Abstract
Mirtrons are a recently described category of microRNA (miRNA) relying on splicing rather than processing by the microprocessor complex to generate pre-miRNA precursors of the RNA interference (RNAi) pathway. Their discovery and subsequent verification provides important information about a distinct class of miRNA and inherent advantages that could be exploited to silence genes of interest. These include micro-processor-independent biogenesis, pol-II-dependent transcription, accurate species generation and the delivery of multiple artificial mirtrons as introns within a single host transcript. Here we determined the sequence motifs required for correct processing of the mmu-miR-1224 mirtron and incorporated these into artificial mirtrons targeting Parkinson’s disease-associated LRRK2 and α-synuclein genes. By incorporating these rules associated with processing and splicing, artificial mirtrons could be designed and made to silence complementary targets either at the mRNA or protein level. We further demonstrate with a LRRK2 targeting artificial mirtron that neuronal-specific silencing can be directed under the control of the human synapsin promoter. Finally, multiple mirtrons were co-delivered within a single host transcript, an eGFP reporter, to allow simultaneous targeting of two or more targets in a combinatorial approach. Thus, the unique characteristics of artificial mirtrons make this an attractive approach for future RNAi applications.
INTRODUCTION
The ability to knockdown genes of interest with RNA interference (RNAi) has revolutionized the way in which gene function has been studied, while exploitation of the highly potent and sequence-specific silencing capabilities of RNAi holds potential as a therapeutic strategy (1). Several RNAi effectors have been developed in recent years. These include, but are not limited to, small interfering RNAs (siRNAs) (2), small-internally segmented interfering RNAs (sisiRNAs) (3), asymmetric interfering RNAs (aiRNAs) (4), short hairpin RNAs (shRNAs) (5), tandem-shRNAs (6) and primary-microRNA (pri-miRNA) mimics (7). While the effector used for each application must be carefully considered, it has been argued by us (8), and others (9), that pri-miRNA mimics are the safest option where long-term DNA-induced expression is desirable. Pol-III transcribed systems such as shRNAs and tandem shRNAs are difficult to spatially restrict, while high levels of expression can lead to toxic accumulation of precursors that compete with endogenous miRNAs for RNAi pathway elements (10). Both problems are limited with pri-miRNA mimics as these can be expressed off pol-II promoters. This approach can restrict expression to only desired cell populations (11), and the typically lower levels of expression result in less competition for the endogenous RNAi pathway (9,10). Further, pri-miRNA mimics can be co-delivered in pri-miRNA cluster mimics to allow combinatorial targeting from the same construct.
Mirtrons are a novel class of miRNA which use a splicing-dependent mode of biogenesis that is distinct from the canonical miRNA pathway, which typically depends on processing of pri-miRNA transcripts by the Drosha/DGCR8 microprocessor complex (12,13). Recently we have experimentally validated the biogenesis of previously predicted mammalian mirtrons and established a mirtron expression system for use in mammalian systems (14). The discovery of mirtrons has further improved our understanding of RNAi biology and added to the complexity of RNAi regulation within the cells. However, mirtrons also provide a novel way of exploiting the RNAi pathway for both research and therapeutic purposes. As with pri-miRNA mimics and pri-miRNA cluster mimics, mirtrons can in theory be expressed from a Pol-II promoter to potentially allow for spatiotemporal control of expression (15). In addition, the potential ability to deliver multiple mirtrons as introns within a host transcript compares with the multiplexing properties of the miRNA cluster mimics (7). However, mirtrons have the additional advantage over these two strategies in that they bypass Drosha processing (12,13,16). The dependence on splicing rather than Drosha processing for pre-miRNA hairpin formation implies that mirtrons would be less dependent on the canonical miRNA biogenesis pathway, which previous research has shown to be capable of saturation (9,10,17). We therefore reasoned that exploiting the mirtron pathway could be an attractive RNAi strategy for future applications.
In this report, we add artificial mirtrons to the RNAi arsenal. Specifically, we describe the characterization and engineering of the mouse miR-1224 mirtron into artificial mirtrons that target Parkinson’s disease (PD)-linked LRRK2 and α-synuclein transcripts. We further demonstrate that artificial mirtrons can initiate RNAi silencing effects comparable to those of shRNAs and have the advantages that they can be expressed from within useful host transcripts and may be paired with pol-II promoters to direct cell-type-specific expression and gene silencing. This work provides a novel gene silencing platform with unique properties that can be exploited for reverse genetics and therapeutic applications.
MATERIALS AND METHODS
Constructs
Mirtron constructs and splicing-deficient variants were designed as previously described, with annealed oligonucleotides containing intronic sequences ligated into the BbsI (NEB, MA, USA) digested pEGFP-Mirt plasmid (14). The dual mirtron variant containing miR-877 and miR-1226 was made by overlapping polymerase chain reaction (PCR) of the 3′-portion of eGFP using the previously reported pEGFP-Mirt-877 plasmid as template (14) and primers containing miR-1226 overhangs. Briefly, the 3′-fragment of eGFP in this plasmid was divided into two smaller fragments by PCR, with a miR-1226 sequence overhang incorporated into the reverse primer of the first PCR and the forward primer of the second PCR. The resultant PCR products were then added to a PCR mixture at a molar ratio of 1:1 for overlapping PCR to create a new eGFP 3′-fragment with miR-1226 inserted as an intron at the site (5′-atcgacttcaag|gaggacggcaaca-3′). This was subsequently sub-cloned back into the pEGFP-Mirt-877 backbone. Mirtrons under the control of the human Synapsin promoter were created by removing mirtron containing eGFP inserts from the pMirt vector using NheI and HindIII restriction enzymes and sub-cloning them into the AAV-6P-SEWB backbone (a kind gift from Dr Sebastian Kügler, Goettingen, Germany) (18) using complementary restriction site overhangs. Pre-miRNA expression plasmids were designed with mirtron stem–loops being transcribed off a U6 promoter. PCR was performed with Pol-III U6 promoter as template and pre-miRNA as 3′-primer as previously reported (14). PCR products were TA-cloned into the pGEM-T Easy vector (Promega) to generate pol-III expressed pre-miRNA plasmids. Target sequences of mirtrons used in the luciferase assay were synthesized as annealed complementary primers and ligated into psiCheck2.2 vector (Promega, USA) with XhoI and NotI restriction sites.
Cell culture, transfections and luciferase assays
HEK-293 cells (ATCC, CRL-1573) and SH-SY5Y cells (ATCC, CRL-2266) were cultured in DMEM supplemented with 10% fetal calf serum. For transfection, cells were grown in 24-well plates to 80% confluence before mirtrons/pre-miRNAs and target plasmid constructs were each transfected at a concentration of 500 ng/ml in HEK-293 cells with Lipofectamine 2000 (Invitrogen, USA). Luciferase activity was assessed using dual-luciferase reporter assay system (Promega, USA) and Wallac-Victor 2 plate reader as per manufacturer’s instructions at 48 h post-transfection. Ratios of renilla luciferase:firefly luciferase were obtained and normalized to specific construct controls utilizing the same promoter. Transfection of NAD and mirt-LRRK2-5NB into SH-SY5Y cells was carried out at 500 ng/ml followed by culture in media containing G418 (Sigma Aldrich, USA) selection antibiotic at a concentration of 50 µg/ml for 3 weeks. Subsequently, cells were maintained in media containing G418 at 10 µg/ml until experiments.
GFP quantification
peGFP-Mirt-transfected HEK-293 cells were lysed at indicated time points and protein content determined using the micro BCA protein assay (Pierce Biotechnology, USA). Fifty micrograms of protein was assayed for GFP fluorescence using a Wallac–Victor plate reader before subtracting background fluorescence determined from cells transfected with target alone.
