Abstract
We searched for new components that are involved in the positive regulation of nuclear gene expression by light by extending a screen for Arabidopsis cue (chlorophyll a/b-binding [CAB] protein-underexpressed) mutants (H.-M. Li, K. Culligan, R.A. Dixon, J. Chory [1995] Plant Cell 7: 1599–1610). cue mutants display reduced expression of the CAB3 gene, which encodes light-harvesting chlorophyll protein, the main chloroplast antenna. The new mutants can be divided into (a) phytochrome-deficient mutants (hy1 and phyB), (b) virescent or delayed-greening mutants (cue3, cue6, and cue8), and (c) uniformly pale mutants (cue4 and cue9). For each of the mutants, the reduction in CAB expression correlates with the visible phenotype, defective chloroplast development, and reduced abundance of the light-harvesting chlorophyll protein. Levels of protochlorophyllide oxidoreductase (POR) were reduced to varying degrees in etiolated mutant seedlings. In the dark, whereas the virescent mutants displayed reduced CAB expression and the lowest levels of POR protein, the other mutants expressed CAB and accumulated POR at near wild-type levels. All of the mutants, with the exception of cue6, were compromised in their ability to derepress CAB expression in response to phytochrome activation. Based on these results, we propose that the previously postulated plastid-derived signal is closely involved in the pathway through which phytochrome regulates the expression of nuclear genes encoding plastid proteins.
The assembly of the photosynthetic machinery in developing leaves of higher plants requires the expression of a set of nuclear and plastidic genes, the products of which will ultimately function in chloroplasts (for review, see Mullet, 1988). The coordinated expression of these genes is regulated by a number of factors, including light (Chory, 1991) and a postulated signal through which the nucleus responds to the functional status of the plastid (Oelmüller et al., 1986; Mayfield, 1990). In dark-grown angiosperms, plastids develop into etioplasts, which accumulate protochlorophyllide and POR (Reinbothe et al., 1996). When plants are exposed to light, profound changes in gene expression occur as the photosynthetic apparatus is assembled and the etioplast develops into a chloroplast. A specific group of photoreceptors, including both phytochromes and cryptochromes (Thompson and White, 1991), play an important role in this transition, but the downstream signaling components involved are only starting to be understood (Terzaghi and Cashmore, 1995).
The developing plastid itself appears to play an important role in the regulation of nuclear gene expression for chloroplast proteins. For instance, the amount of transcripts for several nuclear-encoded chloroplast-localized proteins declines very rapidly following treatments that damage plastid integrity (Oelmüller et al., 1986). These observations suggest a mechanism by which the nucleus can sense the physiological status of the organelle. In support of this hypothesis, Arabidopsis mutants have been isolated in which nuclear gene expression is partially uncoupled from the status of the plastid (Susek et al., 1993).
Various approaches have been used to understand mechanistically the perception and transduction of light signals by plants (Kendrick and Kronenberg, 1994). Genetic screens have been particularly useful (Fankhauser and Chory, 1997). Screens for the deregulation of light responses have identified mutations in the negative elements affecting light signal transduction, notably the DET/COP/FUS class (Chory et al., 1989b; Deng et al., 1992; Miséra et al., 1994). Conversely, positive elements in the light-signaling pathways initiated by phytochromes and cryptochromes have been uncovered in screens for mutants with elongated hypocotyls in various light conditions. These screens have identified the phytochrome apoprotein and chromophore biosynthetic genes (HY1, HY2, PHYA, PHYB, and PHYD), cryptochrome genes (HY4; for review, see Fankhauser and Chory, 1997), and downstream elements involved in the control of elongation or flowering, including ELF3 (Zagotta et al., 1996), FHY1 and FHY3 (Whitelam et al., 1993), HY5 (Oyama et al., 1997), PEF1 (Ahmad and Cashmore, 1996), and RED1 (Wagner et al., 1997). With the exception of phyB and the chromophore mutants hy1 and hy2, none of the other mutants appears to be defective in phytochrome-regulated gene expression or chloroplast development (Chory et al., 1989a; Reed et al., 1994).
In an attempt to identify positive elements specifically involved in phytochrome signaling to nuclear light-regulated promoters, we devised a screen based on selection for the underexpression of a light-regulated promoter, CAB3 (Li et al., 1995). The CAB family of nuclear genes encode the apoproteins of the light-harvesting complex of PSII. Following translation on cytoplasmic ribosomes, the polypeptides encoded (LHCPs) are imported into chloroplasts and are subsequently integrated into the thylakoid membrane, where they form the most abundant chlorophyll-containing complex (Green and Salter, 1996). The expression of the CAB genes is a marker for chloroplast development and is tightly controlled by both light (Karlin-Neumann et al., 1988) and plastid signals. Other factors also control CAB expression, including a circadian clock (Millar and Kay, 1996), hormones (Flores and Tobin, 1986; Bartholomew et al., 1991), and Suc levels (Dijkwel et al., 1997). The interactions between these factors are complex.
Our previous screen identified mutants at two loci that we named cue1 and cue2 (Li et al., 1995). cue1 has been analyzed in detail. It is a reticulate mutant with pale-green mesophyll cells and dark-green bundle-sheath cells aligning the veins (Li et al., 1995). Characterization of this mutant suggested that functional CUE1 is required for phytochrome to derepress CAB expression in the light. This initial screen failed to identify mutants in the genes encoding the photoreceptors, suggesting that an expanded screen might be useful. We report the identification of a series of eight new cue mutants. Two of these mutants are allelic with the well-characterized photomorphogenetic mutants phyB and hy1 (Koornneef et al., 1980; Parks and Quail, 1991; Reed et al., 1993), one is a new allele of cue1, and the other five identify novel loci. All of these mutants appear to have defects in chloroplast development. Analysis of their phenotypes and their responses to red-light pulses suggests a direct role for a previously postulated plastid-derived factor in the pathways through which phytochrome controls nuclear gene expression.
MATERIALS AND METHODS
Genetic Screen and Methods
We used a previously described Arabidopsis line expressing both ADH (alcohol dehydrogenase) and UidA (β-glucuronidase) genes under the control of a CAB3 promoter (Li et al., 1995). The screening procedure was modified as follows: Seeds from approximately 6000 ethyl methanesulfonate-mutagenized plants (M1) were collected in pools of 200 to 300. M2 seeds were grown in liquid medium (Murashige-Skoog salt mixture, Gamborg's vitamin mixture, and 2% Suc) in six-well microtiter plates in batches of 300 seedlings per well with gentle shaking under 150 μmol photons m−2 s−1 white light for 5 d. The Murashige-Skoog medium was then exchanged for medium containing 3.5 mm allyl alcohol for 1 h. These conditions allowed 100% rescue of non-ADH-expressing plants (R002 mutant, Jacobs et al., 1988); therefore, we predicted that mutants with only moderate phenotypes would be rescued.
The CUE loci were mapped using previously described molecular markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994). DNA polymorphisms were scored in a minimum of 35 F2 mutant seedlings (70 chromatids). Double mutants were isolated in the F2 progeny of the respective crosses by searching for novel phenotypes at the expected frequency (1:16). The assignment was confirmed by the absence of segregation in the next generation, as well as by the appearance of similar phenotypes in the progeny of F2 plants identified as single mutants.
