Abstract
The Bmal1 gene is essential for the circadian system, and its promoter has a unique open chromatin structure. We examined the mechanism of topoisomerase I (Top1) to understand the role of the unique chromatin structure in Bmal1 gene regulation. Camptothecin, a Top1 inhibitor, and Top1 small interfering RNA (siRNA) enhanced Baml1 transcription and lengthened its circadian period. Top1 is located at an intermediate region between two ROREs that are critical cis-elements of circadian transcription and the profile of Top1 binding indicated anti-phase circadian oscillation of Bmal1 transcription. Promoter assays showed that the Top1-binding site is required for transcriptional suppression and that it functions cooperatively with the distal RORE, supporting that Bmal1 transcription is upregulated by Top1 inhibition. A DNA fragment between the ROREs, where the Top1-binding site is located, behaved like a right-handed superhelical twist, and modulation of Top1 activity by camptothecin and Top1 siRNA altered the footprint profile, indicating modulation of the chromatin structure. These data indicate that Top1 modulates the chromatin structure of the Bmal1 promoter, regulates Bmal1 transcription and influences the circadian period.
INTRODUCTION
Circadian clocks align behavioral and biochemical process with the day–night cycle. The master clock that generates circadian rhythms in mammals is located in the suprachiasmatic nucleus (SCN) of the hypothalamus where it controls all aspects of physiology such as sleep–wake cycles, body temperature, hormone secretion, blood pressure and metabolism. Coordination among such aspects of physiology by the circadian clock is essential to optimize metabolic responses and strengthen inherent homeostatic regulatory mechanisms (1). The circadian clock generates robust rhythms coupled with changes in the cellular environment. Thus, circadian dysfunction is considered to contribute to the incidence and severity of a wide range of clinical and pathological conditions, including sleep disorders, cancer, depression, metabolic syndrome and inflammation (2).
The molecular mechanism of the circadian oscillator consists of autoregulatory transcriptional and translational feedback loops that have both positive and negative elements. Among the core clock genes, Bmal1 is apparently essential and non-redundant in the mammalian clock and its expression robustly oscillates in the SCN and in peripheral clock cells (3). On the other hand, Bmal1 can be replaced by Bmal2, and in the mouse, Bmal2 is regulated by Bmal1 (4). Therefore, Bmal1 transcription should be closely associated with circadian rhythms. The Bmal1 promoter contains two recognition motifs for retinoic acid receptor-related orphan receptor (ROR) and reverse Erb (REV-ERB) orphan nuclear receptors (ROREs) that are critical elements for Bmal1 oscillatory transcription (5). We previously found that the ROREs are embedded in a unique GC-rich open chromatin structure, with which a nuclear matrix-like structure at the 3′-flanking region cooperates to regulate Bmal1 transcription (6).
DNA topoisomerases regulate the topological status of the DNA double helix and induce either single (type I)- or double (type II)-strand DNA breaks and are thus key enzymes for DNA replication, transcription, recombination and chromatin remodeling (7). Topoisomerase I (type I) is essential for the early development of multicellular eukaryotes and it is also a transcriptional coactivator (8). Topoisomerase activity is essential for the regulation of transcription. Topoisomerase I (Top1) mainly binds the gene promoter at the 3′ parts of transcribed regions and its occupancy is linked to active transcription. Top1 at promoter regions acts directly to stimulate nucleosome disassembly, histone acetylation and gene expression (9). Endogenous glucocorticoid hormones or major cues for circadian rhythms regulate the circadian expression of Top1 (10). The Top1 promoter region also contains an E-box and DBP/E4BP4-binding elements (D-box), where the BMAL1/CLOCK heterodimer and DBP bind, respectively, and thus the clock system controls the autonomous circadian transcription of the Top1 promoter (11). Camptothecin was originally isolated from the Chinese Camptotheca acuminate tree and it is a non-competitive, specific Top1 inhibitor that also induces apoptosis (12). Therefore, many anti-cancer drugs target Top1, and alkaloids such as DNA intercalators also inhibit Top1 activity and exert anti-tumor activity (13). Harmala alkaloids inhibit Top1 activity (14) and we recently found that the harmala alkaloid harmine modulates circadian Bmal1 transcription (15). Although Top1 involvement in circadian transcription is implied as described above, whether or not Top1 directly regulates Bmal1 transcription remains unclear.
Here, we studied the effects of Top1 on Bmal1 transcription and its oscillatory rhythm using camptothecin, a Top1 inhibitor and Top1 small interfering RNA (siRNA). We then analyzed the presence of Top1 on the Bmal1 promoter region and the relationship between Top1 expression and binding to the Bmal1 promoter at the chromatin level. We then investigated how the Top1-binding site on the Bmal1 promoter influences Bmal1 promoter activity. Finally, we characterized the DNA structure around the Top1-binding site on the Bmal1 promoter and confirmed that Top1 changes the chromatin structure of the Bmal1 promoter in vivo.
MATERIALS AND METHODS
Cell culture
Stable cells containing the luciferase reporter gene driven by the Bmal1 promoter region (−197 to +27) were established from NIH3T3 (6) and Sarcoma 180 (16) cells. All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and a mixture of penicillin and streptomycin in a humidified incubator at 37°C under a 5% CO2 atmosphere. RPMI8402 and CPT-K cells were incubated with 1 mM aza-dCTP (Sigma) for 2 days before analyses.
Real-time reporter gene assays
Real-time reporter gene assays proceeded as described (6). Stable reporter cells were stimulated with 100 nM dexamethasone for 2 h and then incubated with DMEM containing 0.1 mM luciferin (Promega), 25 mM HEPES (pH 7.2) and 10% FBS. Bioluminescence was measured and integrated for 1 min at 10-min intervals using a Kronos AB-2500 (ATTO). Data were detrended by subtracting a best fit line followed by subsequent fitting to a sine wave to determine circadian period length as described (15).
Knockdown of Top1 expression
The Stealth RNA interference (RNAi) siRNA duplex for Top1 knockdown and BLOCK-iT Fluorescent Oligo as a negative control were purchased from Invitrogen. The stealth RNAi was a mixture of three siRNAs with the following sequences: 5′-UAUCAAUGAAGUACAGUGCUACAGC-3′ and 5′-GCUGUAGCACUGUACUUCAUUGAUA-3′; 5′-CUUAGUAGUAUAUUCGUGGUCAAGC-3′ and 5′-GCUUGACCACGAAUAUACUACUAAG-3′; 5′-UAUAGUAUCUGAUUGAGUCCUUCCC-3′ and 5′-GGGAAGGACUCAAUCAGAUACUAUA-3′. The siRNAs were introduced into cells using HilyMax (DOJINDO).
Real-time quantitative RT–PCR
Real-time quantitative reverse transcriptase–polymerase chain reaction (RT–PCR) proceeded using a LightCycler (Roche) and the Light Cycler-FastStart DNA Master SYBR Green I kit (Roche) as described (6). The primer sequences were as follows: Actin, 5′-TACGCCAACACAGTGCTGTCTG-3′ and 5′-TTTTCTGCGCAAGTTAGGTTTTGTC-3′; Top1, 5′-TCCCAGATCGAAGCGGATTTC-3′ and 5′-GCTCTAATCTTTTCCTCC-3′; Bmal1, 5′-GGACTTCGCCTCTACCTGTTCA-3′ and 5′-AACCATGTGCGAGTGCAGGCGC-3′. The PCR products cloned into the pGEM-T Easy vector (Promega) served as an authentic template. Relative expression levels were evaluated using Light Cycler software, version 3.5.
Top1-mediated cleavage assays
The Bmal1 promoter region was end-labeled with [γ-32P] ATP using T4 polynucleotide kinase (New England BioLabs) and then incubated with 50 units of Top1 protein (TAKARA) and 0.5 mM camptothecin in 50 mM Tris–HCl (pH 7.5), 120 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol and 30 µg/ml bovine serum albumin at 37°C for 7 min to generate cleavable complexes. The reactions were terminated by the addition of 2 µl of 5% sodium dodecyl sulphate (SDS) and the mixture was treated with 150 µg/ml protenase K for 1 h at 37°C. We purified and resolved DNA by electrophoresis on 8% polyacrylamide–urea gels.
Electrophoretic mobility shift assays
These assays proceeded as described (17). Briefly, a DNA probe (5′-GATTGGTCGGAAAGTAGGTTAGTGGTGCGACATTTAGGGAAGGCAGAAAGTAGGTCAGGGACGGAGG-3′) was end-labeled with [γ-32P] ATP using T4 polynucleotide kinase (New England BioLabs). Portions (10 µl) of the probe suspended in 20 µl of 16 mM HEPES (pH 7.5) containing 150 mM KCl, 16% (v/v) glycerol, 1.6 mM MgCl2, 0.8 mM dithiothreitol, 0.4 mM PMSF, 1 mM EDTA, 0.8 mg/ml of bovine serum albumin, 0.06 mg/ml of poly(dI-dC) and 0.01% NP-40 were incubated with 15 units of Top1 protein (TAKARA). The mixtures were then resolved by electrophoresis on 4% polyacrylamide gels in 40 mM Tris–acetate, 1 mM EDTA and 5% glycerol.