RT–PCR and RT–qPCR
HEK-293 cells were lysed at 48 h post-transfection and RNA harvested using the Trizol protocol (Invitrogen). One milligram of total RNA was reverse transcribed with random hexamers using the Precision Reverse Transcription kit (PrimerDesign, UK) as per the manufacturer’s instructions. Real-time TaqMan PCR was performed with 25 ng of cDNA using TaqMan universal PCR master mix (Applied Biosystems, USA) and TaqMan Gene Expression Assay probes (LRRK2: Hs00417273_m1, Beta-actin: TaqMan beta-actin detection reagents 401 846 Dual Applied Biosystems) as per the manufacturer’s instruction.
Argonaute RNA immuno-precipitation and RT–PCR analysis
AGO1-4 RIP was carried out with modified protocol from Galgano et al. (19). HEK-293 cells were transfected with 1 µg intron variant (NAD, mirt-miR-LRRK2-5NB) together with stated FLAG-tagged AGO protein (20) (Addgene plasmids 19 887-19890) or mock plasmid. At 48 h post-transfection, cells were washed two times with PBS, trypsinized and pelleted. Cells were re-suspended in 500 µl of lysis buffer (50 mM Tris–HCL, 10 mM NaCl, 0.5% Igepal CA-630, 1 mM ethylenediaminetetraacetic acid (EDTA)) before protein concentration was determined with the Bio-Rad Protein assay (Bio-Rad, CA, USA). Subsequently, 200 µg of samples was aliquoted out for further experiments and made up to 500 µl with lysis buffer. Further analysis had subsequently been normalized to sample protein content. AGO proteins were immuno-precipitated for 1 h with 1 µg M2 anti-Flag antibody (Sigma Aldrich, USA) that had been pre-incubated with 30 µl of Dynal protein G magnetic beads (Invitrogen, USA). Beads were separated, and the lysate retained for total RNA extraction using the Trizol extraction method before re-suspension in 30 µl water. Beads were washed three times in wash buffer 1 (50 mM Tris–HCL, 150 mM NaCl, 1 mM MgCl2 0.05% Igepal CA-630) before being re-suspended in 50 µl of sodium dodecyl sulphate (SDS)–EDTA elution buffer (50 mM Tris, pH 8, 100 mM NaCl, 10 mM EDTA, 1% SDS) and heated to 70°C for 10 min. Beads were separated and frozen while elute was retained. A total of 20 µl of one replicate of elute was used for analysis of immuno-precipitation with western blotting, while remaining replicates underwent Trizol extraction of total RNA before re-suspension in 15 µl water.
For small RNA detection, 2 µl of elute or total RNA was used for small RNA stem-loop reverse transcriptase (RT)–PCR using Applied Biosystems TaqMan reverse primers and TaqMan probes against miR-16, mirt-miR-LRRK2-5NB and RNU-24 (Applied Biosystems). Stem–loop reverse transcription of miRNAs was performed with superscript III reverse transcriptase according to the manufacturer’s instructions and as previously described (21). For radioactive PCR and visualization of products, 1.33 µl of cDNA was used in 20 µl reactions with PCR Master Mix (Applied Biosystems) supplemented with 32P-dCTP. Following qPCR analysis to ensure products were in the linear range, 34 cycles were performed for miR-16, 34 cycles for RNU-24 and 39 cycles for mirt-miR-LRRK2-5NB. Samples were run on a 6% TBE-gel, immobilized using GelBond PAG film (Lonza, Switzerland) and autoradiography exposure was carried out.
High throughput sequencing
HEK cells grown in 24-well plates were transfected with 500 ng of stated mirtron. Small RNA libraries were prepared with the Small RNA v1.5 Sample Prep Kit following the manufacturer’s instructions (Illumina). Libraries were sequenced on a Genome Analyzer IIx for 36 cycles following the manufacturer’s protocols. The image analysis and base calling were done using Illumina’s GA Pipeline. Adapters were trimmed with Biopieces remove_adapter script, and remaining sequences were aligned against full-length mirtron hairpins.
Statistical analysis
Statistical significance between control and experimental values was determined using Student’s t-test (paired, two-tailed). All data are expressed as means ± standard deviation unless otherwise stated.
RESULTS
Mmu-miR-1224 is a splicing-dependent mammalian mirtron
Success in RNAi effector design has previously benefited from incorporating antisense sequences targeting genes of interest into endogenous miRNA sequences (7,22). This retains evolutionarily selected recognition sequences for efficient processing of RNAi effectors by RNAi pathway elements such that they maximize likelihood of correct processing and successful silencing. Work in our laboratory has identified the mmu-miR-1224 as a potentially attractive mirtron to engineer to target genes of interest (14). Specifically, this mirtron has a long poly-pyrimidine tract in the 3′-arm that leads to highly efficient splicing from a GFP reporter plasmid and a silencing ability comparable to U6-transcribed pre-miRNA hairpins.
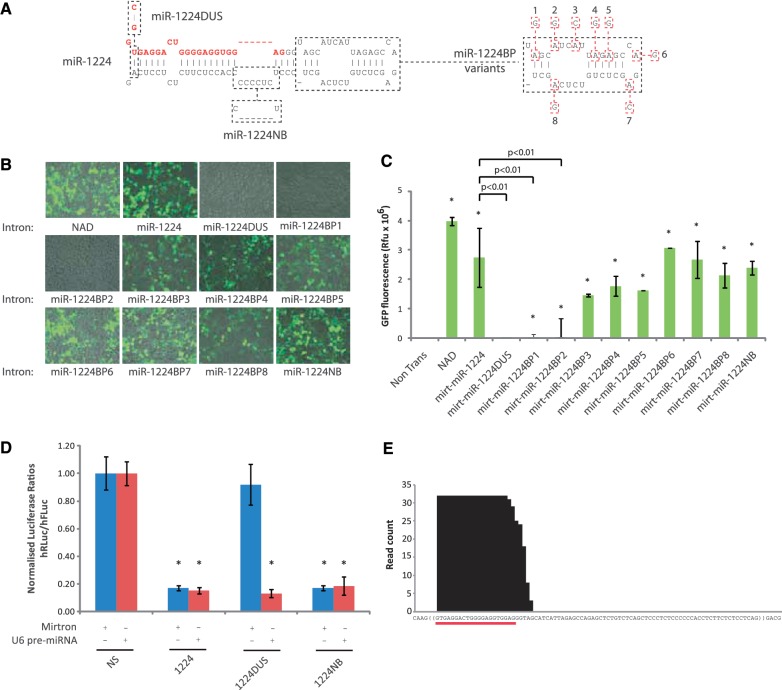
To understand the essential features of this mirtron, the 85-nt long mmu-miR-1224 sequence was placed into a modified eGFP expression plasmid, peGFP-Mirt, as an intron within the eGFP open reading frame (mirt-miR-1224) such that the splicing ability of the intron could be readily determined by the presence of eGFP fluorescence (Figure 1A). Variants of the mirt-miR-1224 mirtron were generated in which mutations were made to key regulatory sequences required for splicing (Figure 1A). Construct mirt-miR-1224DUS (double un-spliceable) included changes to both the 5′-GU and 3′-AG splice site recognition sequences located at the 5′- and 3′-termini of mmu-miR-1224, which are considered essential for splicing (Figure 1A). Constructs mirt-miR-1224BP1-8 (branch point) incorporated mutations to adenosine nucleotides located at sequential positions around the miR-1224 hairpin loop that could represent the site of 2′–5′ diester linkage at the branch point (Figure 1A). This linkage is considered essential for the formation of intermediate branched lariat precursors of the splicing reaction (23). A final construct, mirt-miR-1224NB (no bridge), was generated in which an unpaired row of 6 nt in the 3′-arm of mmu-miR-1224 was excluded in order to see how removal of this secondary structure bulge affected processing and silencing (Figure 1A).
