Summary
Human immunodeficiency virus-1 (HIV-1) envelope protein (Env) and influenza hemagglutinin (HA) are the surface glycoproteins responsible for viral entry into host cells, the first step in the virus life cycle necessary to initiate infection. These glycoproteins exhibit a high degree of sequence variability and glycosylation, which are used as strategies to escape host immune responses. Nonetheless, antibodies with broadly neutralizing activity against these viruses have been isolated that have managed to overcome these barriers. Here, we review recent advances in the structural characterization of these antibodies with their viral antigens that defines a few sites of vulnerability on these viral spikes. These broadly neutralizing antibodies tend to focus their recognition on the sites of similar function between the two viruses: the receptor binding site and membrane fusion machinery. However, some sites of recognition are unique to the virus neutralized, such as the dense shield of oligomannose carbohydrates on HIV-1 Env. These observations are discussed in the context of structure-based design strategies to aid in vaccine design or development of antivirals.
Keywords: HIV-1, Env, influenza, hemagglutinin, antibody, structure
Introduction
The human immunodeficiency virus type 1 (HIV-1) and influenza virus are two pathogens that continue to threaten mankind with epidemics and sporadic pandemics. An estimated 33 million adults and children live with HIV, which corresponds to a worldwide prevalence of 0.8%, with approximately 1.8 million AIDS-related deaths annually (UNAIDS, 2010). Heterosexual transmission now represents on a worldwide basis the most important vehicle for HIV acquisition. However, the infectivity rate of HIV transmission from vaginal intercourse is relatively low and has been estimated to range between approximately 0.0001 and 0.0014 (1). Nevertheless, despite poor infectivity, the HIV-1 epidemic continues due to its high mortality rate in the absence of a cure, lack of world-wide availability of antiviral agents, or a natural mechanism to clear the infection, as well as a nine year delay on average for the progression to acquired immunodeficiency syndrome (AIDS) (2). In stark contrast, 5% to 15% of the world’s population is infected by influenza during a typical seasonal outbreak, which contributes to about 250,000 to 500,000 global deaths annually (WHO). The mortality rate is on the order of about 0.003%, with most deaths occurring in the elderly. In rare instances, influenza virus can become extremely virulent, such as during the 1918 Spanish flu pandemic, when approximately 50 million deaths were recorded, which was roughly 2% of the world’s population at the time. During such pandemics, young adults can be adversely affected, such as in the 1918 and 2009 H1N1 pandemics. Although the pathogenesis of HIV-1 and influenza differ significantly, parallels can be drawn between the mechanisms these two viruses utilize to escape protective immune responses. As such, the development of a universal vaccine remains at the forefront of prevention efforts against these two viruses.
HIV-1 overview and the entry process
HIV-1 is a retrovirus of the Retroviridae family that contains two copies of single-stranded RNA. HIV-1 has a large spherical morphology of approximately 120 nm (3), and electron microscopy studies have revealed that only 9 to 14 irregularly distributed copies of the Env protein are present on the viral membrane, in addition to other proteins acquired from the host cell membrane during budding (4, 5). Env therefore represents the only viral component on the surface of HIV-1 that is accessible for mounting a humoral immune response. Env is expressed as a gp160 precursor and cleaved in the Golgi; the mature Env glycoprotein consists of two non-covalently associated subunits, gp120 and gp41, which assemble into a trimer of heterodimers (6). Env mediates the attachment and fusion of the virus to CD4+ T cells, which is the first step in HIV-1 infection. Obtaining an atomic structure of the full HIV-1 Env trimer has continued to be challenging and so far elusive, but electron microscopy studies have allowed molecular-level characterization of the trimeric protein. Overall, HIV-1 Env adopts a mushroom-shaped structure with the gp120 variable loops 1 and 2 (V1/V2) at its apex and the gp41 trimer inserting in the membrane (5, 7–14). Structural knowledge of the HIV-1 Env has been further enhanced by the atomic structures of individual components. Crystal structures of unliganded gp120 monomer cores (i.e. devoid of the V1/V2 loops, the V3 loop, and truncations at both the N- and C-termini) were recently reported and found to adopt an overall architecture similar to that previously determined for a gp120 core in the CD4-bound state (15, 16). Recently, structural information on the gp120 V1/V2 loops became available from an antibody-scaffold crystal complex structure and revealed that this portion of gp120 can adopt a four antiparallel, disulfide-linked, β-strand topology (17). Although several crystal structures of the gp41 post-fusion, six-helix bundle have been determined, along with crystal and solution structures of the gp41 MPER in various environments, the conformation that gp41 adopts in the pre-fusion state when associated with gp120 remains to be fully elucidated. A recent cryo-EM structure at ~9 Å resolution of a cleaved soluble SOSIP trimer in complex with antibody 17b as a co-receptor mimic has given a tantalizing glimpse of a proposed activated intermediate where the gp41 N-terminal helices are visible in this open conformation (18). Altogether, the various molecular envelopes of HIV-1 Env along with various atomic-level structures of its subunits provide a workable model of the overall structure (Fig. 1A).
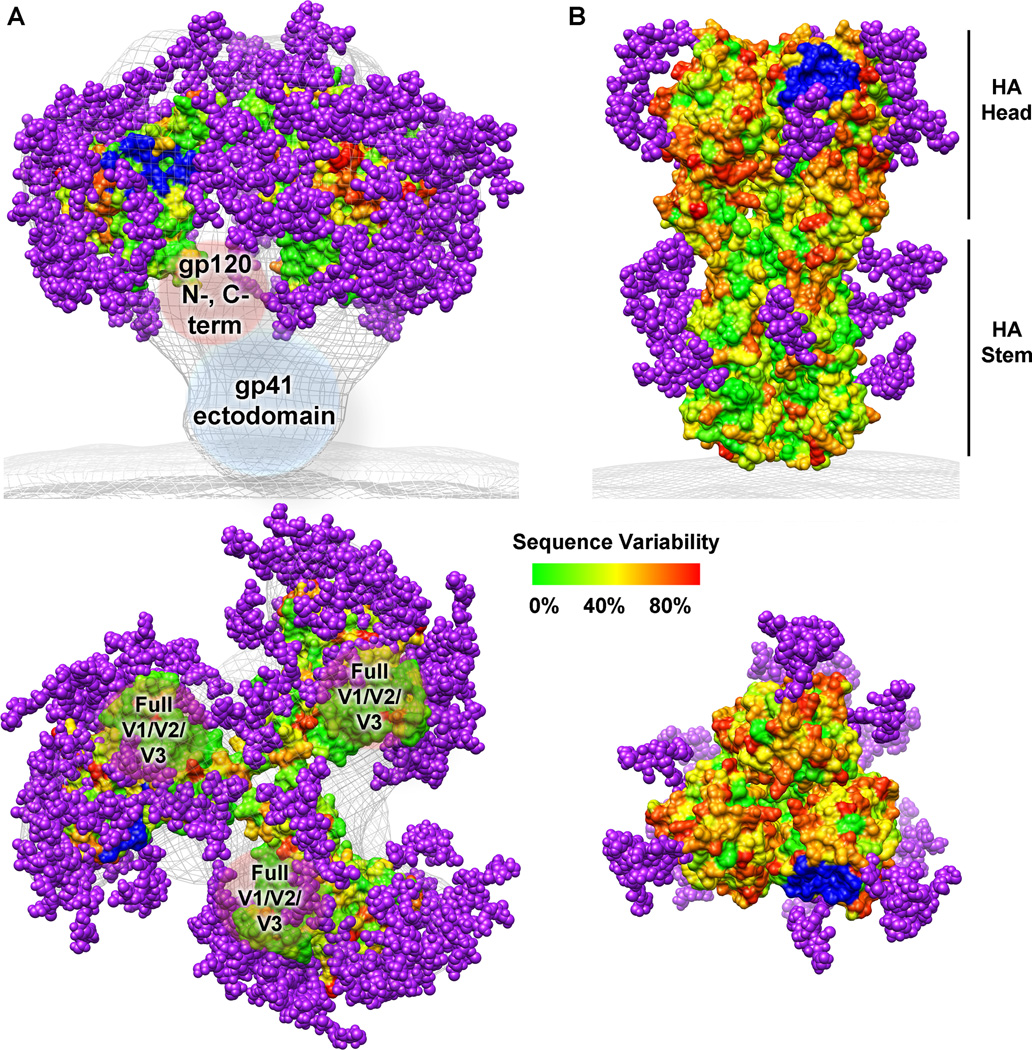
Fig. 1. HIV-1 Env and influenza HA sequence variability and glycosylation.
Sequence variability is represented on the molecular surface as varying colors described on the scale. Potential N-linked glycosylation sites from the consensus sequences are shown as purple spheres. The receptor binding site is colored in blue. (A) As no crystal structure of the full HIV-1 Env trimer is known, a model was generated from the electron microscopy reconstruction of the unliganded HIV-1 Env trimer (gray mesh, EMD ID 5019) (8), the gp120 core structure (PDB ID 3DNN) (8, 15), the gp120 mini-V3 loop (PDB ID 3TYG) (134), and the gp120 V1/V2 loops (PDB ID 3U4E) (17). Missing regions of gp120 (N- and C- termini, and the full V1/V2 and V3 loops) as well as the gp41 ectodomain are labeled inside brown and blue spherical shapes, respectively. (B) The influenza HA trimer structure was rendered using the coordinates from PDB ID 3GBN. This figure was prepared using Chimera (198).