Plant Growth and GUS Assays
Plants were grown in plates on synthetic Murashige-Skoog medium (Murashige-Skoog salt mixture, Gamborg's vitamin mixture, 1% Suc, and 0.8% agar). To ensure uniformity of germination, the plates received a 3-d cold treatment in the dark, followed by 1 h under white light (approximately 100 μmol photons m−2 s−1) prior to transfer to darkness or white light. Except where stated, seedlings were analyzed at 5 d of age. In experiments in which the whole seedling was harvested, seeds were sown on sterile filter paper overlaying the medium. GUS activities (Jefferson, 1987) were measured by fluorescence using 4-methylumbelliferyl β-d-glucuronide (GIBCO-BRL) as a substrate.
Pigments and Protein Analysis
Chlorophylls were extracted in dimethyl formamide at 4°C in the dark, and the concentration of pigments was calculated according to the method of Porra et al. (1989). Anthocyanin accumulation was determined spectrophotometrically as described by Chory et al. (1989b).
Total proteins from greenhouse-grown leaves equivalent to those used for microscopy or 5-d-old dark-grown seedlings were extracted and analyzed as described previously (López-Juez and Hughes, 1995) on PVDF membranes (Merck, Poole, UK); 8 μg of total protein per sample was used in both cases. Quantitation was performed by blotting a dilution series of 8.0, 2.5, and 0.8 μg of protein from each sample. The mouse monoclonal antibodies to LHCPs were a gift from Dr. T. Kunkel (University of Freiburg, Germany). Antiserum recognizing Arabidopsis POR was a gift from Drs. K. Apel and G. Armstrong (ETH Zentrum, Zurich, Switzerland).
Light Treatments and RNA Gel Blots
Plants were grown in 100 μmol photons m−2 s−1 white light in 16-h photoperiods. For single light-pulse experiments, seedlings were grown in the dark for 5 d and treated with the light produced by a tungsten illuminator, filtered through Nikko DIF-BPF-4 induced transmission filters (Vacuum Optics Corp., Tokyo, Japan). For red light, the filter had a maximum A647 and cut-off at 675 nm. The far-red light had a maximum A745 and cut-on at 713 nm. Total fluences were 1,000 μmol photons m−2 for red (20 μmol photons m−2 s−1 for 50 s) and 10,000 μmol photons m−2 s−1 for far-red light (130 μmol photons m−2 s−1 for 75 s). The fluence rate of red light was measured with a PAR meter and that of far-red was estimated using the transmission spectra of the filters and assuming a 20% greater light emission from the tungsten source at 750 than at 650 nm. For experiments with multiple red pulses, light from fluorescent lamps (Gro-Lux, Sylvania) was filtered through a colored glass filter (Li et al., 1995) to provide a narrow-band red peak and attenuated with neutral filters. Pulses of light consisted of 15 min of light of 1.1 μmol photons m−2 s−1 each every 6 h for 7 d.
For RNA extraction about 150 mg of seedlings was added to 150 μL of phenol and 500 μL of extraction buffer (100 mm NaCl, 10 mm Tris, pH 7.5, 1 mm EDTA, and 1% SDS) in a liquid-nitrogen-cooled mortar. The seedlings were ground together with the frozen buffer and phenol into a paste. After the extract was transferred to a microcentrifuge tube, it was extracted with 250 μL of chloroform and spun for 3 min, and the RNA-containing supernatant was precipitated with an equal volume of 4 m LiCl on ice. After the sample was centrifuged, the pellet was resuspended in water. Gel blotting, probe construction, and hybridizations were as previously described (Chory et al., 1989b). The strength of the radioactive signal was quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Light and Electron Microscopy
Rectangular sections extending from the midvein to the leaf edge were cut from the middle part of young, greenhouse-grown Arabidopsis leaves and fixed in 4% formaldehyde plus 3% glutaraldehyde in 0.1 m Pipes buffer, pH 7.2, for 1 h. After the samples were rinsed in Pipes buffer, they were postfixed for 1 h in 1% buffered OsO4. The samples were again washed in buffer, dehydrated in ethanol, and embedded in Spurr's resin following standard procedures. For light microscopy, 0.5-μm sections were stained with 1% toluidine blue in 1% sodium tetraborate. Silver sections were stained with saturated alcoholic uranyl acetate and Reynold's lead stain and viewed on an electron microscope (model EM 109, Zeiss).
RESULTS
Isolation of New cue Mutants
We expanded a previously described mutant screen for identifying light-insensitive mutants of Arabidopsis that monitors for reduced expression of a light-regulated promoter, CAB3 (Li et al., 1995). We screened a mutagenized transgenic line, pOCA108, that is an adh null mutant (R002) carrying two reporter genes: the full-length CAB3 promoter fused to the ADH gene of Arabidopsis and the CAB3 promoter fused to the Escherichia coli GUS (uidA or GUS) gene. ADH activity can be selected against using the substrate allyl alcohol (Li et al., 1995). We first determined conditions that would allow the rescue of mutants with only a slight to moderate deficiency in CAB3 expression. Using these conditions, we screened ethyl methanesulfonate-mutagenized M2 seedlings in pools derived from 200 to 300 M1 plants. A total of 125,000 seedlings were screened, with a minimum of 10 M2 seedlings from each M1 plant. Mutants containing trans-acting mutations that reduced CAB3 promoter activity were distinguished from cis-acting CAB3 promoter mutations using a second reporter, CAB3-GUS.
Of 250 seedlings that survived the allyl alcohol selection, 34 lines had GUS activity of less than 50% of that of the wild-type pOCA108 parent. The mutants were further divided into six classes based on their phenotypes. The first class contained three independently isolated mutants with long hypocotyls, and complementation tests showed them to be alleles of hy1 and phyB. One reticulate mutant, a new allele of cue1, made up the second class. The third class (cue4 and cue9) was uniformly paler than the wild type, with cue9 having a slightly reticulate phenotype. The fourth class (cue3, cue6, and cue8) was virescent, i.e. young leaves or recently expanded tissues (including young inflorescence shoots and the basal margins of older leaves), were pale, whereas more mature tissues were as green as wild-type tissues. The fifth class of mutants included lines with a mild reduction in CAB expression (approximately 50% GUS activity) but no visible phenotype. These will not be described further here. A final group comprised lines that segregated albino seedlings in their progeny. These may correspond to partially dominant albino mutations and were not further investigated. Figure 1 shows the phenotypes of mature hy1, phyB, cue3, cue4, cue6, cue8, and cue9 plants compared with that of pOCA108. These seven mutants are the focus of the studies that follow.
Figure 1.
Visible phenotypes of the cue mutants and the pOCA108 wild type. Plants were grown in vitro for 25 d under long-day growth conditions (16 h of light, 8 h of dark). Smaller plants (cue9, cue6hy1, cue6cue1–3, cue6cue4, and cue6cue8) were photographed at higher magnification for clarity. A, pOCA108 wild type; B, hy1; C, phyB; D, cue3; E, cue6; F, cue8; G, cue4; H, cue9; I, cue1–3; J, cue6hy1; K, cue6phyB; L, cue6cue1–3; M, cue6cue4; and N, cue6cue8. Bars in A to N = 1 cm.