Figure 1.
miR-1224 is splicing-dependent mammalian mirtron processed into small RNA species capable of directing RNAi. (A) Predicted hairpin alignment of murine miR-1224 with boxes indicating variant sequences modified in stated constructs. (B) Representative fluorescent microscopy images of different mirt-miR-1224 variants at 48 h after transfection in HEK-293 cells. (C) Quantification of eGFP fluorescence following 48 h expression of stated miR-1224 variants in HEK-293. Values represent mean fluorescence ±SD from n = 3. *P < 0.05 relative to non-transfected cells. Additionally displayed are P values for differences relative to the mirt-miR-1224 variant. (D) Dual-luciferase reporter assays showing knockdown of a miR-1224 target following co-transfection with indicated mirtron (blue bars) or pre-miRNA (red bars) variants. Values represent mean ratios of Renilla:Firefly luciferase ±SD from n = 6. Mirtron variants are normalized to cells transfected with the NAD variant. Pre-miRNA variants are normalised to cells transfected with a non-specific U6 pre-miRNA hairpin. *P < 0.05 relative to respective normalizing control. (E) Small RNA sequencing from mirt-miR-1224-transfected HEK-293 cells.
Following transfection of mirt-miR-1224 into HEK-293 cells, strong eGFP fluorescence was seen that was qualitatively and quantitatively comparable to that of a 91-nt control NADH-coenzyme Q reductase intron 6 (NAD intron) that lacks hairpin-forming potential (Figure 1B and C). This successful splicing and restoration of a functional eGFP mRNA transcript was abrogated by mutations made to the 5′- and 3′-consensus splice sites of mirt-miR-1224DUS as would be expected if mmu-miR-1224 was a functional intron. Interestingly, while most of the mirt-miR-1224BP variants displayed non-significant effects on eGFP production relative to the wild-type mirt-miR-1224 variant, two of these mutants, mirt-miR-1224BP1 and mirt-miR-1224BP2, almost completely eliminated eGFP production suggesting that these two nucleotides may be located in the predominant branch-point location (Figure 1B and C). These two adenines are within close proximity to one another suggesting they both contribute to the branch-point recognition sequence, although it is unclear which of the two is the site of 2′-phospho-diester linkage in the branched lariat. Collectively the data of these mirt-miR-1224BP variants suggest that maintenance of the miR-1224 loop is likely essential for efficient splicing of this intronic sequence and for the designs of artificial miR-1224 mimics. Finally, expression of mirt-miR-1224NB still resulted in eGFP expression that was quantified as being 60% relative to that of the control NAD intron, compared to 69% for mirt-1224 (Figure 1B and C). This difference was not statistically significant and suggests that removal of the secondary structure bulge that forms part of the poly-pyrimidine tract of miR-1224 may have little, if any, contribution to improving the splicing efficiency of this intronic sequence.
The miRNA derived from the 5′-arm of miR-1224 is enriched in cloning libraries of small RNAs compared to that derived from the 3′-arm indicating that it harbours the mature miRNA species (15). To test RNAi silencing of mirt-1224 variants, a dual-luciferase vector was created that carried a target sequence fully complementary to the miR-1224 5′ sequence in the 3′-untranslated region (UTR) of Renilla luciferase gene. Relative to the NAD intron, mirt-miR-1224 silenced this target by 83% (P < 0.001) (Figure 1D). This silencing was completely abolished by construct mirt-miR-1224DUS to suggest mirt-miR-1224 directed silencing in a splicing-dependent manner (Figure 1D). Despite reduced eGFP fluorescence levels when using construct mirt-miR-1224NB, silencing levels achieved by this construct were directly equivalent to mirt-miR-1224 at 83% (P < 0.001), a result that is consistent with efficient hairpin formation via RNAi processing.
To eliminate the possibility that the mutations introduced into this splicing-deficient variant directly affected knockdown of the target, U6-driven pre-miRNA controls were generated that corresponded to putatively spliced mirtron variants in order to isolate sequence-dependent effects. Expression of the pre-miR-1224DUS variant alongside the miR-1224 luciferase target sequence led to an equivalent silencing of 87% (P < 0.001) compared to the respective wild-type pre-miR-1224 variant, which silenced by 85% (P < 0.001) (Figure 1D). This confirmed that the introduced mutations abrogated silencing of mirtron variants by inhibiting splicing rather than through direct effects on target recognition and RNAi silencing. Despite the slightly reduced splicing efficiency of mirt-1224NB, the pre-miRNA variant of this construct, pre-miR-1224NB, was also able to direct a comparable silencing (81%; P < 0.001) relative to the pre-miR-1224 variant (Figure 1D). These data support a model where a strong secondary structure of the predicted hairpin of mirt-1224NB leads to efficient recognition and/or processing of this modified hairpin by RNAi pathway elements following intron splicing. These effects may compensate for mirt-miR-1224NB’s slightly reduced splicing activity.
Finally, to confirm that an RNAi mechanism was indeed responsible for the silencing effect, small RNA species were deep-sequenced from mirt-miR-1224-transfected HEK-293 cells. Within the 1 766 495 reads that mapped to human miRNAs from four pooled samples, a relatively small total of 32 reads mapped to the hairpin region of the mirt-1224 construct, all of which derived from the 5′-arm from which the predicted mature species derives (Figure 1E). No reads were detected from other regions of the mirt-1224 hairpin. This small number agrees with our previous findings that mirtrons produce guide strands in low abundance despite their relatively potent silencing efficacy when compared to other RNAi effectors (14). Moreover, the 5′-ends of all reads map to the 5′-nucleotide of the intron, thus ensuring accurate guide strand production and minimal exonuclease trimming following mirtron splicing. Taken together, the results suggest miR-1224 intron is both spliced and processed into small RNA species that can direct silencing of a complementary target sequence through an RNAi-like mechanism in a manner that is splicing dependent. The results confirm the classification of miR-1224 as a mammalian mirtron (15) and expand on previous understanding of the sequence constraints of this mirtron (14).
Design of artificial mirtrons targeting the LRRK2 sequence
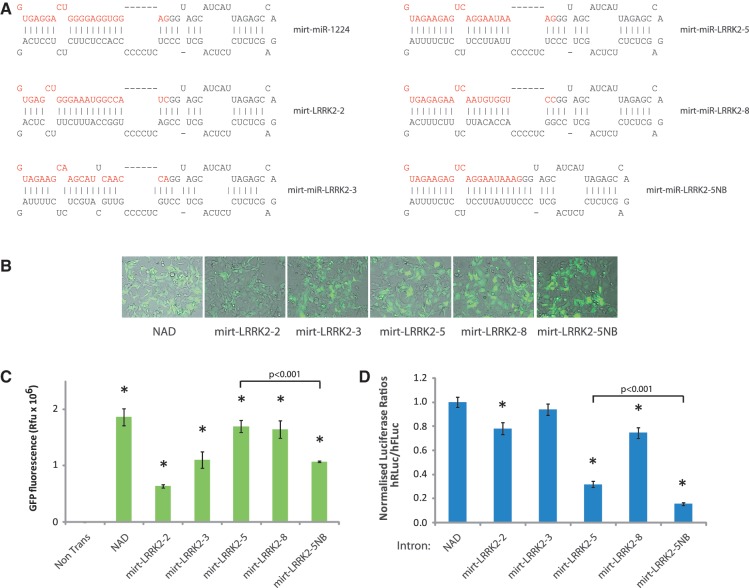
In order to design artificial mirtrons targeting genes of interest, sequences in the PD-associated LRRK2 mRNA sequence were searched for target sequences amenable to both splicing and RNAi requirements for efficient mirtron biogenesis. LRRK2 was selected given that genetic ablation of LRRK2 is protective in PD transgenic mice carrying α-synuclein mutations, suggesting that silencing of LRRK2 could be therapeutically beneficial in PD (24). Specifically, we screened the LRRK2 open reading frame (ORF) for pyrimidine-rich regions, which were complementary to the human consensus splice-site sequence of GURRR, where R is an A or G (25). This rule was applied so that the corresponding antisense species could in theory be inserted into the 5′-arm of the miR-1224 backbone. The list of candidates were then cross-compared to candidate lists defined by publically available RNAi design algorithms in order to rank potential target sequences based on their ability to fulfil both splicing and RNAi requirements. Where possible, complementary sequences to the highest rated target sequences in the RNAi algorithms were subsequently incorporated into the miR-1224 hairpin and inserted into the eGFP ORF. Of the top 10 hits against LRRK2, 4 were chosen to be incorporated into the miR-1224 backbone based on favourable alignments (Figure 2A). A fifth construct was additionally designed based on what was considered to be the most favourable alignment of construct mirt-LRRK2-5. In this construct, named mirt-LRRK2-5-No-Bridge (mirt-LRRK2-5NB), the 6-nt sequence that forms a bulge within the sense arm directly opposite the 3′-end of the antisense species of miR-1224 was removed. This was in order to investigate whether the stronger secondary structure of the hairpin would affect processing in a way similar to that shown previously for mirt-miR-1224NB.