Membrane fusion, as mediated by Env, is a complex process that is only partly understood and has been extensively reviewed elsewhere (19, 20). Briefly, the most accepted view of the HIV-cell membrane fusion process consists of a two-step model that involves first the interaction between the CD4 receptor and gp120, which then induces conformational changes that permit interaction of the CD4-gp120 complex with another cell surface co-receptor (generally CXCR4 or CCR5). The binding between gp120 and co-receptor then triggers further conformational changes in the gp41 transmembrane subunit leading to formation of the extended coil-coil pre-fusion intermediate. At this stage, destabilization of the cell membrane is initiated by membrane insertion of the N-terminal peptide of gp41. Finally, gp41 forms a six-helix bundle that draws the viral and host membranes together. It is this process that commercially available peptide entry inhibitors, such as Enfurvitide, block by inserting an α-helical peptide into a postulated five-helix bundle intermediate and, thus, prevent six-helix bundle formation (6, 21–24). For the cell and viral membranes to be pulled together efficiently, mutational analysis has revealed the importance of the gp41 membrane proximal external region (MPER), a flexible hinge region between the helix bundle and the transmembrane (TM) anchor (25). It is still unclear how many functional Env molecules are required for mediating viral entry; models with anywhere from one to eight Env, CD4, and co-receptor molecules organized in lipid rafts have been implicated in forming the fusion pore (26–31). Although much of the HIV-1 entry process remains to be clearly defined, our current understanding provides strategies to block receptor-mediated HIV-1 membrane fusion, either with inhibitors or monoclonal antibodies that target crucial regions on HIV-1 Env.
Influenza virus overview and the entry process
Influenza A virus is an enveloped virus that uses its major surface glycoprotein hemagglutinin (HA) to mediate viral infection of host cells (32). In contrast to HIV-1 Env, HA is densely clustered on the surface of virus (33) along with another surface antigen the neuraminidase (NA), which is less prevalent than the HA and required to enable progeny release from infected cells. HA is encoded as a single precursor polypeptide, HA0, and three copies assemble into a trimer to form the biological unit. HA0 is subsequently matured extracellularly by proteolysis to generate a pH-dependent metastable trimer, composed of disulfide-linked HA1 and HA2 subunits, which frees the fusion peptide at the N-terminus of HA2 for later fusion events. Generally, HA can be structurally divided into a membrane-distal ‘head’ and a membrane-proximal ‘stem’, each of which plays a distinct role in HA-mediated viral infection (34) (Fig. 1B). The head is composed only from HA1 and contains the binding site for its sialic acid receptor (35). This interaction is fairly weak with millimolar affinity and thus relies on multivalency of binding to achieve sufficient affinity, which leads to endocytosis of the virus and entry into endosomal compartments (36). The stem of HA consists primarily of HA2, along with some HA1 residues, and contains the fusion machinery, which is triggered in the low pH environment of the late endosomes. There, HA undergoes major conformational changes that direct its fusion peptide into the endosomal membranes (37) (Fig. 4C). This pH-dependent activation leads to subsequent fusion of host and virion membranes, which enables release of the influenza genome into the cytoplasm. Thus, prevention of either receptor binding or membrane fusion of HA is key to preventing influenza viral infection.
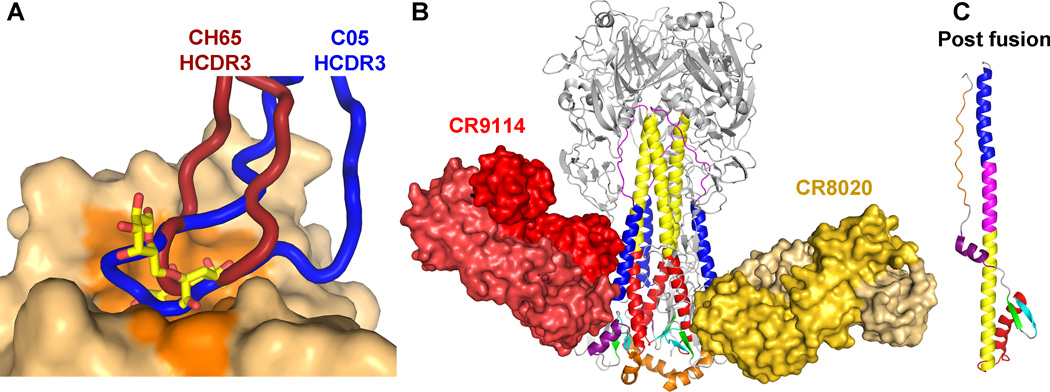
Fig. 4. Recognition of the head and stem subdomains of influenza HA by broadly neutralizing antibodies.
(A) Antibodies CH65 (PDB ID 3SM5) and C05 (PDB ID 4FP8) insert their HCDR3, colored dark red and blue, respectively, into the receptor-binding site to compete with receptor. The HA surface from PDB ID 1MQN is shown, and the residues contacted by receptor are colored in dark orange. The receptor is represented as sticks with carbon in yellow, oxygen in red, and nitrogen in blue. The HA heads of the bnAb complex structures were aligned to that of PDB ID 1MQN. (B) Antibodies CR9114 (PDB ID 4FQI) and CR8020 (PDB ID 3SDY) bind unique epitopes in the stem and inhibit the low pH conformational change shown in (C) (PDB ID 1QU1). The light chains of each antibody are represented in a lighter shade. The HA1 is colored in gray and glycans are not shown. The figure was prepared using Pymol.
HA is the immunodominant protein of influenza virus for the humoral immune response and neutralizing antibodies are rapidly generated against it. There are thousands of unique influenza A strains, which can be divided into two major groups and further classified into 17 HA subtypes established by their uniqueness of reactivity against polyclonal antisera (38, 39). Currently, flu vaccinations are offered annually and are composed of the dominant strains from the circulating influenza A H1 and H3 subtypes as well as influenza B. As influenza viruses constantly undergo antigenic drift that subsequently lead to immune evasion, the vaccine components need to be replaced frequently and almost on an annual basis. Since the efficacy of vaccines depends on the match between the selected vaccine strain(s) and the dominant circulating strain(s), there can be a potential lack of protection if a sufficient discrepancy exists (40, 41). Also, there is concern for pandemic potentials of influenza A by antigenic shift caused by viral reassortment as observed in the 1918 H1N1, 1957 H2N2, 1968 H3N2, and the recent 2009 H1N1 pandemic viruses. Thus, there is a pressing need for a broad-spectrum, cross-reactive vaccine for flu for pandemic preparedness.
Challenges in eliciting protective antibody responses
The presence of HIV-1 Env and influenza HA on the surface of their respective viruses, along with their crucial role for viral infectivity, make them ideal targets for vaccine design. However, it has proven difficult to elicit potent and broad neutralizing antibody responses against these glycoproteins during both the course of natural infection and by vaccination. One mechanism of viral escape common to both HIV-1 and influenza is an elevated genetic diversity of the viruses (Fig. 1) due to an error-prone reverse transcriptase (42). The resulting extensive genetic diversity of HIV-1 and influenza leads to the ability of these viruses to constantly evolve and escape antibody responses capable of neutralizing functional Env and HA variants. For HIV-1 Env, an estimated 35% sequence variation exists between subtypes (commonly called clades) and the evolution rate of HIV-1 is as much as a million times faster than that of animal DNA (43, 44). Although influenza HA strains possess a high degree of sequence variability over time, its diversity is constrained to a given dominant strain at a specific time point (45, 46). On the other hand, the sequence hypervariability of HIV-1 Env is prevalent at all times both geographically and within individuals. As such, it has been estimated that the total HA sequence variability for a given year of an influenza epidemic is equivalent to the degree of HIV-1 Env sequence variability in a single individual (46). Together, these sequence variation comparisons allow an appreciation for why it has been possible to continue to manufacture effective seasonal influenza vaccines, but indicates why creation of a universal influenza vaccine is a more daunting but not impossible task, and perhaps a little more tractable at present than producing an effective HIV-1 vaccine.
Another factor that precludes mounting an effective humoral response against HIV-1 Env and influenza HA is the masking of functional and conserved regions by extensive glycosylation (Fig. 1). Indeed, there is considerable evidence indicating that alterations in the glycosylation pattern of these viral spike proteins have a direct impact on their sensitivity to neutralizing antibodies (47–54). Almost all glycans on Env and HA are N-linked, although some O-linked glycans have also been reported for HIV-1 (55–58). The HIV-1 gp120 has on average 24 potential N-linked glycosylation sites (PNGS) in its consensus sequence, which spans just over 500 residues, and makes it one of the most heavily glycosylated viral proteins ever identified with almost 50% of its molar mass contributed by carbohydrates (Fig. 1A). HIV-1 gp41, on the other hand is much less glycosylated, with four consensus PNGS in its ectodomain. The influenza spike is not nearly as glycosylated as HIV-1 Env, containing a total of six consensus PNGS spanning HA1 and HA2 (59, 60) (Fig. 1B). The total number of glycosylation sites in HAs varies from subtype to subtype and also appears to increase in the years following a pandemic. These additional glycans and alterations in their locations lead to masking and unmasking of epitopes (61–64). In addition to being derived from the host and, hence, poorly immunogenic, the glycan types can be heterogeneous in nature (complex, hybrid, and oligomannose) and glycosylation sites are often differentially utilized in viruses of different clades or subtypes (65–67). These characteristics of the glycans coating the HIV-1 and influenza spike further complicate the task of generating effective broadly neutralizing antibody responses.