Western blotting
Western blotting and SDS–polyacrylamide gel electrophoresis (PAGE) proceeded as described (15). Briefly, nuclear proteins were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) and resolved by 9% SDS–PAGE and transferred to PVDF membranes (Amersham). Non-specific binding was blocked with 3% skim milk in PBS. Proteins were probed with anti-Top1 antibody (Santa Cruz Biotechnology) or anti-lamin A/C antibody (Santa Cruz Biotechnology) and then incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG (Upstate). Immunoreactive proteins were visualized using ECL (Amersham) according to the manufacturer’s instructions.
Chromatin immunoprecipitation assays
The chromatin immunoprecipitation (ChIP) assays proceeded as described (17). Briefly, NIH3T3 cells were incubated with 1% formaldehyde for 10 min at room temperature to cross-link proteins and DNA. The cells were lysed and chromatin was fragmented and immunoprecipitated using the SimpleChIP enzymatic chromatin IP kit (Cell Signaling Technology) and anti-Top1 or anti-Pol II antibodies (Santa Cruz Biotechnology). Purified DNA was analyzed by PCR using the primers 5′-GAACGCGAATTGGTTTGGGTTGTCCG-3′ and 5′-ACACTCACCGTGGCTCGCTGCGAGC-3′.
Transient reporter gene assay
Luciferase reporter gene plasmids and the internal control plasmid, pRL-CMV (Promega) were transfected into NIH3T3 cells. Luciferase was measured using the Dual Luciferase Reporter Assay System (Promega) as described (18). Transcriptional activities were normalized relative to Renilla luciferase activities.
Gel mobility assays
Double-stranded oligonucleotides (AT, dA20/dT20, the Top1-binding region between ROREs, 5′-GGCTGGTGCGACATTTAGGGAA-3′ and the SP1-binding sequence, 5′-GATCGGGGCGGGGC-3′), which were designed to create two-base 5′-protruding ends were ligated to generate concatemers as described (19) and dA20/dT20 served as a migration standard (20). The concatemers were resolved by 6% PAGE. Upon treatment with ethidium bromide, right-handed superhelixes decrease their twist and increase the planarity of the superhelixes, while left-handed superhelixes increase twisting and decrease their degree of planarity (21).
Ligation-mediated PCR
We performed ligation-mediated PCR (LM–PCR) as described (6). Nuclei were isolated and resuspended at a DNA concentration of 1 mg/ml in 15 mM Tris–HCl (pH 7.5) containing 15 mM NaCl, 60 mM KCl, 1 mM CaCl2, 0.34 M sucrose, 15 mM β-mercaptoethanol and 0.5 mM spermidine. Micrococcal nuclease (MNase) (30 units/ml) was added and digestion proceeded for 10 min at 25°C. Genomic DNA was purified and used for first-strand synthesis extending to the cleaved sites using the first synthesis primer (5′-ACACTCACCGTGGCTCGCTGCGAGC-3′). The synthesized DNA was ligated to a double-stranded linker (sense, 5′-GAATTCAGATC-3′; antisense, 5′-GCGGTGACCCGGGAGATCTGAATTC-3′) and amplified by PCR using a linker antisense oligonucleotide and a forward primer (5′-GCACCCGCACTCGGATCCCGCGG-3′). The amplified fragments were linearly amplified using a 32P-end-labeled extension primer (5′-AAGTCCGGCGCGGGTAAACAG-3) and resolved by electrophoresis on 8% denaturing polyacrylamide gels.
RESULTS
The Top1 inhibitor camptothecin elongates circadian period of Bmal1 transcription
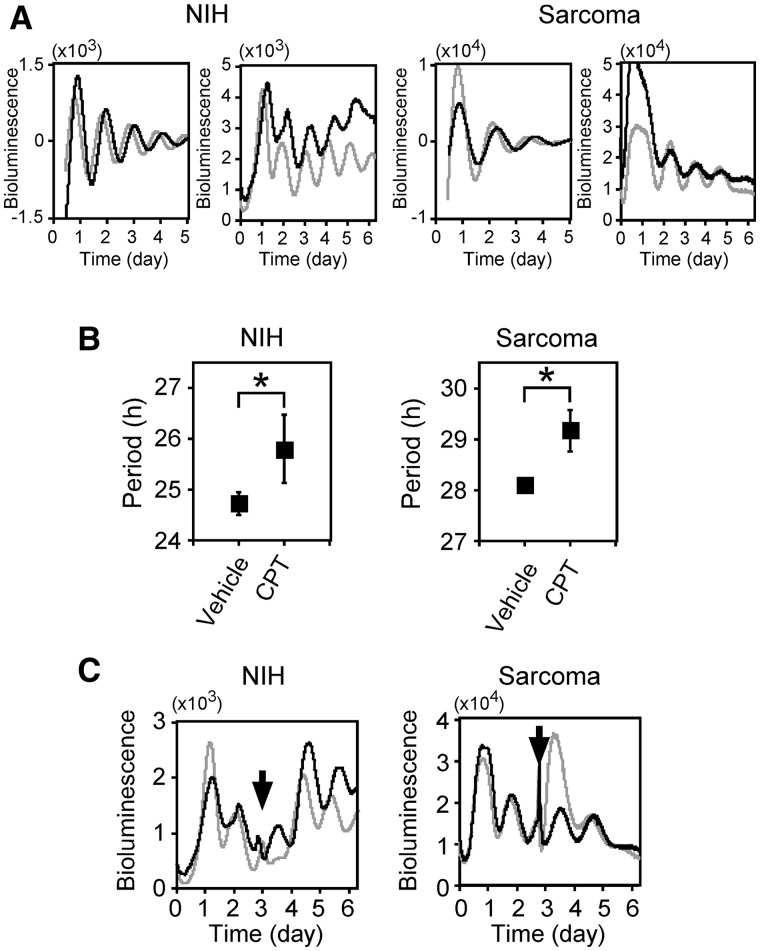
We discovered that a unique chromatin structure on the Bmal1 promoter where SAF-A binds is required for circadian transcriptional regulation of the Bmal1 gene (6). SAF-A interacts with Top1 (22), suggesting the modulation of Top1 activity (23). These findings imply that Top1 is involved in Bmal1 circadian transcription. We examined this notion by investigating whether or not the major Top1 inhibitor camptothecin affects circadian rhythms using real-time reporter gene assays and established stable clones. Figure 1A shows representative results of real-reporter gene assays using stable cells and camptothecin. The period was longer when both NIH3T3-derived and sarcoma 180-derived cell lines were incubated with (black), than without (gray) 5 µM camptothecin. Figure 1B shows that the period lengths of NIH3T3-derived (NIH) cells incubated without and with 5 µM camptothecin were 24.7 ± 0.22 and 25.8 ± 0.66 h, respectively, and those of sarcoma 180-derived (Sarcoma) cells were 28.1 ± 0.06 and 29.2 ± 0.41 h, respectively. Although the period was longer for the sarcoma 180-derived than the NIH3T3-derived cells, camptothecin elongated the period in both lines, indicating that camptothecin elongated the circadian period. Camptothecin can cause apoptosis (12), which might lead to the longer phenotype. Stable reporter cells that have been incubated once with camptothecin and then washed out were used in real-time reporter assays. Both cell lines similarly responded to the camptothecin wash-out with shorter period lengths (NIH, 24.7 ± 1.17 versus 26.7 ± 1.17 h; sarcoma, 28.7 ± 0.59 versus 29.5 ± 0.59 h; Figure 1C). These results suggested that camptothecin induces reversible period elongation and that reporter cells incubated with camptothecin remain healthy. Cell growth assays also revealed that the viability of NIH3T3-derived and sarcoma 180-derived cells incubated with 5 µM of camptothecin for 5 days was 124% and 70%, respectively, showing that both cell lines remained viable after incubation with 5 µM camptothecin, whereas cell growth was slow or arrested (Supplementary Figure S1). These results indicate that camptothecin extends the circadian period of the Bmal1 gene.
Figure 1.
Top1 inhibitor, camptothecin extends period length. (A) Effect of camptothecin on transcriptional oscillation of Bmal1. After NIH3T3 (NIH)- and Sarcoma 180 (Sarcoma)-derived stable clones were stimulated with 100 nM dexamethasone for 2 h, 5 µM camptothecin was added and then bioluminescence was measured. Fit curve data of detrended results (Left) and non-detrended raw data (Right) are representative of at least three independent experiments (control, gray; camptothecin, black). (B) Period analyses. Data are presented as means ± SEM (n > 3, *P < 0.05 by Student’s t-test). (C) Reversible period elongation by camptothecin. Stable clones were stimulated with 100 nM dexamethasone for 2 h, and then bioluminescence was measured with 5 µM camptothecin. Cells were washed and culture medium was replaced with the medium without camptothecin three days later (arrows). Black and gray lines, results with and without cell wash-out and medium change, respectively. Results are representative of triplicate experiments.