Figure 2.
Design of artificial mirtrons targeting LRRK2. (A) Predicted hairpin alignments of LRRK2 targeting artificial mirtrons. (B) Representative fluorescent microscopy images of different artificial mirtron variants targeting LRRK2 48 h after transfection in HEK-293 cells. (C) Quantification of eGFP fluorescence following expression artificial mirtron variants targeting LRRK2 in HEK-293 cells. Values represent mean fluorescence ± SD from n = 3. *P < 0.05 relative to non-transfected cells. (D) Dual-luciferase reporter assays showing knockdown of a LRRK2 target following co-transfection with indicated mirtron variants. Values represent mean ratios of Renilla:Firefly luciferase ± SD from n = 6. *P < 0.05 relative to respective normalizing control.
Transfection into HEK-293 cells revealed that all artificial LRRK2 mirtrons were spliced to levels qualitatively comparable to the previously validated mirt-1224 construct and control NAD intron (Figure 2B). Quantification of fluorescence revealed that mirt-LRRK2-5 and mirt-LRRK2-8 were impressively 91% and 88% as efficient as the NAD intron, respectively, while the removal of the 3′-sequence in mirt-LRRK2-5NB acted to decrease the splicing ability of this construct relative to mirt-LRRK2-5, potentially as a result of a shorter poly-pyrimidine tract (Figure 2C, P < 0.001). It therefore appears that the importance of this bulge is context-dependent, and in the case of this artificial mirtron the bulge contributes to the splicing efficiency of the short intron. Co-transfection with a dual-luciferase reporter plasmid bearing a single large target sequence to all five LRRK2 mirtrons revealed that four out of five variants were able to silence the target. While mirt-LRRK2-2 and mirt-LRRK2-8 silenced the target by ∼20%, both mirt-LRRK2-5 and mirt-LRRK2-5NB were capable of directing 68% (P < 0.001) and 85% (P < 0.001) silencing, respectively (Figure 2D). Thus, while the removal of the 3′-bulge acted to reduce splicing efficiency, it had the desirable benefit of increasing the knockdown efficiency of the mirt-LRRK2-5NB variant (P < 0.001). This could be the result of a thermodynamically more stable hairpin being produced, which is processed more efficiently by the RNA-induced silencing complex. Supporting this hypothesis is the lower ΔG value for the predicted secondary structure of mirt-LRRK2-5NB using the mFold secondary structure prediction algorithm (26): mirt-LRRK2-5 = −7.88 kcal mole−1 and mirt-LRRK2-5NB = −13.76 kcal mole−1. However, comparison of silencing by U6-transcribed pre-miR-LRRK2-5NB and pre-miR-LRRK2-5 revealed near identical levels of silencing of 83% (Supplementary Figure S1). This may be because the high rate of U6-driven transcription of these hairpins masks subtle differences seen when the mature species are produced at lower levels as is the case with the mirtron variants. Irrespective of the reason for this difference, the results suggest that both variants are likely functional artificial mirtrons capable of silencing LRRK2 target sequences.
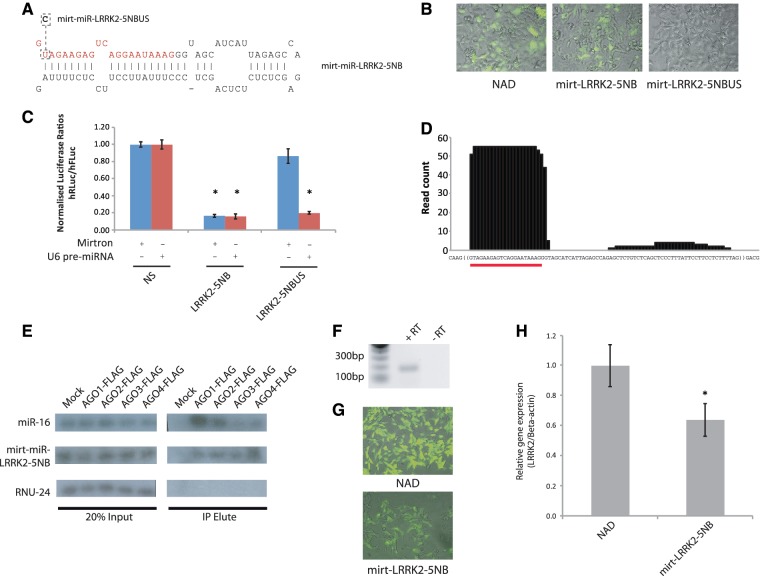
We next decided to further investigate the putative mirtron origins and capabilities of mirt-LRRK2-5NB due to its impressive silencing ability. To test whether LRRK2-5NB was indeed a functional intron, a U→C mutation was made to the 5′-splice site in construct mirt-LRRK2-5NBUS (Figure 3A). Pre-miRNA variants of both LRRK2-5NB (pre-miR-LRRK2-5NB) and LRRK2-5NBUS (pre-miR-LRRK2-5NBUS) were also generated to determine if the mutations made to mirt-LRRK2-5NBUS directly affect the silencing of this construct. As predicted, mutations made to mirt-LRRK2-5NBUS now resulted in no eGFP fluorescence when expressed for 48 h in HEK-293 cells (Figure 3B). Silencing of the LRRK2 target by mirt-LRRK2-5NBUS was also reduced to non-significant levels, suggesting that mirt-LRRK2-5NB is directing silencing in a splicing-dependent manner (Figure 3C). Indeed, both pre-miR-LRRK2-5NB and pre-miR-LRRK2-5NBUS were capable of silencing the target by 84% (P < 0.001) and 80% (P < 0.001), respectively. This means that mutations made to the unspliceable construct do not directly affect the ability of the antisense sequence to direct silencing and instead mediate their effect through abrogation of splicing (Figure 3C).
Figure 3.
mirt-LRRK2-5NB is a splicing-dependent artificial mirtron. (A) Predicted hairpin alignments for mirt-LRRK2-5NB sequences to be incorporated into the pMirt vector. Variant with modifications made to splicing regulatory sequences is indicated by dashed boxes. US = unspliceable. (B) Representative fluorescent microscopy images of different mirt-LRRK2-5NB variants 48 h after transfection in HEK-293 cells. (C) Dual-luciferase reporter assays showing knockdown of a LRRK2 target following co-transfection with indicated mirtron (left panel) and pre-miRNA control (right panel) variants. Values represent mean ratios of Renilla:Firefly luciferase + SD from n = 6. Mirtron variants are normalized to cells transfected with the NAD variant. Pre-miRNA variants are normalised to cells transfected with a non-specific U6 pre-miRNA hairpin. *P < 0.05 relative to respective normalizing control. (D) Small RNA sequencing from mirt-LRRK2-5NB-transfected HEK-293 cells. (E) Radio-labelled RT–PCR of miR-16 (34 cycles), mirt-miR-LRRK2-5NB (39 cycles) and RNU-24 (34 cycles) from total RNA or FLAG immuno-precipitated elutes of cells transfected with mirt-miR-LRRK2-5NB together with FLAG-tagged AGO proteins or a non-FLAG mock transfection. (F) RT–PCR of SH-SY5Y cell total RNA using primers specific for LRRK2. Expected size 191 bp. (G) Representative fluorescent images of SH-SY5Y cells transfected with either the NAD intron or mirt-LRRK2-5NB and maintained in G418 selection marker for four weeks. (H) Relative expression of the LRRK2 transcript determined with RT–qPCR following transfection of SH-SY5Y cells with the NAD intron or mirt-LRRK2-5NB. Values represent mean expression relative to β-actin expression ± SD and n = 6. Transcript levels in the presence of mirt-LRRK2-5NB are normalized to cells transfected with NAD intron.