Whereas HA mainly relies on a high mutation rate and sequence variation, Env has acquired other mechanisms that have allowed it to escape from immune surveillance. These include (i) conformational masking, (ii) the ability of the conserved gp41 MPER to partition to the viral membrane, as well as (iii) a high degree of Env debris displayed on the surface of HIV-1, which is hypothesized to divert and overwhelm the humoral response (51, 68–75).
Strategies for antibody discovery
Over the last couple of decades, discovery of broadly neutralizing monoclonal antibodies (bnAbs) that have activity against a broad range of HIV-1 isolates or influenza subtypes have renewed hope that development of a broadly protective vaccine against these pathogens is indeed possible. In animal experiments, it was demonstrated for HIV-1 that, when some of these bnAbs were passively administered prior to challenge with an HIV-1-like virus, protection of the host was achieved (76–80). These encouraging results suggest that if similar bnAbs can be elicited by vaccination prior to virus exposure, then sterilizing immunity is attainable.
Recent studies have shown that a substantial proportion (between 10% and 25%) of HIV-1 positive individuals who have been seropositive for at least one year possess moderate to broadly neutralizing antibody responses (81–84). In addition, one of these studies estimates that 1% of HIV-infected individuals, termed elite neutralizers, develop sera with unusually high neutralizing breadth and potency (83), and it is from these elite neutralizers that bnAbs have been isolated. In the early to mid-1990s, phage libraries and hybridomas were generated from sera of selected patients and screened for reactivity against Env that allowed for the identification of the first generation of bnAbs: the anti-CD4 binding site (CD4bs) b12 antibody (85, 86), the anti-gp41 MPER 2F5 and 4E10 antibodies (87), and the anti-gp120 glycan coat 2G12 antibody (87). Over the course of many years, these antibodies and their Env epitopes have been the focus of intense investigation, and, as such, crystal structures of these antibodies in complex with their respective epitopes have been determined (88–92). In recent years, novel strategies for the efficient screening of B cells were devised and implemented. These include the gathering of numerous sera of HIV-1 infected individuals from different geographical regions, such as in IAVI Protocol G, the development of novel B-cell stimulation assays, high-throughput micro-neutralization assays, probe development for performing high-throughput binding assays, and 454 pyrosequencing (83, 93–97, reviewed in 98). The use of a combination of these technological advances on different cohorts of HIV-1 patients has recently led to the isolation of new bnAbs that map to common sites of vulnerability on the HIV-1 Env: the CD4bs on gp120, the gp41 MPER, the N160 glycan on gp120 V1/V2, and the N332 glycan on gp120 V3, reviewed in (99–101).
The quest for the discovery of bnAbs against influenza HA, perhaps surprisingly compared to HIV-1, has only recently taken off, despite the establishment and availability of medium-throughput sequence technologies such as phage display antibody libraries in the early 1990s (102). In 1993, the isolation of bnAb C179 from immunized mice marked the first report of an antibody capable of recognizing a conserved antigenic site on HAs of different subtypes (103). In subsequent years, the significant public health concerns surrounding the emergence of the avian and swine flu exposed the need for the discovery of human bnAbs that recognize conserved antigenic sites on HA that could be exploited in the design of a universal or at least a broader spectrum vaccine. Similar techniques to those described for the discovery of anti HIV-1 bnAbs have been utilized to isolate numerous bnAbs targeting conserved regions on the HA receptor binding site and fusion machinery, detailed below. As a result, the recent accumulation of numerous bnAbs converging on the same conserved epitopes has opened up the opportunity to structurally characterize what appear to be the key neutralizing epitopes with an increasing level of detail, which now is greatly informing immunogen design against HIV-1 and influenza.
Antibody responses against HIV-1 Env
The gp120 CD4bs and gp41 MPER as sites of vulnerability
As described previously, key sites of HIV-1 Env vulnerability have been identified from the study of several bnAbs. The best structurally characterized site of vulnerability is the CD4bs, which is recognized by a myriad of bnAbs including b12, VRC01, VRC03, and PGV04, for which co-crystal structures in complex with gp120 core monomers have been determined ((91, 94, 104, reviewed in 105). Structurally, these antibodies mediate their recognition by interacting primarily with a loop in the CD4bs (gp120 residues 365–371 in β15 and α3), particularly via a key salt bridge with gp120 Asp368. Although these bnAbs all contact this gp120 CD4-binding loop, they vary significantly in their neutralization potency and breadth. Non-neutralizing antibodies that interact with the gp120 CD4 binding loop have also been identified, such as b13 and F105, for which co-crystal structures have also been determined (106). From the structural comparisons of neutralizing and non-neutralizing CD4bs antibodies, Kwong and colleagues (106) hypothesize that the angle of approach to CD4bs has to be very precise and compatible with the functional Env trimer configuration. Indeed, visual inspection of the model of the HIV-1 Env in Fig. 1A shows the CD4bs is recessed and located between gp120 protomers as well as shielded by glycans from not only its own gp120 protomer, but also from the heavily glycosylated gp120 outer domain of the adjacent promoter in the Env trimer. As such, for an antibody to gain access to this functionally conserved site, it must avoid significant steric constraints and thus is able only to contact regions of gp120 neighboring the CD4-binding loop that are compatible with a specific angle of approach. For VRC01, VRC03, and PGV04, the broadest bnAbs antibodies isolated to date (up to ~90% neutralization of all circulating HIV-1 viruses), these neighboring interactions include the gp120 loop D (residues 276–283, in addition to the GlcNAc of glycosylated N276) and variable loop 5 (V5, residues 455–462). In support of this idea, a recent study showed by increasing antibody interactions with key regions surrounding the CD4bs using structure-based rational design, antibodies could be developed with increased neutralization breadth and potency (107).
Another site of HIV-1 Env vulnerability is the gp41 MPER against which three major bnAbs have been identified: 2F5, 4E10, and Z13 (87, 108, reviewed in 109, 110). Interestingly, 15 of 25 residues of the gp41 MPER are completely invariant across all HIV-1 sequences and these are at the center of the epitopes recognized by these bnAbs (111). The high degree of sequence conservation in this tryptophan-rich region is best explained by the functional constraints of the Env MPER in various viral processes, such as membrane fusion (25, 109, 112, 113), gp41 oligomerization (114), membrane leakage through pore formation (115–117), as well as binding on epithelial cells to galactosyl ceramide receptors that leads to mucosal infection by transcytosis (118–120). Structurally, the gp41 MPER is predicted to adopt an α-helical secondary structure with amphipathic character, which allows it to partition with the viral membrane (121, 122). As revealed in the co-crystal structures, the residues buried upon 2F5, Z13, and 4E10 binding are spanned by Leu661-Trp670, Trp670-Thr676, and Phe673-Trp680, respectively (90, 92, 123). Although 2F5, Z13, and 4E10 recognize core epitope regions of the MPER that are overlapping, the structure that this gp41 region adopts when in complex with each antibody differs significantly. As revealed in the co-crystal structures with peptide epitopes, 2F5, Z13, and 4E10 recognize their core epitopes using a type-1 β-turn, two perpendicular α-helical turns, and an α-helical secondary structure, respectively. These crystal structures therefore clearly highlight the structurally dynamic nature of the gp41 MPER and how each structure represents a distinct site of vulnerability for HIV-1. Recently, extensive biophysical techniques were utilized to shed light into the neutralization mechanism of these anti-MPER directed bnAbs (122, 124, 125). Interestingly, it appears that 2F5, Z13, and 4E10, to different extents, are able to access and possibly extract their conserved epitopes from a membrane environment thereby halting viral entry by restricting the motion and orientation of important gp41 MPER structural elements (122, 124, 125).
HIV-1 glycans and broadly neutralizing antibodies
Long thought solely as a shield covering conserved regions on HIV-1 Env, the glycan coat of the gp120 silent face has recently emerged as a vulnerable entity amenable to recognition by bnAbs. Indeed, analysis of humoral responses in infected subjects has revealed that the HIV-1 Env glycan coat possesses characteristics that make it immunogenic and lead to the development and amplification of B cells to mediate its recognition (97, 126, 127). More specifically, HIV-1 Env glycans are packed very densely, which drastically limits the processing of individual carbohydrates by the host cell glycosylation machinery during viral maturation and renders a high proportion of glycans on the HIV-1 Env as being immature oligomannose (IOM) type. Various studies have estimated the contribution of these IOM glycans to be 46% to 98% of the total carbohydrates on the surface of Env, depending on viral isolates and production systems (58, 67, 128). These IOM glycans are predominantly localized on the intrinsic patch, a densely arranged glycan cluster on the gp120 outer domain monomer face opposite to the CD4bs, as well as on regions involved in gp120 trimerization, such as V1/V2, where closely interacting quaternary structural elements are thought to hinder access to glycan-processing enzymes (58, 129) (Fig. 2A). Glycosylation sites, as analyzed by mass spectroscopy, that have a propensity to harbor primarily glycans of the IOM type include N230, N234, N241, N262, N289, N332, N339, N386, and N392 (65, 66). Biochemical and structural characterizations suggest that positions N156, N160, N173, and N295 likely also have carbohydrates of the IOM type (17, 130). The gp120 IOM are relatively well conserved because of their role in protein folding (131) and facilitating entry into CD4+ T cells via the interaction with ancillary receptors, such as DC-SIGN (132). Even if these IOM glycans are derived from the host’s endoplasmic reticulum and Golgi pathways during the HIV-1 life cycle, they appear as ‘non-self’ to the immune system because of their dense clustering and IOM nature, thereby representing motifs not usually observed in host glycoproteins. These clustered IOM patches located on gp120 have now been revealed to be the target of many bnAbs, including 2G12, PGT128, and PG9 (93, 97, 133) (Fig. 2A).
