Abstract
H3K4me3 is a histone modification that accumulates at the transcription-start site (TSS) of active genes and is known to be important for transcription activation. The way in which H3K4me3 is regulated at TSS and the actual molecular basis of its contribution to transcription remain largely unanswered. To address these questions, we have analyzed the contribution of dKDM5/LID, the main H3K4me3 demethylase in Drosophila, to the regulation of the pattern of H3K4me3. ChIP-seq results show that, at developmental genes, dKDM5/LID localizes at TSS and regulates H3K4me3. dKDM5/LID target genes are highly transcribed and enriched in active RNApol II and H3K36me3, suggesting a positive contribution to transcription. Expression-profiling show that, though weakly, dKDM5/LID target genes are significantly downregulated upon dKDM5/LID depletion. Furthermore, dKDM5/LID depletion results in decreased RNApol II occupancy, particularly by the promoter-proximal Pol lloser5 form. Our results also show that ASH2, an evolutionarily conserved factor that locates at TSS and is required for H3K4me3, binds and positively regulates dKDM5/LID target genes. However, dKDM5/LID and ASH2 do not bind simultaneously and recognize different chromatin states, enriched in H3K4me3 and not, respectively. These results indicate that, at developmental genes, dKDM5/LID and ASH2 coordinately regulate H3K4me3 at TSS and that this dynamic regulation contributes to transcription.
INTRODUCTION
Covalent post-translational modification of core histones constitutes a principal regulatory mechanism in eukaryotic chromatin. Histone modifications are diverse, involve multiple residues and contribute to the regulation of most genomic processes, from RNA transcription and processing, to DNA replication, recombination and repair, and chromosome segregation [reviewed in (1,2)].
In this context, methylation of lysine 4 in histone H3 (H3K4) constitutes a well-documented case where a specific histone modification influences the functional state of chromatin [reviewed in (3,4)]. H3K4-methylation preferentially occurs at transcriptionally active chromatin domains. In particular, tri-methyl H3K4 (H3K4me3) occurs at the transcription-start site (TSS) of active genes and is important for transcription activation (5–13). Similarly, di-methyl H3K4 (H3K4me2) is also enriched at TSS, showing a broader distribution than H3K4me3. On the other hand, mono-methyl H3K4 (H3K4me) preferentially locates at transcriptional enhancers (14).
How is the pattern of H3K4-methylation established/maintained, as well as the molecular basis of its contribution to transcription regulation, are not yet fully understood. Specific methyl-transferases (KMTs) and demethylases (KDMs) are known to regulate H3K4-methylation [reviewed in (15,16)]. Although most species contain several H3K4-KMT2s, H3K4-methylation is largely mediated by ASH2, an evolutionarily conserved component of H3K4–KMT2 complexes that, lacking methyltransferase activity, is required for H3K4me3 in yeast, Drosophila and mammalian cells (17–25). In addition, in Drosophila, ASH2 was recently shown to localize at TSS (26). On the other hand, two H3K4 KDMs have been identified in Drosophila, dKDM1/LSD1/SU(VAR)3-3 that demethylates H3K4me and H3K4me2, but not H3K4me3 (27), and the Jumonji-domain containing protein dKDM5/LID, which effectively demethylates H3K4me3 (28–31).
In this work, we analyze the contribution of dKDM5/LID to the regulation of the pattern of H3K4me3. Previous reports showed that dKDM5/LID is a component of various co-repressor complexes (32,33), which play a role in repression of NOTCH target genes (32,34). Similarly, several mammalian KDM5 isoforms associate with components of co-repressor complexes (35) and mediate repression (36–41). On the other hand, dKDM5/LID has also been implicated in activating transcription, as lid was originally identified as a trxG gene (42), which is required for optimal Ubx expression (29,30), and antagonizes heterochromatin-mediated gene silencing (30,34).
Here, we report that dKDM5/LID localizes at TSS of developmental genes and regulates H3K4me3. dKDM5/LID target genes are highly transcribed and, opposite to what would be expected from its demethylase activity, dKDM5/LID positively contributes to their expression. Here, we also show that dKDM5/LID target genes are bound, and positively regulated, by ASH2. However, dKDM5/LID and ASH2 recognize two different chromatin states at TSS, enriched in H3K4me3 and not, respectively. These results indicate that dKDM5/LID and ASH2 cooperate to dynamically regulate H3K4me3 at TSS of developmental genes for their efficient transcription.
MATERIALS AND METHODS
Fly stocks and cell lines
lidRNAi (9088R2) was obtained from NIG-FLY. dKDM5/LID depletion by lidRNAi was determined by qRT-PCR (Figure 2A), western blot (Figure 2A and Supplementary Figure S1A), immunostaining (Supplementary Figure S2) and ChIP-qPCR (Supplementary Figure S1D). lidk06801 is described in (42). GAL4 lines used in this study were yw;Act5CGAL4, w;enGAL4-UASGFP and w;ptcGAL4-UASGFP (described in Bloomington Stock Centre). Stable S2 cell line expressing ASH2-HA is described in (43).
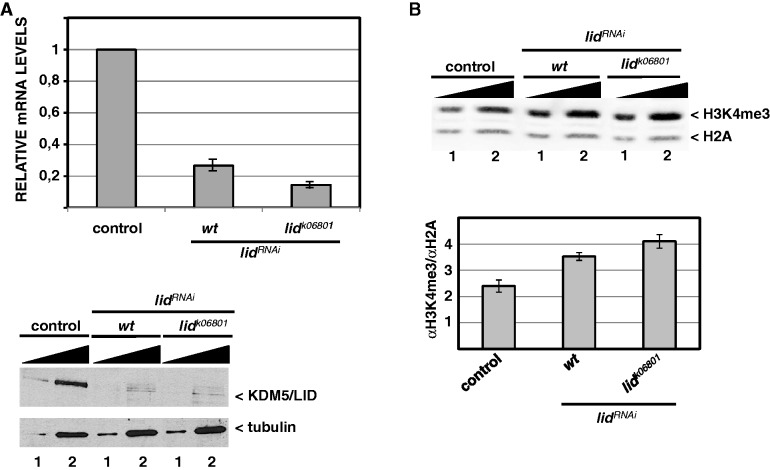
Figure 2.