Next, a codon-modified, scrambled target to mirt-LRRK2-5NB was generated in order to test the sequence specificity of the interaction between LRRK2-5NB and its target sequence (Supplementary Figure S2). Specifically, the third nucleotide of each codon in the target site was changed to an alternate nucleotide such that the scrambled sequence would still be transcribed into the same amino-acid sequence due to redundancy in the triplet code. As expected, both pre-miRNA variants and mirt-LRRK2-5NB showed a decreased ability to silence the scrambled target implying that silencing is a sequence-specific effect that strongly resembles an RNAi-like mechanism (Supplementary Figure S2). This was further confirmed by the detection of small RNA species corresponding predominantly to the 5′-arm of mirt-LRRK2-5NB following deep sequencing of transfected HEK-293 cells (Figure 3D). As with miR-1224, a relatively modest 55 reads out of the 1 766 495 small RNA reads from four pooled samples mapped to mirt-miR-LRRK2-5NB. Importantly, using RNA immuno-precipitation of FLAG-tagged Argonaute (AGO) proteins revealed that these mature small RNA species were enriched in FLAG-tagged AGO1-4 elutes relative to mock-transfected cells in a similar way to the canonical miRNA, miR-16, but not the small nucleolar RNA, RNU-24 (Figure 3E). Small RNA detection of miR-16 and mirt-miR-LRRK2-5NB in total RNA fractions is expected to be due to mature RNA present in endogenous AGO1-4 proteins that is not immuno-precipitated by the FLAG antibody. Collectively, these results provide compelling evidence that artificial mirtrons function through the RNAi pathway to silence cognate target sequences.
We next investigated how artificial mirtrons compared to existing RNAi methodologies. To do this, identical 2.5ƒm amounts of the LRRK2-5NB hairpin sequence in either a U6-transcribed pre-miRNA or mirtron context were transfected with the dual-luciferase target. Encouragingly, the silencing by these two constructs at these equivalent concentrations was near identical (91% pre-miR-LRRK2-5NB, 87% mirt-miR-LRRK2-5NB, P < 0.01) when compared to the target alone condition, suggesting that mirtrons have comparable efficacy to pre-miRNA/shRNA effectors (Supplementary Figure S3).
As a final test of LRRK2-5NBs potential, attempts were made to detect knockdown of endogenous, full-length LRRK2 transcripts by this artificial mirtron. RT–PCR was initially used to determine that LRRK2 was expressed in the human dopaminergic SH-SY5Y cell line (Figure 3F). Subsequently, constructs containing the NAD intron and mirt-LRRK2-5NB were transfected into SH-SY5Y cells and cells grown in G418 selection marker to enrich for transfection for 4 weeks. Analysis of eGFP-fluorescence from the NAD intron and LRRK2-5NB constructs confirmed that nearly all surviving cells at this time produced eGFP-fluorescence (Figure 3G). Furthermore, endogenous LRRK2 transcripts were reduced 36% (P < 0.05) to confirm that artificial mirtrons may also effectively direct silencing of endogenous gene targets (Figure 3H). The reduction in silencing relative to the dual-luciferase assays may be due to a less accessible target site within the secondary structure of the full-length LRRK2 transcript. Alternatively, 3′-UTR targeting for the dual-luciferase reporter biases incorporation of guide strands into AGOs 1–4, whereas only AGO2 can function when targeting the coding sequence. Arguing against this second point and favouring the first is the fact that we have previously designed shRNAs against 3′-UTR luciferase targets that work well against full-length open reading frames (27,28), suggesting that AGO protein availability is not the limiting factor in the silencing effect. Attempts were made to transiently transfect U6 hairpins into SH-SY5Y to test whether the pre-miRNA variants also have limited silencing of LRRK2. However, the limited transfection efficiency in these cells precluded detection of a silencing effect.
Targeting of α-synuclein with miR-1224 mimics
To confirm that miR-1224 can be used as a structural backbone for artificial mirtron mimics targeting other genes of interest, we applied the previously described approach to design artificial mirtrons to the ORF of human α-synuclein that is also linked to PD. Like LRRK2, the silencing of α-synuclein has potential as a therapy for PD (29). Six artificial mirtron sequences were designed and tested against this gene, and a single variant found to be spliced and silence a corresponding luciferase and full-length target. Construct α-syn-mirt-1 was shown to splice to levels 26% of the NAD intron, and silence a fully complementary luciferase target by 26% (P < 0.05) and a full-length mCherry-tagged transcript by 27% (P < 0.05, Supplementary Figure S4). This modest level of silencing could be due to the poor splicing efficiency or alternatively due to an inefficient RNAi effector being generated following splicing. Despite the limited eGFP expression, the latter explanation is favoured since a pre-miRNA control variant of α-syn-mirt-1 was shown to silence the dual-luciferase target by a comparable 33% (P < 0.05) to the mirtron variant (Supplementary Figure S4), indicating that the mature hairpin produced is not a strong RNAi effector. Collectively, the results demonstrate that mirt-LRRK2-5NB is not unique and that artificial mirtrons based on miR-1224 can be designed targeting other genes of interest.
LRRK2-5NB can direct cell type-specific silencing when expressed off neuron-specific pol-II promoters
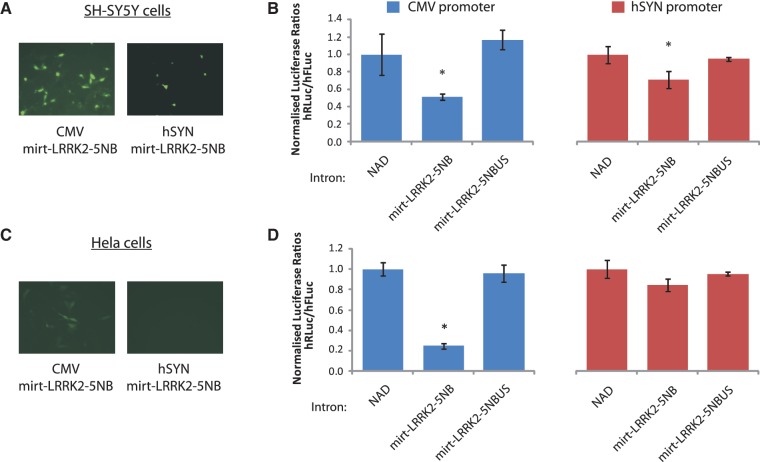
Having demonstrated that functional artificial mirtrons could be designed, we next investigated some of the putative advantages of such an artificial mirtron system. One potential benefit of using artificial mirtrons is that silencing could be directed to specific cell types of interest. To investigate this, a previously validated promoter was sourced with properties, making it suitable for pairing with the LRRK2-5NB mirtron. The human synapsin-1 (hSYN) minimal promoter has previously been demonstrated to direct neuronal-specific expression of eGFP when incorporated into an AAV-vector and delivered into the rat brain (18).
