Abstract
Polyuridylation is emerging as a ubiquitous post-translational modification with important roles in multiple aspects of RNA metabolism. These poly (U) tails are added by poly (U) polymerases with homology to poly (A) polymerases; nevertheless, the selection for UTP over ATP remains enigmatic. We report the structures of poly (U) polymerase Cid1 from Schizoscaccharomyces pombe alone and in complex with UTP, CTP, GTP and 3′-dATP. These structures reveal that each of the 4 nt can be accommodated at the active site; however, differences exist that suggest how the polymerase selects UTP over the other nucleotides. Furthermore, we find that Cid1 shares a number of common UTP recognition features with the kinetoplastid terminal uridyltransferases. Kinetic analysis of Cid1’s activity for its preferred substrates, UTP and ATP, reveal a clear preference for UTP over ATP. Ultimately, we show that a single histidine in the active site plays a pivotal role for poly (U) activity. Notably, this residue is typically replaced by an asparagine residue in Cid1-family poly (A) polymerases. By mutating this histidine to an asparagine residue in Cid1, we diminished Cid1’s activity for UTP addition and improved ATP incorporation, supporting that this residue is important for UTP selectivity.
INTRODUCTION
Many RNA metabolic pathways depend on nucleotidyl transferases to catalyze the template-independent addition of nucleotide monophosphates (NMP) at the 3′-end of RNA targets (1). These non-template-encoded nucleotides provide an additional layer of control in determining the fate of these RNAs. The best studied members of these polymerases, the poly (A) polymerases (PAPs), add poly (A) tails to the 3′-ends of RNA. In addition to the PAPs, there have been a number of polymerases identified that catalyze the addition of poly (U) tails to 3′-ends of RNAs. Over the past few years, these poly (U) polymerases (PUPs) have emerged as a potentially widespread class of polymerases involved in a previously under-appreciated mechanism of RNA metabolism (2).
Poly (U) tails have been found on a number of different RNA substrates. An early example of a poly (U) tail being added post-transcriptionally came from work showing that micro-RNA (miRNA) decay intermediates had been uridylated at their 3′-ends (3). It was also demonstrated that kinetoplastid RNAs are heavily edited by the insertion and deletion of uridine nucleotides by a group of terminal uridyl transferases (TUTases) (4). More recently, uridylation has been expanded to include a number of other targets including miRNAs, histone mRNAs and polyadenylated mRNAs (5–7). With the advent of deep sequencing techniques, it is becoming apparent that oligouridylation is likely a conserved, widespread mechanism of RNA decay (2).
One of the first described PUPs was Schizosaccharomyces pombe Cid1. Cid1 was initially identified as a PAP with residual PUP activity (8,9). However, later work demonstrated that Cid1 is specific for poly (U) incorporation in vitro and in vivo, thus reclassifying it as a PUP (10,11). This was followed by the discovery that Cid1 plays a role in the degradation of polyadenylated mRNAs, and its poly (U) activity forms the basis of a novel mRNA decay pathway in fission yeast that may be conserved in higher eukaryotes (7).
The discovery of Cid1 led to the identification of a group of six homologs in S. pombe (9), which were later shown to be conserved across eukaryotes (12). Cid1 family proteins contain conserved signature motifs placing them within the DNA polymerase β superfamily (1). A feature that appears to be unique to Cid1 family proteins is their ability to be PUPs, PAPs or potentially both depending on their context (10,13–16). A number of Cid1 family PUPs have been identified, but sequence analysis was not able to determine what makes particular Cid1 family members PUPs while others are PAPs (10). In the case of Cid1, there is a demonstrated preference for UTP incorporation, but the protein can add poly (A) tails when provided exclusively with ATP. Thus, several key questions surrounding Cid1’s selectivity remain unanswered: (i) How can Cid1 selectively incorporate UTP over ATP? (ii) How can Cid1 incorporate ATP and UTP, but not GTP and CTP?
To address these questions, we have determined crystal structures of S. pombe Cid1 alone and in complex with each ribonucleotide triphosphate (NTP) to resolutions between 2.0 and 2.7 Å (Table 1). The structures reveal that Cid1 can accommodate each of the 4 nt at the active site. However, differences in ribose and base recognition suggest a mechanism for selection of UTP and ATP over GTP and CTP. Furthermore, comparison of Cid1 with structures from trypanosomal TUTases revealed that these proteins share several common features for recognizing UTP. In order to better understand Cid1 selectivity for UTP over ATP, we measured the in vitro kinetic activity of Cid1 for UTP and ATP and found that UTP is the kinetically preferred substrate. Finally, we have identified several residues that appear to be important for UTP selectivity. In particular, we identified a histidine residue in the active site that is commonly found in confirmed Cid1 family PUPs, but this residue is replaced by an asparagine residue in confirmed Cid1 family PAPs. Mutation of this residue in Cid1 to an asparagine residue results in a decrease in PUP activity, but PAP activity is significantly improved.
Table 1.