Depletion of dKDM5/LID increases global H3K4me3. (A) lidRNAi efficiently depletes dKDM5/LID. At the top, the relative levels of dKDM5/LID mRNA determined by qRT-PCR are presented for control wild-type and lidRNAi flies. At the bottom, dKDM5/LID protein levels are analyzed by western blot in control wild-type and lidRNAi flies. dKDM5/LID depletion was induced by the Actin5C-GAL4 driver in either wt or heterozygous lidk06801/+ mutant flies. RNA was extracted from wing imaginal discs. Protein extracts were prepared from a mixture of imaginal discs, and increasing amounts of extract were analyzed: 1X (lanes 1) and 5X (lanes 2). Antibodies used were rat polyclonal αLID (1:10000) and, as loading control, α-β-tubulin (1:2000). (B) Global H3K4me3 levels are determined by western blot in control wt (left) and lidRNAi knockdown flies, where dKDM5/LID depletion was induced by the Actin5C-GAL4 driver in either wt (center) or heterozygous lidk06801/+ mutant flies (right). Protein extracts were prepared as in (A) and increasing amounts of extract were analyzed: 1X (lanes 1) and 2X (lanes 2). Results are presented for a single exposure of the same gel blotted simultaneously with αH3K4me3 (1:2000) and, for normalization, αH2A (1:2500) antibodies. Quantitative analyses of the results, carried out with Odyssey scanner, are shown at the bottom. Errors bars are standard deviation. Results are the average of two independent experiments.
Antibodies
Rat αLID polyclonal antibodies were raised from a mixture of two truncated GST-fusions harboring amino acids 1–888 and 1295–1826, respectively. Rabbit αLID polyclonal antibodies used in ChIP-experiments were raised against polypeptide 1295–1826 and IgGs were purified using Econo-Pac IgG purification columns (Bio-Rad). Specificity of the antibodies was determined by western blot using whole cell extracts prepared from a mixture of imaginal discs obtained from wt and lidRNAi flies where ubiquitous dKDM5/LID depletion was induced by the Actin5C-GAL4 driver. As seen in Supplementary Figure S1A and B, both αLID antibodies recognize a single major band in wt flies or S2 cells, which is barely detectable in lidRNAi flies. Immunostaining experiments also show high specificity of the antibodies (Supplementary Figure S2). Performance of rabbit αLID polyclonal antibodies in ChIP-experiments was determined by western blot. As shown in Supplementary Figure S1C, both whole αLID antiserum and the purified IgGs specifically immunoprecipitate dKDM5/LID very efficiently both from whole cell extracts and crosslinked chromatin. In addition, as judged by ChIP-qPCR, binding of dKDM5/LID to sites detected in wt flies is strongly reduced in lidRNAi flies (Supplementary Figure S1D).
The rest of antibodies used in these experiments were commercially available: αH3K4me3 (Abcam/ab8580), αH2A (Abcam/ab13923), αH3 (Cell Signaling, 9715), αHA (Roche, 12CA5), αPol IIoser5 (Abcam/ab5131), αPol IIoser2 (Abcam/ab5095) and α-β-tubulin (Millipore/MAB3408).
Determination of H3K4me3 levels by western blot
Protein extracts were prepared from imaginal discs dissected in PBS containing 0.05% Igepal (5 µl per larvae). After addition of an equal volume of loading buffer, discs were homogenized with a pestle, boiled with 10% β-mercaptoethanol and centrifuged. To avoid any interference of slight differences in loadings and detection conditions, samples were analyzed by western blot using simultaneously both αH3K4me3 (1:2000) and, for normalization, αH2A (1:2500) antibodies. Quantitative analyses were carried out with Odyssey scanner using infrared conjugated secondary antibodies (1:10000) (LI-COR) and LI-COR odyssey software (v3.0).
ChIP experiments
For ChIP, chromatin was prepared according to (44) from pools of about 300–500 wing imaginal discs or from cultured S2 cells, and sonicated to obtain fragments ranging from 200 to 500 bp. Immunoprecipitations (IPs) were basically performed as described in (45), using 1–2 µg of specific αH3K4me3, rabbit polyclonal αLID (IgG purified), αHA, αRNApol II, αPol IIoser5 and αPol IIoser2 antibodies. For ChIP-qPCR, triplicates from two independent biological replicates were analyzed following the ΔCt method (see Supplementary Table S1 for primers used in these experiments). For ChIP-seq, library construction, cluster generation and sequencing analysis using the Illumina GAII Genome Analyzer were performed following manufacture’s protocols (www.illumina.com). In brief, libraries, prepared using Illumina’s ChIP-Seq Sample Prep Kit from 10 ng of ChIP/input DNA, were size selected to ∼300 bp on an agarose gel. Adaptor-modified DNA fragments were subjected to limited PCR amplification (18 cycles). Using Illumina’s Cluster Generation Kit, libraries were subject to cluster generation at 8 pM concentration as one sample per lane. Sequencing-by-synthesis was performed for 38 cycles. ChIP-seq experiments for H3K36me3, Pol IIoser5, Pol IIoser2 and ASH2 in wt flies are described in (26).
Bioinformatics analysis of ChIP-seq data
Except where otherwise indicated, all analyses were performed with the Bioconductor software (46). Solexa/Illumina sequencing data for dKDM5/LID and H3K4me3 in both wt and lidRNAi flies, and for Pol IIoser5 and Pol IIoser2 in lidRNAi flies were pre-processed with the standard Illumina pipeline version 1.5.1 and sequences were aligned to the Drosophila melanogaster genome (UCSC dm3 version) with the Bowtie software 0.12.5 (47). We kept sequences mapping to a unique location in the genome, allowing up to 2 mismatches in the first 28 bases, and used the Bowtie default values for filtering low-quality sequences. For H3K36me3, Pol IIoser5, Pol IIoser2 and ASH2 in wt flies, aligned sequencing data were obtained from (26) (GSE24115). As PCR over-amplification artifacts typically result in a single sequence being repeated a large number of times, only the first 100 appearances of a given sequence were considered for analysis. We removed strand specific biases using alignPeaks in the htSeqTools package (48), and binding sites were determined with a two-step procedure. First, we used enrichedRegions in htSeqTools to find genomic regions with coverage above 10 and showing high accumulation of sequences in the IP sample compared with the control. We selected regions with coverage above 10 and compared the proportion of reads inside/outside of each region between the IP sample and its corresponding control with a logistic regression likelihood-ratio test. We defined enriched regions as those with a Benjamini–Yekutieli adjusted P-value below 0.05. In a second step, we used enrichedPeaks from htSeqTools to define peaks (i.e. putative binding sites) as locations within the enriched regions with coverage above 50. When combining sequences from two replicated experiments, the coverage cut-off for peak calling was 100. For Pol IIoser2, H3K36me3 and ASH2, which show a moderate enrichment along relatively large genomic regions, we used a coverage cut-off of 5 in the first step and skipped the peak-calling step. ChIP-seq profiles and binding sites were deposited in the NCBI Gene Expression Omnibus repository under accession number GSE27081. Coverage plots at selected genomic regions were generated with the Integrative Genomics Viewer (IGV) (49). Binding sites were assigned to the closest gene using the UCSC refflat gene annotations (http://hgdownload.cse.ucsc.edu/goldenPath/dm3/database/ refFlat.txt.gz). We found the closest transcript to a binding site by computing the distance between the midpoint of the site and the midpoint of all transcripts. Binding sites with no transcript 1000 bp upstream/downstream of their start/end locations (respectively) were left unannotated.