Top1 modulates Bmal1 expression and the circadian period
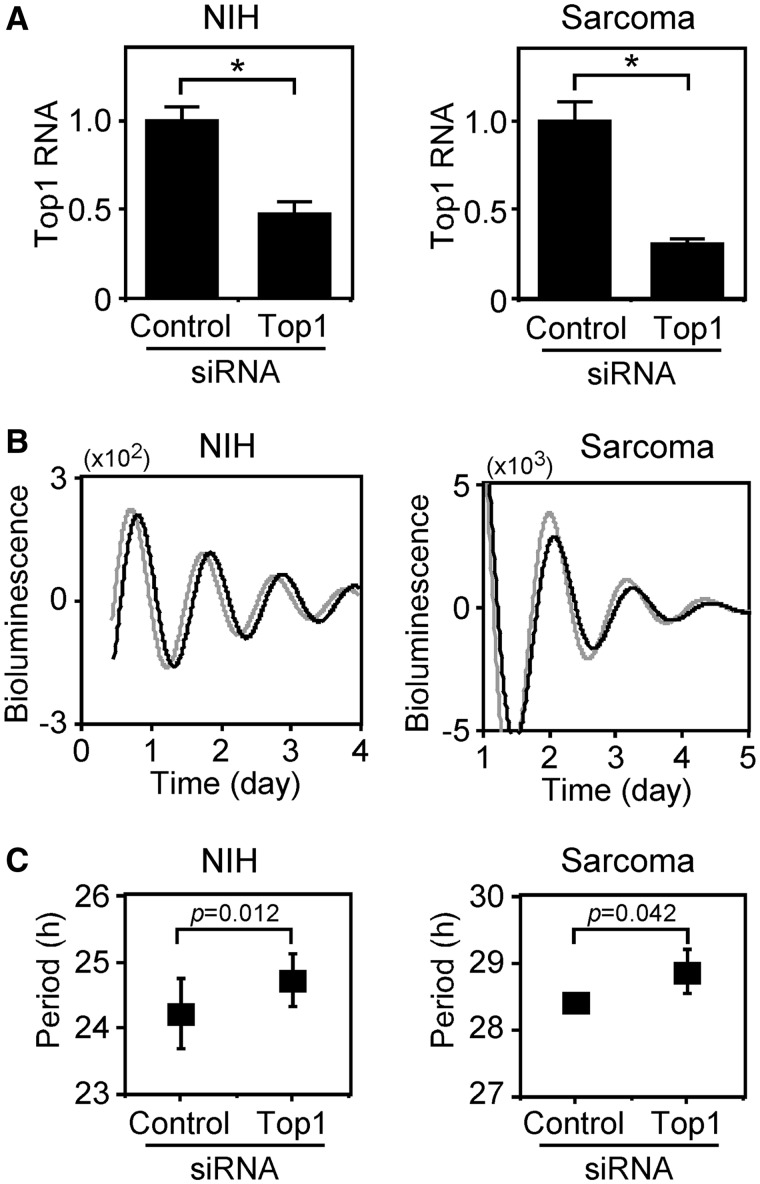
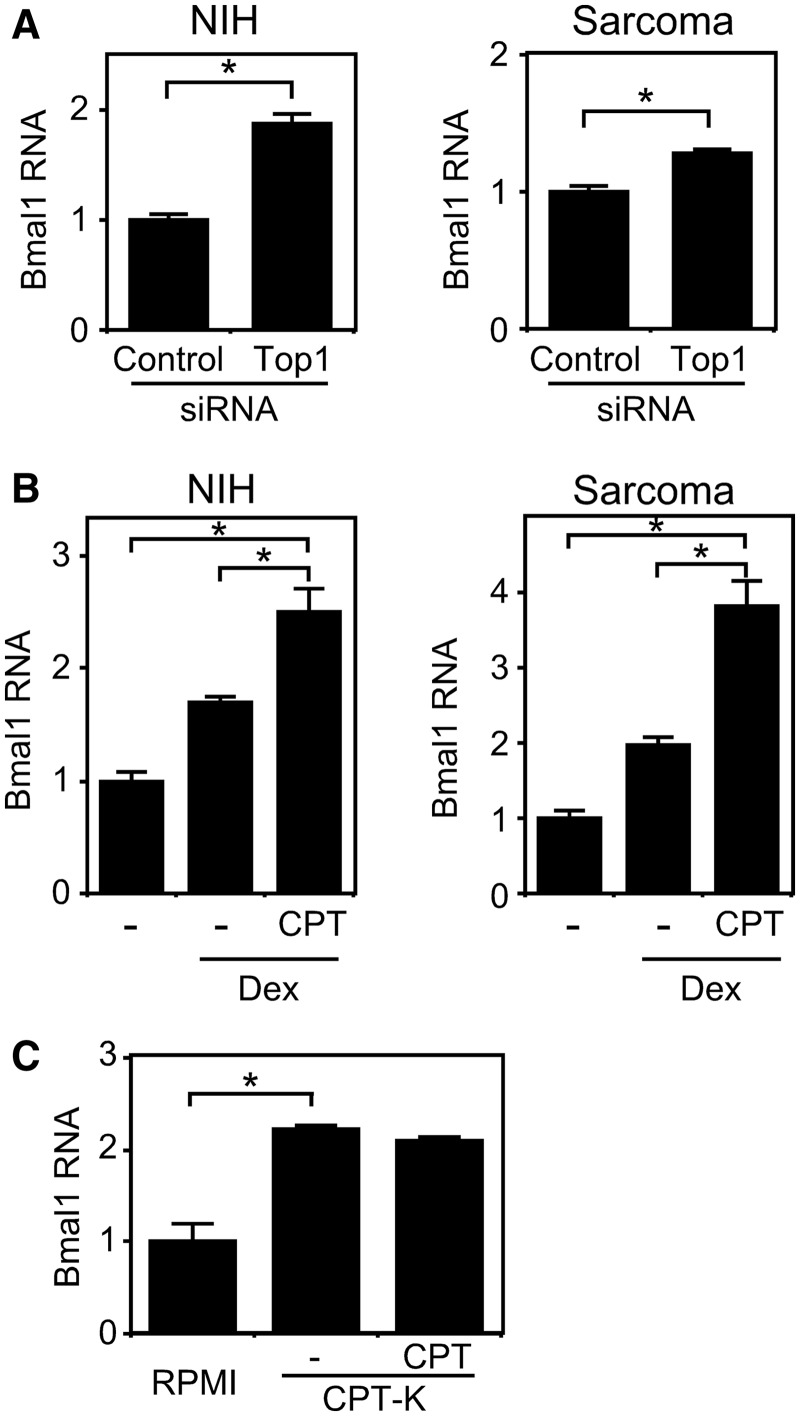
We evaluated the relationship between the amount of Top1 gene expression and period length. Figure 2A shows that knockdown using siRNA reduced Top1 expression in both cell lines. Real-time reporter gene assays with siRNA revealed that the circadian periods of the NIH3T3-based cells (NIH) harboring control and Top1 siRNAs were 24.2 and 24.7 h, respectively, and that those of the sarcoma 180-derived cells harboring control and Top1 siRNAs were 28.4 and 28.9 h, respectively (Figure 2B). These results indicate that the downregulation of Top1 gene expression extends the circadian period of the Bmal1 gene. We (24) and others (25) have recently reported that the amount of Bmal1 expression is related to circadian rhythms. Here, we evaluated the relationship between levels of Bmal1 and Top1 gene expression. Knockdown of Top1 expression by siRNA enhanced Bmal1 transcription in both cell lines (Figure 3A), suggesting that Top1 expression is related to Bmal1 transcription. After resetting Bmal1 expression with dexamethasone, the cells were incubated with 5 µM camptothecin for 24 h and then transcripts were analyzed by real-time quantitative RT–PCR. Figure 3B shows that camptothecin enhanced Bmal1 transcription in both cell lines. We also found lower levels of Bmal1 expression in the RPMI8402 cell line than in its Top1 mutant line CPT-K that has reduced Top1 activity (26) (Figure 3C). In addition, Bmal1 transcription did not respond to camptothecin in CPT-K cells (Figure 3C), which is consistent with the report by Kjeldsen et al. that the catalytic activity of Top1 in CPT-K cells is insensitive to camptothecin (27). These findings suggest that reducing Top1 activity induces Bmal1 transcription and therefore, Top1 is closely associated with Bmal1 transcriptional regulation. These results together with previous findings suggest that Top1 participates not only in Bmal1 at the transcriptional level but also in its circadian period.
Figure 2.
Knockdown of Top1 expression affects period length. Stealth RNAi siRNA duplex for Top1 knockdown (Top1) and BLOCK-iT Fluorescent Oligo as a negative control (Control) were transfected into NIH3T3 (NIH)- and Sarcoma 180 (Sarcoma)-derived stable clones. Expression of Top1 analyzed 24 h after transfection using real-time quantitative RT–PCR. Levels of RNA were normalized to Actin expression (A). Expression levels are indicated relative to that transfected with BLOCK-iT Fluorescent Oligo. Values are means ± SEM of triplicate assays. *P < 0.05 (Student’s t-test). At 24 h after starting to analyze transfection circadian periods using real-time reporter gene assays. Fit curve data of detrended results (B) are representative of at least five independent experiments (control, gray; camptothecin, black). Period analyses (C) are plotted as means ± SEM (n > 5). P-values against control siRNA data were assessed by Student’s t-test.
Figure 3.