Data collection and refinement statistics for Cid1 crystals
| Apo | MgUTP | Mg 3′-dATP | MgCTP | MgGTP | CaUTP | |||
|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||
| Wavelength (Å) | 0.9786 | 0.9786 | 0.9718 | 0.9786 | 0.9786 | 0.9786 | ||
| Space group | P212121 | |||||||
| Cell dimensions | ||||||||
| a, b, c (Å) | 53.87, 62.57, 111.16 | 53.58, 62.86, 111.50 | 53.68, 62.52, 111.21 | 53.64, 62.72, 111.84 | 53.41, 62.9, 111.44 | 53.49, 62.37, 111.21 | ||
| Resolution (Å)a | 2.0 (2.1–2.0)* | 2.3 (2.36–2.3) | 2.7 (2.770–2.7) | 2.1 (2.154–2.1) | 2.5 (2.565–2.5) | 2.5 (2.565–2.5) | ||
| Rmeasa,b | 10.1 (64.7) | 11 (38.8) | 17.7 (68.1) | 8.5 (26.5) | 11.1 (38.5) | 14.1 (53.2) | ||
| I/σIa | 12.7 (2.4) | 14.5 (4.4) | 11.28 (2.44) | 16.3 (6.9) | 13.8 (4.43) | 10.8 (3.23) | ||
| Completeness (%)a | 99.3 (97.8) | 99.4 (99.6) | 99.6 (99.7) | 99.7 (99.3) | 99.9 (99.9) | 99.7 (99.7) | ||
| Redundancya | 5 (3) | 5 (5.6) | 5.1 (5.3) | 6.5 (6.2) | 6.4 (6.6) | 5.9 (6.0) | ||
| Refinement | ||||||||
| Resolution (Å)a | 48.3–2.0 | 48.3–2.3 | 41.6–2.7 | 48.3–2.1 | 48.2–2.5 | 48.2–2.5 | ||
| No. reflections | 25923 | 17243 | 10732 | 22623 | 13540 | 13382 | ||
| Rworkc/Rfreed | 19.5/23.7 | 20.2/24.4 | 21.2/26.9 | 20.0/24.2 | 19.7/24.9 | 19.2/24.5 | ||
| No. atoms | ||||||||
| Protein | 2622 | |||||||
| NTP | N/A | 29 | 30 | 29 | 32 | 29 | ||
| Ions (Mg+2, Ca+2) | N/A | 1 | 1 | 1 | 1 | 1 | ||
| No. water molecules | 166 | 100 | 44 | 164 | 74 | 69 | ||
| Avg B-factors (Å2) | ||||||||
| Protein | 24.3 | 28.6 | 29.3 | 18.0 | 28.9 | 29.7 | ||
| NTP | N.A. | 22.8 | 52.6 | 48.6 | 63 | 18.6 | ||
| Ion (Mg+2, Ca+2) | N.A. | 28.8 | 40.15 | 38.0 | 47.5 | 23.8 | ||
| Water | 32.5 | 34.6 | 26.3 | 27.8 | 32.0 | 29.1 | ||
| R.m.s. deviationse | ||||||||
| Bond lengths (Å) | 0.009 | 0.009 | 0.009 | 0.008 | 0.009 | 0.009 | ||
| Bond angles (°) | 1.082 | 1.185 | 1.222 | 1.098 | 1.169 | 1.220 | ||
| Ramachandran favored (%) | 98.43 | 98.12 | 96.47 | 98.12 | 98.12 | 98.75 | ||
| Ramachandran disallowed (%) | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Molprobity score [percentile] | 1.23 [99th] | 1.67 [98th] | 2.11 [97th] | 1.3 [99th] | 1.56 [99th] | 1.56 [99th] | ||
aValues in parentheses are for highest resolution shell. bRmeas = ∑h[nh/(nh − 1)]1/2∑i|Ihl − Ih|/∑h∑i Ih,i, where Ih is the mean intensity of symmetry-equivalent reflections and nh is the redundancy. cRwork=∑||Fo| − |Fc||/∑|Fo|. dRfree is the same as Rwork, but calculated based on 5% of the data excluded from refinement. eR.m.s. deviations from target geometries.
MATERIALS AND METHODS
Cid1 construct design
Full-length and Cid1 ΔN 31 were PCR amplified from S. pombe cDNA using the following primers: cid1_fl_ndeI; cid1_dN31_ndeI; cid1_xhoI and the PCR products were inserted between the NdeI/XhoI sites of pET28b (Novagen) resulting in an N-terminal 6X His fusion tag. The C-terminally truncated version, Cid1 (31–377), was prepared by using the QuikChange mutagenesis protocol with the primers: cid1_378stop_f and cid1_378stop_r. Additional mutants were prepared using the QuikChange mutagenesis protocol. All constructs were verified by DNA sequencing. Primers are listed in Supplementary Table S1.
Cid1 expression and purification
Escherichia coli BL-21 (DE3) RIL cells (Stratagene) transformed with the pET28b Cid1 constructs were grown at 310 K in LB medium containing kanamycin and chloramphenicol until reaching an OD600 ∼0.6. The temperature was reduced to 291 K and protein expression was induced by addition of 1 mM isopropyl β-d-thiogalactoside (IPTG). Cells were harvested 16 h after induction by centrifugation. Cell pellets were resuspended in Buffer A (50 mM HEPES pH 6.8, 500 mM NaCl and 5 mM 2-mercaptoethanol). Cell walls were broken by sonication and the cell debris was clarified by centrifugation at 20 000g. All subsequent purification steps were performed at 277 K. The supernatant was loaded onto Talon affinity resin (Clontech) equilibrated with Buffer A. Following a wash with Buffer B (50 mM HEPES pH 6.8, 1 M NaCl and 5 mM 2-mercaptoethanol), the bound proteins were eluted in Buffer C (50 mM HEPES pH 6.8, 500 mM NaCl, 150 mM imidazole and 5 mM 2-mercaptoethanol). The N-terminal hexahistidine tag was removed by thrombin digestion in Buffer D [50 mM MES pH 6.3, 200 mM NaCl, 2 mM dithioerythritol (DTE) and 2 mM CaCl2] overnight at 277 K. The cleaved protein was loaded directly onto a HiTrap Heparin HP column (GE Healthcare) equilibrated in Buffer D lacking CaCl2. The bound protein was eluted using a linear gradient of 30 column volumes to Buffer E (50 mM MES pH 6.3, 1 M NaCl and 2 mM DTE). Cid1 proteins were further purified by size exclusion chromatography on a Superdex™ 75 column (GE Healthcare) equilibrated with Buffer F (50 mM MES pH 6.5, 250 mM NaCl and 2 mM DTE). Cid1 proteins were subsequently concentrated to ∼5–10 mg/ml and stored at 277 K.
Cid1 crystallization and NTP soaks
Crystals for Cid1 (31–377) grew at 293 K in a sitting-drop vapor diffusion setup using a 1:2 ratio of protein to reservoir within 3–4 days. The reservoir solution contained 100 mM CHES buffer pH 9.5 and PEG 3350 at concentrations between 5% and 10% (w/v). Single crystals were transferred to a cryoprotectant solution containing the mother liquor supplemented with 25% (v/v) glycerol and subsequently flash cooled in liquid nitrogen.