To generate hSYN-pMirt constructs, eGFP variants carrying the NAD intron, LRRK2-5NB or LRRK2-5NBUS were excised from respective pMirt vectors and ligated downstream of the hSYN promoter in the AAV-6P-SEWB expression plasmid (18). Transfection of the LRRK2-5NB dual-luciferase target together with each of the hSYN constructs was carried out in the unrelated HeLa and SH-SY5Y cell lines to test for cell specificity of expression and silencing. At the same time, CMV promoter-driven constructs were transfected under identical conditions for comparison of fluorescence and silencing efficacy between the identical constructs under the control of the different promoters. While levels of eGFP expression were somewhat smaller than in HEK-293 cells largely due to transfection efficiency, the use of a CMV promoter successfully resulted in detectable eGFP expression for the NAD and mirt-LRRK2-5NB constructs but not the mirt-LRRK2-5NBUS in both cell-lines, with ratios between the two comparable to that seen previously in HEK-293 cells (Figure 4A and C; Supplementary Figure S5). This agrees with the ubiquitous expression patterns expected of the CMV promoter. In contrast, hSYN-directed low-level expression of eGFP from NAD and mirt-LRRK2-5NB constructs in the neuronal SH-SY5Y but not in HeLa cell line (Figure 4A and C; Supplementary Figure S5). This confirms a neuronal-specific expression pattern of the hSYN promoter as reported previously.
Figure 4.
LRRK2-5NB can be expressed in a cell-type specific manner using the human synapsin promoter. (A) Representative fluorescent images of mirt-LRRK2-5NB expressed off the CMV or human synapsin promoter in SH-SY5Y cells. (B) Dual-luciferase reporter assays showing knockdown of a LRRK2 target following co-transfection with indicated mirtron variants expressed off the CMV (left panel) or human synapsin (right panel) promoter in SH-SY5Y cells. Values represent mean ratios of Renilla:Firefly luciferase ± SD from n = 3. Mirtron variants are normalized to cells transfected with the NAD variant. Pre-miRNA variants are normalised to cells transfected with a non-specific U6 pre-miRNA hairpin. *P < 0.05 relative to respective normalizing control. (C) Representative fluorescent images of mirt-LRRK2-5NB expressed off the CMV or human synapsin promoter in Hela cells. (D) Dual-luciferase reporter assays showing knockdown of a LRRK2 target following co-transfection with indicated mirtron variants expressed off the CMV (left panel) or human synapsin (right panel) promoter in Hela cells. Values represent mean ratios of Renilla:Firefly luciferase ± SD from n = 3. Mirtron variants are normalized to cells transfected with the NAD variant. Pre-miRNA variants are normalized to cells transfected with a non-specific U6 pre-miRNA hairpin. *P < 0.05 relative to respective normalizing control.
To determine if gene silencing was also directed in a cell-type-specific manner, dual-luciferase assays were performed at 48 h post-transfection. Use of the CMV promoter resulted in significant levels of silencing of the LRRK2-5NB target by 75% (P < 0.05) and 48% (P < 0.05) in Hela and SH-SY5Y cells, respectively, with LRRK2-5NBUS failing to silence the target (Figure 4B and D). However, expression with the hSYN promoter resulted in significant silencing of the target by mirt-LRRK2-5NB in SH-SY5Y cells by 29% (P < 0.05), respectively, but not in HeLa cells (Figure 4B and D). While improved promoter activity is desirable, collectively the data imply that silencing by mirtrons can be directed in a cell-type-specific manner under the hSYN promoter.
Co-delivery of multiple mirtrons within one host transcript
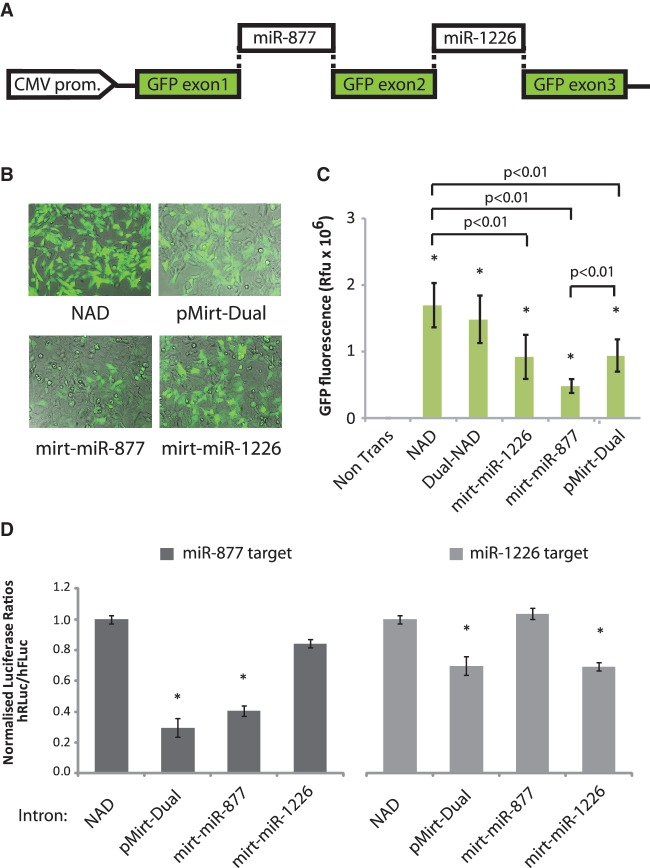
An additional advantage of using artificial mirtrons over several other RNAi strategies is that multiple mirtrons can in theory be included within a single host transcript to direct combinatorial RNAi. This can either be used to enhance silencing of the desired target by including multiple copies of the same mirtron or different mirtrons targeting the same transcript, or alternatively it could be used to silence multiple gene targets by incorporating different mirtrons. As a preliminary step towards developing this application for future studies, a dual-construct, pMirt-Dual, was produced which contained both previously validated mirtrons, mirt-miR-877 and mirt-miR-1226 (14), as introns within the eGFP transcript separated by a 90-nt exon (Figure 5A). Fluorescence microscopy and eGFP fluorescence quantification after expression of the construct in HEK-293 cells showed that splicing to produce a functional eGFP transcript was evident (Figure 5B and C). Interestingly, the pMirt-Dual construct generated fluorescence at 55% of the level of the NAD intron which is significantly greater than the single mirt-miR-877 construct (P < 0.01) and equivalent to that of mirt-miR-1226. This suggests that the splicing efficiency of mirt-miR-877 can be improved through incorporation of a stronger intron. Importantly, co-transfection with either the miR-877 or miR-1226 targets demonstrated that the dual-mirtron construct was able to silence both reporter constructs to near identical levels to each of the respective single mirtron constructs, while the single mirt-miR-877 or mirt-miR-1226 constructs were only able to silence their own respective gene targets (Figure 5D). The results confirm that multiple mirtrons can be incorporated into a single host gene to effectively silence target transcripts and suggests that mirtrons could be used for future combinatorial RNAi applications.
Figure 5.
Combinatorial delivery of multiple artificial mirtrons in a single host transcript. (A) Schematic of pMirt-dual which carries both the mirt-miR-877 and mirt-miR-1226 mirtrons. (B) Representative fluorescent images of HEK-293 cells expressing indicated mirtron variants at 48 h post-transfection. (C) Quantification of eGFP fluorescence following expression indicated intron variants in HEK-293 cells. Values represent mean fluorescence ± SD from n = 3. *P < 0.05 relative to non-transfected cells. (D) Dual-luciferase reporter assays showing knockdown of a target complementary to the 5′-arm of mirt-miR-877 (left panel) or the 3′-arm of mirt-miR-1226 (right panel) following co-transfection with indicated mirtron variants in HEK-293 cells. Values represent mean ratios of Renilla:Firefly luciferase ± SD from n = 3. Mirtron variants are normalized to cells transfected with the NAD variant. *P < 0.05 relative to respective normalizing control.
DISCUSSION
In this report, we show that artificial mirtrons represent a new type of expressed RNAi trigger to be added to the ever-growing arsenal of RNAi effectors. Specifically, we have identified key sequence elements required for splicing in the miR-1224 mirtron. These have then been incorporated into artificial miR-1224 mimics placed within the eGFP ORF, which successfully direct silencing of the human LRRK2 and α-synuclein target sequences in a splicing and RNAi-dependent manner. Finally, we have demonstrated advantages of using artificial mirtron mimics such as pol-II-regulated silencing and combinatorial delivery of multiple mirtron effectors. This work opens the use of artificial mirtrons as a new method for targeted silencing of genes-of-interest in which advantages of the mirtron system can be exploited for therapeutic benefit.