The seven mutations selected for further study were recessive when backcrossed to the pOCA108 parent. For each mutant, the pale phenotype segregated in a manner consistent with a mutation at a single locus (data not shown). cue3 and cue4 exhibited mild seedling lethality (χ2 tests indicated significant differences from a 3:1 F2 ratio for cue4). Pairwise complementation tests confirmed that these seven lines define distinct loci (results not shown). We mapped the CUE loci and Table I shows their chromosomal positions and approximate distance (in centimorgans) to nearby molecular markers.
Table I.
Mapping summary
| Mutation | Chromosome | Marker
|
|
|---|---|---|---|
| North | South | ||
| cue3 | 3 | nga126 (4.5) | nga162 (2.1) |
| cue4a | 1 | g2395 (4.8) | m235 (0) |
| cue6 | 5 | DFR (15.8) | nga129 (6.3) |
| cue8 | 5 | nga106 (6.3) | nga139 (3.8) |
| cue9 | 1 | nga63 (6.8) | g2395 (3.7) |
Numbers in parentheses indicate centimorgans.
No recombinants were found between m235 and cue4 among 82 chromatids.
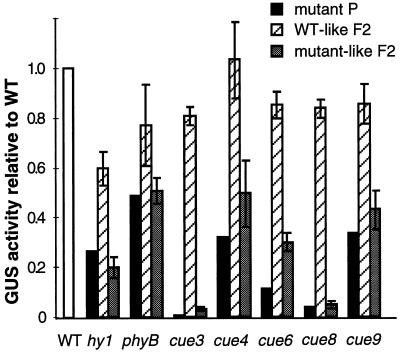
We monitored the expression of the endogenous CAB genes, as well as the genes encoding the ribulose-1,5-bisphosphate carboxylase small subunit (RBCS) in white-light-grown cue seedlings. Figure 2 shows that all of the mutations resulted in a moderate reduction of the total amount of CAB mRNAs. In most cases, although not appreciably for hy1 and cue4, the RBCS transcripts were also affected to a comparable extent. The visible phenotype and reduced gene expression phenotypes were linked. Figure 3 shows the cosegregation of the reduced CAB expression phenotype with the visible greening defect.
Figure 2.
Steady-state mRNA levels for the CAB and RBCS gene families are reduced in cue mutant seedlings under white light (WL). Seedlings were grown for 5 d under 100 μmol photons m−2 s−1 light in 16-h photoperiods, and harvested for RNA extraction 5 h into the photoperiod of d 5. RNA gel blots were hybridized to the corresponding probes and normalized to the signal from rRNA using a phosphor imager. WT, Wild type.
Figure 3.
Cosegregation of the visible and the CAB underexpression phenotypes of each cue mutant. After each mutant was backcrossed, the segregating F2 progeny were scored as mutant or wild type (WT). Three samples of three to five seedlings each were used for GUS measurements. For each mutant values are shown relative to the wild type for the parental line (black bars), wild type-like (striped bars), and visibly mutant (gray bars) F2 seedlings. For phyB, seedlings of intermediate length (presumably heterozygous), not shown here, showed wild-type GUS activities. Error bars = sd.
cue Mutants Are Defective in Greening and in the Expression of Genes for Photosynthetic Proteins
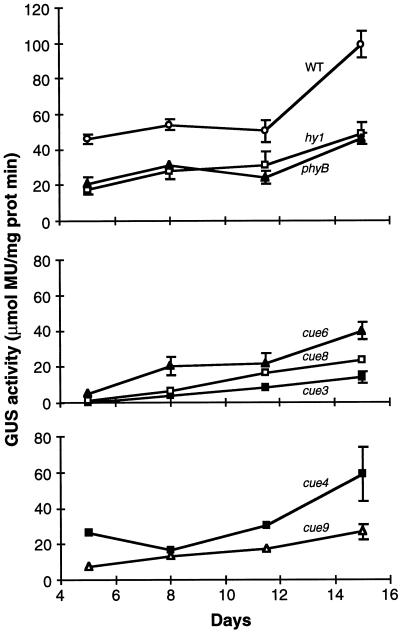
To quantify the CAB expression defect, a time course of GUS accumulation was performed. A close inspection of Figure 4 shows that the three classes of visible phenotypes—virescent, yellow-green, and long hypocotyl—correspond to different patterns of CAB3-driven GUS expression. The phytochrome mutants showed a general reduction in reporter activity throughout the period examined, during which time the activity increased about 2-fold in both the wild type and the mutants. The slow-greening mutants had a much more pronounced defect early in development than later, with GUS increasing at least 5-fold over the period analyzed. cue4, a uniformly pale mutant, showed an overall defect similar to hy1 or phyB; however, cue9 was intermediate between cue4 and the slow-greening class.
Figure 4.
Kinetics of CAB3 promoter-driven GUS accumulation in cue mutant seedlings in the light. Seedlings were grown for 15 d under the conditions described in Figure 2 and harvested, and GUS activity was measured. The curves are grouped according to the type of visible phenotype, either wild type (WT) or long-hypocotyl mutants showing overall reductions in GUS activity (top), slow-greening mutants showing very low early activities and subsequent steep increases (middle), and uniformly pale or reticulate mutants (bottom) appearing similar to those in the top panel or intermediate between the upper and middle ones. Three samples of 1 to 10 seedlings depending on age are shown; error bars = sd. MU, 4-Methylumbelliferyl β-d-glucuronide; prot, protein.
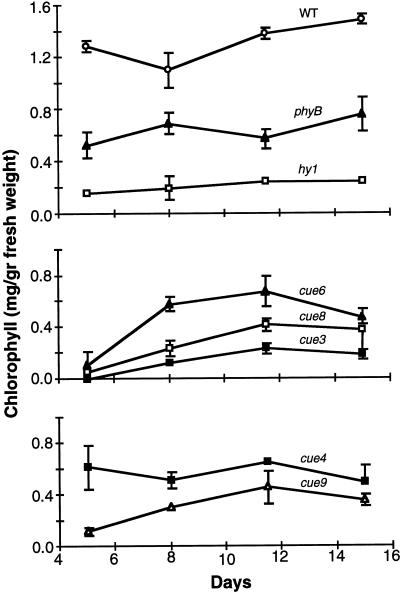
We also measured chlorophyll accumulation throughout this same developmental period. The data presented in Figure 5 show that chlorophyll accumulation correlates well with the pale phenotypes and the CAB expression patterns described in Figure 4. Phytochrome-deficient and uniformly pale mutants showed an increase in chlorophyll content of 2- to 3-fold during this period. In the virescent mutants, chlorophyll levels increased between 7- and 60-fold. To further analyze this relationship between CAB expression and chlorophyll accumulation, we examined leaves at two stages of development of the cue6 mutant (which has the most pronounced virescent phenotype). Compared with mature cue6 leaves, the youngest, palest cue6 tissues (leaves less than 5 mm long) had both a very high chlorophyll a/b ratio, indicative of relatively low amounts of LHCP (13.1 versus 3.0, the wild-type value being 3.4), and a greatly reduced level of CAB3-GUS activity (8% versus 33% of the wild type).