For NTP soaks, crystals were transferred to a drop containing 100 mM CHES buffer pH 9.5, 10% (w/v) PEG 3350 and 20% (v/v) glycerol with 10 mM ATP, UTP, CTP or GTP and 20 mM MgCl2. Generally, crystals were soaked for 30 min to 1 h and then flash cooled in liquid nitrogen. However, crystals soaked with MgUTP showed significant degradation upon transfer to the soaking solution. These crystals were harvested after 15 min of soaking. Synchrotron diffraction data were collected at the beamline X10SA at the Swiss Light Source (Switzerland) at 100 K and processed with XDS (17).
Structure determination
Initial phases for free Cid1 (31–377) were obtained by molecular replacement using PHASER(18) with the Trypanosoma brucei (Tb) Tut4 structure (PDB ID: 2Q0C) as a search model. NTP-bound structures used the free Cid1 structure as a search model. Model building was performed in cycles of manual building with COOT (19) and refinement using REFMAC (20) and the simulated annealing protocol from CNS (21). Model quality was evaluated with MolProbity (22). All figures were prepared using PYMOL (23).
Cid1 gel-based activity assays
Purified, recombinant Cid1 variants were assayed for polymerase activity in a buffer containing 20 mM HEPES–KOH pH 7.5, 100 mM KCl, 1 mM MgCl2 and 0.5 mM DTE. 20 µM penta-adenosine (A5) RNA or 20 µM of 30-mer RNA (5′-CACUUAAGGAGGUAUACUAUGUUCACGAUC-3′) was mixed with 5 µM protein and 1 mM nucleotide and the assay was allowed to proceed for 1 h at 303 K to run a long-term reaction and 15 min at 310 K for a short-term elongation reaction, respectively. The reaction was stopped by the addition of an equal volume of 8 M urea and 2 µl loading dye. Products were analyzed on 20% denaturing polyacrylamide gels run at 35 W for 45 min. RNA bands were visualized by staining with Methylene Blue. The A5 RNA was purchased HPLC purified from Integrated DNA Technologies. The 30-mer RNA was purchased HPLC purified from IBA life sciences.
Cid1 coupled activity assays
Cid1 polymerization activity was monitored by coupling the production of pyrophosphate during the polymerization reaction to production of inorganic phosphate by yeast inorganic pyrophosphatase (New England Biolabs) and subsequently to cleavage of fluorescent 7-methylguanosine (Sigma) by bacterial purine nucleoside phosphorylase (Sigma) (24,25). Each 400 -µl reaction mixture contained 0.5 U purine nucleoside phosphorylase, 0.2 U yeast inorganic pyrophosphatase, 100 µM 7-methylguanosine, 5 µM RNA (30-mer) substrate, 0.2 µM Cid1 and various concentrations of NTP in a buffer containing 10 mM HEPES–NaOH pH 7.5, 150 mM KCl, 5 mM MgCl2 and 0.5 mM DTE. Reactions were started by addition of NTP. Reaction progress was monitored by the decrease in fluorescence at 400 nm with excitation wavelength set to 300 nm in a FP-8500 Spectrofluorometer (Jasco) with excitation and emission bandwidths of 5 nm. All reactions were temperature controlled at 310 K. Kinetic parameters, apparent KM and Vmax, were obtained by fitting the initial velocities as a function of NTP concentration into a standard Michaelis–Menten kinetics model using the software Anemona.xlt. NTPs for crystallization were purchased from Sigma, and those for the activity measurements were purchased from Jena Biosciences.
RESULTS AND DISCUSSION
Structure of Cid1
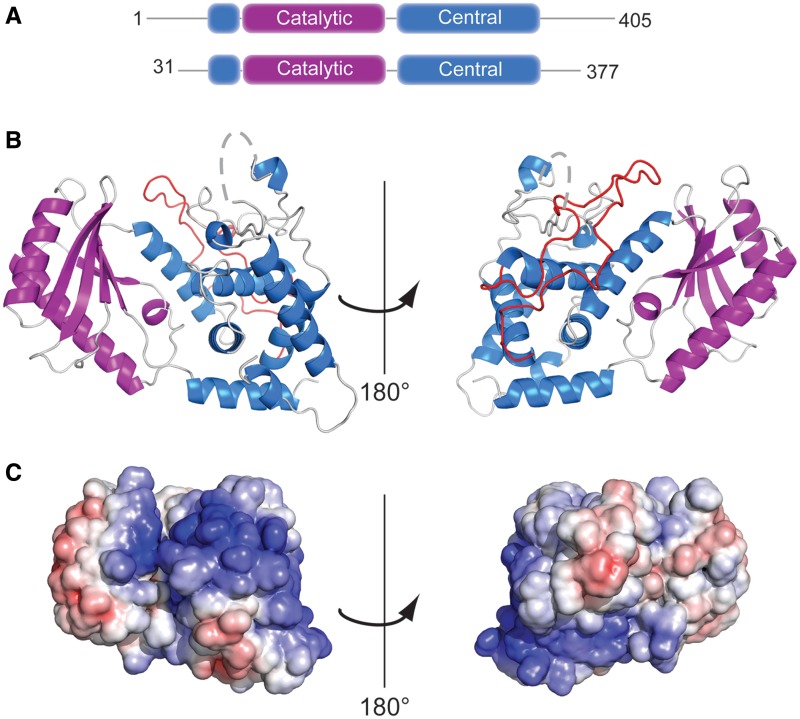
Attempts to crystallize the full length or a previously identified (11) N-terminally truncated variant, Cid1 ΔN31, were unsuccessful. However, a variant of Cid1-containing residues 31–377 was identified through partial proteolysis and secondary structure prediction. Crystallization trials using this variant produced crystals that diffracted to 2.0 Å resolution. This variant retains full enzymatic activity and represents the functional core of the Cid1 protein, containing both the catalytic and nucleotide triphosphate-binding motifs (Figure 1A, Supplementary Figure S1). The structure of the apoprotein was determined by molecular replacement using the structure of T. brucei (Tb) Tut4 (PDB ID: 2Q0C, 26% sequence identity) as the initial search model. In all, nine residues from the N-terminus (residues 31–39) and 15 residues from a loop in the central domain (residues 308–322) were disordered and were not incorporated into the structures. Details of the structure determination and model quality are given in Table 1.
Figure 1.