We measured abundance by computing the reads per kilobase per million (RPKM) (50). RPKM is computed as 106R/(ML), where R is the number of reads mapped to a given gene, M is the total number of reads and L is the distance between TSS and TES in kb. We assessed the distribution of ChIP-seq reads around the TSS by plotting the average read coverage. We used the function plotMeanCoverage in the Bioconductor package htSeqTools (48). Shortly, in order to render data from different experiments comparable, the software normalizes the coverage in each gene dividing by the mean coverage in that gene. We removed local irregularities in the average coverage by applying a loess smoothing (span parameter set to 0.1).
We compared genes with binding sites both for H3K4me3 and dKDM5/LID versus genes with only H3K4me3 in terms of Gene Ontology (GO) and KEGG pathway enrichment. We assessed statistical significance with Fisher's exact test, with Benjamini–Yekutieli multiple testing adjustment.
Genome-wide associations in peak calls between ChIP-seq experiments were assessed by cross-tabulating the presence/absence of a peak in each gene and experiment. Statistical significance was assessed via chi-square tests, with P-values obtained from 10 000 permutations to take into account a possible lack of independence between genes.
Expression profiling
Expression profiling analyses were performed using wing imaginal discs from control wt and lidRNAi knockdown flies where ubiquitous depletion was induced by crossing to flies carrying an Actin5C-GAL4 driver. For each condition, around 30 discs were dissected in PBS and RNA was extracted using a combination of Trizol (Invitrogen) and RNeasy minikit (Quiagen). Duplicates were processed for each genotype. Hybridization targets were prepared from 25 ng total RNA using isothermal amplification SPIA Biotin System v2 (NuGEN Technologies, Inc.). 2.2 μg of cDNA was hybridized per Drosophila Genome 2.0 GeneChip (Affymetrix). GeneChips were scanned in a GeneChip Scanner 3000 (Affymetrix). CEL files were generated from DAT files using GCOS software (Affymetrix). Microarray data were preprocessed via quantile normalization and RMA summarization (51). To assess differential expression, we used the limma moderated t-test statistics to compute posterior probabilities of differential expression (PDE), following the empirical Bayes semi-parametric procedure described in (52) and setting the FDR at 0.05. The PDE is a measure of statistical uncertainty: PDE = 1 indicates that the corresponding gene is differentially expressed and PDE = 0 that it is not. Microarray data for lid mutants were deposited in the NCBI Gene Expression Omnibus repository under accession number GSE27081. Microarray data for ash2I1 mutants were obtained from (17) (GPL3797).
When the effects of dKDM5/LID depletion on the expression of individual genes was determined by qRT-PCR, 20 wing imaginal discs from control wt and lidRNAi flies were dissected and RNA extraction was performed with Zymo Research RNA extraction kit, following the instructions of the manufacturer. Two independent replicates were performed and analyzed following the ΔΔCt method. Briefly, candidate genes were first normalized against the control gene Sply for each sample and then represented as a fold change of RNA level in lidRNAi mutants relative to control wt flies. Primers used in these experiments are summarized in Supplementary Table S1.
RESULTS
dKDM5/LID localizes at TSS of developmental genes and regulates H3K4me3
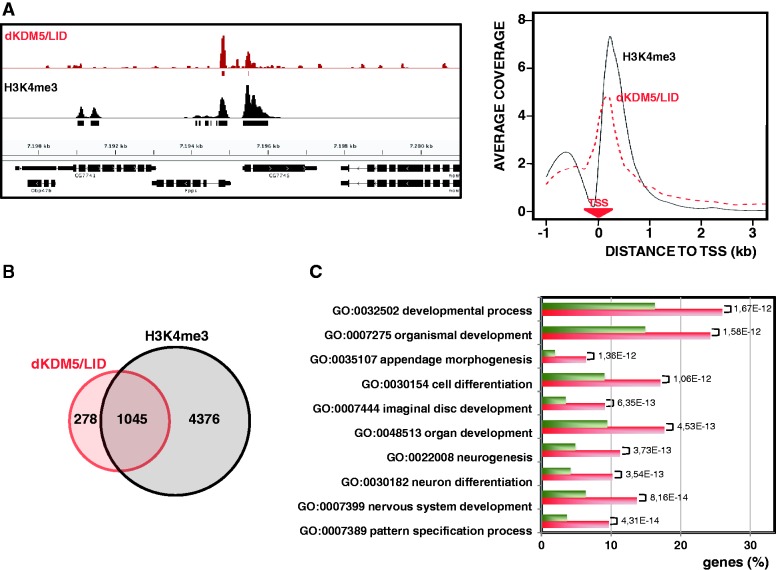
ChIP-seq analyses were performed to determine the genomic distribution of dKDM5/LID in the wing imaginal disc (see ‘Materials and Methods’ section, and Supplementary Figures S1 and S2, for characterization of the αLID antibodies). Approximately 65% of dKDM5/LID binding sites lay within gene coding regions accumulating at TSS sites (Figure 1A), the rest mapping at upstream regulatory regions (22%) and intergenic regions (13%). dKDM5/LID binds TSS regions that are marked with H3K4me3 (Figure 1A), as ∼80% of dKDM5/LID target genes carry H3K4me3 at TSS (Figure 1B). In addition, dKDM5/LID and H3K4me3 show similar distributions at TSS (Figure 1A, right). However, dKDM5/LID target genes account for only 19% of all the genes containing H3K4me3 at TSS (Figure 1B), indicating that dKDM5/LID is present only in a subset of H3K4me3 target genes. GO analysis indicates that the subset of genes containing both H3K4me3 and dKDM5/LID is enriched in specific functions related to developmental processes, morphogenesis and differentiation when compared with genes containing only H3K4me3 (Figure 1C). These results indicate that dKDM5/LID preferentially locates at TSS of developmentally regulated genes containing H3K4me3.
Figure 1.
dKDM5/LID localizes at TSS of developmental genes containing H3K4me3. (A) ChIP-seq coverage profiles of dKDM5/LID (red) and H3K4me3 (black) across a representative region are presented. Rectangles underneath each profile indicate the position of the corresponding peaks/binding sites. Genomic organization of the region is indicated. On the right, the distribution around TSS is presented for dKDM5/LID (red) and H3K4me3 (black). For each gene, the coverage profile was normalized dividing by the average coverage in that gene. The position of the TSS is indicated. (B) Venn diagram showing the intersection between dKDM5/LID (red) and H3K4me3 (black) target genes. (C) The percentage of genes containing both dKDM5/LID and H3K4me3 (red) and genes containing only H3K4me3 (green) are shown for the twenty most enriched functions. Statistical significance of the differences (Benjamini–Yekutieli adjusted P-value) is indicated.