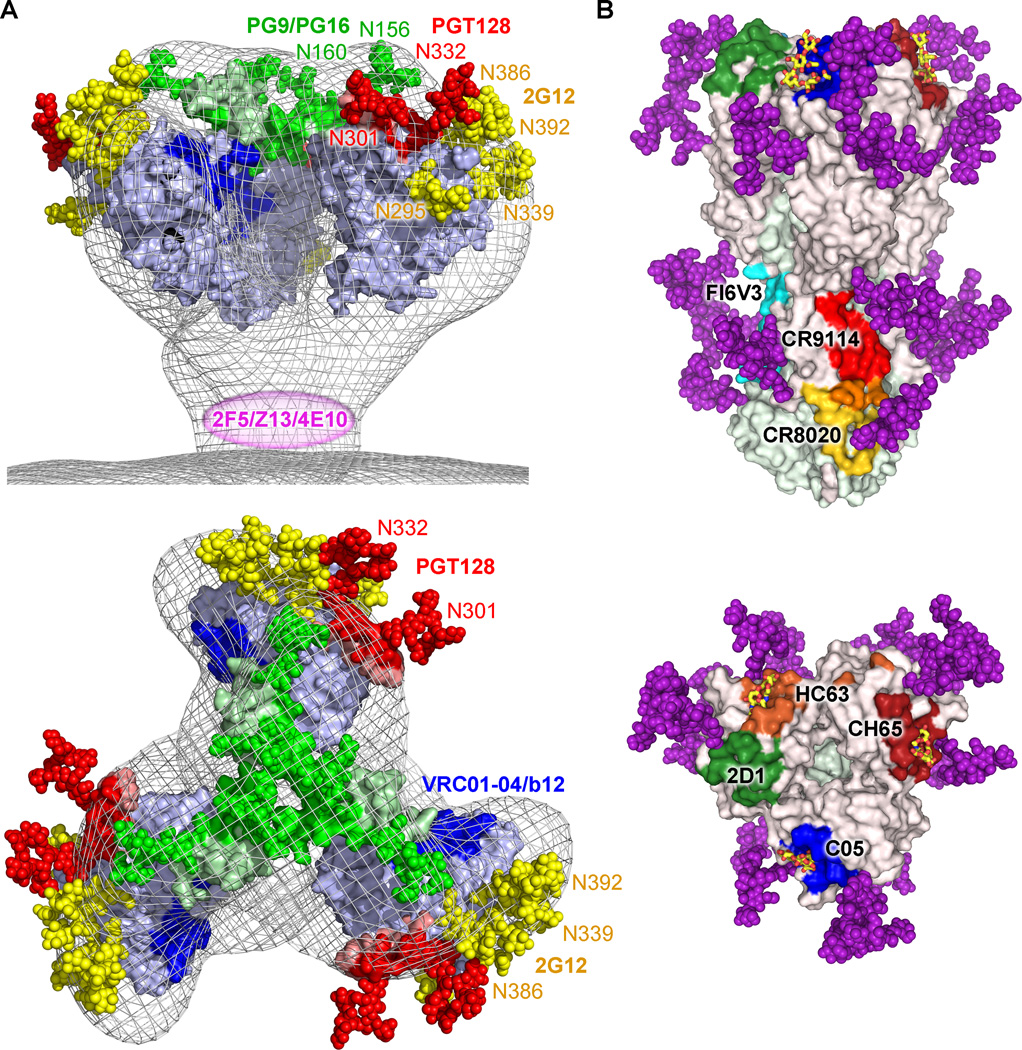
Fig. 2. HIV-1 Env and influenza HA sites of vulnerability.
(A) Crucial oligomannose carbohydrates of the HIV-1 glycan coat are part of epitopes recognized by broadly neutralizing antibodies PGT128 (N301 and N332, red), PG9 (N156 and N160, green), and 2G12 (N295, N339, N386 and N392, yellow + N332, red). Other sites of vulnerability recognized by broadly neutralizing antibody include the CD4bs (blue) and the gp41 membrane proximal external region (MPER) (magenta). (B) Comparison of structurally characterized neutralizing antibody epitopes against influenza HA. For each antibody-HA complex, contacting residues were determined by Contacsym (199) and the equivalent positions are mapped onto the surface representation of A/Brevig Mission/1/1918 (H1N1) HA from PDB ID 3GBN: red, CR9114 (PDB ID 4FQI); yellow, CR8020 (PDB ID 3SDY); cyan, FI6V3 (PDB ID 3ZTN); blue, C05 (PDB ID 4FP8); dark red, CH65 (PDB ID 3SM5); green, 2D1 (PDB ID 3LZF); orange, HC63 (PDB ID 1KEN). Overlapping residues between the CR9114 and CR8020 footprints are colored in orange. The α2-6 sialic acid receptor analog from PDB ID 3UBE is shown with carbon in yellow, oxygen in red, and nitrogen in blue. Glycans are illustrated as in Fig. 1. This figure was prepared using Pymol.
2G12
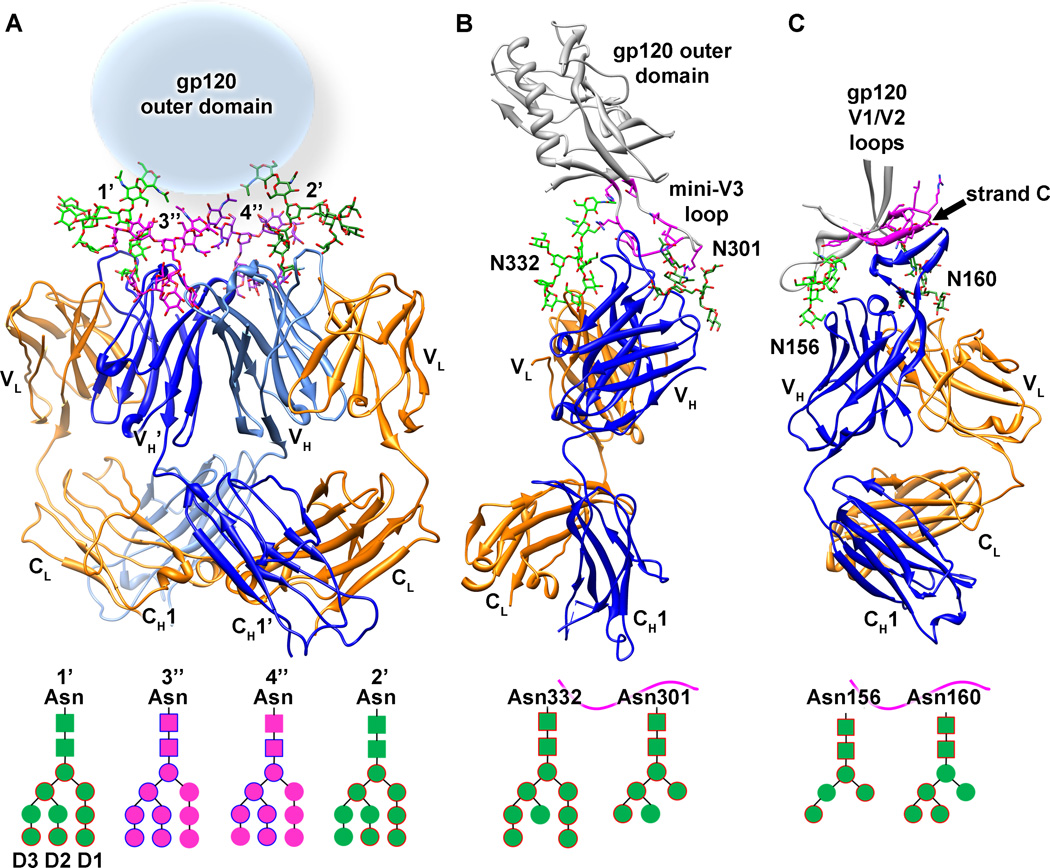
In the early 1990s, bnAb 2G12 was isolated from a HIV-1-positive individual possessing a lymphocyte count of at least 500 cells/mm3as well as a CD4+ T cell:CD8+ T-cell ratio greater than one (87). Subsequent structural and biochemical studies of 2G12 revealed that it adopts an unusual domain exchanged dimer configuration, where the variable heavy chains are swapped on adjacent Fab arms, both when unliganded and when bound to a soluble trimeric Env construct (88) (Fig. 3A). By somatic mutations of key residues during B-cell affinity maturation, domain swapping in this unique antibody becomes possible by (i) weakening the contacts in the VH/VL interface, (ii) structural rearrangement of the Fab elbow region located between the VH and CH1-regions including incorporation of a proline, and (iii) formation of a complementary VH/VH’ interface (88). This unusual linear architecture of the Fab portion of the IgG expands the antibody paratope to 20 Å × 60 Å, with a distance between the two conventional VH/VL combining sites of approximately 35 Å. The crystal structure of 2G12 in complex with Man9GlcNAc2 moieties demonstrates how four carbohydrates can simultaneously interact with the expanded paratope via two types of interactions (Fig. 3A). The high affinity site of glycan recognition is located in the conventional combining sites (sites 1’ and 2’ in Fig. 3A), and although several glycan residues are contacted by 2G12 in these two sites, recognition of the terminal Manα1-2Man disaccharide on the D1 arm accounts for 85% of the interaction, which contains 12 hydrogen bonds (88). The secondary sites of glycan interaction (sites 3” and 4” in Fig. 3A) reside at unique VH/VH’ interface created by domain swapping. Although the specificity for glycan recognition at this site appears less than that of terminal Manα1-2Man disaccharides in primary sites, the secondary combining site nonetheless forms between eight and nine hydrogen bonds with the D2 arm of Man9GlcNAc2 moieties and primarily interacts with the central mannose residue. Despite the intrinsic low affinity of isolated protein-glycan interactions, it is the increased multivalency by clustering of several specific protein-glycan interactions on the 2G12 domain-swapped dimer paratope that is thought to result in recognition of gp120 with a binding affinity in the nanomolar range (88). Mutagenesis studies have revealed that the most important IOM glycans required for 2G12 neutralization are located at glycosylation sites N295, N332, N339, N386, and N392 (130). Based on the gp120 core and the 2G12:Man9GlcNAc2 structures, it becomes possible to hypothesize that 2G12 neutralizes HIV-1 Env by recognition of glycans at positions N332 and N392 in its primary high affinity combining sites, whereas proximal glycans at positions N295, N339, and N386 either reside in the secondary combining sites or play an indirect role in limiting glycan processing of the key carbohydrate moieties by clustering effects (88).