Structure of Cid1 PUP. (A) Schematic representation of the domain organization of Cid1 and the construct used for crystallization. (B) Overall structure of Cid1 shown as ribbon representation with the catalytic and central domains shown in purple and blue, respectively. The dashed gray line indicates missing loop region between residues 307–323. The long loop connecting helices H7 and H8 is colored in red and most likely serves as a protein docking site. Right panel shows Cid1 rotated by 180°. (C) Cid1 shown as solvent accessible surface representation. Coloring is according to the electrostatic surface potential over the range from +7 kT/e (blue) to −7 kT/e (red).
Cid1 adopts a fold similar to previously characterized DNA Pol β family members containing a catalytic domain and a central domain. The N-terminal catalytic domain is packed against the C-terminal central domain burying 1578 Å2 solvent accessible surface area. The catalytic domain (residues 56–165) contains a mixed, 5-stranded β-sheet (S1–S5) packed against two α-helices, H2 and H3. The non-contiguous central domain (residues 40–55 and 166–377) adopts a primarily helical structure with six α-helices and two short β-strands. The catalytic domain and central domain form a deep cleft with the catalytic domain’s β-sheet forming one wall of the cleft and helices from the central domain forming the other wall (Figure 1A and Supplementary Figure S1). Within this cleft, the catalytic triad of aspartate residues is found on strands 2 and 5 of the catalytic domain, and residues from the nucleotide recognition motif (residues 327–339) are found in a mostly unstructured region of the central domain (Supplementary Figure S1). Comparison with previously solved structures in the PDB demonstrates that Cid1 most closely resembles the structure of human PAPD1 (CαRMSD 1.75 Å) (26) and the terminal uridyltransferases RET2 (CαRMSD 2.07 Å) (27), MEAT1 (CαRMSD 2.06 Å) (28) and TUT4 (CαRMSD 1.84 Å) (29) from T. brucei. Structural comparison with the Cid1-like protein Trf4 from Saccharomyces cerevisiae (30) revealed a 3.1 Å CαRMSD. A primary structural difference between Cid1 and these polymerases is the loop connecting helices H7 and H8 in the central domain. In general, this loop appears to represent a unique feature in each of these polymerases. In Cid1, this loop (residues 226–270) forms a long insertion between helices H7 and H8 adopting an unstructured conformation that extends along the back of the central domain and nearly reaches the active site (Figure 1B). In TbMEAT1 this loop forms the so-called bridging domain which was proposed to function as a protein-binding site (28). In Trf4 this loop is only five residues long (residues 364–368), but several residues within this loop interact with the Air2p zinc knuckles (30). Thus, it is tempting to speculate that Cid1 may also use this loop for protein–protein interactions.
Cid1 members generally lack an additional RNA-binding domain, instead they rely on additional RNA-binding proteins to target them to particular RNAs for polyadenylation (12). Despite the lack of an RNA-binding domain, Cid1 is able to act on a variety of RNA targets (7,10,11). Analysis of the electrostatic surface potential of Cid1 reveals that there is an extended positively charged patch running along the surface of the Cid1 molecule. This positively charged region runs from the NTP-binding cleft and wraps around the central domain. In the absence of a protein-binding partner, this site is likely to act as a non-specific RNA-binding surface of the protein (Figure 1C). The presence of such an extended region of positive charge explains how Cid1 is able to polymerize various RNA targets without the requirement for additional binding partners, unlike some other Cid1 polymerases which show no activity in the absence of their binding partner (16).
Cid1–UTP recognition
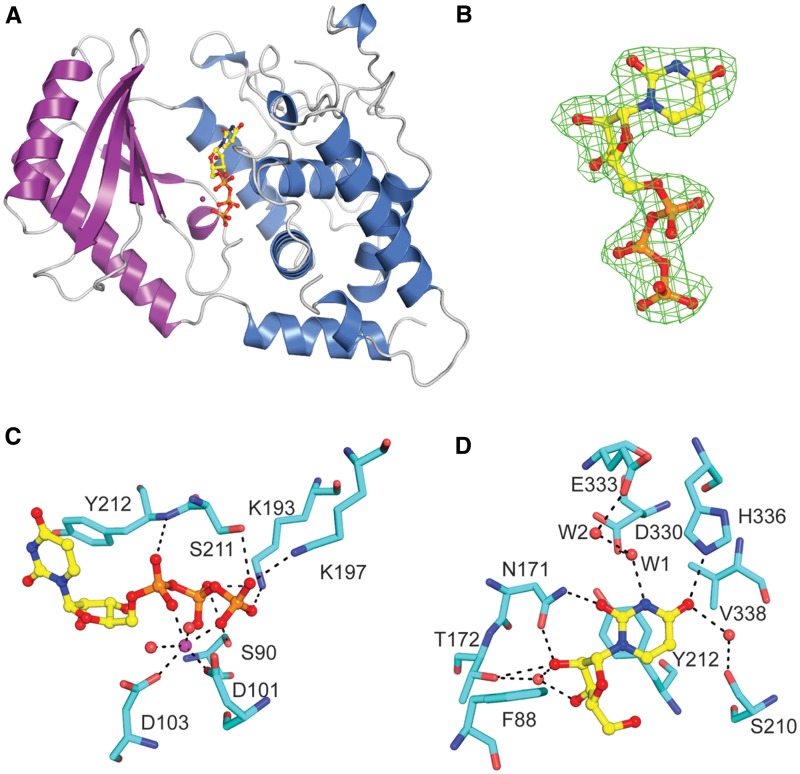
In vitro Cid1 uses both ATP and UTP as substrates for polymerization, and it displays some activity for CTP and GTP (8,10,11). However, the protein has been shown to preferentially use UTP as its substrate both in vivo and in vitro (10,11). In order to better understand Cid1’s UTP specificity, we determined the crystal structure of Cid1 bound to UTP with either Mg2+ or Ca2+ divalent cations (Figure 2, Table 1 and Supplementary Figure S2).
Figure 2.
Structure of Cid1–MgUTP complex. (A) Overview of Cid1–MgUTP structure. UTP binds into a cleft between the catalytic and central domains of Cid1. UTP is shown in ball and stick representation, Cid1 is shown as ribbon representation. (B) UTP molecule from Cid1–MgUTP structure with a Fo − Fc omitted map (green mesh) contoured at 3 σ. (C) Cid1 recognition of Mg2+ and triphosphate moieties of UTP. Mg2+shown as sphere. (D) Key ribose/base interactions between Cid1 and UTP.