Figure 5.
Chlorophyll accumulation is reduced in cue mutants grown in the light. Total chlorophyll was measured following extraction from seedlings identical to those used for GUS accumulation kinetics shown in Figure 4. The seedlings shown are long-hypocotyl mutants (top), virescent mutants (middle), and uniformly pale or reticulate mutants (bottom). Each measurement represents three samples of one to five seedlings; error bars = sd. WT, Wild type.
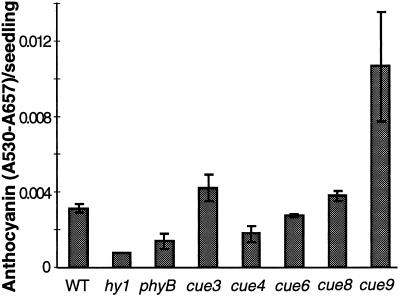
To examine a different light-regulated response in the mutants, we measured anthocyanin accumulation. Anthocyanins accumulated in response to phytochrome in nonphotosynthetic cells. Figure 6 indicates that anthocyanin was not reduced in any of the new cue mutants. (Anthocyanin levels in cue4, although lower in this individual experiment, were comparable overall to the wild type when analyzed over a series of ages.) Anthocyanin content was actually higher than wild-type levels in cue9. This is in contrast to the photoreceptor mutants, which exhibited a 2- to 4-fold reduction in anthocyanin levels. As such, the cue mutations appear to affect specifically chlorophyll accumulation and the expression of photosynthetic genes.
Figure 6.
Anthocyanin content of cue mutants does not correlate with CAB expression or chlorophyll reduction. Anthocyanin was extracted from seedlings 3.5 d after the transfer of seeds into light, as described for Figure 2. This time was shown in preliminary experiments to correspond approximately to the peak of anthocyanin accumulation in the wild type (WT). Some of the cue mutants were delayed in their growth, but the pattern shown here was maintained when examined at d 6. Three samples are shown; error bars = sd.
Some cue Mutants Are Affected in Both Basal Levels and Phytochrome Induction of CAB Expression
Results of many studies have underscored the requirement for chloroplast integrity (functional chloroplast transcription/translation) for proper expression of nuclear-encoded photosynthetic proteins. The reduced photosynthetic gene expression in the new cue mutants might be due to defects in photosynthetic physiology or chloroplast development rather than being a direct consequence of a defect in phytochrome signaling. In an attempt to discriminate between these possibilities, experiments were performed in which red-light pulses were given to etiolated seedlings. The light-pulse treatments can induce (derepress) gene expression, with only modest changes in seedling development or physiology, more accurately reflecting the primary effects of photoreceptor activation. This induction is mediated at least in part by phytochrome (Karlin-Neumann et al., 1988; Reed et al., 1994).
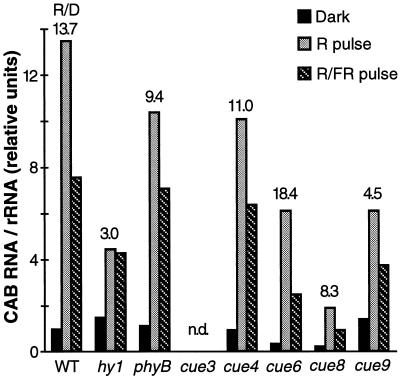
Figure 7 shows the effect of a single pulse of red light on the derepression of CAB gene expression: a 14-fold increase in levels of total CAB mRNAs in the wild type. As reported previously and as shown in Figure 7, CAB accumulation was slightly reduced in phyB and about 80% reduced in hy1 (Chory et al., 1989a; Reed et al., 1994). In these mutants, the dark basal levels of CAB mRNA accumulation were similar to the wild type. In contrast, the new cue mutants differed from the wild type in two ways: First, clear reductions in the basal levels of CAB mRNA in dark-grown seedlings were observed in the virescent mutants cue3, cue6, and cue8. Second, moderate decreases in the ratio of light to basal levels of CAB were seen in cue3, cue4, cue8, and cue9 but not in cue6. The amount of CAB mRNA accumulated in response to the red-light pulse was reduced in all cases. A far-red pulse was able to reverse the effect of a red-light pulse about 50% in all genotypes, with the exception of hy1, indicating involvement of phytochrome.
Figure 7.
Induction by a single red-light (R) pulse or reversion by far-red (FR) light following red light of the accumulation of CAB mRNA in etiolated cue mutant seedlings. Five-day-old etiolated seedlings were treated (or not) with a pulse of light, as indicated in “Materials and Methods,” and returned to darkness for 4 h before harvesting. The number above each bar represents the derepression by a red-light pulse or the ratio of red light to the basal dark (D) level. CAB mRNA was not detected in the cue3 samples. n.d., None detected.
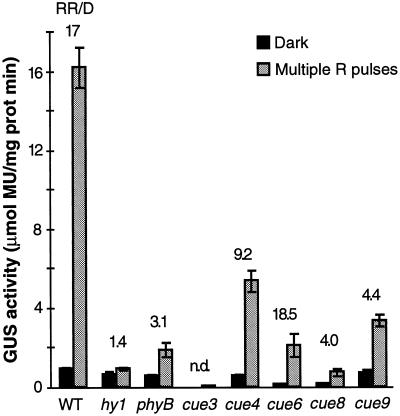
As a control, the expression of eIF4A, a translation initiation factor, was shown to be unaffected by the red-light pulse in the wild type and in the mutants (data not shown). Similar trends were observed in the cue mutants when GUS accumulation was monitored by repeated red-light pulses (Fig. 8). Although this assay measures essentially the same phenomenon as the single pulse treatment, in preliminary experiments we found it to be a more highly reproducible, quantitative measurement of the effect of photoreceptor activation. These studies demonstrated a clear reduction in the basal dark levels of GUS activity in the virescent mutants and a moderate defect in the derepression by light of the CAB3 promoter in all of the mutants except cue6.
Figure 8.
Induction by repeated red-light (R) pulses of the accumulation of CAB3-driven GUS activity in cue mutant seedlings. A pulse of light of 11 μmol photons m−2 s−1 was given for 15 min every 6 h from germination. Otherwise the seedlings remained in the dark. GUS activity was measured after 7 d, in three samples of six seedlings each. The number above each genotype represents the derepression by the pulses or the ratio of GUS activity following a red-light pulse to the dark (D) value. Error bars = sd. MU, 4-Methylumbelliferyl β-d-glucuronide; prot, protein; WT, wild type; n.d., none detected.
cue Mutants Have Defective Plastids
To examine chloroplast development in the cue mutants, we performed both light and electron microscopic analyses of leaf sections and assayed amounts of LHCP and POR proteins in light-grown and etiolated seedlings. Together, the analyses indicate a correlation among reduced greening, defects in mesophyll structure, and delayed differentiation of chloroplasts.
