatment group using Statistica software (StatSoft, Inc.), and P ≤ 0.05 was considered significant. A significant inhibition of development of metastases was observed in mice treated with P-Ad-101F6 (P = 0.002), P-Ad-NPRL2 (P = 0.001), P-Ad-FUS1 (P = 0.002), and P-Ad-HYAL2 (P = 0.014), respectively, compared with mice treated with PBS, P-Ad-EV, or P-Ad-LacZ but no significant inhibition in mice treated with P-Ad-BLU (P = 0.818) or P-Ad-HYAL1 (P = 0.904).
DISCUSSION
In this study, we used recombinant adenoviral vectors to introduce individual WT 3p21.3 genes into 3p-deficient tumor xenograft or tumor cell lines. The ectopic expression of WT 3p21.3 genes 101F6, FUS1, and NPRL2 effectively inhibited the growth of 3p-deficient NSCLC A549, H1299, and H460 cells in vitro but had no effect on the growth of H358 cells (which remains heterozygous for multiple polymorphic markers in the 3p21.3 650-kb homozygous deletion region) or on the growth of normal HBECs, suggesting the specificity of the exogenous WT 3p21.3 genes in inhibiting tumor cell growth. These findings also indicate the possibility that exogenous expression of 3p21.3 genes will be safe as cancer gene therapy agents because they caused no generalized cytotoxicity to normal cells or in mice treated systemically. The tumor suppressing effects of some TSGs under normal physiological conditions are generally mediated by increased levels of TSG expression in response to the oncogenic and environmental stimuli. These include p53, WAF1, BAX, and BAK (20), e.g., expression of both endogenous WT p53 gene and protein increased 6–8-fold after heat treatment of myeloblastic leukemia cells and DNA-binding activity of p53 increased >17-fold after γ-irradiation of human glioblastoma cells (21, 22). The level of 3p21.3 gene expression by the adenoviral vector-mediated 3p21.3 gene transfer in normal HBEC cells was increased ~10–15-fold (data not shown) and is close to the elevated levels of TSG expression induced by environmental stimuli under physiological conditions shown by that of p53.
In most cases, there is loss of heterozygosity at the 3p21.2 locus with no mutations detected in the remaining allele. However, haplo-insufficiency can be associated with abrogation of tumor suppressor activity, e.g., the p27 gene is haploinsufficient for tumor suppression with tumor suppressor activity critically dependent on the absolute level of p27 protein expression (23). Elevated p27 expression inhibits cell cycle progression and promotes apoptosis in human glioma, colon, NSCLC, and mantle cell lymphoma, suggesting that p27 acts as a rheostat rather than as an on/off switch tumor suppressor in suppressing neoplasia (24). Similar to p27, some of the 3p21.3 genes are possibly inactivated by haploinsufficincy (10), and the modulation of protein expression may play an important role for their tumor suppressor activities. Furthermore, the overexpression of these 3p21.3 genes at higher MOIs may be pharmaceutically appropriate for enhancing their function as cancer therapeutics and may be necessary for proper TSG function to overcome degradation pathways and inactive pathways in the cancer cell. The selectivity of the vectors with respect to growth inhibition and induction of the specific pathway of apoptosis in cancer compared with normal cells further supports their physiological role.
Inhibition of cell proliferation and induction of cell death by activated TSGs, such as retinoblastoma (Rb) and p53, are attributed primarily to these genes’ ability to mediate cell cycle arrest and apoptosis (25–27). Because apoptosis is a genetically programmed cellular response to environmental stresses or stimuli, inactivation of TSGs involved in the apoptotic pathways could result in deregulated cell proliferation and tumorigenesis. To elucidate the mechanism governing the inhibition of NSCLC cell growth by 3p21.3 genes, we studied the effects of exogenously expressed 3p21.3 genes on apoptosis mediated by adenoviral vector transduction. Introduction of WT 3p21.3 genes 101F6, FUS1, and NPRL2 into the 3p-deficeint A549, H1299, and H460 cells induced apoptosis. However, this was not a generalized feature of 3p21.3 gene overexpression, as the HYAL2, HYAL1, and BLU genes from this same 120-kb region did not induce a significant increase in apoptosis in the same lung cancer cells. The time and the magnitude of the induction of apoptosis by these 3p21.3 genes were also well correlated with those of the inhibition of growth observed in vitro. These observations suggest that the tumor suppressing function mediated by the 3p21.3 genes is through induction of apoptosis.