The Cid1–UTP structure reveals that UTP is bound within the cleft formed between the catalytic domain and the central domain (Figure 2A and B). Unlike other polymerases (28,31), the Cid1–UTP structure revealed only minor conformational rearrangements when comparing the apo- and UTP-bound structures (Cα RMSD 0.26 Å). UTP recognition is achieved using residues from both the catalytic and central domains with residues from each domain participating in both sugar/base and triphosphate recognition. In the UTP-bound structures, a single divalent cation is coordinated by the triphosphate moiety (either Mg2+ in the active form or Ca2+ in the inhibited form), by the side chain carboxyl groups of the conserved aspartate residues 101 and 103 and by water molecules (Figure 2C). The γ-phosphate group is bound through contacts to the side chains of K193,K197, S211 and S90. The β-phosphate group is bound by the side chains of K193 and S90 and the α-phosphate group is bound by a direct contact to the main chain amide of Y212 (Figure 2C).
Ribose recognition is achieved using a combination of hydrophobic and hydrogen bonding interactions. The ribose, in the C3′-endo conformation, is packed against the aromatic side chains of Y212 and F88. The 2′-hydroxyl group forms a hydrogen bond with the side chain carbonyl group of N171 and a nearby water molecule. This water molecule bridges by additional hydrogen bonds the 3′-hydroxyl group of the ribose with the side chain hydroxyl group of T172. Noteworthy, T172 does also form a hydrogen bond to the 2′-hydroxyl group of the ribose (Figure 2D). This direct ribose recognition by N171 side chain group and the direct and water-mediated contacts to T172 are likely to be important for discriminating between UTP and dUTP. A similar interaction has been observed in the terminal uridyltransferase structures from T. brucei; however, in these structures, T172 is replaced by a serine residue that makes direct contact to both the 2′- and 3′-hydroxyls (27,28 and Supplementary Figure S2). The 3′-hydroxyl group of the ribose forms several additional hydrogen bonds with surrounding water molecules, including a water-mediated contact to the β-phosphate group and to the side chain hydroxyl group of S93.
The uridine base is bound in an anti-conformation and forms a hydrophobic stacking interaction against Y212 with an additional hydrophobic interaction to V338. Each of the Watson–Crick hydrogen bonding partners of the uridine base is satisfied through direct or water-mediated contacts to the protein. The O4 carbonyl group is recognized by a direct hydrogen bond to the Nε of H336. The O4 carbonyl group also forms a water-mediated contact to S210. The N3 imino proton forms a hydrogen bond to a water molecule (W1) that also forms hydrogen bonds with the O2 carbonyl group and the side chain carboxylate of D330. This water is additionally bound to an adjacent water molecule (W2) that interacts with the carboxylate group of E333. The O2 carbonyl group is additionally recognized by the side chain amine of N171 (Figure 2D).
Comparison of Cid1–UTP to the Trypanosmal uridyltransferases
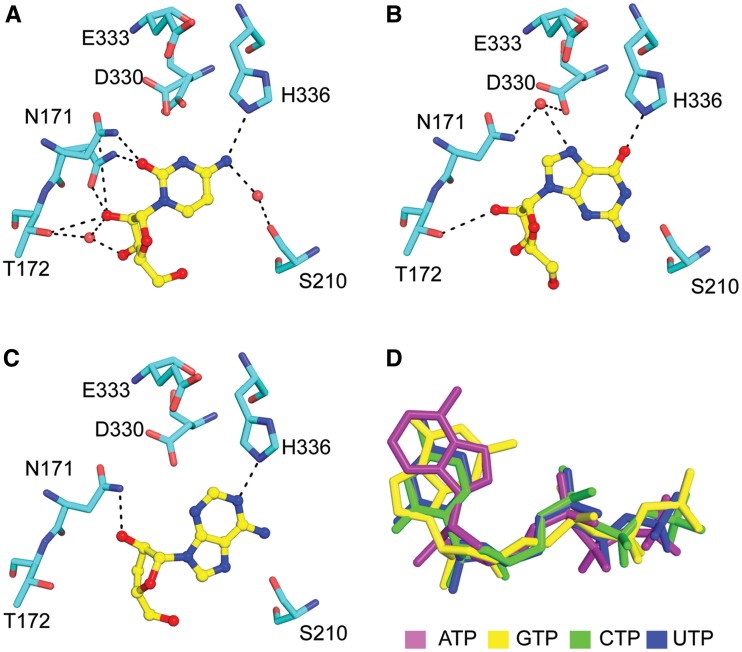
The Cid1–UTP structure shares a number of interactions also found in the UTP-bound Trypanosomal uridyltransferase structures. In all of the structures, the base is stacked against the aromatic side chain of a tyrosine (Cid1, RET2, TUT4) or phenylalanine (MEAT1) residue. The dual sugar–base interaction observed between N171 and the 2′-hydroxyl group of the ribose and the O2 carbonyl group of the uridine base is observed in all structures of uridyltransferases to date (Figure 2D and Supplementary Figure S2). This interaction is critical because it is able to specifically distinguish dUTP from UTP and it serves as a pyrimidine sensor by forming a hydrogen bond to the O2 carbonyl group of the UTP base. In soaking experiments with each of the other three NTPs, we find that the side chain of N171 changes conformation in response to the presence of UTP and CTP, but remains in a similar conformation observed for the apo structures when ATP or GTP are present in the soaking buffer (Figure 3). This conformational change was previously observed in soaks of TbTUT4 with dUTP (29), but not in the presence of ATP or GTP (32). Thus, in Cid1 it appears that N171 may function as a pyrimidine sensor. Further similarities are found with the recognition of the N3 imino hydrogen, which is recognized through a water-mediated contact to an acidic amino acid side chain (Figure 2D and Supplementary Figure S2). In the case of TbRET2, this water molecule was proposed to be selective for UTP because it is oriented by two acidic side chains in such a way that only the lone pair electrons of oxygen could accept a hydrogen bond donor from the N3 imino of the uridine base (Supplementary Figure S2A) (27).
Figure 3.
NTP recognition by Cid1. Key ribose–base interactions observed for (A) Cid1–CTP recognition; (B) Cid1–GTP recognition and (C) Cid1-3′-dATP recognition. (D) Relative positioning of NTPs in the active site after superpositioning each of the Cid1–NTP structures. For panels A–C coloring is same as Figure 2, coloring for NTPs in panel D is indicated in the figure.