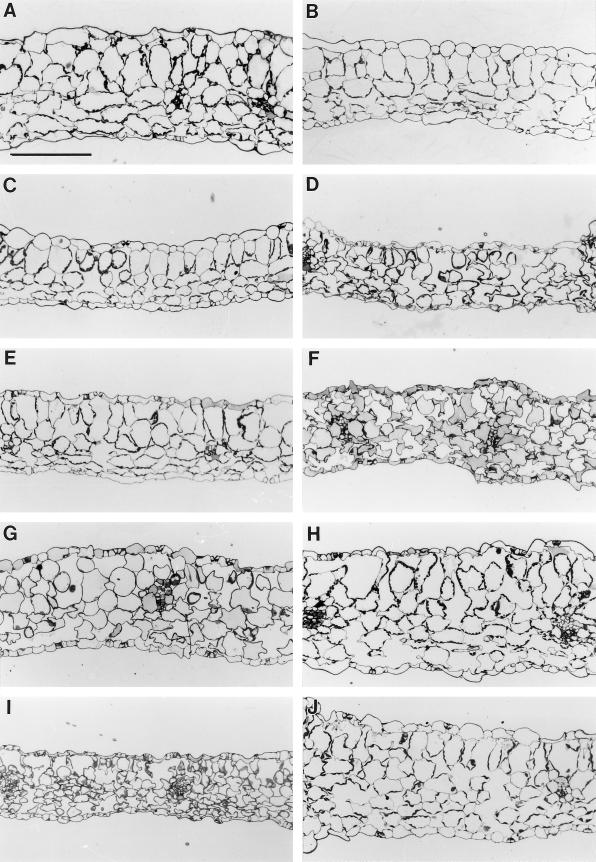
Cross-sections of leaves from the phytochrome-deficient and cue4 mutants (Fig. 9) indicate a reduced leaf thickness compared with the wild type (Fig. 9, B, C, and E). In other respects, these leaves appear normal. In contrast, cue3, cue6, cue8, and cue9 showed areas in which air spaces appear in the palisade mesophyll (Fig. 9, D, F, I, and J). This phenomenon, which is also observed in some mutants with a reticulate phenotype, has been interpreted to be a result of reduced growth and division rates of mesophyll cells, resulting in hollow spaces when the epidermis and vascular bundles expand (E. Kinsman and K. Pyke, personal communication). The virescent cue6 mutant showed a gradual transition from abnormal, underdeveloped mesophyll in young leaves to almost wild-type tissue in the center of mature leaves (Fig. 9, F–H).
Figure 9.
Light microscopy of leaf sections of the different cue mutants. A, Wild type; B, hy1-6.2; C, phyB-17.6; D, cue3; E, cue4; F, cue6 young leaf, toward the margin (mostly pale tissue); G, cue6 young leaf, toward the midvein (increased greening); H, cue6 mature leaf (greenest tissue for this mutant); I, cue8; J, cue9 (section bordering the midvein, to the left). Bar in A = 100 μm; all panels are to same scale.
The electron micrographs shown in Figure 10 indicate that the greening defect is associated with a reduction in both plastid size and the size of the granal stacks in the various mutants. Although detailed quantitation was not carried out, Figure 10 shows chloroplasts representative of 10 to 60 recorded chloroplasts (and a much greater number visually inspected). As previously described (Chory et al., 1989a), thin grana were found in hy1 (Fig. 10B). The three virescent mutants, cue3, cue6, and cue8, had the least developed grana (Fig. 10, D, G, and I). cue4 had the mildest phenotype and the greatest area of appressed thylakoids (Fig. 10E). The defect in organelle development was restricted to chloroplasts, since mitochondria appeared normal in all cases (Fig. 10, insets). The delayed greening in cue6 appeared to correlate with a transition from proplastids at the margins of young leaves to mature chloroplasts in green tissues (Fig. 10, F–H). A similar transition was observed for cue3 and cue8 leaves (data not shown). We also noticed that chloroplasts from the cue9 mutant, which exhibits some reticulation in the leaves, were more differentiated in cells close to the midvein than in the mesophyll (Fig. 10, compare J and K).
Figure 10.
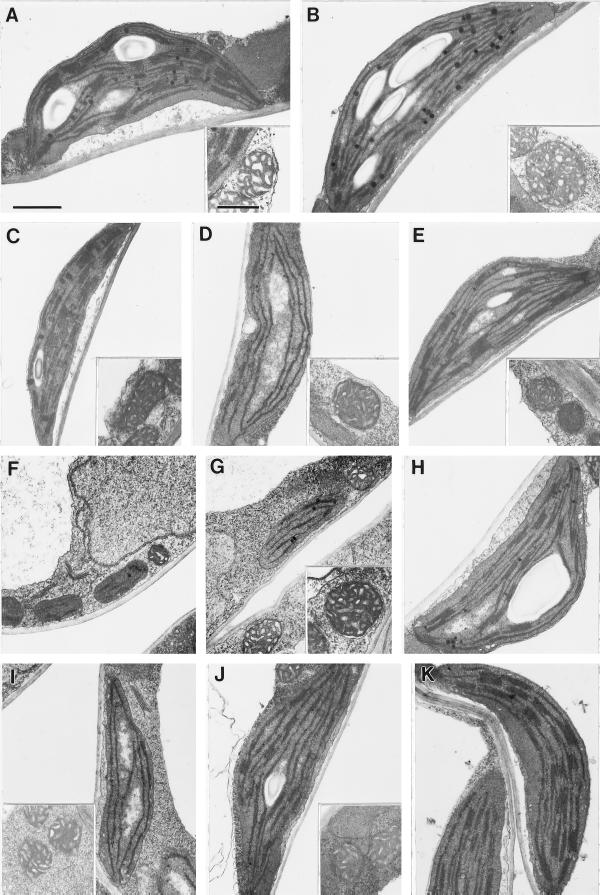
Ultrastructure of plastids from each of the cue mutants at stages of intermediate greening and at different stages for cue6. Mitochondria shown for reference in each case (inset, except for cue6). A, Wild type; B, hy1-6.2; C, phyB-17.6; D, cue3, from young leaf, toward midvein; E, cue4; F, cue6, proplastids from margin of young leaf; G, cue6, from young leaf toward the midvein (intermediate greening); H, cue6, chloroplast from mature leaf; I, cue8, from young leaf, toward midvein; J, cue9, from mesophyll cell not close to the midvein; K, cue9, from cell close to central vascular bundle. Bar in A = 1 μm; all panels are to same scale. Bar in A, inset = 0.5 μm; all insets are to same scale.
To further assess chloroplast development, the accumulation of LHCP was quantified by western analysis using total proteins from leaf tissue. Figure 11A shows that the amounts of LHCP were reduced in all of the cue mutants to an extent that was related to the accumulation of chlorophyll (Fig. 5) and the degree of chloroplast development (Fig. 10).
Figure 11.
Accumulation of chloroplast (LHCP) and etioplast (POR) marker proteins in each cue mutant. Total protein was extracted initially in SDS-urea solution at 80°C, followed by acetone precipitation and SDS-PAGE. The top membrane contained 8 μg per sample of protein from green leaves analyzed with an antibody against LHCP. The bottom membrane contained the same amount of protein from etiolated seedlings analyzed with an antiserum against total POR. WT, Wild type.