Levels of Top1 activity affect Bmal1 transcription. Knockdown of Top1 enhances Bmal1 transcription. Stealth RNAi siRNA duplex for Top1 knockdown (Top1) and BLOCK-iT Fluorescent Oligo as a negative control (Control) were transfected into NIH3T3 (NIH)- and Sarcoma 180 (Sarcoma)-derived stable clones and then Bmal1 expression was analyzed by real-time quantitative RT–PCR 24 h later (A). Expression levels are indicated relative to that transfected with BLOCK-iT Fluorescent Oligo. Inhibition of Top1 activity by camptothecin enhances Bmal1 transcription (B). NIH3T3 (NIH)- and Sarcoma 180 (Sarcoma)-derived stable cells were stimulated with 100 nM dexamethasone (Dex) for 2 h, incubated with 5 µM camptothecin for 24 h and then Bmal1 expression was analyzed using real-time quantitative RT–PCR. Expression levels are indicated relative to that without either dexamethasone or camptothecin. Reduced Top1 activity enhances Bmal1 expression (C). Bmal1 expression was assessed in RPMI8402 cells and its Top1 mutant line, CPT-K using real-time quantitative RT–PCR. Expression levels are indicated relative to that in RPMI8402 cells. Levels of RNA were normalized to Actin expression. Values are means ± SEM of triplicate assays. *P < 0.05 (Student’s t-test). CPT, 5 µM camptothecin for 12 h.
Top1 is located at the intermediate region between the ROREs in the Bmal1 promoter
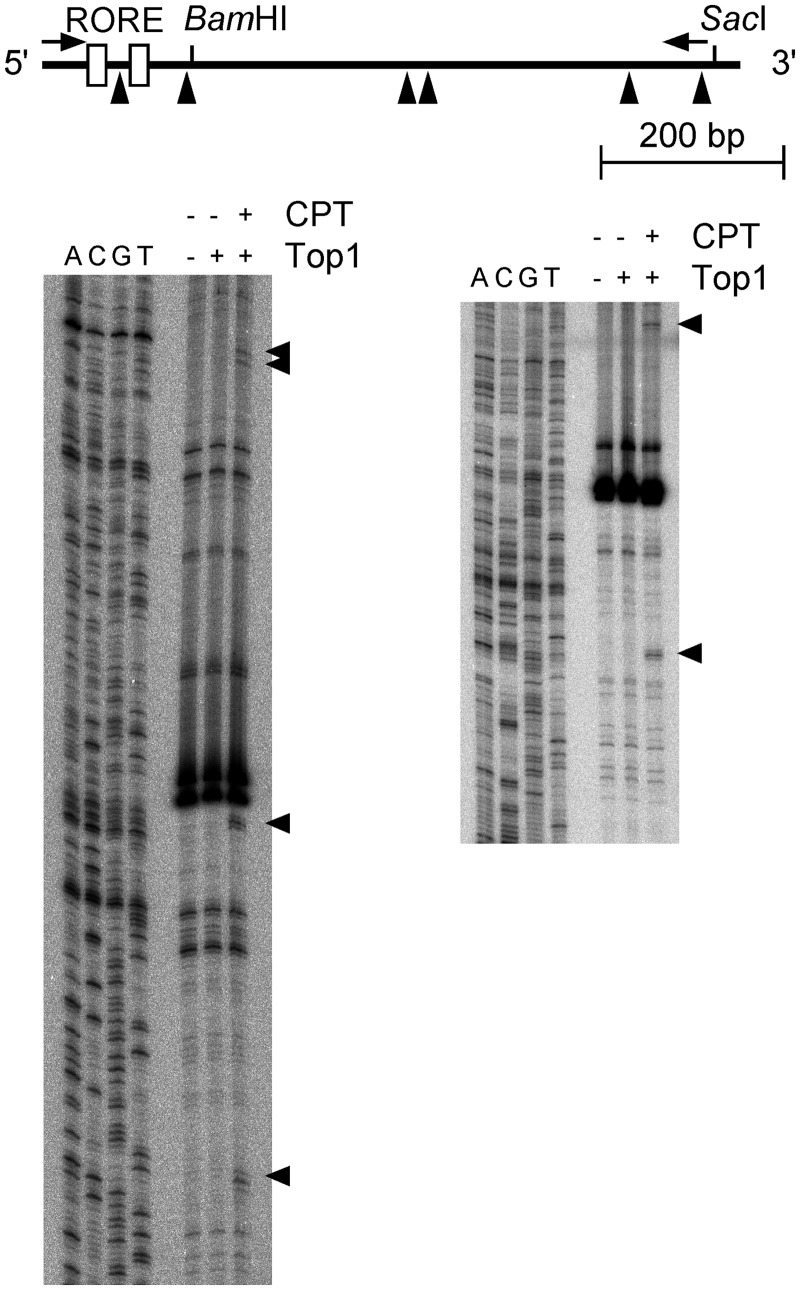
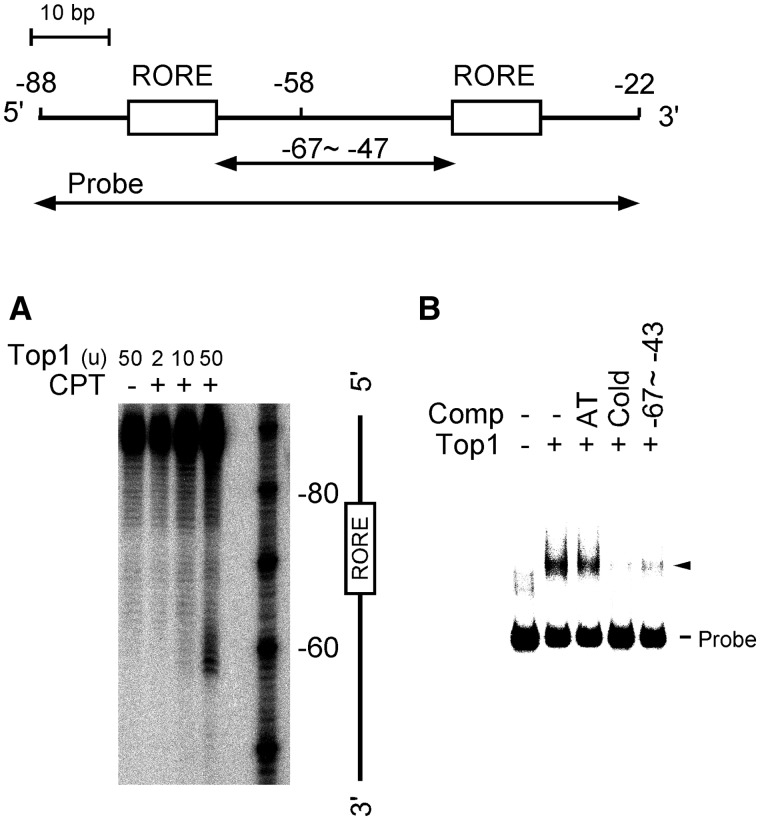
We examined the presence of Top1 at the Bmal1 promoter region. The results of Top1-mediated cleavage assays of the Bmal1 promoter fragment indicated that bands at positions −58, +10, +201, +210, +357 and +450 (Figure 4; triangles) were specifically cleaved to cleavable complexes only after incubation with the TOP1 inhibitor, camptothecin. These findings suggested that Top1-binding sites are located on the Bmal1 promoter region in vitro and that one of them is located between the ROREs, which are critical cis-elements for the circadian oscillation of Bmal1 transcription (Figure 4). To confirm that a Top1-binding site is indeed located between the ROREs, we performed Top1-mediated cleavage assays and EMSA using a DNA fragment around the ROREs (nucleotides −88 to −22). The results showed that specific cleavage of a site at −58 depended on the amount of Top1 (Figure 5A). Figure 5B shows that a Top1–DNA complex was diminished by adding a full-length unlabeled DNA probe or the intermediate region of the ROREs (nucleotides −67 to −43), whereas the control competitor (dA30/dT30) exerted little effect. These results suggest that Top1 binds to the intermediate region (position at −58) between the ROREs, which are critical cis-elements for circadian Bmal1 transcription, in the Bmal1 promoter region.
Figure 4.
Screening of Top1-binding sites in the Bmal1 promoter region. Top1-binding sites were determined by Top1-mediated cleavage assays. Bmal1 promoter region was amplified by PCR using sense (5′-TAACGGGAAGAGGCAGGTATC-3′) and antisense (5′-TCCAGAAGCTCCCGGGGATGTATC-3′) DNA probes. DNA fragments of the Bmal1 promoter region that had been end-labeled with [32P] on either sense (Left panel) or antisense (Right panel) strand was reacted with either 50 units of Top1 alone or 50 units of Top1 plus 0.5 mM camptothecin. Purified DNA was resolved on 8% polyacrylamide–urea gels adjacent to a sequencing ladder primed with the same 32P-labeled oligonucleotide as a position marker. Top1-mediated cleavage sites are indicated by triangles and summarized in map. Open boxes and arrows, RORE and labeled probes, respectively. CPT, camptothecin.
Figure 5.
Top1 binds to intermediate region between ROREs in Bmal1 promoter region. Top1-mediated cleavage assay (A). DNA fragment around the ROREs (nucleotides −88 to −22: 5′-GATTGGTCGGAAAGTAGGTTAGTGGTGCGACATTTAGGGAAGGCAGAAAGTAGGTCAGGGACGGAGG-3′) was end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. DNA fragment was reacted with either 50 units of Top1 alone or with 2–50 units of Top1 plus 0.5 mM camptothecin. Purified DNA was resolved on 8% polyacrylamide–urea gels. CPT, camptothecin. EMSA using probe, nucleotides −88 to −22, 15 units of Top1 protein and a 100-fold molar excess of the following competitors (Comp): AT, control oligonucleotides, (dA)30 and (dT)30; unlabeled probe, nucleotides −88 to −22; −67 to −43, nucleotides −67 to −4 (B). Arrowhead, shifted band.