Despite the many similarities, differences are evident how Cid1 recognizes the uridine O4 carbonyl group. First, Cid1 directly recognizes the O4 carbonyl group through a hydrogen bond to H336. In contrast, in the T. brucei structures, there is a leucine or valine residue at the equivalent position, thus, no specific protein–base contact occurs at this position. Instead, interaction with the O4 carbonyl group is usually through a water-mediated contact to an invariant arginine residue. This arginine residue is also present in Cid1 (R340), but does not interact with the O4 carbonyl group. The only exception is the structure of TbMEAT1 where an asparagine side chain (N181) amine forms a hydrogen bond with the O4 carbonyl (Supplementary Figure S2B). In Cid1 and RET2, the O4 group forms a water-mediated interaction to the hydroxyl group of S210 (Cid1) or T317 (RET2) residue (Figure 2D and Supplementary Figure S2A).
Structures of Cid1 with ATP, CTP and GTP
In addition to the UTP-bound structure, we also determined structures of Cid1 bound to each of the other NTP substrates by soaking the apo crystals with cordycepin triphosphate (3′-dATP), GTP, or CTP in the presence of Mg2+ (Table 1, Figure 3 and Supplementary Figure S3). We used 3′-dATP as a mimic for ATP, because both soaking and cocrystallization experiments in which ATP was used, revealed only weak electron density for the triphosphate group.
Comparison of the nucleotide-bound structures revealed that triphosphate binding was very similar in each of the different nucleotide structures (not shown), but differences exist in ribose/base recognition that may explain some of the differences in nucleotide incorporation observed in vitro. The first difference is that N171, which forms a dual ribose–base interaction with UTP, is capable of forming the same interaction with CTP (Figures 2D and 3A), but not with ATP or GTP (Figure 3B and C). In the ATP and GTP structures, the side chain of N171 is turned away and makes no contact to the ribose in the GTP structure (Figure 3B) and a single hydrogen bond with the O2′-hydroxyl group in the 3′-dATP structure (Figure 3C). Second, in the 3′-dATP-bound structure, the ribose position is shifted in comparison to CTP, GTP or UTP structures (Figure 3D). Third, the structures of UTP, CTP and ATP all have the base in the anti-conformation, but in the GTP structure the GTP is bound in the syn-conformation (Figure 3B).
How do these differences relate to the in vitro activity observed for Cid1? In the UTP-, GTP- and ATP-bound structures, the Nε atom of H336 acts as donor for a hydrogen bond which is required for base recognition. For the CTP-bound structure, the Nε of H336 would act as an acceptor to interact with the amine at position 4 of the cytidine base. In our in vitro experiments, we observed activity for ATP, UTP and GTP, but no activity for CTP (Figure 4A). This suggests that the preferred H336 tautomer is protonated on Nε; therefore, only ATP, UTP and GTP can serve as substrates for polymerization. We surmised that it might be possible to influence NTP incorporation by changing the pH and, hence, the protonation state of H336. However, we observed no activity for CTP incorporation at any pH tested (pH 6–9), and we observed that ATP, GTP and UTP incorporation peaked near pH 8.0 (data not shown). Thus, while the protonation state of H336 may influence ATP, UTP and GTP incorporation, it is likely that additional contacts are important for selecting against CTP. The most likely candidate is the water molecule that forms a hydrogen bond to the imino of the uridine base, which is not present in the Cid1–CTP structure (Figure 3A). This contact was previously argued to have a role in selecting for UTP over CTP in the case of TbRET2 (27), and it may be that Cid1 uses a similar mechanism to select for UTP.
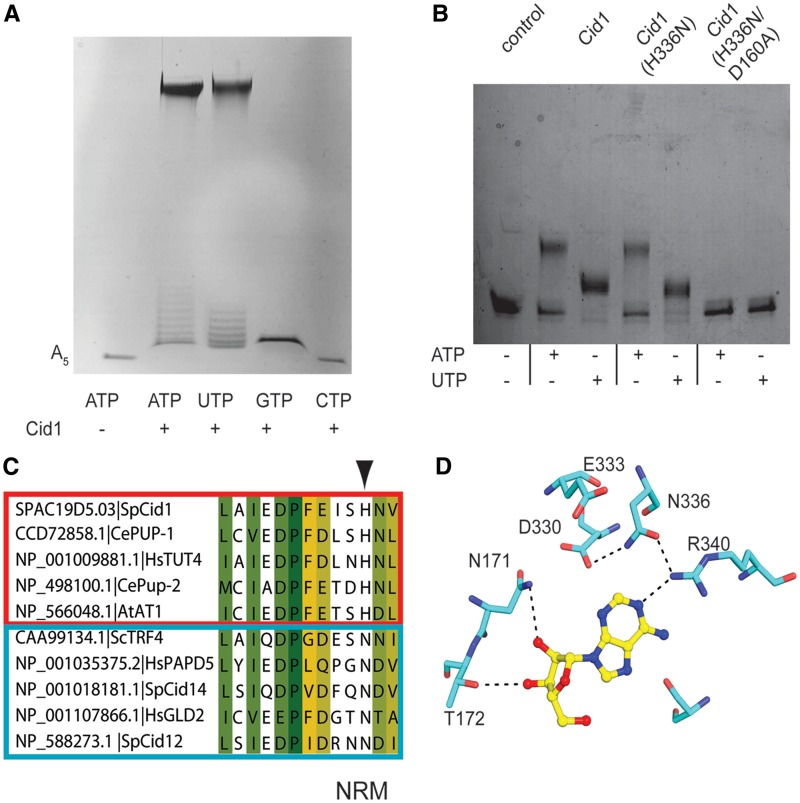
Figure 4.