To examine whether the reduced dark CAB expression phenotypes in the virescent cue mutants correlated with a defect in etioplast development, levels of POR protein were quantified by western analysis. POR accumulates in etioplasts and is the most abundant constituent of the prolamellar body. We found a parallel between POR accumulation and the defect in CAB expression in etiolated seedlings. For example, Figure 11B shows that there is a large reduction in POR accumulation in the virescent mutants cue3, cue6, and cue8, which also show reduced basal levels of CAB. Virtually no POR was detected in cue3. Unexpectedly, dark-grown hy1 appeared to have a slightly lower POR content than phyB, which was confirmed by a dilution series (results not shown).
Double-Mutant Studies
Genetic epistasis studies were performed to determine whether the genes defined by the cue mutations (or processes depending on them) play a role in light-signal transduction. We crossed cue6 with representatives of each class of mutants identified in the screen, including the phytochrome mutants. It should be noted that the nature of the mutations in the CUE alleles is unknown; however, the phyB mutation appears to be a null allele, because no PHYB protein was detected on western blots (E. López-Juez and M. Furuya, unpublished observations). Figure 1 shows the visible phenotypes of the double mutants. cue6hy1 and cue6phyB show the long hypocotyl and petiole defects of the phytochrome-deficient mutants and the virescent phenotype of cue6. This argues against a role for CUE6 in light-controlled cell elongation. However, Table II shows that, although the cue6phyB double mutant had delayed greening, the reduction in chlorophyll levels was not additive with phyB (levels of 29% of wild type would have been expected if these mutations were additive). In contrast, cue8 appeared to be fully epistatic to cue6, whereas cue6 and cue1 resulted in fully additive chlorophyll deficiencies. The phenotype of cue6cue4 appeared by visual inspection to be closer to that of cue6. This lack of additive effects suggests that the pathways affected by cue6 and phyB or cue6 and cue8 (and perhaps cue4) do overlap at least partially.
Table II.
Greening phenotype of double mutants, measured as microgram chlorophyll per gram fresh weight in 7-d-old seedlings
| Genotype | Total Chlorophyll (% of wild type) |
|---|---|
| pOCA108 | 100 |
| phyB | 55 |
| cue1 | 52 |
| cue6 | 53 |
| cue8 | 13 |
| cue6:phyB | 43a |
| cue6:cue1 | 29 |
| cue6:cue8 | 14a |
Indicates a lack of additivity of the reduction in chlorophyll content.
DISCUSSION
The cue Phenotype Defines a Specific Subset of Chloroplast Development Mutants
We demonstrated in this study that the screen for Arabidopsis cue mutants can be used to successfully identify mutants in light signaling. However, the cue phenotype probably defines a relatively broad class of mutants, as indicated by the number of loci identified and the fact that within related phenotypes we did not find multiple alleles at a single locus.
We did not anticipate that most of the cue mutants would have defects in greening, since it has been shown that large reductions in CAB expression do not necessarily result in defective antenna accumulation (Flackmann and Kuhlbrandt, 1995). Likewise, reductions in LHCP are not necessarily correlated with defects in CAB mRNA accumulation (Zhang et al., 1992). That the cue mutants isolated have visible pale phenotypes suggests that the loci identified may encode products that function in the plastid. However, the screen has probably defined a specific subset of plastid defects, since many pale mutants do not have reduced levels of CAB. For instance, we obtained several mutants with pale phenotypes in which CAB expression was very close to normal.
The Arabidopsis pale cress (Reiter et al., 1994), reticulata (Li et al., 1995), and chlorina3 (Carol et al., 1996) mutants also have very pale or albino phenotypes but close to wild-type levels of CAB expression. In addition, Arabidopsis transgenic plants with reduced expression of an ankyrin-repeat-containing gene possess a phenotype remarkably similar to that of cue6, and yet CAB expression is normal in the chlorotic tissues (Zhang et al., 1992). Furthermore, a large number of mutants that have pale or albino phenotypes have a defect in carotenoid biosynthesis (Robertson, 1975). None of the cue mutants so far examined (cue4, cue6, and cue8) are carotenoid deficient (H.P. Mock and B. Grimm, personal communication). No dark phenotype would be expected of mutants affected in carotenoid biosynthesis. The cue mutants thus belong to a third class of chloroplast-defective mutants, including, as extreme cases, Arabidopsis cla1 (Mandel et al., 1996) and Antirrhinum dag (Chatterjee et al., 1996), which show fully arrested plastid development and absence of CAB expression. Fortunately, the fact that the cue mutants described result only in partial defects in plastid development allows the interaction between phytochrome and plastid signals to be uncovered. It is also possible that some of the cue mutations define genes directly required for the control of CAB expression and that the greening defect is secondary to this effect.
The cue Phenotype Can Be Explained by a Close Association between Plastid and Phytochrome Signal Transduction
A central issue in the characterization of the cue mutants has been to determine the basis for the cue phenotype in each case. Three possible interpretations can be envisaged: (a) CAB expression is affected directly through a failure to transduce light signals, (b) CAB expression is affected directly through a regulatory pathway unrelated to light, or (c) CAB expression is affected indirectly through a defect in plastid development. Assuming that plastid function and light-signal transduction are independent, we predicted that light-signal transduction mutants would have wild-type basal levels of CAB mRNA, with a defect specifically in derepression of CAB expression by light pulses.
Conversely, the expectation was that CAB expression would be derepressed normally by light pulses in plastid function mutants. The results for two of the five new cue mutants indicated both a dark phenotype and a light induction defect. This fact, together with the evidence for defective chloroplast and etioplast development, forced us to revise our basic assumption: plastid function and phytochrome signal transduction may in fact be closely related. Whereas functional CUE proteins are required prior to the perception of light pulses, the same proteins or their products must be present during or after the light pulse for a full response to take place. This leads us to propose a role for plastids in both the expression of CAB in the dark and its induction by phytochrome.
Do Plastids Play a Direct Role in Phytochrome Signal Transduction?
The issue of whether plastid signaling is related to the control of nuclear gene expression by light has been addressed in the past. It has been argued that, although light and plastid control of promoter activity were presumed to be separate processes, part of the overall effect of light on chloroplast development might be due not to the direct effect of light on transcription but to the subsequent increase in plastid-signaling capacity associated with the developing chloroplast (Mayfield, 1990). One study addressed the possibility of a direct relation on the control of transcript accumulation using the albostrians mutant of barley. This nuclear mutation results in the lack of plastid ribosomes in cells of “white” tissue and in a very reduced expression of nuclear genes for photosynthetic proteins. The remaining expression was shown to be light regulated (Hess et al., 1994), and it was therefore proposed that light regulation was independent of the plastid signal. However, the fact that the mutant accumulates some chlorophyll in very pale tissues indicates that the deficiency in plastid translation is not complete. Since the induction by light in pale and green tissues was not quantified, the possibility that a partial defect exists in both plastid function and light induction cannot be ruled out.