Results reported above suggest that, in developmental genes, dKDM5/LID regulates H3K4me3 at TSS. To test this hypothesis, we performed ChIP-seq analyses in wing imaginal discs from lid mutants. For this purpose, we used lidRNAi knockdown flies that carry a UASGAL4 construct expressing a synthetic hairpin from the coding region of lid that, upon crossing to flies expressing GAL4, generates siRNAs to silence lid expression. lidRNAi efficiently depletes dKDM5/LID when expressed both in wt and heterozygous lid k06801/+ mutant flies, so that, when crossed to flies carrying a ubiquitous Actin5C-GAL4 driver, lid mRNA levels are reduced with respect to original control levels by 70–85%, respectively (Figure 2A, top). Concomitantly, protein levels are strongly reduced (Figure 2A, bottom), and global H3K4me3 levels are significantly increased (Figure 2B). Increased global H3K4me3 is also readily detectable by immunostaining (Supplementary Figure S2). For the rest of the experiments, ubiquitous dKDM5/LID depletion was always performed in heterozygous mutant flies using Actin5C-GAL4 driver, except in expression profiling experiments, where ubiquitous depletion was performed in wt flies.
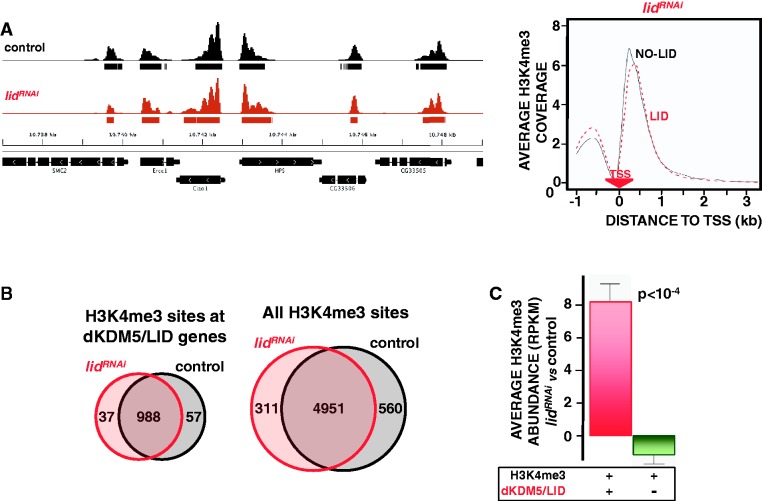
As shown in Figure 3A, dKDM5/LID depletion does not alter the pattern of H3K4me3 that, like in control wt flies, maps to TSS in lidRNAi flies, showing a remarkably similar distribution both in genes containing dKDM5/LID or not (Figure 3A, right). Furthermore, no additional H3K4me3 sites are observed, as 96% of H3K4me3 sites detected at dKDM5/LID target genes in lidRNAi flies are also present in control wt flies (Figure 3B, left), and a similar percentage (94%) is observed when all H3K4me3 sites are considered (Figure 3B, right). On the other hand, in lidRNAi flies, H3K4me3 abundance specifically increases at TSS of dKDM5/LID target genes, whereas non-target genes are not significantly affected (Figure 3C). Increased H3K4me3 was also detected when individual genes were analyzed by ChIP-qPCR (Figure 5C). Altogether, these results show that dKDM5/LID depletion increases H3K4me3 levels at TSS without noticeably affecting the actual number and location of H3K4me3 sites.
Figure 3.
dKDM5/LID depletion increases H3K4me3 at TSS. (A) ChIP-seq coverage profiles of H3K4me3 across a representative region are presented both in control wt flies (black) and lidRNAi knockdown flies (red), where dKDM5/LID depletion was induced by the Actin5C-GAL4 driver in heterozygous lidk06801/+ mutant flies. Rectangles underneath each profile indicate the position of the corresponding peaks/binding sites. Genomic organization of the region is indicated. On the right, H3K4me3 distribution around TSS in lidRNAi knockdown flies is presented for genes containing dKDM5/LID (red) or not (black). For each gene, the coverage profile was normalized dividing by the average coverage in that gene. The position of the TSS is indicated. (B) Venn diagrams showing the intersection between H3K4me3 sites in control wt (black) and lidRNAi knockdown flies (red) detected at dKDM5/LID target genes (left) and when all H3K4me3 sites are considered (right). (C) Relative H3K4me3 abundance in lidRNAi knockdown flies versus control wt flies is presented for genes containing both dKDM5/LID and H3K4me3 (red) and genes containing only H3K4me3 (green). Statistical significance of the difference (Kruskal–Wallis P-value) is indicated.
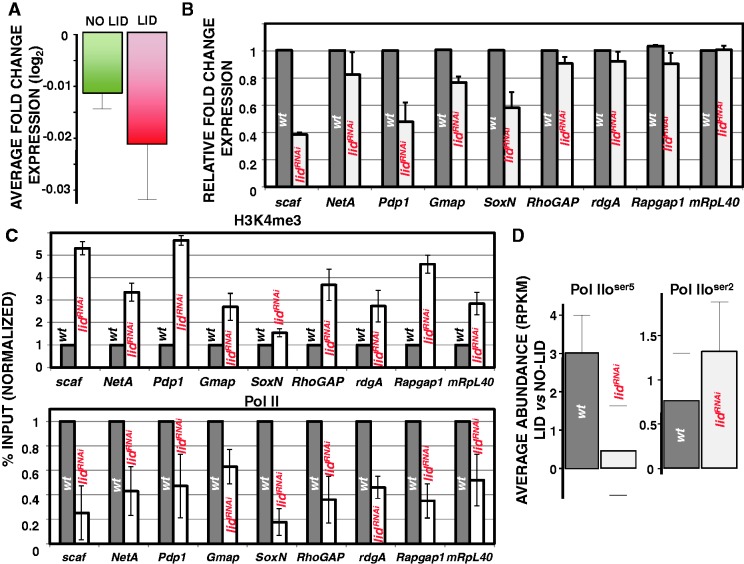
Figure 5.
dKDM5/LID target genes are weakly downregulated in lidRNAi flies. (A) Average fold change of expression in lidRNAi flies is presented for dKDM5/LID target genes (red) or non-target genes (green). (B) mRNA levels of nine dKDM5/LID target genes in lidRNAi knockdown (white) and control wt (black) flies is determined by qRT-PCR. mRNA levels were determined in relation to a non-target gene (Sply) and normalized respect to those observed in control wt flies. (C) H3K4me3 and RNApol II levels at the dKDM5/LID target genes shown in panel B are determined by ChIP-qPCR in lidRNAi knockdown (white) and control wt (black) flies. Results are presented as percentage of input normalized with respect to wt. (D) Relative Pol IIoser5 (left) and Pol IIoser2 (center) abundance at dKDM5/LID target genes versus non-target genes is presented in lidRNAi knockdown (white) and control wt (black) flies.