PUP and PAP activity of Cid1. (A) Long-term in vitro polymerase assay with Cid1 construct used for structure determination. Cid1 was incubated in the presence of A5 RNA with indicated NTP. Reaction products were then separated on a 20% urea–polyacrylamide gel. RNA products were visualized by staining with methylene blue. (B) Short-term in vitro polymerase assay comparing activity of wild-type Cid1 in the presence of 30-mer RNA compared with the variants H336N and H336N/D160A. Detection and staining of reaction products were performed as described for (A). (C) Sequence alignment of nucleotide recognition motifs (NRM) from various Cid1-family polymerases. Boxed in red are verified PUPs and in blue are verified PAPs (10). Arrowhead indicates the position of H336 in Cid1. Accession numbers and common names are given. Abbreviations are as follows: Schizosaccharomyce pombe (Sp), Saccharomyces cerevisiae (Sc), Caenorhabditis elegans (Ce), Homo sapiens (Hs) and Arabidopsis thaliana (At). (D) Structure of Cid1 (H336N) showing recognition of ATP. Coloring is the same as in Figure 2.
One difference in purine recognition by Cid1 is that GTP is bound in a syn-conformation while ATP, UTP and CTP are all bound in the anti-conformation (Figure 3). Our in vitro results demonstrate that Cid1 incorporates around one or two GTP residues (confirmed by mass spectrometry, data not shown), but then polymerization is stopped. For TbTUT4, it was previously shown that stacking interactions between the RNA and the nucleotide base are important for the formation of a productive complex (32). Thus, it is tempting to speculate that in the ternary complex, the presence of syn-GTP in the active site may result in a reduced stacking interaction with the 3′-base of the RNA, and thus poor nucleotide addition.
UTP versus ATP selection
In order to confirm the activity of our crystallized construct, we used a gel-based assay to monitor nucleotide addition to a short poly(A) RNA (sequence: AAAAA) primer. In this assay, Cid1 demonstrated robust poly(A) and poly(U) incorporation for both full-length protein (data not shown) and the truncated Cid1 construct used for crystallization (Figure 4A). This assay also demonstrated weak or no activity for GTP and CTP, respectively (Figure 4A), similar as described previously (8,11). We also tested a catalytically inactive variant where one of the three catalytic aspartates was mutated to alanine (D160A), and could not observe any nucleotide addition, excluding that the activity was due to any copurifying contaminant (data not shown). In contrast to the previously reported results (11), we observed two different kinds of reaction products after long-term incubation (Figure 4A). It seemed, that Cid1 added single nucleotides to the short RNA primer and subsequently became more processive and elongated long RNA tails in our assays. We surmised that the initial non-processive nucleotide addition is due to inefficient binding to RNA and that longer RNA primers should be elongated much more processively. Indeed, once a longer RNA species was used in our assay and the elongation time was reduced, Cid1 appeared to be fully processive since we do not observe the RNA ladder any more (Figure 4B). Furthermore, whereas the original RNA primer was fully depleted after 15 min of poly-uridylation by Cid1, residual substrate was observed for a poly-adenylation reaction.
Whereas during long elongation times, Cid1 seemed to be as effective in adding poly(A) tails when compared with the production of poly(U) tails (Figure 4A) the enzyme showed some preference for UTP addition under limited elongation times (Figure 4B), similar as described previously (11). However, our qualitative gel-based assay could not explain Cid1’s selectivity for UTP against ATP and suggested robust activity for ATP in the absence of UTP (Figure 4A). In order to address this issue, we used a kinetic assay to determine the kinetic parameters of ATP and UTP addition by Cid1 on longer RNA species. With this assay, we determined that the apparent KM values for UTP and ATP are ∼30-fold higher for ATP than for UTP. Additionally, the rate of catalysis and catalytic efficiency of Cid1 with UTP was higher than what was observed for Cid1 with ATP (Table 2 and Supplementary Figure S4). All of which point to a kinetic preference for UTP over ATP as the preferred Cid1 substrate.
Table 2.
Kinetic parameters for Cid1 with ATP and UTP and Cid1 H336N with ATP and UTP
| Protein + NTP | KM (µM) ± SE | kcat (s−1) | kcat/ KM(M−1 s−1) |
|---|---|---|---|
| Cid1 + ATP | 312 ± 0.09 | 0.14 | 4.5 × 104 |
| Cid1 + UTP | 12 ± 0.16 | 0.80 | 6.6 × 102 |
| Cid1 (H336N) + ATP | 65 ± 0.10 | 0.52 | 7.9 × 103 |
| Cid1 (H336N) + UTP | 41 ± 0.15 | 0.35 | 8.5 × 103 |
SE—standard error.
A key difference between Cid1 and trypanosomal TUTa-ses in the presence of H336 which makes direct contact to the O4 of the uridine base. A sequence alignment of Cid1-related polymerases from higher eukaryotes revealed that a subset of these polymerases have histidine at this position while in others this position is usually occupied by asparagine residue. This difference was intriguing, and upon sorting these polymerases based on demonstrated PUP or PAP activity (10), we found that all of the confirmed PUPs have a histidine at this position while PAPs generally have asparagine residue at this position (Figure 4C). Thus, we suspected that it might be possible to re-engineer Cid1 to act similar to PAPs by mutating H336 to asparagine. We created this variant of Cid1, Cid1 (H336N), and we found that Cid1 (H336N) had a significantly decreased apparent KM for ATP (5-fold reduced) and the rate of catalysis had improved. In contrast, the apparent KM for UTP had increased and the rate of catalysis was slightly reduced (Table 2). Apparently, the mutation of H336 to an asparagine residue dramatically improved Cid1’s ability to bind and elongate ATP on RNA primers and it is tempting to speculate that Cid1 PUPs have evolved from PAPs by losing their capability of using ATP. Similar as observed for the wild-type protein, the in vitro gel-based assay did not reveal a significant difference in ATP and UTP incorporation by Cid1 (H336N) (Figure 4B). We also tested a catalytically inactive variant Cid1 (D160A/H336N) to ensure that the activity we observed was not due to a copurifying polymerase (Figure 4B).
To better understand the altered selectivity for this mutant on a molecular level, we crystallized Cid1 (H336N) with ATP (Supplementary Table S2). The structure reveals that the histidine to asparagine mutation indirectly facilitates base recognition by forming a hydrogen bond between its side chain carbonyl group to R340 which directly contacts the N1 position of the base. Also, the asparagine side chain amine forms a hydrogen bond to the side chain of D330, which may potentially disrupt recognition of the N3 imino of a uridine base (Figure 4D).