Fig. 3. Recognition of carbohydrate-containing HIV-1 Env epitopes by three broadly neutralizing antibodies.
Protein components are rendered as secondary structure cartoons, whereas glycans are represented as sticks. The light chain and heavy chain of the antibodies are colored in orange and shades of blue, respectively. The glycans recognized by the antibodies are represented as cartoon models below the crystal structures with squares and circles representing GlcNAc and mannose moieties, respectively. For each glycan, the moieties are circled if they have at least 10 Å2 of surface area buried in the antibody paratope, a threshold used for defining interacting regions. Regions on gp120 that form the primary site of interaction are colored green, whereas secondary binding sites are colored magenta. (A) The crystal structure of the domain-swapped 2G12 antibody dimer in complex with Man9GlcNAc2 sugar moieties reveals an interaction with up to four oligomannose glycans in its multivalent combining site (PDB ID 1OP5). Labeled 1’ and 2’ are the glycans interacting in the primary combining sites, whereas the glycans contacting the secondary combining sites are labeled 3” and 4”. (B) As seen in the crystal structure of the complex with an engineered gp120 outer domain (eODm3), antibody PGT128 recognizes two glycans (N301 and N332) and a β-strand (base of gp120 V3) (PDB ID 3TYG). (C) Antibody PG9 also recognizes two glycans (N156 and N160) and a β-strand (gp120 V2 loop) in a crystal structure of a complex with a gp120 V1/V2 scaffold (PDB ID 3U4E). The figure was prepared using Chimera (198).
PGT128
A multitude of new bnAbs with increased potency and breadth has been isolated recently from the International AIDS Vaccine Initiative (IAVI) Protocol G initiative. This extensive study yielded antibodies of the PGT 125-131 family (97). Neutralization and binding experiments with HIV-1 Env mutants suggested that recognition of gp120 by this antibody family was mediated by interactions with IOM carbohydrates, especially the glycan at position N332 (97). Recently, a crystal structure of PGT128 in complex with a gp120 outer domain construct (eODmV3) revealed the sites of recognition at atomic detail (134). PGT128, a non-domain swapped antibody, recognizes three main features on gp120: two different glycans (Man8/9GlcNAc2 at position N332 and Man5GlcNAc2 at position N301), and a β-strand at the base of the V3 loop (Fig. 3B). The main component of the interaction is the Man8/9 moiety derived from the glycosylation site at position N332, particularly its D1 and D3 arms with which PGT128 makes 11 of the 16 total hydrogen bonds. To mediate this interaction with high affinity, antibodies of the PGT125-128 family have evolved an unusual six-residue insertion in HCDR2, which effectively promotes interactions with the N332 D1 arm of the Man8/9 moiety. The secondary glycan contacted by PGT128 in the co-crystal structure is located at position N301 and recognition is centered on the core of the Man5GlcNAc2 moiety that is visible for this glycan in the crystal structure. Finally, the 19-residue PGT128 HCDR3 apex makes mostly backbone contacts with C-terminal residues of the mini-V3 loop. The HCDR3 of PGT128 forms a β-strand secondary structure, suggesting that it could potentially form a β-strand-type structure with the full-length V3 loop in the context of the Env trimer. In the same report, electron microscopy studies of a PGT128-soluble Env SOSIP construct demonstrated that three Fab molecules were bound to the three gp120 outer domains on the face opposite to the CD4bs and oriented at an approximately 120° angle relative to the viral membrane (134). Because PGT128 has one of the highest potencies (lowest concentration to achieve neutralization) of any anti-HIV-1 bnAb isolated to date, and because of its relatively high neutralization breadth (~70% of circulating strains), it makes its epitope an attractive target for structure-based vaccine design.
PG9
Two other anti-HIV-1 bnAbs that have recently seen significant progress in the structural characterization of their epitopes are PG9 and PG16, which were also isolated from the IAVI Protocol G initiative (83, 93, 135). Crystal structures of the unliganded PG16 antibody revealed that the 28-residue HCDR3 adopts a hammerhead-like structure with sulfated tyrosines, and paratope mutagenesis experiments showed that these sulfated tyrosines at the top surface of this extended hammerhead loop play an important role in mediating HIV-1 neutralization (136, 137). Neutralization and binding studies indicate that PG9 and PG16 recognize a glycan-dependent N160 epitope located in the gp120 V1/V2 loop that is preferentially displayed on surface-anchored gp160 trimers (93). However, extensive screening of monomeric gp120 constructs from different HIV-1 isolates led to the discovery of a few gp120 monomeric constructs that are efficiently bound by PG9, PG16, or both (17). PG9 was recently co-crystallized in complex with the V1/V2 strands of two HIV-1 isolates, CAP45 and ZM109, inserted into a protein scaffold. Similar to PGT128, the structure revealed that the PG9 epitope also consists of two different glycans and a β-strand (17)(Fig. 3C). In both cases, a Man5GlcNAc2 glycan at position N160 is the predominant element recognized by PG9, with up to 11 hydrogen bonds at the interface. Interestingly, the secondary glycosylation site is occupied by a Man5GlcNAc2 glycan from either position N156 in the CAP45 isolate or from position N173 in the ZM109 isolate, indicating some promiscuity in the glycan recognition. However, although deriving from different Asn residues in the two different isolates, these glycans are seen to reside in the same spatial location. PG9 is therefore dependent on recognition of the N160 glycan but can adapt to either interaction with a N156 or N173 glycan in its secondary site, depending on which HIV-1 isolate it neutralizes. Finally, the extended PG9 HCDR3 hammerhead penetrates through the glycans to form intermolecular parallel β-sheet interactions with backbone residues of the V1/V2 strand C. Cationic gp120 residues also interact with anionic residues from the PG9 HCDR3, including two sulfated-tyrosine residues. Interestingly, other V1/V2 recognizing bnAbs for which unliganded crystal structures are also reported in this study (CH04 and PGT145) show similar extended anionic HCDR3 loops capable of penetrating the glycan shield (17). Although informative, these crystal structures do not directly explain why these bnAbs have a preference for a quaternary epitope.
Antibody responses against influenza HA
Antibodies against the HA head
The membrane-distal head subdomain of HA is the immunodominant element of the HA, and antibodies are rapidly generated against this hypervariable region. Clusters of antigenic sites have been identified in the head, indicating that it contains an almost continuous carpet of epitopes that can be targeted by antibodies; however, some of these sites seem to be more immunodominant than others and undergo constant antigenic drift to escape immune pressure and can vary quite substantially between strains even from the same subtype (61, 138). Therefore, due to the ability of the virus to rapidly mutate and evade immune detection, it has long been believed that antibodies against the head are merely strain specific and, thus, have a narrow breadth of neutralization, which gives rise to the need for nearly annual vaccine strain reformulation. However, despite the sequence variation in the head, all HAs have a functionally conserved receptor binding site for host cell recognition and, thus, residues involved in receptor binding have limited mutational freedom as they are constrained to maintain viral fitness (Fig. 1B).