The design of artificial mirtrons is a challenging task since both splicing and then RNAi requirements must be met in order for success. Here, screening ORFs for pyrimidine-rich regions with complementarity to a theoretical purine-rich 5′ splice site based on the human consensus sequence of GURRR [where R is an A or G (25)] was used to narrow down potential candidate target sites of artificial miR-1224 mimics, which would use the 5′-arm as the mature species. Candidates were then compared against results of the same ORFs filtered through RNAi design algorithms to identify the highest scoring RNAi effectors that match the desired splicing requirements. Using this simple approach, we were able to design artificial mirtrons that successfully silenced the LRRK2 and α-synuclein sequences.
An impressive 85% silencing was achieved by one artificial mirtron, mirt-LRRK2-5NB, against a dual-luciferase LRRK2 target sequence that was directly comparable to the silencing by a U6-transcribed pre-miRNA hairpin (shRNA) targeting the same sequence. This is promising since it suggests that artificial mirtrons can silence on a par with one of the most commonly used RNAi approaches, and to levels desirable to RNAi researchers. However, this level of silencing was not seen when targeting the full-length LRRK2 endogenous transcript where 36% knockdown was demonstrated. Despite this, the difference should not detract from the proof-of-principle of using artificial mirtrons since the reduced silencing of the endogenous target can be explained by features specific to this transcript. This most likely involves a secondary structure of the endogenous LRRK2 transcript that reduces accessibility to the target site relative to the 3′-UTR of the Renilla luciferase gene. Indeed, no reduction in silencing was seen between dual-luciferase targeting and full-length targeting of α-synuclein by α-syn-mirt-1, despite the modest levels of silencing achieved, while artificial mirtrons currently in early stages of development in our lab have demonstrated >75% knockdown against full-length targets (data not shown).
A range of novel strategies for producing RNA-mediated silencing, such as small internally segmented interfering RNAs (3), asymmetric interfering RNAs (4), pri-miRNA mimics (9,28) and pri-miRNA cluster mimics (7), have been developed in recent years that address many of the issues associated with employing RNAi. This includes toxicity due to competition with the endogenous pathway, toxicity due to off-target effects, delivery specificity and prolonged duration of silencing (8). Current trends in expressed RNAi triggers, with which artificial mirtrons should be directly compared, suggest that pri-miRNA mimics are the most suitable DNA-encoded approach for therapeutic gene silencing (9,28). Alternatives of shRNAs and tandem shRNAs rely on use of ubiquitous pol-III promoters to drive expression and are therefore difficult to spatially restrict to cell types of interest using viral tropism alone. This lack of promoter-regulated specificity is a major limitation of pol-III transcribed approaches for therapeutic application and indeed of siRNAs (30,31). Furthermore, high levels of expression also lead to toxicity in various pre-clinical models due to competition with the endogenous pathway (9,10). In contrast, pri-miRNA mimics can be expressed off pol-II promoters that have potential to direct cell-type-specific silencing and drive expression at much lower levels (9,11). Both characteristics can reduce the off-target effects in heterogeneous populations of cells, the first by restricting exogenous antisense production to only the cell populations desired, and the second by providing less competition for the endogenous RNAi pathway that can be saturated (10).
It is demonstrated here that artificial mirtrons and pri-miRNA mimics share the ability to be expressed in a cell-type-specific manner using pol-II transcription. We have shown that the mirt-LRRK2-5NB mirtron could be driven by the human synapsin promoter to direct silencing in the neuronal SH-SY5Y cell line but not the cervical cancer-derived HeLa cell line. Silencing in SH-SY5Y cells was reduced from 48%% to 29% when using the human synapsin promoter rather than the CMV promoter, and it is expected that this is because either the 495-bp synapsin promoter that was used (18) was a minimal promoter variant with incomplete activity or alternatively that the synapsin promoter is not strongly activated in this cell line. Irrespective, this desirable quality of cell-type-specific silencing is possible since artificial mirtrons can be expressed within host mRNA transcripts that have elaborate initiation and termination signals bearing recognition sequences for cell-type-specific transcription factors. As such, the potential for off-target effects of artificial mirtrons within heterogeneous populations of cells is likely to be restricted both by limiting aberrant expression and expression levels, although this last point remains to be fully determined experimentally. Importantly, despite few having been exploited for RNAi at present, numerous cell-type-specific promoters have been identified, which can now be placed at the disposal of artificial mirtrons and pri-miRNA mimics (32,33).
Another promising RNAi approach being investigated for several therapeutic settings is the combination of gene silencing with other therapies. This may include delivery of multiple RNAi effectors when it is beneficial to target biological pathways rather than single targets in cancers or complex genetic diseases, or where high rates of viral mutation dictate that silencing multiple targets is required to avoid viral tolerance of silencing (34–36). Alternatively, effectors have been delivered in combination with other beneficial therapeutics such as genetically corrected target genes as a means of non-allele specific correction of disease-linked mutations (37,38). Delivery of multiple RNAi effectors and supplementary transgenes has been hampered by the difficulty of co-delivery of the multiple constructs with their respective promoters in a single vector. RNAi approaches have been devised, which can release multiple antisense species from a single transcript to limit this problem such as tandem shRNAs and pri-miRNA cluster mimics (6,7). Yet tandem shRNAs would be expected to suffer the same disadvantages as pol-III transcribed shRNA constructs, while the potency of an antisense species is lost the further it is placed away from the base of the hairpin stem to limit the number of effectors that can be delivered in one construct (39,40). Given the advantages of pri-miRNA mimics, the pri-miRNA cluster mimics are considered the optimal combinatorial RNAi delivery mechanism, although these have been scarcely investigated at present (7).
Here we show that artificial mirtrons can now be included alongside the existing approaches being used in combination therapies since two previously validated mirtrons, miR-1226 and miR-877 (14), were successfully delivered within one host construct without any loss of silencing ability in this study. The incorporation of artificial mirtrons into the ORF of eGFP also suggests that it will also be possible for mirtrons to be inserted into alternate ORFs of therapeutically beneficial transgenes to allow combinatorial therapies driven by a single promoter. Importantly, the short length of artificial mirtrons will not add significantly to the packaging size of the delivery of the host transcript even when multiple mirtrons are incorporated, making it a useful combinatorial approach where restrictions on packaging size are present such as when using certain viral approaches.
Finally, the small RNA sequencing reads of mirt-miR-1224 and the mirt-LRRK2-5NB artificial mirtron importantly show that the 5′-terminal nucleotide of the mature species generated is highly reproducible and corresponds to the first nucleotide of these short introns within eGFP. This was expected since the splicing machinery is initiated and directed by the presence of essential sequence requirements within introns (25,41,42). Recognition allows splicing to be carried out reproducibly at the same positions of a pre-mRNA transcript such that identical spliced introns are produced in each reaction. When considered in the mirtron pathway, in which the 5′- and 3′-ends of the removed intron define the termini of the pre-miRNA like substrate, it implies that accurate pre-miRNA species generation and seed-sequence definition will occur when the 5′-arm represents the mature strand. Indeed these results were reflected in the deep sequencing reads we obtained for both the natural mirtron, miR-1224, and our artificial mirtron, mirt-LRRK2-5NB. This is important since even shifts of just 1 nt of the position of the seed region can either lead to abrogated activity or aberrant off-target effects as new gene pools are targeted (43). Non-homogenous populations of both mature and pre-miRNAs have been seen following pri-miRNA processing implying that Drosha fidelity is not 100% reproducible (43). Together it suggests that this may be limiting in therapeutic approaches utilizing pri-miRNA mimics since the potential for abrogated activity or increased off-target effects would be enhanced. Collectively this point could be used to argue that artificial mirtrons should be preferable to pri-miRNA mimics for targeting genes of interest, although further supporting evidence needs to be collected before this can be concluded.