To demonstrate whether the observed inhibitory effects of these 3p21.3 genes on tumor cell growth in vitro could be reproduced in vivo, we evaluated the efficacy of 3p21.3 genes in suppressing tumor growth by directly injecting Ad-3p vectors into A549 or H1299 tumor xenografts in nu/nu mice. Growth of both A549 and H1299 tumors was suppressed significantly by treatments with Ad-101F6, Ad-FUS1, and Ad-NPRL2. Furthermore, we explored the tumor suppressing potential of 3p21.3 genes in inhibiting experimental metastases in vivo by systemic administration of protamine-Ad-3p complexes. The novel protamine-Ad-3p complexes developed as part of this study allowed us to deliver 3p21.3 genes efficiently to the lung by systemic injection. The development of metastases was inhibited effectively by the protamine-adenovirus complex-mediated transfer of the 101F6, FUS1, HYAL2, and NPRL2 genes. These in vivo data are consistent with the in vitro data, further supporting the roles of 3p21.3 genes as TSGs.
Two of the 3p21.3 genes, HYAL1 and RASSF1C, showed neither tumor suppressor activity in vitro nor in vivo nor apoptosis-inducing activity in vitro in all cell lines tested. Recently, one splicing isoform of one of the genes, RASSF1A, has been shown to undergo epigenetic inactivation by promoter region hypermethylation acquired in tumors. This isoform, but not the expressed RASSF1C isoform, also exhibits functional tumor suppressing activity. Consistent with these results, we also found no significant effects on growth of NSCLC cells and induction of apoptosis in these NSCLC cells in vitro and in vivo in our experiments using the Ad-RASSF1C vector (data not shown). The results with RASSF1A indicate that it will be important to study all of the genes in the region for loss of expression via tumor-acquired promoter hypermethylation. Alternatively, with 3p allele loss, haploinsufficiency of one or more of these 3p21.3 genes may play a role in tumorigenesis. On the basis of the evidence that multiple contiguous 3p21.3 genes, including 101F6, NPRL2, RASSF1A, and FUS1, exhibited varied degrees of tumor suppressing activity, we propose that genes in this 3p21.3 120-kb region act together as part of a tumor suppressor region to suppress tumor growth through their functional activation of tumor suppressing pathways. Likewise, their inactivation may contribute directly to the development of cancer because of haploinsufficiency, loss of expression, or rarely mutations. In summary, we have demonstrated here for the first time that introduction of several WT 3p21.3 genes (101F6, NPRL2, and FUS1) contiguously located within a 120-kb region by recombinant adenoviral vector-mediated gene transfer into 3p-deficient tumor xenografts and tumor cell lines efficiently suppressed tumor cell growth and metastases and induced apoptosis in vitro and in vivo. These results suggest the role of these genes as TSGs. A better understanding of the biological function of genes in this region may result in the development of new strategies for the prevention, early detection, diagnosis, and treatment for lung cancer and other human cancers.
Acknowledgments
The Minna’s Lab thanks Eva Forgacs, Gena Mele, and Adrin Avila for assistance with this work and Dr. Yoshitaka Sekido for his invaluable assistance in identifying and characterizing the genes in this 3p21.3 region.
Footnotes
Supported in part by grants from the National Cancer Institute and the NIH SPORE (2P50-CA70970-04, to J. D. M. and J. A. R.; P01 CA78778–01A1, to J. A. R.; and CA71618, to J. D. M.), gifts to the Division of Surgery from Tenneco and Exxon for the Core Laboratory Facility, the UT M. D. Anderson Cancer Center Support Core Grant (CA 16672), a grant from the Tobacco Settlement Funds as appropriated by the Texas State Legislature (Project 8), a W. M. Keck Gene Therapy Career Development Grant (L. J.), and a sponsored research agreement with Introgen Therapeutics, Inc. (SR93-004-1).