The receptor-binding site is a shallow groove that is framed by the 130 loop, the 150 loop, the 190 helix, and the 220 loop. The sequences of these loops are relatively conserved compared to the rest of the head in order to bind sialic acid receptors (Fig. 1B). Recent reports have described receptor binding site-targeted bnAbs that overlap with the natural receptor, effectively neutralizing virus by antagonizing receptor binding (139, 140) (Fig. 2B). These antibodies have considerable binding breadth, although not nearly as broad as the stem antibodies, and illustrate that eliciting a broad neutralizing antibody response against the hypervariable HA head is possible.
CH65 and receptor mimicry
One such antibody that targets residues conserved for receptor binding is CH65 (139). It was isolated from peripheral blood mononuclear cells from a subject vaccinated with the 2007 trivalent inactivated vaccine (141). CH65 was discovered to have broad neutralization activity against a large number of HAs specific to the H1 subtype, spanning across strains isolated from 1986 to 2007. The crystal structure reveals that CH65 targets residues conserved in receptor binding by inserting HCDR3 into the receptor binding site (Fig. 4A) but also relies on the use of four additional CDR loops to recognize variable residues. Although the interaction of HCDR3 with HA is proposed to resemble the interaction with receptor, it does not quite achieve true mimicry, as it does not extend to make key interactions with residues in the 220 loop. Moreover, CH65 binds a large HA footprint and, thus, contacts variable residues adjacent to and outside of the receptor binding site, which accounts for its limited breadth.
C05 and binding with a single loop
Another example of an antibody that targets the receptor binding site is C05 (140). It is a human antibody identified from seasonal influenza infection survivors through phage display. This antibody possesses heterosubtypic reactivity and crosses the HA group barrier and targets certain strains from the H1, H2, H3, H9, and H12 subtypes. C05 binds HA primarily through the use of its 24 amino-acid HCDR3 and contacts a compact and conserved HA footprint in the receptor binding site. In addition, the HCDR3 loop extends to make additional contacts with the receptor binding loops in comparison to CH65 (Fig. 4A). Although C05 possesses no clear receptor mimicry, its binding is quite remarkable, because it demonstrates that the receptor binding site can be bound with nanomolar affinity by essentially a single loop, in comparison to the very weak millimolar affinity of sialylated glycan receptors. In addition, avidity appears to play a major role for its expanded subtype recognition in that bivalent binding of IgG is much greater than that of monovalent Fab, likely due to the accessibility of the receptor binding site at the apex of HA on the viral surface for crosslinking HAs. The multiplicity of binding of IgG in combination with the conserved contacted residues in the receptor binding site likely extend C05’s breadth of binding and neutralization.
Recent antibodies such as CH65 and C05 are remarkable in that they both specifically target the relatively small footprint of the shallow receptor binding site. Not surprisingly, the cross-reactivity of an antibody is highly related to the conservation of the residues that it targets. Discovery of antibodies such as these and their binding modes may prove critical in deciding how to better select vaccine strains. Whether or not avidity is a general trend for other receptor binding site-targeted antibodies is suggestive but remains to be seen.
HA stem-targeted antibodies
The membrane proximal stem subdomain of HA, in contrast to the head, has much less sequence variability and, thus, is highly desirable for being targeted by the immune response. The stem region is not as highly exposed as the head and, hence, not as accessible to antibodies or as immunogenic. Many of the residues in the stem are highly conserved, as they contribute to the fusion machinery and are likely to be constrained from mutation. Antibodies that target this site could inhibit influenza viral infection by blocking HA maturation or preventing the low pH conformational change that triggers the fusion process (reviewed in 142) (Fig. 2B). For many years, it was believed that the stem subdomain could not be targeted by antibodies despite the fact that a mouse monoclonal antibody C179 was shown in 1993 to target this region and neutralize H1 and H2 subtypes (103). Nevertheless, perhaps because these were not human antibodies and that bnAbs to influenza were not at the forefront of vaccine efforts, not much attention was paid to bnAbs against influenza virus. However, all that changed about four years ago with the discovery of a number of human bnAbs that were shown to target the HA stem region (143–146). The discovery of these antibodies reinvigorated the field and illustrated that a universal flu vaccine may indeed be achievable. Furthermore, it is now clear that individuals do produce stem antibodies in the natural course of infection (147, 148) or through vaccination (149), but these have been overlooked in large part because all of the tests for vaccine efficacy are based on hemagglutination inhibition assays (HAI) that do not detect neutralization by stem antibodies.
Influenza group 1 specific antibodies
In 2008–2009, three groups reported discovery of antibodies that neutralized virtually all 10 subtypes of the group 1 family of HAs (143, 144, 146). These antibodies, as exemplified by CR6261 and F10, are very similar in sequence and structure and all derive from the VH1-69 germline gene family (145, 146). They bind the HA stem using only their heavy chain VH Ig domain and insert their hydrophobic HCDR2, the hallmark of the VH1-69-encoded germline gene family, into a hydrophobic pocket between the HA helices in the stem region (145, 146). These antibodies are protective in mice from lethal challenge of influenza virus from H1 and H5 subtypes. Furthermore, these antibodies are not highly mutated from their putative germline precursor and, hence, could potentially be relatively easy to elicit by vaccination or infection. CR6261 also was shown to protect against the pH-dependent conformational change in the HA and, thereby, provide a mechanism for its neutralization ability (145).
Influenza group 2 specific antibodies
The team from Crucell extensively screened memory B cells, which led to the identification of bnAb CR8020 with reactivity against group 2, the remaining 6 of the known 17 HA subtypes. These antibodies recognized a conserved epitope lower down the stem close to the viral membrane (150)(Fig. 4B) than that of the previously described group 1 specific antibodies and uncovered another highly conserved site of vulnerability in the HA stem. The epitope for CR8020 only partially overlaps with the CR6261 epitope. CR8020 is derived from a completely different VH family (VH1-18) and uses five out of its six total CDRs for interaction with the HA. CR8020 not only blocks the pH-dependent conformational change, but also maturation of the HA0 into HA1 and HA2, providing yet another possible mechanism of neutralization (150).
Pan-influenza A antibodies
In a further development, other antibodies have now been discovered that neutralize all influenza A subtypes from group 1 and group 2. The first of these to be reported was bnAb FI6V3 that was also not from the VH1-69 encoded family but rather from VH3-30 (151). FI6V3 binds to a similar epitope to CR6261-like Abs but is oriented very differently, such that it approaches the stem from a near orthogonal direction parallel to the viral membrane, and uses both its light and heavy chain for interaction. FI6V3 manages to avoid the group 2 specific glycans and, hence, broadens its specificity compared to the group 1 specific stem antibodies. Another even more remarkable antibody has been found that neutralizes not only all tested influenza A subtypes but also influenza B viruses of both lineages (152). CR9114 is a VH1-69 germline encoded antibody, discovered by phage display, and recognizes nearly the same epitope as the other VH1-69 germline encoded antibodies, such as CR6261 and F10, using only its heavy chain (152)(Fig. 4B). CR9114 differs only slightly in orientation and angle of approach from the VH1-69 group 1 specific antibodies, but is rotated by about 90° in relation to FI6V3. Whereas CR6261 and F10 are sensitive to larger residues that are present in group 2 HAs, CR9114 and FI6V3 can accommodate larger residues at these positions and also avoid the glycans present in different groups and subtypes. These structures also show that these glycans can have some conformational flexibility and can be gently pushed out of the way for the bnAbs to contact the highly conserved protein surface (151, 152).
Influenza B antibodies
To truly have a universal flu vaccine, influenza B viruses must be taken into account. These viruses are ever present and cause considerable infection with substantial economic consequences and thus an influenza B strain is represented in the trivalent seasonal flu vaccine. Influenza B viruses can be divided into two antigenically distinct linages (Victoria and Yamagata) and bnAbs should be able to neutralize viruses from both linages. Three bnAbs have recently been reported that protect mice from lethal challenge by viruses from both lineages (152). We have previously described CR9114, which binds to the highly conserved stem region in both flu A and flu B HAs. The two other antibodies CR8033 and CR8071 bind to conserved regions in the head around the receptor binding site and the vestigial esterase region in the center of the HA, respectively. In yet another potential mechanism of neutralization, these antibodies appear to inhibit egress of progeny virus from infected cells (152).