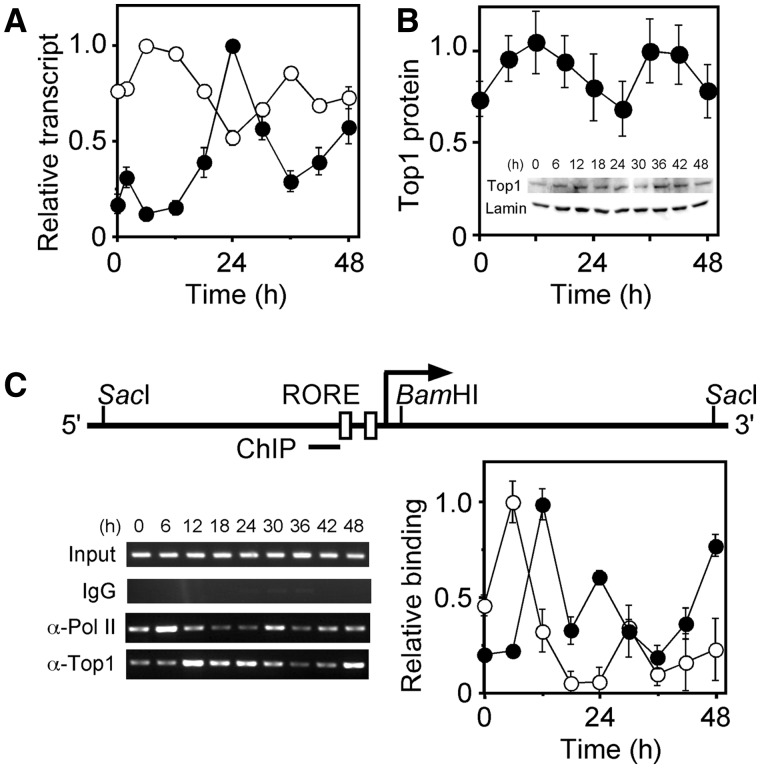
NIH3T3 cells stimulated with dexamethasone express Top1 with circadian oscillation (11). The profile of Top1 expression was oscillatory with a peak at 24 h and troughs at 12 and 36 h, which was anti-phase to the Bmal1 profile (Figure 6A). Western blots using an anti-Top1 antibody revealed that Top1 protein showed an anti-phase profile to Top1 transcription though the circadian rhythm did not show the statistic significance, which was consistent with previous report by Kuramoto et al. (10). We next confirmed Top1 binding around the ROREs in vivo using ChIP assays and an anti-Top1 antibody in NIH3T3 cells stimulated with dexamethasone. Figure 6C shows that the ROREs region was amplified by the ChIP assays using anti-Top1 antibody, indicating Top1 binding between the ROREs in vivo. Furthermore, the profile of Top1 binding was oscillatory with peaks at 12, 24 and 48 h (Figure 6C, filled circles). ChIP assays using an anti-RNA polymerase II (Pol II) antibody revealed similar circadian profiles between Pol II binding to the Bmal1 promoter and of Bmal1 transcription, whereas the oscillatory phase of Pol II binding was advanced by 6 h compared with that of Bmal1 transcription (Figure 6C, unfilled circles). The peaks at 24 and 48 h of the Top1-binding profile were the circadian oscillation, which conserved the periodic relationship of the oscillatory phase among Top1 and Bmal1 transcriptions and Pol II binding. These results indicate that Top1 binds between the ROREs in vivo and implies that the Top1 binding impacts on Bmal1 transcription.
Figure 6.
Top1 in NIH3T3 cells. (A) Oscillatory transcription of Top1 gene. NIH3T3 cells were stimulated with 100 nM dexamethasone and then transcripts were analyzed by real-time quantitative RT–PCR. Unfilled and filled circles respectively indicate Bmal1 and Top1 expression. Levels of RNA were normalized to Actin expression, and peak value was set at 1. Values are means ± SEM of triplicate assays. (B) Western blot of Top1 protein. NIH3T3 cells were stimulated with 100 nM dexamethasone for 2 h, incubated for the indicated period, and then Top1 protein was analyzed by western blotting. Levels of Top1 protein were normalized to those of Lamin, and peak value was set at 1. Values are means ± SEM of triplicate assays. Representative results are provided in the inset. Top1, anti-Top1 antibody; Lamin, anti-lamin A/C antibody. (C) Top1-and Pol II-binding on Bmal1 promoter in NIH3T3 cells stimulated with dexamethasone and then analyzed by ChIP assays followed by real-time quantitative PCR. Amplified DNA region was indicated as a horizontal bar (ChIP). Unfilled and filled circles respectively indicate Pol II and Top1 binding. Values for binding were normalized to input, and peak value was set at 1. Values are means ± SEM of triplicate assays. PCR products were also resolved on 2% agarose gels. IgG, mouse immunoglobulin; α-Pol II, anti-Pol II antibody; α-Top1, anti-Top1 antibody.
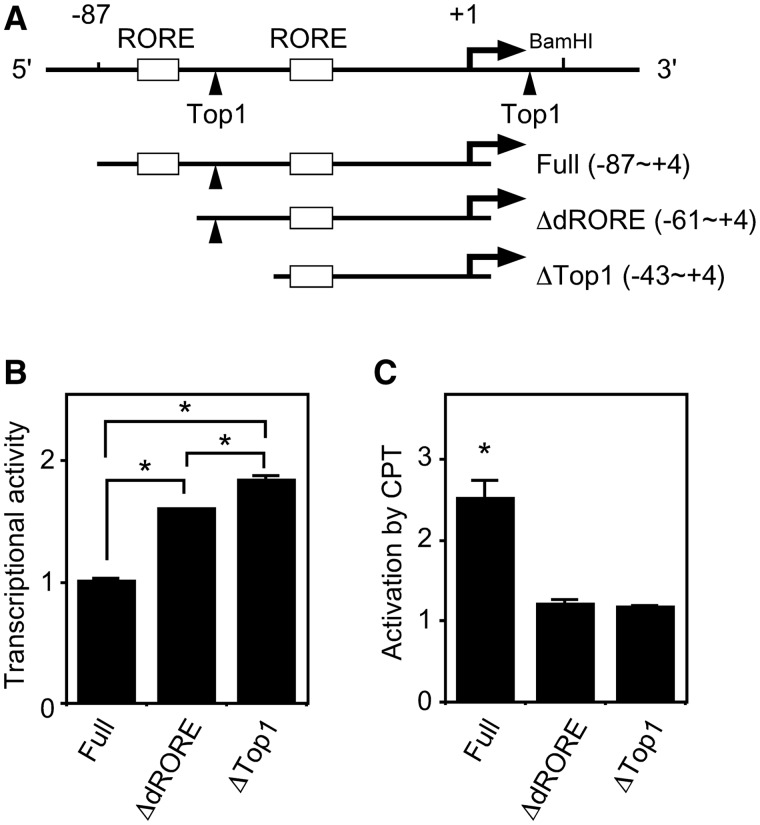
Top1 between the ROREs modulates promoter activity
We evaluated the effect of Top1 between the ROREs on Bmal1 transcription using transient reporter assays and deletion constructs of the Bmal1 promoter (Figure 7A). Deleting the distal RORE (ΔdRORE) enhanced transcriptional activity, suggesting that the distal RORE functions in transcriptional suppression (Figure 7B). In comparison, deleting the Top1-binding site (ΔTop1) resulted in transcriptional activation, suggesting that the Top1-binding site is required for transcriptional suppression (Figure 7B). We then included camptothecin in reporter assays to determine the effect of Top1 activity on Bmal1 transcription. Camptothecin activated Bmal1 promoter activity ∼2.5-fold in the full construct (nucleotides −87 to +4) (Figure 7C), which was consistent with the results shown in Figure 7B as well as the knockdown of Top1 expression by siRNA (Figure 3A) and by camptothecin (Figure 3B), which enhanced Bmal1 transcription. Camptothecin did not activate transcription in the construct with a deleted Top1-binding site (ΔTop1), and moreover, ΔdRORE also could not be activated by camptothecin, suggesting that Top1 cooperates with the distal RORE in Bmal1 transcription.
Figure 7.
Top1-binding site between ROREs is involved in Bmal1 gene transcriptional activity. Schematic representation of deletion constructs of Bmal1 promoter (A). Arrow and arrowheads, transcription start site and Top1-binding sites, respectively. Open boxes, ROREs. Top1-binding site is involved in transcriptional suppression of Bmal1 gene (B). Reporter plasmid, pGL3-promoter containing the indicated Bmal1 promoter region was transfected into NIH3T3 cells. Normalized expression levels were calculated relative to luciferase activity of full construct (nucleotides −88 to +4). Values are means ± SEM of triplicate assays. *P < 0.05 (Student’s t-test). (C) Activation of Bmal1 transcription by CPT requires distal RORE. Reporter plasmids were transfected into NIH3T3 cells, 5 µM camptothecin (CPT) was added 24 h later and then cells were incubated for a further 24 h. Normalized expression levels were calculated relative to luciferase activity of each construct without camptothecin. Values are means ± SEM of triplicate assays. *P < 0.05 (Student’s t-test).