dKDM5/LID target genes are actively transcribed
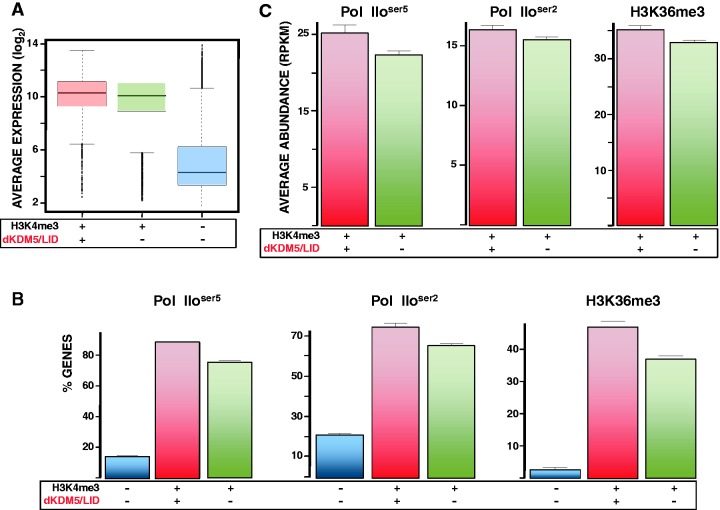
Genes marked with H3K4me3 at TSS are generally expressed to high levels. However, considering that dKDM5/LID demethylates H3K4me3 at TSS, it was possible that dKDM5/LID target genes would be expressed only to low levels. To test this hypothesis, we performed expression-profiling experiments in wt wing imaginal discs to determine gene expression as a function of presence or not of dKDM5/LID. As shown in Figure 4A, genes containing both dKDM5/LID and H3K4me3 are as highly expressed as genes containing only H3K4me3. In addition, ChIP-seq analyses detected binding of both the promoter-proximal Pol IIoser5 and the elongating Pol IIoser2 active RNApol II forms at most dKDM5/LID target genes (Figure 4B). As a matter of fact, in comparison to genes containing only H3K4me3, dKDM5/LID target genes are enriched in active RNApol ll, particularly Pol IIoser5 (Figure 4C). Similarly, dKDM5/LID target genes are also enriched in H3K36me3 (Figure 4B and C), a modification that is deposited during elongation (53). Altogether, these results indicate that dKDM5/LID target genes are actively engaged in transcription.
Figure 4.
dKDM5/LID target genes are actively transcribed. (A) Box plot showing the expression of genes carrying both dKDM5/LID and H3K4me3 (red), only H3K4me3 (green) or lacking H3K4me3 (blue). (B) Binding of Pol IIoser5 (left), Pol IIoser2 (center) and H3K36me3 (left) to genes carrying both dKDM5/LID and H3K4me3 (red), only H3K4me3 (green) or lacking H3K4me3 (blue) is presented as the percentage (%) of genes under each category that contain Pol IIoser5, Pol IIoser2 and H3K36me3, respectively. (C) Average Pol IIoser5 (left), Pol IIoser2 (center) and H3K36me3 (left) abundance is presented at genes carrying both dKDM5/LID and H3K4me3 (red) and only H3K4me3 (green).
The association of dKDM5/LID with actively transcribed genes suggests a positive contribution to transcription. To address this question, we performed expression-profiling experiments in wing imaginal discs from lidRNAi flies where ubiquitous depletion was induced by the Actin5C-GAL4 driver. As shown in Table 1, dKDM5/LID depletion induces only a weak effect on transcription, as <1% of dKDM5/LID target genes are detected differentially expressed with high probability (PDE > 0.95) (see ‘Materials and Methods’ section for details). This set of genes is highly enriched (10-fold) in downregulated genes, showing an average fold-change expression of −2.91. This unbalance towards downregulated genes is also observed with genes detected differentially expressed at lower confidences (PDE > 0.9 to PDE > 0.5) (Table 1) or, even, when all dKDM5/LID target genes are considered (down/up ratio, 1.15) (Supplementary Table S2). As a matter of fact, though weakly (∼3%), average expression of all dKMD5/LID target genes is significantly decreased in lidRNAi flies with respect to non-target genes (P = 0.0077) (Figure 5A). It must also be noticed that genes detected differentially expressed with increasing confidence do not differ in H3K4me3 content or expression in wt flies (Supplementary Figure S3). Altogether, these results strongly suggest that, though weakly, dKDM5/LID depletion downregulates expression of target genes. qRT-PCR analyses confirm these results, as 8 out of 9 dKDM5/LID target genes tested (PDEs ranging from 0 to 0.96) were found downregulated to different extents in lidRNAi flies (Figure 5B). In addition, ChIP-qPCR analyses confirm increased H3K4me3 and reduced RNApol ll occupancy at these genes (Figure 5C). The effect of dKDM5/LID depletion on RNApol II occupancy was further analyzed by ChIP-seq. In these experiments, we determined Pol IIoser5 and Pol IIoser2 occupancy at dKDM5/LID target genes in comparison to non-target genes both in wt and lidRNAi flies. As shown in Figure 5D, Pol IIoser5 occupancy at dKDM5/LID target genes, which is higher than at non-target genes in wt flies (see also Figure 4C), is strongly reduced in lidRNAi flies, while Pol IIoser2 occupancy increases slightly. Reporter-expression experiments are also consistent with a positive contribution to transcription, as expression of a vestigial(vg)-LacZ reporter construct in the wing imaginal disc was strongly reduced upon dKDM5/LID depletion or in null lidk06801/lidk06801 mutant clones (Supplementary Figure S4).
Table 1.
dKDM5/LID depletion downregulates expression of target genes
| PDE | Average fold-change expression | Upregulated genes | Downregulated genes | Down/up ratio | P-value |
|---|---|---|---|---|---|
| >0.95 | −2.91 | 1 | 10 | 10 | 0.0117 |
| >0.90 | −1.49 | 11 | 31 | 2.82 | 0.0029 |
| >0.80 | −1.26 | 34 | 85 | 2.50 | P <0.0001 |
| >0.70 | −1.16 | 71 | 133 | 1.87 | P <0.0001 |
| >0.60 | −1.11 | 110 | 178 | 1.62 | P <0.0001 |
| >0.50 | −1.07 | 175 | 235 | 1.34 | P = 0.0035 |
The number of dKDM5/LID target genes upregulated and downregulated in lidRNAi flies is presented for several cut-offs on the PDE (see ‘Materials and Methods’ section for details). The down/up ratio and the average fold-change expression are indicated. Statistical significance of the differences is also indicated (Binomial test for 50/50 up/down proportions).