In summary, the characteristics of the mammalian mirtron pathway make the artificial mirtrons developed in this study a highly desirable RNAi effector to be added alongside existing RNAi technologies. Specifically, we have shown that artificial mirtrons can be designed that direct comparable levels of silencing to existing RNAi technologies such as U6-driven pre-miRNAs/shRNAs, silencing can be restricted in a cell-type-specific manner and that combinatorial delivery of multiple mirtrons in one host construct is possible. If the design of artificial mirtrons can be optimized further in future, then their advantages could make them one of the leading synthetic RNAi effectors available to researchers.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Medical Research Council Studentship (to C.S.); A*STAR scholarship (to Y.S.); Medical Research Council and Parkinson’s UK (to M.J.A.W.); Oppenheimer Trust and National Research Foundation (South Africa) (to M.S.W.). Funding for open access charge: Parkinson's UK research grant.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
C.R.S., Y.S. and M.S.W. designed the experiments. C.R.S. and Y.S. generated the constructs. C.R.S. performed the experiments with contributions from H.C. and Y.S. C.R.S. analysed the data. C.R.S. and M.J.A.W. wrote the article.
REFERENCES
- 1.Lares MR, Rossi JJ, Ouellet DL. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010;28:570–579. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Bramsen JB, Laursen MB, Damgaard CK, Lena SW, Babu BR, Wengel J, Kjems J. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res. 2007;35:5886–5897. doi: 10.1093/nar/gkm548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun X, Rogoff HA, Li CJ. Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat. Biotechnol. 2008;26:1379–1382. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- 5.McManus MT, Petersen CP, Haines BB, Chen J, Sharp PA. Gene silencing using micro-RNA designed hairpins. RNA. 2002;8:842–850. doi: 10.1017/s1355838202024032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saayman S, Arbuthnot P, Weinberg MS. Deriving four functional anti-HIV siRNAs from a single Pol III-generated transcript comprising two adjacent long hairpin RNA precursors. Nucleic Acids Res. 2010;38:6652–6663. doi: 10.1093/nar/gkq460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aagaard LA, Zhang J, von Eije KJ, Li H, Saetrom P, Amarzguioui M, Rossi JJ. Engineering and optimization of the miR-106b cluster for ectopic expression of multiplexed anti-HIV RNAs. Gene Ther. 2008;15:1536–1549. doi: 10.1038/gt.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sibley CR, Seow Y, Wood MJ. Novel RNA-based strategies for therapeutic gene silencing. Mol. Ther. 2010;18:466–476. doi: 10.1038/mt.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl Acad. Sci. USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen TT, Marion I, Hasholt L, Lundberg C. Neuron-specific RNA interference using lentiviral vectors. J. Gene Med. 2009;11:559–569. doi: 10.1002/jgm.1333. [DOI] [PubMed] [Google Scholar]
- 12.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 14.Sibley CR, Seow Y, Saayman S, Dijkstra KK, El Andaloussi S, Weinberg MS, Wood MJ. The biogenesis and characterization of mammalian microRNAs of mirtron origin. Nucleic Acids Res. 2011;40:438–448. doi: 10.1093/nar/gkr722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boudreau RL, McBride JL, Martins I, Shen S, Xing Y, Carter BJ, Davidson BL. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kugler S, Kilic E, Bahr M. Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area. Gene Ther. 2003;10:337–347. doi: 10.1038/sj.gt.3301905. [DOI] [PubMed] [Google Scholar]
- 19.Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010;6:e1001063. doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, Rebres R, Roach T, Seaman W, Simon MI, et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc. Natl Acad. Sci. USA. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao K, Masuda A, Matsuura T, Ohno K. Human branch point consensus sequence is yUnAy. Nucleic Acids Res. 2008;36:2257–2267. doi: 10.1093/nar/gkn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin X, Parisiadou L, Gu XL, Wang L, Shim H, Sun L, Xie C, Long CX, Yang WJ, Ding J, et al. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson's-disease-related mutant alpha-synuclein. Neuron. 2009;64:807–827. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 26.Jacobson AB, Zuker M. Structural analysis by energy dot plot of a large mRNA. J. Mol. Biol. 1993;233:261–269. doi: 10.1006/jmbi.1993.1504. [DOI] [PubMed] [Google Scholar]
- 27.Sibley CR, Wood MJ. Identification of allele-specific RNAi effectors targeting genetic forms of Parkinson's disease. PLoS One. 2011;6:e26194. doi: 10.1371/journal.pone.0026194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scholefield J, Greenberg LJ, Weinberg MS, Arbuthnot PB, Abdelgany A, Wood MJ. Design of RNAi hairpins for mutation-specific silencing of ataxin-7 and correction of a SCA7 phenotype. PLoS One. 2009;4:e7232. doi: 10.1371/journal.pone.0007232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCormack AL, Mak SK, Henderson JM, Bumcrot D, Farrer MJ, Di Monte DA. Alpha-synuclein suppression by targeted small interfering RNA in the primate substantia nigra. PLoS One. 2010;5:e12122. doi: 10.1371/journal.pone.0012122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim TE, Park MJ, Choi EJ, Lee HS, Lee SH, Yoon SH, Oh CK, Lee BJ, Kim SU, Lee YS, et al. Cloning and cell type-specific regulation of the human tyrosine hydroxylase gene promoter. Biochem. Biophys. Res. Commun. 2003;312:1123–1131. doi: 10.1016/j.bbrc.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 33.Dziedzicka-Wasylewska M, Solich J. Neuronal cell lines transfected with the dopamine D2 receptor gene promoter as a model for studying the effects of antidepressant drugs. Brain Res. Mol. Brain Res. 2004;128:75–82. doi: 10.1016/j.molbrainres.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 34.O'Grady M, Raha D, Hanson BJ, Bunting M, Hanson GT. Combining RNA interference and kinase inhibitors against cell signalling components involved in cancer. BMC Cancer. 2005;5:125. doi: 10.1186/1471-2407-5-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Grimm D, Kay MA. Combinatorial RNAi: a winning strategy for the race against evolving targets? Mol Ther. 2007;15:878–888. doi: 10.1038/sj.mt.6300116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubodera T, Yokota T, Ishikawa K, Mizusawa H. New RNAi strategy for selective suppression of a mutant allele in polyglutamine disease. Oligonucleotides. 2005;15:298–302. doi: 10.1089/oli.2005.15.298. [DOI] [PubMed] [Google Scholar]
- 38.O'Reilly M, Palfi A, Chadderton N, Millington-Ward S, Ader M, Cronin T, Tuohy T, Auricchio A, Hildinger M, Tivnan A, et al. RNA interference-mediated suppression and replacement of human rhodopsin in vivo. Am. J. Hum. Genet. 2007;81:127–135. doi: 10.1086/519025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu YP, von Eije KJ, Schopman NC, Westerink JT, ter Brake O, Haasnoot J, Berkhout B. Combinatorial RNAi against HIV-1 using extended short hairpin RNAs. Mol. Ther. 2009;17:1712–1723. doi: 10.1038/mt.2009.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saayman S, Barichievy S, Capovilla A, Morris KV, Arbuthnot P, Weinberg MS. The efficacy of generating three independent anti-HIV-1 siRNAs from a single U6 RNA Pol III-expressed long hairpin RNA. PLoS One. 2008;3:e2602. doi: 10.1371/journal.pone.0002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 42.Guth S, Martinez C, Gaur RK, Valcarcel J. Evidence for substrate-specific requirement of the splicing factor U2AF(35) and for its function after polypyrimidine tract recognition by U2AF(65) Mol. Cell Biol. 1999;19:8263–8271. doi: 10.1128/mcb.19.12.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H, Ye C, Ramirez D, Manjunath N. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA. PLoS One. 2009;4:e7566. doi: 10.1371/journal.pone.0007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.