We have previously described evidence suggesting that translationally functional plastids are required for normal phytochrome derepression of CAB expression. Growth of seedlings in the dark in the presence of the organellar protein-biosynthesis inhibitor chloramphenicol results in a gene-expression phenotype very similar to that observed in cue3, cue4, cue8, and cue9: the basal level of CAB mRNA is reduced and the induction by light is also impaired (López-Juez et al., 1996). In agreement with the features of the previously characterized plastid signal in mustard (Oelmüller et al., 1986), basal and light-induced mRNA levels were affected only when organellar translation was inhibited early but not after 48 h of seedling growth. When seedlings are grown in the dark on medium containing the carotenoid-biosynthesis inhibitor norflurazon, CAB expression is still normally derepressed by a light pulse; however, a high-fluence, high-intensity pulse capable of driving the synthesis and subsequent photooxidation of some amount of chlorophyll results in reduced derepression compared with a low-fluence-rate pulse (E. López-Juez and J. Chory, unpublished results). In the absence of norflurazon it is the high-fluence pulse that leads to higher CAB expression.
Two alternative models could account for these observations (López-Juez et al., 1996). In the first, a plastid signal would control the amplitude of the phytochrome-induced signal (or vice versa), with both signals being required simultaneously but acting through separate intermediates. In a second model, the primary target of the phytochrome-induced signal would be the plastid rather than nuclear genes such as CAB. This signal, modulating plastid activity, would subsequently be relayed to the nucleus from the organelle. Several sites of action for the CUE gene products are possible in both models. If the plastid-derived and phytochrome-induced signals are distinct but simultaneously required, we predict that the cue mutations primarily affect plastid function. This would be the most likely explanation for their altered etioplast phenotype. If the plastids actually mediate phytochrome signaling, then the CUE gene products could play a role either inside the plastid or upstream of it, in a signaling pathway between phytochrome and the plastid. A direct role for some of the CUE genes in light signal transduction in spite of their mutant phenotype in the dark is also a possibility if one assumes a residual flow through the phytochrome signal transduction system in dark-grown seedlings. Either model predicts a close interaction between the phytochrome-induced and plastid-derived signals and could explain the observed epistasis (lack of additivity) between the phytochrome and cue mutations.
Similarities and Differences among the cue Mutants
Each mutant described here shows unique features, making it likely that different primary processes are affected in each case. The cue3 mutation results in the most dramatic plastid defect, since POR protein levels were barely detectable. CAB expression was also undetectable in young cue3 seedlings, although the expression of a control gene (eIF4A) was not affected. The reduction in CAB expression in cue4 can be ameliorated under lower light (40 μmol photons m−2 s−1; data not shown), and the expression in the dark was also similar to that in the wild type.
Like cue3, cue6 and cue8 are probably involved in processes required for normal plastid development, even in the dark. cue6 is unique because it is the only mutation that does not reduce the effectiveness of light pulses in derepressing CAB expression. cue9 shows the highest level of seedling anthocyanin and the greatest reduction in CAB derepression by light pulses, making its gene product particularly interesting. A phenomenon of negative reciprocal control has been proposed between the separate signal transduction pathways inducing photosynthetic gene expression and anthocyanin biosynthetic gene expression (Bowler et al., 1994). The CUE9 gene product could be an element in the former pathway prior to the source of the negative crosstalk signal. Only the molecular identification of the mutated genes will allow the assignment of functions to each of the CUE genes.
Greening-defective mutants have been widely characterized in other species, particularly in the cereals. As in Arabidopsis, one abundant class affects chloroplast structural components or chlorophyll biosynthesis itself without affecting CAB gene expression (Taylor, 1989; Knoetzel and Simpson, 1991), and another common class is primarily defective in carotenoid biosynthesis (Robertson, 1975). A nuclear mutation in the grass Lolium temulentum results in a slow-to-green phenotype and aberrant plastids (Oughan et al., 1992), but the nature of the mutation and its effect on the expression of genes such as CAB are not known.
A slow-greening phenotype is displayed by cr88, a recently described Arabidopsis mutant defective in the light regulation of nitrate reductase (Lin and Cheng, 1997). This mutant does in fact show reduced CAB expression, as well as a long-hypocotyl phenotype. The mutant maps to a position clearly distinct from any of the cue mutations described here (C.L. Cheng, personal communication). CR88 appears to play a role upstream of the CUE gene products, controlling both morphological and photosynthetic gene expression responses.
The proposed notion that phytochrome regulation of nuclear photosynthetic gene expression could be a manifestation of the underlying plastid-nuclear-signaling mechanism would explain in a simple way the multiplicity of regulatory mechanisms observed for the CAB genes. Remarkably, studies published to date that have analyzed short promoter elements required for light or plastid response of photosynthetic genes have so far failed to distinguish between light- and plastid-responsive sequences (Argüello-Astorga and Herrera-Estrella, 1996; Bolle et al., 1996). It would be interesting to assay light-responsive photosynthetic gene expression in cells devoid of plastids. The existence of plastid-division mutants in which some cells can be found to be completely devoid of plastids (Robertson et al., 1995) would make these kinds of experiments possible.
ACKNOWLEDGMENTS
We thank the members of the Chory laboratory, in particular Drs. R. Larkin, S. Streatfield, and M. Surpin, as well as Dr. K. Pyke and Prof. J. Bowyer at Royal Holloway, for their helpful discussions in the course of this investigation.
Abbreviations:
- CAB
chlorophyll a/b-binding protein
- LHCP
light-harvesting chlorophyll protein
- POR
protochlorophyllide oxidoreductase
Footnotes
This work was supported by a grant from the U.S. Department of Energy (no. ER13993 to J.C.). While at The Salk Institute, E.L.-J. was a fellow of the Spanish Ministry of Education and of the North Atlantic Treaty Organization. R.P.J. is a long-term fellow of the International Human Frontier Science Program Organization. J.C. is an associate investigator at the Howard Hughes Medical Institute.