ASH2 regulates dKDM5/LID target genes
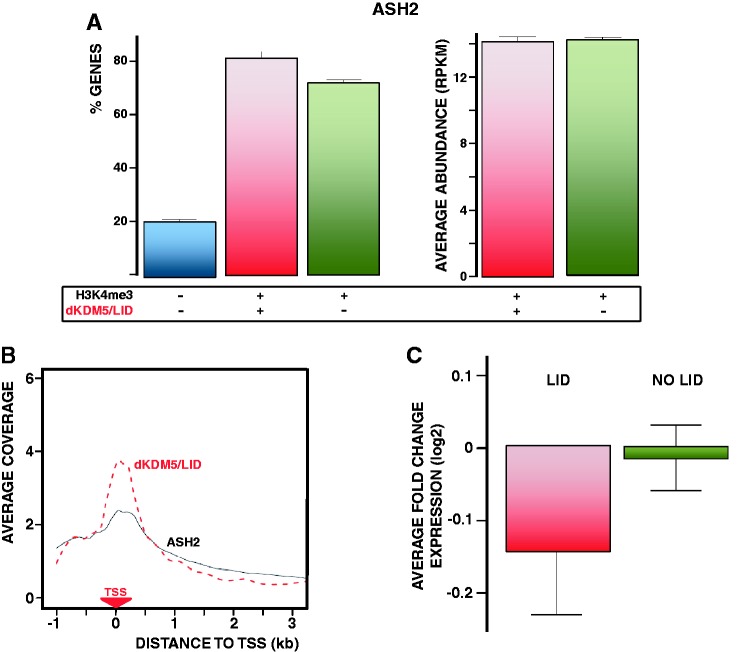
It was shown earlier that, in Drosophila, ASH2 binds at TSS of most genes containing H3K4me3 (26), and is required for H3K4me3 (17). In this context, considering that dKDM5/LID only binds 19% of total H3K4me3-containing genes (Figure 1B), we determined whether dKDM5/LID target genes are also bound/regulated by ASH2. As shown in Figure 6A (left), ∼80% of dKDM5/LID target genes are bound by ASH2. As a matter of fact, ASH2 occupancy at dKDM5/LID target genes is as high as at genes containing only H3K4me3 (Figure 6A, right). In addition, the distributions of dKDM5/LID and ASH2 at TSS are highly overlapping (Figure 6B). ASH2 has a dual effect on transcription, as ∼60% of ASH2 target genes found differentially expressed in ash2I1 mutants are downregulated, the rest being significantly upregulated (26). Both sets of genes, which display differential structural and functional features, contain H3K4me3 at TSS. In this context, dKDM5/LID preferentially localizes at genes downregulated in ash2I1 mutants, as average expression of dKDM5/LID target genes is significantly decreased in ash2I1 mutants, whereas expression of genes containing only H3K4me3 is not (P < 0.0001) (Figure 6C). Furthermore, like dKDM5/LID target genes, genes downregulated in ash2I1 mutants are enriched in developmental functions, whereas upregulated ones are enriched in ubiquitous/house keeping functions (26).
Figure 6.
dKDM5/LID target genes are regulated by ASH2. (A) On the left, binding of ASH2 to genes carrying both dKDM5/LID and H3K4me3 (red), only H3K4me3 (green) or lacking H3K4me3 (blue) is presented as the percentage (%) of genes under each category that contain ASH2. On the right, average ASH2 abundance is presented at genes carrying both dKDM5/LID and H3K4me3 (red) and only H3K4me3 (green). (B) The distribution of dKDM5/LID (red) and ASH2 (black) around TSS is presented for the genes containing both factors. For each gene, the coverage profile was normalized dividing by the average coverage in that gene. The position of TSS is indicated. (C) Average fold change of expression in ash2I1 mutant flies is presented for genes carrying both dKDM5/LID and H3K4me3 (red) or only H3K4me3 (green).
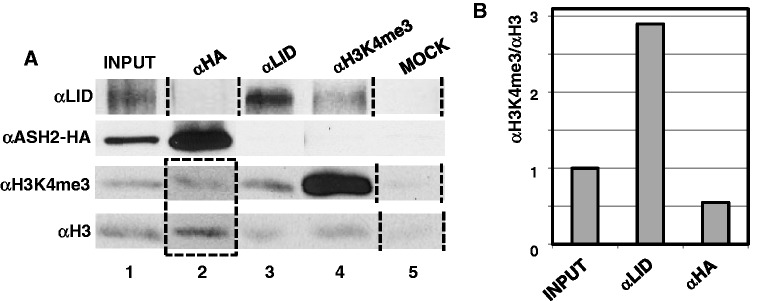
Altogether, these results show that genes regulated by dKDM5/LID are also regulated by ASH2. Both factors bind at TSS, regulate H3K4me3 and positively contribute to their expression. Next, we asked whether dKDM5/LID and ASH2 bind simultaneously. To address this question, we performed ChIP-experiments in in vitro cultured cells, which constitute a homogeneous cell population, and, to overcome the lack of efficient αASH2 antibodies, we used a stable S2 cell line expressing an ASH2-HA tagged construct. In these experiments, crosslinked chromatin was subjected to IP with αLID and αHA antibodies. As shown in Figure 7, when chromatin is immunoprecipitated with αHA antibodies, dKDM5/LID cannot be detected (Figure 7A, lane 2, row αLID) and, vice versa, no ASH2-HA is detected after IP with αLID antibodies (Figure 7A, lane 3, row αASH2-HA). In addition, chromatin immunoprecipitated with αLID antibodies is significantly enriched in H3K4me3 (Figure 7A, lane 3, rows αH3K4me3 and αH3, and Figure 7B), whereas chromatin immunoprecipitated with αHA has a low H3K4me3 content (Figure 7A, lane 2, rows αH3K4me3 and αH3, and Figure 7B). Altogether, these results indicate that dKDM5/LID and ASH2 do not bind simultaneously and recognize different chromatin states at TSS, enriched in H3K4me3 and not, respectively. Consistent with this hypothesis, when chromatin was immunoprecipitated with αH3K4me3 antibodies, dKDM5/LID was detected but ASH2 was not (Figure 7A, lane 4, rows αLID and αASH2-HA).
Figure 7.
dKDM5/LID and ASH2 do not bind simultaneously and recognize different chromatin states. (A) Crosslinked chromatin was immunoprecipitated with αHA (lane 2), αLID (lane 3), αH3K4me3 (lane 4) or no (lane 5) antibodies and the presence of dKDM5/LID (row αLID), ASH2-HA (row αASH2-HA), H3K4me3 (row αH3K4me3) and H3 (row αH3) determined by western blot using rat polyclonal αLID (1:10000), αHA (1:500), αH3K4me3 (1:2000) and αH3 (1:1000) antibodies. Notice that to present data in the same order some lanes have been rearranged (indicated by the dotted lines). Notice also that analyses with αH3K4me3 and αH3 antibodies of the ChIP performed with αASH2-HA (lane 2, rows αH3K4me3 and αH3; indicated by the dotted rectangle) was performed in a different set of gels together with the input and mock for proper quantification and comparison to the results obtained in the ChIP performed with αLID. (B) Quantitative analysis of the enrichment in H3K4me3 versus H3 in the ChIPs performed with αLID (lane 3, rows αH3K4me3 and αH3 in panel A) and αASH2-HA (lane 2, rows αH3K4me3 and αH3 in panel A).