Unique chromatin structure around the Top1-binding site in the Bmal1 promoter
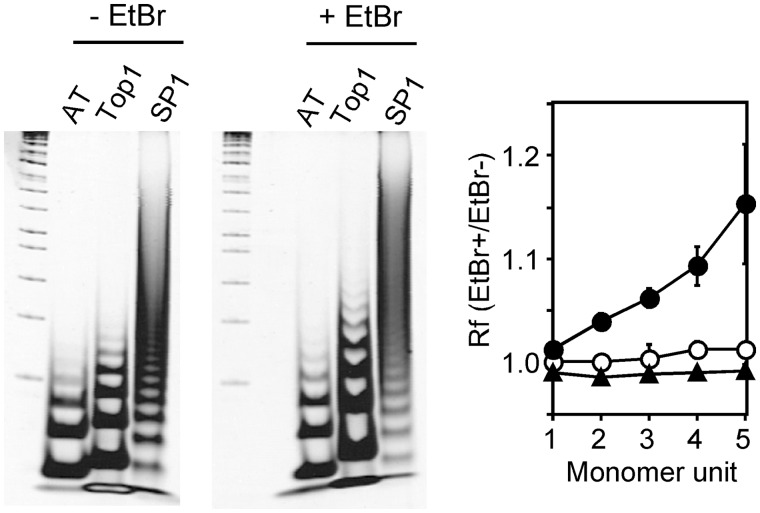
Because the contribution of Top1 to Bmal1 transcription suggests that the DNA structure around the Top1-binding site is important, we evaluated the DNA configuration of the Top1-binding region between the ROREs using gel mobility assays (Figure 8). Homologous regions of an SP1-binding sequence were often located around the Bmal1 promoter (positions at −157, −113, −107, +141 and +225), and all of them were independent of the Top1-binding region. Thereafter, we used A20/T20 and SP1-binding fragments as the standards for a superhelical twist (20) and for comparable DNA fragments, respectively. Ethidium bromide exerted little effect on both the A20/T20 and SP1-binding fragments (Figure 8) because their Rf values were almost 1, indicating that they behave like left-handed superhelical twists (21). On the other hand, the Top1-binding region between the ROREs was more electrophoretically retarded in the presence, than in the absence of ethidium bromide, indicating behavior similar to that of a right-handed superhelical twist (21). These results indicate that the DNA structure around the ROREs is unique and strengthens the notion that Top1 is involved in Bmal1 transcription by altering DNA topology on the Bmal1 promoter.
Figure 8.
Unique conformation of DNA strand at Top1-binding region between ROREs. Oligonucleotides were unidirectionally concatemerized and assayed in 6% polyacrylamide gels with or without 5 µg/ml of ethidium bromide (EtBr). AT, dA20/dT20; Top1, Top1-binding region between ROREs; SP1, SP1-binding sequence. Data are representative of triplicate experiments. Ratios of relative mobility (Rf) with or without ethidium bromide are plotted against their oligonucleotide units. Values are means ± SEM of triplicate assays. Unfilled and filled circles, Top1-binding region between ROREs and dA20/dT20, respectively. Triangles, SP1-binding sequence.
Camptothecin alters chromatin structure around ROREs
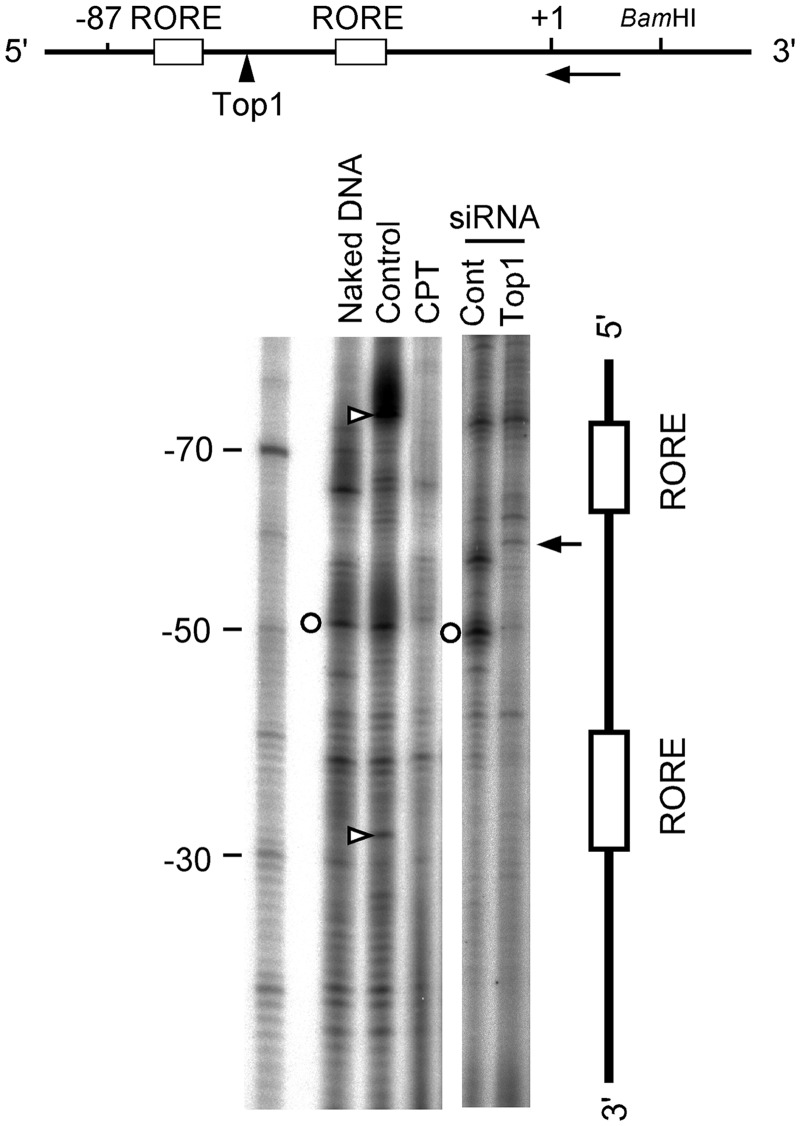
The functional inhibition of Top1 binding around the ROREs suggested modulation in chromatin structure. We evaluated these changes using LM–PCR and NIH3T3 cells incubated with the Top1 inhibitor, camptothecin (Figure 9). The bands at the ROREs in the control sample or NIH3T3 cells (Figure 9, open triangles) were undetectable in naked DNA, indicating that these bands reflect a chromatin-specific structure that generates a micrococcal nuclease-sensitive region. Camptothecin caused the disappearance of these bands, indicating that the chromatin structure was released and assumed a state like naked DNA because these digested bands were not evident in naked DNA. Such a state was not evident in the sample treated with Top1 siRNA, in which cleaved bands (Figure 9, arrow at −58) appeared, suggesting the release of Top1 binding. The encircled band between the ROREs appeared in both the naked and control samples but not in cells incubated with either camptothecin or Top1 siRNA, suggesting that a change in Top1 modulates the chromatin structure between the ROREs in vivo (Figure 9). These results indicate that Top1 plays an important role in modulating the chromatin structure around the ROREs on the Bmal1 promoter in vivo.
Figure 9.
Top1 contributes to footprinting profiles around ROREs. NIH3T3 cells were incubated with 5 µM camptothecin or siRNAs (Cont, negative control; Top1, topoisomerase I) and analyzed by LM–PCR 12 h later. Arrow and arrowhead in the map of the Baml1 promoter, labeled probe and Top1-binding sites, respectively. Circles and triangles, bands that disappeared in presence of camptothecin plus Top1 siRNA, and in that of camptothecin alone, respectively. Arrow, Top1-binding site at position −58. Numbers beside marker indicate nucleotide position of Bmal1 gene.
Overall, these results indicate that Top1 alters the chromatin structure around the ROREs on the Bmal1 promoter to regulate Bmal1 transcription and is thus involved in circadian period regulation.
DISCUSSION
Effect of Top1 inhibitors on circadian period
Camptothecin, a major Top1 inhibitor, and Top1 siRNA both prolonged the circadian period of the Bmal1 gene in both NIH3T3-derived and sarcoma 180-derived cell lines (Figures 1 and 2). We recently found that the harmala alkaloid, harmine which inhibits Top1 activity (28), extends the circadian period of Bmal1 transcription by enhancing RORα function (15). Lee et al. recently reported that BMAL1 overexpression elongates the circadian period by ∼30 min (25). We also confirmed using real-time reporter gene assays that knockdown and overexpression of the Bmal1 gene result in the short- and long-period phenotype, respectively (24). Both camptothecin and Top1 siRNA enhanced Bmal1 transcription in both NIH3T3-derived and sarcoma 180-derived cell lines (Figure 3A and B) and the Top1 mutant expressed more Bmal1 (Figure 3C), suggesting that a decrease in Top1 activity enhances Bmal1 expression. Our results (Figures 1–3) are consistent with the reported relationship between Bmal1 expression and its circadian period.
The activity of Top1 is inhibited after modification by poly(ADP-ribosyl)ation (29), which reportedly contributes to setting the period length of the Arabidopsis central oscillator and its inhibition shortens period length (30). The results of thist study are consistent with the notion that Top1 inhibition lengthens the circadian period.