The abbreviations used are: TSG, tumor suppressor gene; NSCLC, non-small cell lung cancer; TUNEL, terminal deoxynucleotidyl transferase-mediated nick end labeling; WT, wild type; HBEC, human bronchial epithelial cell; GFP, green fluorescence protein; pfu, plaque-forming unit(s); vp, viral particle; FACS, fluorescence-activated cell sorter; MOI, multiplicities of infection.
References
- 1.Zochbauer-Muller S, Gazdar AF, Minna JD. Molecular pathogenesis of lung cancer. Ann Rev Physiol. 2002;64:681–708. doi: 10.1146/annurev.physiol.64.081501.155828. [DOI] [PubMed] [Google Scholar]
- 2.Fong K, Sekido Y, Minna JD. The molecular basis of lung carcinogenesis. In: Coleman WB, Tsongalis G, editors. The Molecular Basis of Human Cancer. Totowa, NJ: Humana Press; 2001. pp. 379–405. [Google Scholar]
- 3.Park IW, Wistuba II, Maitra A, Milchgrub S, Virmani AK, Minna JD, Gazdar AF. Multiple clonal abnormalities in the bronchial epithelium of patients with lung cancer. J Natl Cancer Inst (Bethesda) 1999;91:1863–1868. doi: 10.1093/jnci/91.21.1863. [DOI] [PubMed] [Google Scholar]
- 4.Wistuba II, Lam S, Behrens C, Virmani AK, Fong KM, LeRiche J, Samet JM, Srivastava S, Minna JD, Gazdar AF. Molecular damage in the bronchial epithelium of current and former smokers. J Natl Cancer Inst (Bethesda) 1997;89:1366–1373. doi: 10.1093/jnci/89.18.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wistuba II, Berry J, Behrens C, Maitra A, Shivapurkar N, Milchgrub S, Mackay B, Minna JD, Gazdar AF. Molecular changes in the bronchial epithelium of patients with small cell lung cancer. Clin Cancer Res. 2000;6:2604–2610. [PMC free article] [PubMed] [Google Scholar]
- 6.Wistuba II, Behrens C, Virmani AK, Mele G, Milchgrub S, Girard L, Fondon JW, Garner HR, McKay B, Latif F, Lerman MI, Lam S, Gazdar AF, Minna JD. High resolution chromosome 3p allelotyping of human lung cancer and preneoplastic/preinvasive bronchial epithelium reveals multiple, discontinuous sites of 3p allele loss and three regions of frequent breakpoints. Cancer Res. 2000;60:1949–1960. [PubMed] [Google Scholar]
- 7.Girard L, Zochbauer-Muller S, Virmani AK, Gazdar AF, Minna JD. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 2000;60:4894–4906. [PubMed] [Google Scholar]
- 8.Maitra A, Wistuba II, Washington C, Virmani AK, Ashfaq R, Milchgrub S, Gazdar AF, Minna JD. High-resolution chromosome 3p allelotyping of breast carcinomas and precursor lesions demonstrates frequent loss of heterozygosity and a discontinuous pattern of allele loss. Am J Pathol. 2001;159:119–130. doi: 10.1016/S0002-9440(10)61679-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wistuba II, Behrens C, Milchgrub S, Bryant D, Hung J, Minna JD, Gazdar AF. Sequential molecular abnormalities are involved in the multistage development of squamous cell lung carcinoma. Oncogene. 1999;18:643–650. doi: 10.1038/sj.onc.1202349. [DOI] [PubMed] [Google Scholar]
- 10.Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. Cancer Res. 2000;60:6116–6133. [PubMed] [Google Scholar]
- 11.Sekido Y, Ahmadian M, Wistuba II, Latif F, Bader S, Wei MH, Duh FM, Gazdar AF, Lerman MI, Minna JD. Cloning of a breast cancer homozygous deletion junction narrows the region of search for a 3p21.3 tumor suppressor gene. Oncogene. 1998;16:3151–3157. doi: 10.1038/sj.onc.1201858. [DOI] [PubMed] [Google Scholar]
- 12.Wu X, Zhao Y, Honn SE, Tomlinson GE, Minna JD, Hong WK, Spitz MR. Benzo[a]pyrene diol epoxide-induced 3p21.3 aberrations and genetic predisposition to lung cancer. Cancer Res. 1998;58:1605–1608. [PubMed] [Google Scholar]