DISCUSSION
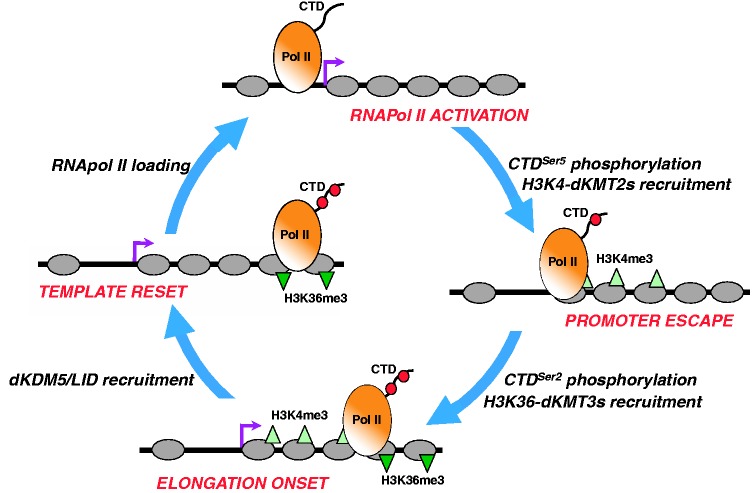
Here, we report that dKDM5/LID localizes at TSS of developmental genes and regulates H3K4me3. dKDM5/LID target genes are actively transcribed and, though weakly, they are significantly downregulated in lidRNAi mutant flies. Previous reports already suggested a positive contribution of dKDM5/LID to transcription (29,30,42). Our results also show that dKDM5/LID target genes are bound by ASH2, an evolutionarily conserved component of H3K4-KMT2 complexes that localizes at TSS and is required for H3K4me3 (17–25). In addition, dKDM5/LID target genes are strongly downregulated in ash2 mutant flies. These observations indicate that dKDM5/LID and ASH2 act coordinately to regulate H3K4me3 at TSS of developmental genes for their efficient transcription. dKDM5/LID and ASH2, however, do not bind chromatin simultaneously, indicating that they act at different moments during transcription. These observations strongly favor a model by which ASH2 and dKDM5/LID act sequentially during transcription to facilitate its progression (Figure 8). On this regard, work performed in budding yeast links chromatin modification events to sequential RNApol II activation. At a first step, TFIIH-mediated phosphorylation of CTDSer5 recruits scKMT2/SET1 to methylate H3K4 (54), and induces promoter escape. Later, the onset of productive transcription involves phosphorylation of CTDSer2, which results in recruitment of H3K36 KMT3/SET2 both in budding yeast and mammals (55,56). dKDM5/LID recruitment might also be regulated during transcription cycle progression. In this context, it is possible that, after RNApol II activation and subsequent H3K4-methylation, dKDM5/LID is recruited and transient demethylation resets chromatin to the original ‘unmethylated’ state, facilitating the next RNApol II molecule to initiate progression through the transcription cycle. Consistent with this model, it was shown that the C-terminal PHD-finger of dKDM5/LID, or the mammalian homolog KDM5A/JARID1A, specifically binds H3K4me2,3 (57,58) and, furthermore, we have shown that dKDM5/LID binds chromatin enriched in H3K4me3, whereas chromatin bound by ASH2 is poor in H3K4me3. Finally, our results also show that dKDM5/LID depletion significantly reduces RNApol ll occupancy, in particular by the promoter-proximal Pol IIoser5 active form, providing a basis for the positive contribution of dKDM5/LID to transcription. In contrast, occupancy by the elongating Pol IIoser2 form is not similarly affected, showing a tendency to be slightly increased. It is possible that, in the absence of dKDM5/LID, constitutive/increased H3K4me3 at TSS affects RNApol II pausing and, hence, transcription efficiency. Actually, it has been shown that depletion of NELF, a factor required for RNApol II pausing, results in a general downregulation of its target genes both in Drosophila and human cells (59,60).
Figure 8.
A model for the coordinated action of dKDM5/LID and ASH2-associated dKMT2s in transcription regulation. ASH2/dKMT2s and dKDM5/LID might act at different steps of the transcription cycle to facilitate its progression. Phosphorylation of CTDSer5 results in recruitment of ASH2/dKMT2s to methylate H3K4 and promotes promoter escape. Later, phosphorylation of CTDSer2 recruits H3K36 dKMT3s and marks the onset of productive elongation. Next, transient H3K4me3 demethylation by dKDM5/LID resets chromatin to the original ‘unmethylated’ state, so that a new transcription cycle starts with loading/activation of the next RNApol II molecule.
Several reasons could account for the weakness of the observed effect of dKDM5/LID depletion on gene expression. On one hand, though dKDM5/LID content is strongly reduced in lidRNAi, depletion is not complete. Note that null lid mutations could not be used, as they are lethal during late embryo/early larvae development. Second, although dKDM5/LID is the only enzyme known to specifically demethylate H3K4me3 in Drosophila, additional KDMs might exist capable of playing a similar function. At this respect, it was reported that dKDM2, which was originally found to demethylate H3K36me2 (61), might also be capable of demethylating H3K4me3 (62). Thus, it is possible that loss of dKDM5/LID is partially compensated by dKDM2. As a matter of fact, a genetic interaction was recently reported between dKDM5/lid and dKDM2 (57).
The proposed function of dKDM5/LID in the regulation of transcription is likely conserved, as it was recently reported that mammalian KDM5B/JARID1B preferentially localizes at TSS of developmental genes and regulates H3K4me3 (41). Mammalian KDM5C/JARID1C has also been shown to bind at TSS (63). Interestingly, although KDM5B/JARID1B is required to efficiently silence stem and germ cell specific genes during neuronal differentiation, its depletion in ESCs shows also a weak downregulation of target genes (41). In Drosophila, dKDM5/LID has also been shown to be involved in repression of some developmental genes (32). In fact, in the wing imaginal disc, ∼20% of dKDM5/LID target genes show no detectable H3K4me3. Altogether, these observations suggest that dKDM5/LID might play a dual function; repressing specific genes during development and, in differentiated cells, regulating H3K4me3 dynamics at TSS during transcription.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–2 and Supplementary Figures 1–4.