Lessons from the structural studies of anti-Env and anti-HA bnAbs
The structural characterization of both HIV-1 Env and influenza HA in complex with a plethora of different and diverse antibodies has allowed definition of the key sites of vulnerability on these viruses. Interestingly, despite significant differences in the biology of the viruses, the key functional regions of each viral spike are targeted by these bnAbs, resulting in potent and broad neutralization (reviewed in 101, 153). On both Env and HA, the receptor binding site and a region proximal to the viral membrane involved in the fusion machinery are targets of broadly neutralizing antibodies. Whereas high-affinity binding to the receptor binding site on the functional trimer most likely leads to potent neutralization by directly preventing receptor engagement on host cells, binding of neutralizing antibodies to a region proximal to the viral membrane appears to prevent membrane fusion. To access these sites of functional importance on the viral spikes, antibody responses need to evolve mechanisms to by-pass masking and decoy strategies employed by the viruses around these sites of vulnerabilities, such as extensive glycosylation, loops of high sequence variability, and steric occlusion from the viral membrane. Two other strategies for antigen recognition by broadly neutralizing antibodies against HIV-1 Env and influenza HA is the recognition of a quaternary epitope, such as for PG9, PG16, and PGT145 against Env (93, 97) and FI6V3 and HC63 against HA (151, 154). There have also been indications that some antibodies, such as b12 IgG, 4E10 IgG, and PGT128 IgG against HIV-1 Env and C05 IgG against influenza HA are capable of cross-linking spikes as a mechanism of increasing neutralization avidity (134, 140, 155, 156). In addition to the receptor-binding sites and the membrane proximal external region, the IOM glycans on the gp120 have recently been directly implicated in recognition by numerous antibodies including 2G12, PGT128, and PG9 for which structural information describing their interaction with the antigen is now available. The two glycans at the center of these newly discovered epitopes are a Man8/9 glycan at position N332 near the base of the V3 loop and a Man5 moiety at position N160 on strand C of the V2 loop. As highlighted in Fig. 3, the exact interaction of each antibody with the IOM glycans is highly specific including interaction with the less accessible GlcNAc residues, as well as the mannoses of the D1, D2, and D3 arms. It remains to be fully determined how recognition of these elements translates into extremely potent HIV-1 neutralization. Although the influenza HA has also been characterized to contain at least two IOM sugars for certain subtypes, one at position N165 and the other at position N289 of HA1 (157, 158), no broadly neutralizing antibodies have yet been identified that interact directly with these glycans other than minor skirmishes in accessing the underlying conserved protein epitopes. As such, direct recognition of IOM glycans by antibodies to achieve broad neutralization seems at present to be a property unique to HIV-1 but may become an avenue of exploration for influenza and other highly glycosylated viruses.
Structure-based vaccine and small inhibitor design
The amalgamation of structural information of the sites of vulnerability on HIV-1 Env and influenza HA has led to the possibility of designing antigens and inhibitors by structure-based processes with the ultimate goal to re-elicit neutralizing antibodies by vaccination that block or ameliorate infection (reviewed in 159, 160). Not surprisingly, the receptor binding site on both viruses has received significant attention in this regard. Various recent gp120-resurfaced proteins and scaffold molecules have been generated in immunogen design efforts, in some instances with alteration of glycosylation sites attempting to mask immunodominant epitopes and emphasize a better presentation of the CD4bs (161–166). In the case where these molecules were tested as immunogens, none so far have been able to generate a neutralizing antibody response possessing both breadth and potency (161, 162, 164, 165). A recent structural study comparing neutralizing and non-neutralizing CD4bs antibodies has suggested that the angle of approach on the functional HIV-1 trimer is critical in determining the neutralizing capacity of these antibodies (106). As such, monomeric gp120 antigens might be limited in their ability to adequately present the CD4 binding site as an effective immunogen. Similarly, antigens presenting the conserved gp41 MPER in a scaffolded environment very successfully displayed the desired structural epitope but failed to elicit neutralizing antibodies when tested as immunogens (111, 167–169). Interestingly, in one instance, the structural characterization of an antibody isolated from a vaccinated animal in complex with the immunogen revealed that it recognized the desired structure while not being able to neutralize the virus (167). This result and others suggests that the presentation of the gp41 MPER in a trimeric environment, and possibly surrounded by phospholipids to represent the membrane, might be necessary in order to re-elicit antibodies that possess all of the required components to achieve neutralization (170–174). Finally, several attempts to generate synthetic, microbial, and recombinant glycoconjugates as immunogens have recently been performed to mimic the IOM cluster recognized by bnAb 2G12 (175–180). In some cases, the glycoconjugates were successful in eliciting antibodies that bound gp120, but none were capable of eliciting broad neutralization. In part, this lack of success can be attributed to the fact that the exact 2G12 epitope on gp120 has not yet been structurally revealed, and therefore efforts in structure-based design of glycoconjugates are somewhat hindered. Without question, the recent structural description of the PGT128 and PG9 carbohydrate epitopes will lead to increased activity in the structure-based design of glycan-targeting immunogens looking to re-elicit broadly neutralizing antibodies against gp120.
Most bnAbs against HIV-1 that have been isolated from elite neutralizers possess unusual characteristics, reviewed in (153), such as a high degree of somatic hypermutation. However, it remains unclear whether all of these mutations are required to achieve broad and potent neutralization, or whether they are a mere byproduct of accumulated somatic mutations from antibody maturation during the course of a prolonged infection. For example, 2G12 was recently found to only require a few substitutions from its germline precursor sequence to achieve a domain-exchanged configuration (181). Knowledge on the degree of somatic hypermutation required to broadly and potently neutralize HIV-1 is of utmost importance as it will directly impact vaccination strategies that will look to generate bnAbs from germline precursors present in the B-cell repertoire of naive individuals (182–184).
Headless HAs and HA chimeric constructs as alternative vaccine strategies
As an alternative vaccination strategy against influenza, headless HA constructs were created to elicit an antibody response against the stem. These constructs omitted the immunodominant hypervariable head so that only the highly conserved HA stem is presented, and immunization with these constructs does indeed protect mice in vivo (185, 186). Although these first studies provide some optimism that an influenza vaccine can be based upon truncated HA or simplified epitope, other considerations must still be addressed, such as presenting the epitopes in a similar context to that presented in the HA trimer.
It has recently been shown that individuals infected with the 2009 H1N1 pandemic virus generate high titers of stem-specific antibodies (148, 187). The sequence of the HA head from this virus differs substantially from the previously circulating seasonal H1 strain, but is remarkably similar to that from the 1918 virus (64). Thus, the elderly population had preexisting immunity to the 2009 pandemic virus. However, it seems that sudden incorporation of an antigenically distinct head can direct the immune response against the conserved stem. An alternative form of immunogen design may then be to create HA chimeras, where the head subdomains are swapped on the background of a constant stem (188), and provide yet another means to pandemic preparedness through HA stem antibody elicitation.
Designer proteins for neutralizing viruses
As the number of solved bnAb structures increases, trends in antigen recognition may emerge that can be utilized to aid the design of small molecule or protein scaffold inhibitors of viral pathogens. This idea has already been successfully applied to the CR6261 epitope, where a de novo protein was designed to mimic the residues that CR6261 uses to contact the hydrophobic groove in the HA stem (189, 190). Disembodied side chains were modeled onto ‘hot spot’ cavities on the HA surface, and a suitable protein scaffold was identified to display these key side chains for HA interaction. These designs, HB36 and HB80, also inhibit the low pH-induced conformational change in HA and display at least equivalent, if not better, binding and neutralization breadth than CR6261. Crystal structures of these scaffolds in complex with HA show that the designs were successfully predicted even down to side-chain rotamer conformations. This raises the possibility that this method may be further applied to other sites of vulnerability on HA and possibly even be extended to the HIV Env-1 and other viruses. There are significant advantages of creating non-immunoglobulin designed proteins as therapies for influenza as they are cheaper and faster to produce in yeast fermenters in comparison to vaccines, which are produced in chicken eggs and can take at least six months to produce in enough quantity. Also, the delivery mode can be engineered to be oral bioavailable for facilitated administration and distribution.
Much of the effort in designing small molecules against the receptor binding site has been attempted by using sialic acid as a scaffold; however, sialic acid binds weakly with millimolar affinity (36) and is thus a poor starting point. For the first time, high affinity recognition against the receptor binding site by a single loop has been observed by Fab C05 (140). Though there is no obvious receptor mimicry, this structure may also inform on the development of small molecules, whether proteins or drug-like molecules that target the receptor binding site.
Despite the functional conservation of receptor binding of HA, sequence variability within the receptor-binding site will remain a significant hurdle for immune or small molecule recognition. Different subtypes use different residues to recognize their sialic acid receptor. For instance, changes can be incorporated that do not influence receptor binding, most notably by amino-acid insertions or deletions, that alter the shape and size of the receptor binding site and influence antibody recognition and binding. Also, the receptor does not make direct contacts with residues along the tip of the 150 loop, which is at the apex of the HA spike; thus, these positions are not constrained for receptor binding and can be quite variable. Despite these hurdles that limit the binding potential, C05 uses multivalent binding in the context of IgG to neutralize strains that the weak affinity monovalent Fab cannot (140). Weaker affinity arising from reduced specificity and less affinity maturation may be required to neutralize the more variable receptor binding site compared to the much more highly conserved stem region. The increased affinity by avidity raises the possibility that variable regions in the top of the HA head, such as the 150 loop, can be tolerated and this concept can be extended to isolate or elicit antibodies similar to C05.
There have been several other reports of antibodies that also map at the apex of the head of HA near the receptor binding site. These consist of H1-specific antibodies that appear to complement CH65 so as to cover all H1 strains (191, 192), pan-H2 specific antibodies (193), a whole class of pan-H3 antibodies (194–196), and another that is remarkably heterosubtypic (197). Although structures of these Abs have not yet been reported, structural information from these antibodies may also aid in development of vaccines or antibody and small molecule therapies as they target HAs from those viruses that have infected or are currently circulating in humans.