LITERATURE CITED
- Ahmad M, Cashmore AR. The pef mutants of Arabidopsisthaliana define lesions early in the phytochrome signaling pathway. Plant J. 1996;10:1103–1110. doi: 10.1046/j.1365-313x.1996.10061103.x. [DOI] [PubMed] [Google Scholar]
- Argüello-Astorga GR, Herrera-Estrella LR. Ancestral multipartite units in light-responsive plant promoters have structural features correlating with specific phototransduction pathways. Plant Physiol. 1996;112:1151–1166. doi: 10.1104/pp.112.3.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew DM, Bartley GE, Scolnik PA. Abscisic acid control of RBCS and Cab transcription in tomato leaves. Plant Physiol. 1991;96:291–296. doi: 10.1104/pp.96.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bolle C, Kusnetsov VV, Herrmann RG, Oelmüller R. The spinach AtpC and AtpD genes contain elements for light-regulated, plastid-dependent and organ-specific expression in the vicinity of the transcription start sites. Plant J. 1996;9:21–30. doi: 10.1046/j.1365-313x.1996.09010021.x. [DOI] [PubMed] [Google Scholar]
- Bowler C, Yamagata H, Neuhaus G, Chua N-H. Phytochrome signal transduction pathways are regulated by reciprocal control mechanisms. Genes Dev. 1994;8:2188–2202. doi: 10.1101/gad.8.18.2188. [DOI] [PubMed] [Google Scholar]
- Carol P, Perez P, Begot L, Seyer C, Rocipon M, Mache R (1996) Transposon tagging of genes important for chloroplast function. 7th International Conference on Arabidopsis Research (abstract). Norwich, UK, p 212
- Chatterjee M, Sparvoli S, Edmunds C, Garosi P, Findlay K, Martin C. DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J. 1996;15:4194–4207. [PMC free article] [PubMed] [Google Scholar]
- Chory J. Light signals in leaf and chloroplast development: photoreceptors and downstream responses in search of a transduction pathway. New Biol. 1991;3:538–548. [PubMed] [Google Scholar]
- Chory J, Peto CA, Ashbaugh M, Saganich R, Pratt LH, Ausubel FM. Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell. 1989a;1:867–880. doi: 10.1105/tpc.1.9.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto CA, Feinbaum R, Pratt LH, Ausubel FM. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell. 1989b;58:991–999. doi: 10.1016/0092-8674(89)90950-1. [DOI] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβ homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- Dijkwel PP, Huijser C, Weisbeek PJ, Chua N-H, Smeekens SCM. Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell. 1997;9:583–595. doi: 10.1105/tpc.9.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. Light control of plant development. Annu Rev Cell Dev Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- Flackmann R, Kuhlbrandt W. Accumulation of plant antenna complexes is regulated by posttranscriptional mechanisms in tobacco. Plant Cell. 1995;7:149–160. doi: 10.1105/tpc.7.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores S, Tobin EM. Cytokinin modulation of Lhcp messenger-RNA levels—the involvement of post-transcriptional regulation. Plant Mol Biol. 1986;11:409–415. doi: 10.1007/BF00039021. [DOI] [PubMed] [Google Scholar]
- Green BR, Salter AH. Light regulation of nuclear-encoded thylakoid proteins. In: Andersson B, Salter AH, Barber J, editors. Molecular Genetics of Photosynthesis. Oxford, UK: Oxford University Press; 1996. pp. 75–103. [Google Scholar]
- Hess WR, Muller A, Nagy F, Borner T. Ribosome-deficient plastids affect transcription of light-induced nuclear genes: genetic evidence for a plastid-derived signal. Mol Gen Genet. 1994;242:305–312. doi: 10.1007/BF00280420. [DOI] [PubMed] [Google Scholar]
- Jacobs M, Dolferus R, van den Bossche D. Isolation and biochemical analysis of ethyl methanesulfonate-induced alcohol dehydrogenase null mutations of Arabidopsis thaliana (L.) Heynh. Biochem Genet. 1988;26:105–122. doi: 10.1007/BF00555492. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Karlin-Neumann GA, Sun L, Tobin EM. Expression of light-harvesting chlorophyll a/b-protein genes is phytochrome-regulated in etiolated Arabidopsisthaliana seedlings. Plant Physiol. 1988;88:1323–1331. doi: 10.1104/pp.88.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM, eds (1994) Photomorphogenesis in Plants, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Knoetzel J, Simpson D. Expression and organisation of antenna proteins in the light-sensitive and temperature-sensitive barley mutant chlorina-104. Planta. 1991;185:111–123. doi: 10.1007/BF00194522. [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using codominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- Li H-M, Culligan K, Dixon RA, Chory J. CUE1: a mesophyll cell-specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell. 1995;7:1599–1610. doi: 10.1105/tpc.7.10.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Cheng CL. A chlorate-resistant mutant defective in the regulation of nitrate reductase gene expression in Arabidopsis defines a new HY locus. Plant Cell. 1997;9:21–35. doi: 10.1105/tpc.9.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Juez E, Hughes M. Effect of blue light and red light on the control of chloroplast acclimation of light-grown pea leaves to increased fluence rates. Photochem Photobiol. 1995;61:106–111. [Google Scholar]
- López-Juez E, Streatfield S, Chory J (1996) Light signals and autoregulated chloroplast development. In W Briggs, RL Heath, EM Tobin, eds, Regulation of Plant Growth and Development by Light, ASPP Symposium Series. American Society of Plant Physiologists, Rockville, MD, pp 144–152
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, Leon P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 1996;9:649–658. doi: 10.1046/j.1365-313x.1996.9050649.x. [DOI] [PubMed] [Google Scholar]
- Mayfield SP. Chloroplast gene regulation: interaction of the nuclear and chloroplast genomes in the expression of photosynthetic proteins. Curr Opin Cell Biol. 1990;2:509–513. doi: 10.1016/0955-0674(90)90135-2. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Kay S. Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:15491–15496. doi: 10.1073/pnas.93.26.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miséra S, Müller AJ, Weiland-Heidecker U, Jürgens G. The FUSCA genes of Arabidopsis: negative regulators of light responses. Mol Gen Genet. 1994;244:242–252. doi: 10.1007/BF00285451. [DOI] [PubMed] [Google Scholar]
- Mullet J. Chloroplast development and gene-expression. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:475–502. [Google Scholar]
- Oelmüller R, Levitan I, Bergfeld R, Rajasekhar VK, Mohr H. Expression of nuclear genes as affected by treatments acting on the plastids. Planta. 1986;168:482–492. doi: 10.1007/BF00392267. [DOI] [PubMed] [Google Scholar]
- Ougan HJ, Thomas AM, Thomas BJ, Roberts PC, Mutinda C, Hayward MD, Dalton SJ. Leaf development in Lolium temulentum L.: characterisation of a slow-to-green mutant. New Phytol. 1992;122:261–272. doi: 10.1111/j.1469-8137.1992.tb04230.x. [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Quail PH. Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell. 1991;3:1177. doi: 10.1105/tpc.3.11.1177. 1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]
- Reed JW, Nagatani A, Elich T, Fagan M, Chory J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S, Reinbothe C, Lebedev N, Apel K. PORA and PORB, two light-dependent protochlorophyllide-reducing enzymes of angiosperm chlorophyll biosynthesis. Plant Cell. 1996;8:763–769. doi: 10.1105/tpc.8.5.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter RS, Coomber SA, Bourett TM, Bartley GE, Scolnik PA. Plant Cell. 1994;6:1253–1264. doi: 10.1105/tpc.6.9.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DS. Survey of the albino and white endosperm mutants of maize: their phenotypes and gene symbols. J Hered. 1975;66:67–74. [Google Scholar]
- Robertson EJ, Pyke K, Leech RM. arc6, an extreme chloroplast division mutant of Arabidopsis also alters proplastid proliferation and morphology in shoot and root apices. J Cell Sci. 1995;108:2937–2944. doi: 10.1242/jcs.108.9.2937. [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- Taylor WC. Regulatory interactions between nuclear and plastid genomes. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:211–233. [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Thompson WF, White MJ. Physiological and molecular studies of light-regulated nuclear genes in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:423–466. [Google Scholar]
- Wagner D, Hoecker U, Quail PH. RED1 is necessary for phytochrome B-mediated red light-specific signal transduction in Arabidopsis. Plant Cell. 1997;9:731–743. doi: 10.1105/tpc.9.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng JR, Caron P, Anderson ML, Cowl JS, Harberd NP. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR. The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 1996;10:691–702. doi: 10.1046/j.1365-313x.1996.10040691.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Scheirer DC, Fowle WH, Goodman HM. Expression of antisense or sense RNA of an ankyrin repeat-containing gene blocks chloroplast differentiation in Arabidopsis. Plant Cell. 1992;4:1575–1588. doi: 10.1105/tpc.4.12.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]