FUNDING
MICINN [CSD2006-49 and BFU2009-07111 to F.A.], [ACI2009-0903 to M.C.]; CSIC [200420E583 and 201120E001]; Generalitat de Catalunya [SGR2009-1023 to F.A.]; I3P predoctoral fellowship (to M.L.L). This work was carried out within the framework of the ‘Centre de Referència en Biotecnologia’ of the ‘Generalitat de Catalunya’. Funding for open access charge: MICINN [BFU2009-07111].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are thankful to Dr A. Shilatifard for the S2 cell line expressing ASH2-HA and to Dr M. Milán for fly stocks, advice and reading of the manuscript. We are also thankful to Mrs E. Fuentes and E. Freire for technical assistance.
REFERENCES
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Ruthenburg AJ, Li H, Patel DJ, Allis DC. Mutivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 2008;20:1–8. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 8.Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl Acad. Sci. USA. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science. 2002;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- 10.Noma K, Allis CD, Grewal S. Transitions in distinct histone H3 methylation patterns at heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 11.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 12.Schneider R, Bannister AJ, Myers FA, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 13.Schübeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarez-Venegas R, Avramova Z. SET-domain proteins of the Su(var)3-9, E(z) and trithorax families. Gene. 2002;285:25–37. doi: 10.1016/s0378-1119(02)00401-8. [DOI] [PubMed] [Google Scholar]
- 16.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 17.Beltran S, Angulo M, Pignatelli M, Serras F, Corominas M. Functional dissection of the ash2 and ash1 transcriptomes provides insights into the transcriptional basis of wing phenotypes and reveals conserved protein interactions. Genome Biol. 2007;8:R67. doi: 10.1186/gb-2007-8-4-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl Acad. Sci. USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl Acad. Sci. USA. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 22.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder R. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 23.Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, Verdine GL. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 2006;13:704–712. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sims R, Jr, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pérez-Lluch S, Blanco E, Carbonell A, Raha D, Snyder M, Serras F, Corominas M. Genome-wide chromatin occupancy analysis reveals a role for ASH2 in transcriptional pausing. Nucleic Acids Res. 2011;39:4628–4639. doi: 10.1093/nar/gkq1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schäfer C, Phalke S, Walther M, Schmidt A, Jenuwein T, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol. Cell. 2007;26:103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 28.Eissenberg JC, Lee MG, Schneider J, Ilvarsonn A, Shiekhattar R, Shilatifard A. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat. Struct. Mol. Biol. 2007;14:344–346. doi: 10.1038/nsmb1217. [DOI] [PubMed] [Google Scholar]
- 29.Lee N, Zhang J, Klose RJ, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat. Struct. Mol. Biol. 2007;14:341–343. doi: 10.1038/nsmb1216. [DOI] [PubMed] [Google Scholar]
- 30.Lloret-Llinares M, Carré C, Vaquero A, de Olano N, Azorín F. Characterisation of Drosophila melanogaster JmjC+N histone demethylases. Nucleic Acids Res. 2008;36:2852–2863. doi: 10.1093/nar/gkn098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, Karch F, Bray SJ, Demmers JAA, Verrijzer CP. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol. Cell. 2009;35:782–793. doi: 10.1016/j.molcel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 33.Lee N, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. The H3K4 demethylase Lid associates with and inhibits histone deacetylase Rpd3. Mol. Cell. Biol. 2009;29:1401–1410. doi: 10.1128/MCB.01643-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Stefano L, Walker JA, Burgio G, Corona DF, Mulligan P, Näär AM, Dyson NJ. Functional antagonism between histone H3K4 demethylases in vivo. Genes Dev. 2011;25:17–28. doi: 10.1101/gad.1983711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 36.Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Rin6a/MBLR, a polycomb-like protein. Cell. 2007;128:877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Liefke R, Oswald F, Alvarado C, Ferres-Marco D, Mittler G, Rodriguez P, Dominguez M, Borggrefe T. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010;24:590–601. doi: 10.1101/gad.563210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Bigas N, Kisiel TA, Dewaal DC, Holmes KB, Volkert TL, Gupta S, Love J, Murray HL, Young RA, Benevolenskaya EV. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol. Cell. 2008;31:520–530. doi: 10.1016/j.molcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, Impey S. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011;30:1473–1484. doi: 10.1038/emboj.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitz SU, Albert M, Malatesta M, Morey L, Johansen JV, Bak M, Tommerup N, Abarrategui I, Helin K. Jarid1b targets gene regulating development and is involved in neural differentiation. EMBO J. 2011;30:4586–4600. doi: 10.1038/emboj.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gildea JJ, Lopez R, Shearn A. A screen for new trithorax group genes identified little imaginal dics, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics. 2000;156:645–663. doi: 10.1093/genetics/156.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A. The COMPASS family of H3K4 methylases in Drosophila. Mol. Cell Biol. 2011;31:4310–4318. doi: 10.1128/MCB.06092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papp B, Müller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 46.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Planet E, Stephan-Otto Attolini C, Reina O, Flores O, Rossell D. htSeqTools: high-throughput sequencing quality control, processing and visualization in R. Bioinformatics. 2011;28:589–590. doi: 10.1093/bioinformatics/btr700. [DOI] [PubMed] [Google Scholar]
- 49.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat. Biotech. 2010;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by rna-seq. Nat. Methods. 2008;7:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 51.Irizarry R, Hobbs B, Collin B, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TS. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 52.Rossell D, Guerra R, Scott C. Semi-parametric differential expression analysis via partial mixture estimation. Stat. Appl. Genet. Mol. Biol. 2008;7:1–15. doi: 10.2202/1544-6115.1333. [DOI] [PubMed] [Google Scholar]
- 53.Bell O, Conrad T, Kind J, Wirbelauer C, Akhtar A, Schübeler D. Transcription-coupled methylation of histone H3 at lysine 36 regulates dosage compensation by enhancing recruitment of the MSL complex in Drosophila melanogaster. Mol. Cell Biol. 2008;28:3401–3409. doi: 10.1128/MCB.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ng HH, Robert F, Young TA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 55.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P. Solution structure of the Set2-Rpb1 interacting domain of Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc. Natl Acad. Sci. USA. 2005;102:17636–17641. doi: 10.1073/pnas.0506350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L, Greer C, Eisenman RN, Secombe J. Essential functions of the histone demethylase Lid. PLoS Genet. 2010;6:e1001221. doi: 10.1371/journal.pgen.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo J-L, Patel DJ, Allis CD. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun J, Li R. Human negative elongation factor activates transcription and regulates alternative transcription initiation. J. Biol. Chem. 2010;285:6443–6452. doi: 10.1074/jbc.M109.084285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Verrijzer CP. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 2008;22:2799–2810. doi: 10.1101/gad.484208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kavi HH, Birchler JA. Drosophila KDM2 is an H3K4me3 demethylase regulating nucleolar organization. BMC Res. Notes. 2009;2:217. doi: 10.1186/1756-0500-2-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]