Top1 in the Bmal1 promoter
Our Top1-mediated cleavage assays identified several candidate Top1-binding sites in the Bmal1 promoter region (Figure 4) and confirmed that the Top1-binding site is located among ROREs that are critical elements for circadian transcription of the Bmal1 gene (Figures 4 and 5). The consensus sequence for Top1 is not so clearly defined and a couple of sequences are proposed (31,32). The nucleotide sequence 5′-CGACATT-3′ around the Top1-mediated cleavage site among the ROREs followed the consensus Top1 recognition sequence, 5′-NRRYRNN-3′, in which R is purine, Y is pyrimidine and N is any nucleotide (33). We previously discovered that the Bmal1 promoter region has a unique chromatin structure with G/C-rich sequences (6) and therefore, SP1-binding sequences are often identified around the Bmal1 promoter region. Although G/C-rich sequences reportedly affect stiffness and curvature stability (34), SP-1-binding sites and the DNA region containing the Top1-binding site among the ROREs behaved like left- and right-handed superhelical twists, respectively (Figure 8). This finding reinforced the importance of the chromatin structure around the ROREs because right-handed superhelices upstream of a promoter enhance transcription more than planar or left-handed superhelices (35). These results also suggest that changes in DNA topology among the ROREs induced by Top1 play an important role in Bmal1 transcription.
Top1 itself was expressed with circadian oscillation (Figure 6A) as reported (11). As shown in Figure 6B, Top1 protein showed an anti-phase profile to Top1 transcription though the circadian rhythm did not show the statistic significance, which is consistent with a previous report by Kuramoto et al. (10). On the other hand, the profile of Top1 binding (Figure 6C) was different from that of Top1 protein (Figure 6C). These results suggest the presence of post-transcriptional regulation of Top1 for its binding. Top1 is post-transcriptionally modulated by proteolysis (36), phosphorylation (37) and SUMO-1 modification (38). The Top1 modulations for binding between the ROREs remain unclear and this issue requires further investigation. The profile of Top1 binding was oscillatory with peaks at 12, 24 and 48 h (Figure 6C). To reset the circadian rhythm, the cells were stimulated with dexamethasone that modulates not only Top1 transcription (10) but also Top1 activity on the phosphatidylinositol 3-kinase-Akt-signaling pathway (39). Furthermore, the circadian profile of Top1 protein delays by 12 h compared with that of glucocorticoid (10). Taken together, these results suggest that the first peak at 12 h after dexamethasone stimulation might be brought by the simple response of Top1 to dexamethasone. On the other hand, the peaks at 24 and 48 h conserved the periodic relationship of the oscillatory phase among Top1 and Bmal1 transcriptions and Pol II binding, implying that the Top1 binding impacts on Bmal1 transcription.
Top1 rhythmically bound to the site among the ROREs (Figure 6C), suggesting a functional commitment to circadian Bmal1 transcription. The oscillation phase of Top1 binding differed from that of Pol II (Figure 6C), implying that Top1 is required to relax the supercoils generated by transcription in an active promoter (40) and that Top1 regulates open chromatin and controls gene expression (9). We found that the region around the ROREs comprises an open chromatin structure (6), which is consistent with our notion that Top1 alters the chromatin structure around the ROREs and modulates Bmal1 transcription.
Changes in the chromatin structure around the ROREs are important for circadian Bmal1 transcription (6), and Top1 inhibition using camptothecin or Top1 siRNA induced changes in the chromatin configuration around the ROREs (Figure 9) and enhanced Bmal1 transcription (Figure 3). On the other hand, camptothecin inhibits gene transcription by increasing Pol II escape from promoter-proximal pausing sites (41) and Pol II constitutively binds to the per promoter (42), implying that camptothecin sustains the promoter-proximal pausing of the per gene which represses Bmal1 transcription.
Effect of Top1 on Bmal1 promoter activity
Transient reporter assays showed that the distal RORE was required to repress Bmal1 gene expression (Figure 7B), which is consistent with the previous finding that both ROREs are required for Bmal1 repression by Rev-erbα [43]. The intermediate region between the ROREs containing the Top1-binding site was also required for repression (Figure 7B), supporting our hypothesis that Top1 is involved in transcriptional regulation of the Bmal1 gene. On the other hand, camptothecin activated Bmal1 promoter activity when the distal RORE and the Top1-binding site remained intact (Figure 7C), suggesting that Top1 cooperates with the distal RORE in Bmal1 transcription. Akashi and Takumi proposed that the distal RORE is essential to drive transcriptional oscillation, whereas the proximal RORE is necessary for high oscillation amplitude (44). Both ROREs are required for Bmal1 oscillation, homodimer formation with Rev-erbα and recruitment of the nuclear receptor corepressor complex (43). Because the ROREs are separated by a 24-bp gap, DNA alterations around them play an important role in processes such as homodimerization through modulating formation of the chromatin structure, thus indicating the importance of the Top1-binding site in the 24-bp gap. Furthermore, the intermediate region represented a unique DNA structure, which would strengthen the involvement of Top1 in its active modification (Figure 8). In fact, disrupting Top1 in the intermediate region between the ROREs with either camptothecin or Top1 siRNA changed the footprints (Figure 9), suggesting that Top1 is involved in Bmal1 transcription by altering the chromatin structure around the ROREs.
Chronotherapeutic insight into phyto-alkaloids
Camptothecin has powerful anti-tumor activity. Biological events and drug metabolism are both under the control of the circadian clock and therefore not only circadian rhythms but also drug and chronopharmaceutical (pharmaceuticals that focus on biological rhythms) pharmacokinetics, effects and safety should be considered in terms of risk of disease (45). For example, tolerance for CPT-11, a camptothecin analog that is largely administered to cancer patients, is subject to circadian rhythms (46) and the target of CPT-11, Top1, is expressed in a circadian manner (Figure 6A). Therefore, knowledge of chronotherapy using CPT-11 is advancing (47). Here, we showed that camptothecin can modulate Bmal1 expression and the circadian period (Figures 1, 3 and 7), which provides new insight into the mechanisms of chronopharmaceutical actions.
Circadian regulation of Bmal1 transcription
Physiological circadian rhythms are features of organisms ranging from bacteria to humans and represent vigor in the face of environmental changes. To adapt circadian rhythms to various environments, lower organisms regulate gene expression by arranging chromosomal compaction (48) and by modifying DNA topology (49). We previously reported that chromatin dynamics are involved in circadian Bmal1 transcription (6). Here, we showed that Top1 alters DNA topology around ROREs and modulates Bmal1 transcription as well as the circadian period, which might be a vestige of prehistoric adaptation. The circadian clock in mammals is defined as a negative feedback loop based on reciprocal interaction between the activators BMAL1 and CLOCK, and the repressors PER and CRY, and the stoichiometric relationship among the clock components is critical for robust circadian rhythms (25). Our findings provide new insights into the robustness of circadian rhythms and indicate that the chromatin architecture of clock genes including DNA topology is a determinant of these rhythms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1.
FUNDING
KAKENHI (23592756) (JSPS) and operational subsidies from AIST (METI). Funding for open access charge: AIST (METI).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Yoshimitsu Yamazaki (AIST) for valuable discussions.