CONCLUSIONS
Structures of Cid1 free and bound to each of the ribonucleotide triphosphates reveal that the protein can accommodate each of the individual nucleotides at the active site. However, there are a number of differences in nucleotide recognition that are likely to contribute to selectivity for UTP over ATP, GTP and CTP. First, selectivity for UTP relies on multiple direct and water-mediated contacts to fully satisfy the hydrogen bonding requirements of the Watson–Crick face of the uridine base. Many of these contacts appear to be shared among uridyltransferases (27–29). The structures of Cid1 with CTP, GTP and ATP show that the Watson–Crick hydrogen bonding requirements are not fully satisfied for any of these nucleotides. One unique feature of Cid1 is the presence of a histidine in the nucleotide recognition motif. This histidine makes direct contact to each of the 4 nt, and it may act to select against CTP based on its protonation state. Sequence alignments of Cid1-family proteins with confirmed poly(A) or poly(U) activity revealed that this histidine is found solely in confirmed PUPs while confirmed PAPs tend to have an asparagine residue at this position. In order to test if this residue is important for defining poly(A) or poly(U) activity, we mutated H336 to asparagine. With this variant, we found that activity for UTP was slightly decreased but we observed a significant improvement in ATP activity, suggesting that this residue could play an important role in determining whether a Cid1 family member is a PUP or a PAP.
Overall, this work demonstrates that Cid1 is able to accommodate all 4 nt in the active site, but there are differences in recognition that suggest UTP is the preferred NTP. Previous work on Cid1 demonstrated that Cid1 nucleotide selectivity is dependent on the incoming RNA substrate (11). Thus, a key question that we have not addressed with this work is to explain the role of the RNA substrate in selecting the incoming nucleotide. Thus, while our structures represent the first key step in elucidating Cid1’s UTP selectivity, it is likely that the full specificity requirements will only be revealed with structures of the ternary complex.
ACCESSION NUMBERS
Atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank for each of the structures listed: Cid1 apo (4FH3), Cid1-MgUTP (4FH5), Cid1-CaUTP (4FHP), Cid1-Mg 3′dATP (4FHY), Cid1-MgGTP (4FHW), Cid1-MgCTP (4FHV), Cid1(H336N)-MgATP (4FHX) structures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–4.
FUNDING
The Deutsche Forschungsgemeinschaft [ME 3135/2-1 to A.M.] and A.M. is member of CellNetworks—Cluster of Excellence (EXC81). Funding for open access charge: Max-Planck-Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Hannes Mutschler and Jochen Reinstein for advice on the coupled assays and Christina Sendlmeier and all members of the Meinhart group for stimulating discussions. The authors are grateful to Ilme Schlichting for continuous encouragement and support. The authors also thank the Heidelberg/Dortmund data collection teams, the scientific staff at the beamline X10SA, Paul Scherrer Institute (Villigen, Switzerland) for help with data collection and Chris Roome and Ingrid Vetter for support of the crystallographic software.
REFERENCES
- 1.Martin G, Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi YS, Patena W, Leavitt AD, McManus MT. Widespread RNA 3′-end oligouridylation in mammals. RNA. 2012;18:394–401. doi: 10.1261/rna.029306.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 4.Aphasizhev R. RNA uridylyltransferases. Cell Mol. Life Sci. 2005;62:2194–2203. doi: 10.1007/s00018-005-5198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol. 2009;16:616–623. doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Read RL, Martinho RG, Wang SW, Carr AM, Norbury CJ. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl Acad. Sci. USA. 2002;99:12079–12084. doi: 10.1073/pnas.192467799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang SW, Toda T, MacCallum R, Harris AL, Norbury C. Cid1, a fission yeast protein required for S-M checkpoint control when DNA polymerase delta or epsilon is inactivated. Mol. Cell Biol. 2000;20:3234–3244. doi: 10.1128/mcb.20.9.3234-3244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak JE, Wickens M. A family of poly(U) polymerases. Rna-a Publicat. RNA Soc. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell. Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevenson AL, Norbury CJ. The Cid1 family of non-canonical poly(A) polymerases. Yeast. 2006;23:991–1000. doi: 10.1002/yea.1408. [DOI] [PubMed] [Google Scholar]
- 13.Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- 14.Wickens M, Kwak JE. Molecular biology. A tail tale for U. Science. 2008;319:1344–1345. doi: 10.1126/science.1154946. [DOI] [PubMed] [Google Scholar]
- 15.Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- 17.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 18.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 20.(1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 21.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. The PyMOL Molecular Graphics System, Version 1.5.0.1 Schrödinger, LLC. [Google Scholar]
- 24.Mutschler H, Gebhardt M, Shoeman RL, Meinhart A. A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 2011;9:e1001033. doi: 10.1371/journal.pbio.1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulikowska E, Bzowska A, Wierzchowski J, Shugar D. Properties of two unusual, and fluorescent, substrates of purine-nucleoside phosphorylase: 7-methylguanosine and 7-methylinosine. Biochim. Biophys. Acta. 1986;874:355–363. doi: 10.1016/0167-4838(86)90035-x. [DOI] [PubMed] [Google Scholar]
- 26.Bai Y, Srivastava SK, Chang JH, Manley JL, Tong L. Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase. Mol. Cell. 2011;41:311–320. doi: 10.1016/j.molcel.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng J, Ernst NL, Turley S, Stuart KD, Hol WG. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stagno J, Aphasizheva I, Bruystens J, Luecke H, Aphasizhev R. Structure of the mitochondrial editosome-like complex associated TUTase 1 reveals divergent mechanisms of UTP selection and domain organization. J. Mol. Biol. 2010;399:464–475. doi: 10.1016/j.jmb.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. J. Mol. Biol. 2007;366:882–899. doi: 10.1016/j.jmb.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamill S, Wolin SL, Reinisch KM. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc. Natl Acad. Sci. USA. 2010;107:15045–15050. doi: 10.1073/pnas.1003505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balbo PB, Bohm A. Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis. Structure. 2007;15:1117–1131. doi: 10.1016/j.str.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stagno J, Aphasizheva I, Aphasizhev R, Luecke H. Dual role of the RNA substrate in selectivity and catalysis by terminal uridylyl transferases. Proc. Natl Acad. Sci. USA. 2007;104:14634–14639. doi: 10.1073/pnas.0704259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.