- 13.Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S, Sekido Y, Latif F, Milchgrub S, Toyooka S, Gazdar AF, Lerman MI, Zabarovsky E, White M, Minna JD. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst (Bethesda) 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 15.Kondo M, Ji L, Kamibayashi C, Tomizawa Y, Randle D, Sekido Y, Yakota J, Kashuba V, Zabarovsky E, Kuzmin I, Lerman M, Roth JA, Minna JD. Overexpression of candidate tumor suppressor gene FUS1 isolated from the 3p21.3 homozygous deletion region leads to G1 arrest and growth inhibition of lung cancer cells. Oncogene. 2001;20:6258–6262. doi: 10.1038/sj.onc.1204832. [DOI] [PubMed] [Google Scholar]
- 16.Fondon JW, Mele GM, Brezinschek RI, Cummings D, Pande A, Wren J, O’Brien KM, Kupfer KC, Wei MH, Lerman M, Minna JD, Garner HR. Computerized polymorphic marker identification: experimental validation and a predicted human polymorphism catalog. Proc Natl Acad Sci USA. 1998;95:7514–7519. doi: 10.1073/pnas.95.13.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roehm NW, Rodgers GH, Hatfield SM, Glasebrook AL. An improved colorimetric assay for cell proliferation and viability utilizing the tetrazolium salt XTT. J Immunol Methods. 1991;142:257–265. doi: 10.1016/0022-1759(91)90114-u. [DOI] [PubMed] [Google Scholar]
- 18.Nishizaki M, Meyn RE, Atkinson EN, White RA, Roth JA, Ji L. Synergistic inhibition of human lung cancer cell growth by adenovirus-mediated wild-type p53 gene transfer in combination with docetaxel and radiation therapeutics in vitro and in vivo. Clin Cancer Res. 2001;7:2683–2689. [PubMed] [Google Scholar]
- 19.Ji L, Fang B, Yen N, Fong K, Minna JD, Roth JA. Induction of apoptosis and inhibition of tumorigenicity and tumor growth by adenovirus vector-mediated fragile histidine triad (FHIT) gene overexpression. Cancer Res. 1999;59:3333–3339. [PubMed] [Google Scholar]
- 20.Bishay K, Ory K, Lebeau J, Levalois C, Olivier MF, Chevillard S. DNA damage-related gene expression as biomarkers to assess cellular response after γ irradiation of a human lymphoblastoid cell line. Oncogene. 2000;19:916–923. doi: 10.1038/sj.onc.1203405. [DOI] [PubMed] [Google Scholar]
- 21.Kastan MB, Radin AI, Kuerbitz SJ, Onyekwere O, Wolkow CA, Civin CI, Stone KD, Woo T, Ravindranath Y, Craig RW. Levels of p53 protein increase with maturation in human hematopoietic cells. Cancer Res. 1991;51:4279–4286. [PubMed] [Google Scholar]
- 22.Ohnishi T, Wang X, Ohnishi K, Matsumoto H, Takahashi A. p53-dependent induction of WAF1 by heat treatment in human glioblastoma cells. J Biol Chem. 1996;271:14510–14513. doi: 10.1074/jbc.271.24.14510. [DOI] [PubMed] [Google Scholar]
- 23.Fero ML, Randel E, Gurley KE, Roberts JM, Kemp CJ. The murine gene p27Kip1 is haplo-insufficient for tumour suppression. Nature (Lond) 1998;396:177–180. doi: 10.1038/24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kemp CJ, Sun S, Gurley KE. p53 induction and apoptosis in response to radio- and chemotherapy in vivo is tumor-type-dependent. Cancer Res. 2001;61:327–332. [PubMed] [Google Scholar]
- 25.Evan G, Littlewood T. A matter of life and cell death. Science (Wash DC) 1998;281:1317–1322. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- 26.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 27.Vousden KH. p53: death star. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]