REFERENCES
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Bechtold DA, Gibbs JE, Loudon AS. Circadian dysfunction in disease. Trends Pharmacol. Sci. 2010;31:191–198. doi: 10.1016/j.tips.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Bunger MK, Wilbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi S, Hida A, McGuinness OP, Wasserman DH, Yamazaki S, Johnson CH. Circadian clock gene Bmal1 is not essential; functional replacement with its paralog, Bmal2. Curr. Biol. 2010;20:316–321. doi: 10.1016/j.cub.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 6.Onishi Y, Hanai S, Ohno T, Hara Y, Ishida N. Rhythmic SAF-A binding underlies circadian transcription of the Bmal1 gene. Mol. Cell. Biol. 2008;28:3477–3488. doi: 10.1128/MCB.02227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 8.Kretzschmar M, Meisterernst M, Roeder RG. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc. Natl Acad. Sci. USA. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durand-Dubief M, Persson J, Norman U, Hartsuiker E, Ekwall K. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J. 2010;29:2126–2134. doi: 10.1038/emboj.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuramoto Y, Hata K, Koyanagi S, Ohdo S, Shimeno H, Soeda S. Circadian regulation of mouse topoisomerase I gene expression by glucocorticoid hormones. Biochem. Pharmacol. 2006;71:1155–1161. doi: 10.1016/j.bcp.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 11.Yang F, Nakajima Y, Kumagai M, Ohmiya Y, Ikeda M. The molecular mechanism regulating the autonomous circadian expression of Topoisomerase I in NIH3T3 cells. Biochem. Biophys. Res. Commun. 2009;380:22–27. doi: 10.1016/j.bbrc.2008.12.186. [DOI] [PubMed] [Google Scholar]
- 12.Onishi Y, Azuma Y, Sato Y, Mizuno Y, Tadakuma T, Kizaki H. Topoisomerase inhibitors induce apoptosis in thymocytes. Biochim. Biophys. Acta. 1993;1175:147–154. doi: 10.1016/0167-4889(93)90017-j. [DOI] [PubMed] [Google Scholar]
- 13.Pommier Y, Pourquier P, Fan Y, Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998;1400:83–105. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 14.Funayama Y, Nishio K, Wakabayashi K, Nagao M, Shimoi K, Ohira T, Hasegawa S, Saijo N. Effects of β- and γ-carboline derivatives of DNA topoisomerase activities. Mutat. Res. 1996;349:183–191. doi: 10.1016/0027-5107(95)00176-x. [DOI] [PubMed] [Google Scholar]
- 15.Onishi Y, Oishi K, Kawano Y, Yamazaki Y. The harmala alkaloid, harmine is a modulator of circadian Bmal1 transcription. Biosci. Rep. 2011;32:45–52. doi: 10.1042/BSR20110002. [DOI] [PubMed] [Google Scholar]
- 16.Hara Y, Onishi Y, Oishi K, Miyazaki K, Fukamizu A, Ishida N. Molecular characterization of Mybbp1a as a co-repressor on the Period2 promoter. Nucleic Acids Res. 2009;37:1115–1126. doi: 10.1093/nar/gkn1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onishi Y, Kiyama R. Interaction of NF-E2 in the human β-globin locus control region before chromatin remodeling. J. Biol. Chem. 2003;278:8163–8171. doi: 10.1074/jbc.M209612200. [DOI] [PubMed] [Google Scholar]
- 18.Onishi Y, Kiyama R. Enhancer activity of HS2 of the human β-LCR is modulated by distance from the key nucleosome. Nucleic Acids Res. 2001;29:3448–3457. doi: 10.1093/nar/29.16.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Onishi Y, Wada-Kiyama Y, Kiyama R. Expression-dependent perturbation of nucleosomal phases at HS2 of the human β-LCR: possible correlation with periodic bent DNA. J. Mol. Biol. 1998;284:989–1004. doi: 10.1006/jmbi.1998.2244. [DOI] [PubMed] [Google Scholar]
- 20.Wanapirak C, Onishi Y, Wada-Kiyama Y, Ohyama T, Kiyama R. Conservation of DNA bend sites with identical superhelical twists among the human, mouse, bovine, rabbit and chicken β-globin genes. DNA Res. 2000;7:253–259. doi: 10.1093/dnares/7.4.253. [DOI] [PubMed] [Google Scholar]
- 21.Brukner I, Belmaaza A, Chartrand P. Differential behavior of curved DNA upon untwisting. Proc. Natl Acad. Sci. USA. 1997;94:403–406. doi: 10.1073/pnas.94.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czubaty A, Girstun A, Kowalska-Loth B, Trzcinska AM, Purta E, Winczura A, Grajkowski W, Staron K. Proteomic analysis of complexes formed by human topoisomerase I. Biochim. Biophys. Acta. 2005;1749:133–141. doi: 10.1016/j.bbapap.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Trzcinska-Daneluti AM, Gorecki A, Czubaty A, Kowalska-Loth B, Girstun A, Murawska M, Lesyng B, Staron K. RRM proteins interacting with the cap region of topoisomerase I. J. Mol. Biol. 2007;369:1098–1112. doi: 10.1016/j.jmb.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 24.Onishi Y, Kawano Y, Yamazaki Y. Lycorine, a candidate for the control of period length in mammalian cells. Cell. Physiol. Biochem. 2012;29:407–416. doi: 10.1159/000338495. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y, Chen R, Lee HM, Lee C. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J. Biol. Chem. 2011;286:7033–7042. doi: 10.1074/jbc.M110.207217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andoh T, Ishii K, Suzuki Y, Ikegami Y, Kusunoki Y, Takemoto Y, Okada K. Characterization of a mammalian mutant with a camptothecin-resistant DNA topoisomerase I. Proc. Natl Acad. Sci. USA. 1987;84:5565–5569. doi: 10.1073/pnas.84.16.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kjeldsen E, Bonven BJ, Andoh T, Ishii K, Okada K, Bolund L, Westergaard O. Characterization of a camptothecin-resistant human DNA topoisomerase I. J. Biol. Chem. 1988;263:3912–3916. [PubMed] [Google Scholar]
- 28.Sobhani AM, Ebrahimi SA, Mahmoudian M. An in vitro evaluation of human DNA topoisomerase I inhibition by Peganum harmala L. seeds extract and its beta-carboline alkaloids. J. Pharm. Pharm. Sci. 2002;5:19–23. [PubMed] [Google Scholar]
- 29.Ferro AM, Olivera BM. Poly(ADP-ribosylation) of DNA topoisomerase I from calf thymus. J. Biol. Chem. 1984;259:547–554. [PubMed] [Google Scholar]
- 30.Panda S, Poirier GG, Kay SA. tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the arabidopsis circadian oscillator. Dev. Cell. 2002;3:51–61. doi: 10.1016/s1534-5807(02)00200-9. [DOI] [PubMed] [Google Scholar]
- 31.Jaxel C, Kohn KW, Pommier Y. Topoisomerase I interaction with SV40 DNA in the presence and absence of camptothecin. Nucleic Acids Res. 1988;16:11157–11169. doi: 10.1093/nar/16.23.11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker LH, Champoux JJ. Analysis of the biased distribution of topoisomerase I break sites on replicating Simian Virus 40 DNA. J. Mol. Biol. 1993;231:6–18. doi: 10.1006/jmbi.1993.1252. [DOI] [PubMed] [Google Scholar]
- 33.Shen CC, Shen CK. Specificity and flexibility of the recognition of DNA helical structure by eukaryotic topoisomerase I. J. Mol. Biol. 1990;212:67–78. doi: 10.1016/0022-2836(90)90305-6. [DOI] [PubMed] [Google Scholar]
- 34.Dlakic M, Harrington RE. Bending and torsional flexibility of G/C-rich sequences as determined by cyclization assays. J. Biol. Chem. 1995;270:29945–29952. doi: 10.1074/jbc.270.50.29945. [DOI] [PubMed] [Google Scholar]
- 35.Hirota Y, Ohyama T. Adjacent upstream superhelical writhe influences an Escherichia coli promoter as measured by in vivo strength and in vitro open complex formation. J. Mol. Biol. 1995;254:566–578. doi: 10.1006/jmbi.1995.0639. [DOI] [PubMed] [Google Scholar]
- 36.Schmitt B, Buhre U, Vosberg HP. Characterisation of size variants of type I DNA topoisomerase isolated from calf thymus. Eur. J. Biochem. 1984;144:127–134. doi: 10.1111/j.1432-1033.1984.tb08440.x. [DOI] [PubMed] [Google Scholar]
- 37.Pommier Y, Kerrigan D, Hartman KD, Glazer RI. Phosphorylation of mammalian DNA topoisomerase I and activation by protein kinase C. J. Biol. Chem. 1990;265:9418–9422. [PubMed] [Google Scholar]
- 38.Mao Y, Sun M, Desai SD, Liu LF. SUMO-1 conjugation to topoisomerase I: A possible repair response to topoisomerase-mediated DNA damage. Proc. Natl Acad. Sci. USA. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akagi T, Fukagawa T, Kage Y, To H, Matsunaga N, Koyanagi S, Uchida A, Fujii A, Iba H, Ikemura T. Role of glucocorticoid receptor in the regulation of cellular sensitivity to irinotecan hydrochloride. J. Pharmacol. Sci. 2009;109:265–274. doi: 10.1254/jphs.08219fp. [DOI] [PubMed] [Google Scholar]
- 40.Capranico G, Marinello J, Baranello L. Dissecting the transcriptional functions of human DNA topoisomerase I by selective inhibitors: implications for physiological and therapeutic modulation of enzyme activity. Biochim. Biophys. Acta. 2010;1806:240–250. doi: 10.1016/j.bbcan.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 41.Baranello L, Bertozzi D, Fogli MV, Pommier Y, Capranico G. DNA topoisomerase I inhibition by camptothecin induces escape of RNA polymerase II from promoter-proximal pause site, antisense transcription and histone acetylation at the human HIF-1α gene locus. Nucleic Acids Res. 2010;38:159–171. doi: 10.1093/nar/gkp817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor P, Hardin PE. Rhythmic E-box binding by CLK-CYC controls daily cycles in per and tim transcription and chromatin modifications. Mol. Cell. Biol. 2008;28:4642–4652. doi: 10.1128/MCB.01612-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbα recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol. Endocrinol. 2005;19:1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 44.Akashi M, Takumi T. The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- 45.Ohdo S. Chronopharmaceutics: pharmaceutics focused on biological rhythm. Biol. Pharm. Bull. 2010;33:159–167. doi: 10.1248/bpb.33.159. [DOI] [PubMed] [Google Scholar]
- 46.Filipski E, Lemaigre G, Liu XH, Mery-Mignard D, Mahjoubi M, Levi F. Circadian rhythm of irinotecan tolerability in mice. Chronobiol. Int. 2004;21:613–630. doi: 10.1081/cbi-120040183. [DOI] [PubMed] [Google Scholar]
- 47.Innominato PF, Levi FA, Bjarnason GA. Chronotherapy and the molecular clock: Clinical implications in oncology. Adv. Drug Deliv. Rev. 2010;62:979–1001. doi: 10.1016/j.addr.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc. Natl Acad. Sci. USA. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salvador ML, Klein U, Bogorad L. Endogenous fluctuations of DNA topology in the chloroplast of Chlamydomonas reinhardtii. Mol. Cell. Biol. 1998;18:7235–7242. doi: 10.1128/mcb.18.12.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]