Long-standing challenges and concluding remarks
Despite significant advances in the structural characterization of the HIV-1 Env and the influenza HA in the last decades, some major milestones still remain to be attained to complete the molecular understanding of these viral glycoproteins, which in turn would lead to better guiding structure-based design strategies. In the case of HIV-1, it is unequivocal that the most pressing need is for a structure of the HIV-1 trimer at atomic resolution. It is becoming increasingly clear that the steric constraints present in the membrane-anchored trimeric Env greatly affect its antigenicity, such as the angle of approach of CD4bs antibodies, the glycosylation profile leading to an increase in IOM sugars, as well as the formation of quaternary epitopes located at the spike apex. As such, a detailed structural understanding of the HIV-1 Env trimer would enable the creation of constrained immunogens that possess all the characteristics necessary to present the epitopes of broadly neutralizing antibodies in a more native context.
For influenza, the future seems very bright for the design of a universal broad-spectrum vaccine due to the current structural arsenal of antibodies that cover influenza A and influenza B. As there are now multiple structures of antibodies in complex with HA that target the same functionally constrained epitopes in the receptor binding site and stem, the real challenge now is how to collectively take this information and apply it towards the design of immunogens or small molecules. The design of vaccines or small molecules as therapies based on the known bnAbs may be more tractable in the short-term, in large part due to not requiring sterilizing immunity as for retroviruses.
Acknowledgments
We thank R.L. Stanfield, C. Dreyfus, and D.C. Ekiert for data analysis and helpful discussions. This work was supported by the International AIDS Vaccine Initiative Neutralizing Antibody Center, NIH grants AI84817 (I.A.W.), Canadian Institutes of Health Research fellowship (J.-P.J.) and the NIH Molecular Evolution Training Program (P.S.L.). This is manuscript XXX from The Scripps Research Institute.
Footnotes
The authors state to have no financial or personal relationships that could be viewed as a potential conflict of interest.
References
- 1.Gray RH, et al. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet. 2001;357:1149–1153. doi: 10.1016/S0140-6736(00)04331-2. [DOI] [PubMed] [Google Scholar]
- 2.Forsman A, Weiss RA. Why is HIV a pathogen? Trends Microbiol. 2008;16:555–560. doi: 10.1016/j.tim.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Gelderblom HR, Hausmann EH, Ozel M, Pauli G, Koch MA. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 4.Zhu P, et al. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci USA. 2003;100:15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 6.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 7.Zhu P, Winkler H, Chertova E, Taylor KA, Roux KH. Cryoelectron tomography of HIV-1 envelope spikes: further evidence for tripod-like legs. PLoS Pathog. 2008;4:e1000203. doi: 10.1371/journal.ppat.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White TA, et al. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 2010;6:e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu SR, et al. Single-particle cryoelectron microscopy analysis reveals the HIV-1 spike as a tripod structure. Proc Natl Acad Sci USA. 2010;107:18844–18849. doi: 10.1073/pnas.1007227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White TA, et al. Three-dimensional structures of soluble CD4-bound states of trimeric simian immunodeficiency virus envelope glycoproteins determined by using cryo-electron tomography. J Virol. 2011;85:12114–12123. doi: 10.1128/JVI.05297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris A, et al. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc Natl Acad Sci USA. 2011;108:11440–11445. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moscoso CG, et al. Quaternary structures of HIV Env immunogen exhibit conformational vicissitudes and interface diminution elicited by ligand binding. Proc Natl Acad Sci USA. 2011;108:6091–6096. doi: 10.1073/pnas.1016113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu G, Liu J, Taylor KA, Roux KH. Structural comparison of HIV-1 envelope spikes with and without the V1/V2 loop. J Virol. 2011;85:2741–2750. doi: 10.1128/JVI.01612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon YD, et al. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci USA. 2012;109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran EE, et al. Structural Mechanism of Trimeric HIV-1 Envelope Glycoprotein Activation. PLoS Pathog. 2012;8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallo SA, et al. The HIV Env-mediated fusion reaction. Biochim Biophys Acta. 2003;1614:36–50. doi: 10.1016/s0005-2736(03)00161-5. [DOI] [PubMed] [Google Scholar]
- 20.Markovic I, Clouse KA. Recent advances in understanding the molecular mechanisms of HIV-1 entry and fusion: revisiting current targets and considering new options for therapeutic intervention. Curr HIV Res. 2004;2:223–234. doi: 10.2174/1570162043351327. [DOI] [PubMed] [Google Scholar]
- 21.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 22.Kilby JM, et al. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 23.Shu W, Liu J, Ji H, Radigen L, Jiang S, Lu M. Helical interactions in the HIV-1 gp41 core reveal structural basis for the inhibitory activity of gp41 peptides. Biochemistry. 2000;39:1634–1642. doi: 10.1021/bi9921687. [DOI] [PubMed] [Google Scholar]
- 24.Sia SK, Carr PA, Cochran AG, Malashkevich VN, Kim PS. Short constrained peptides that inhibit HIV-1 entry. Proc Natl Acad Sci USA. 2002;99:14664–14669. doi: 10.1073/pnas.232566599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munoz-Barroso I, Salzwedel K, Hunter E, Blumenthal R. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J Virol. 1999;73:6089–6092. doi: 10.1128/jvi.73.7.6089-6092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhmann SE, Platt EJ, Kozak SL, Kabat D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J Virol. 2000;74:7005–7015. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierson TC, Doms RW. HIV-1 entry and its inhibition. Curr Top Microbiol Immunol. 2003;281:1–27. doi: 10.1007/978-3-642-19012-4_1. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, Kurteva S, Ren X, Lee S, Sodroski J. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J Virol. 2005;79:12132–12147. doi: 10.1128/JVI.79.19.12132-12147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klasse PJ. Modeling how many envelope glycoprotein trimers per virion participate in human immunodeficiency virus infectivity and its neutralization by antibody. Virology. 2007;369:245–262. doi: 10.1016/j.virol.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magnus C, Rusert P, Bonhoeffer S, Trkola A, Regoes RR. Estimating the stoichiometry of human immunodeficiency virus entry. J Virol. 2009;83:1523–1531. doi: 10.1128/JVI.01764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magnus C, Regoes RR. Analysis of the subunit stoichiometries in viral entry. PLoS One. 2012;7:e33441. doi: 10.1371/journal.pone.0033441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 33.Harris A, et al. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci USA. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 35.Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 36.Sauter NK, et al. Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: a 500-MHz proton nuclear magnetic resonance study. Biochemistry. 1989;28:8388–8396. doi: 10.1021/bi00447a018. [DOI] [PubMed] [Google Scholar]
- 37.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 38.Air GM. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc Natl Acad Sci USA. 1981;78:7639–7643. doi: 10.1073/pnas.78.12.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 40.Salzberg S. The contents of the syringe. Nature. 2008;454:160–161. doi: 10.1038/454160a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Centers for Disease Control and Prevention (CDC) Interim within-season estimate of the effectiveness of trivalent inactivated influenza vaccine--Marshfield, Wisconsin, 2007–08 influenza season. MMWR Morb Mortal Wkly Rep. 2008;57:393–398. [PubMed] [Google Scholar]
- 42.Parvin JD, Moscona A, Pan WT, Leider JM, Palese P. Measurement of the mutation rates of animal viruses: influenza A virus and poliovirus type 1. J Virol. 1986;59:377–383. doi: 10.1128/jvi.59.2.377-383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korber B, et al. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 44.Gaschen B, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 45.Karlsson Hedestam GB, Fouchier RA, Phogat S, Burton DR, Sodroski J, Wyatt RT. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat Rev Microbiol. 2008;6:143–155. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- 46.Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull. 2001;58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- 47.Back NK, et al. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology. 1994;199:431–438. doi: 10.1006/viro.1994.1141. [DOI] [PubMed] [Google Scholar]
- 48.Kolchinsky P, Kiprilov E, Sodroski J. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J Virol. 2001;75:2041–2050. doi: 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCaffrey RA, Saunders C, Hensel M, Stamatatos L. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J Virol. 2004;78:3279–3295. doi: 10.1128/JVI.78.7.3279-3295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang SM, et al. Modified HIV envelope proteins with enhanced binding to neutralizing monoclonal antibodies. Virology. 2005;331:20–32. doi: 10.1016/j.virol.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, et al. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J Virol. 2008;82:638–651. doi: 10.1128/JVI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang CC, et al. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc Natl Acad Sci USA. 2009;106:18137–18142. doi: 10.1073/pnas.0909696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Binley JM, et al. Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. J Virol. 2010;84:5637–5655. doi: 10.1128/JVI.00105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wanzeck K, Boyd KL, McCullers JA. Glycan shielding of the influenza virus hemagglutinin contributes to immunopathology in mice. Am J Respir Crit Care Med. 2011;183:767–773. doi: 10.1164/rccm.201007-1184OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geyer H, Holschbach C, Hunsmann G, Schneider J. Carbohydrates of human immunodeficiency virus. Structures of oligosaccharides linked to the envelope glycoprotein 120. J Biol Chem. 1988;263:11760–11767. [PubMed] [Google Scholar]
- 56.Hansen JE, Clausen H, Hu SL, Nielsen JO, Olofsson S. An O-linked carbohydrate neutralization epitope of HIV-1 gp 120 is expressed by HIV-1 env gene recombinant vaccinia virus. Arch Virol. 1992;126:11–20. doi: 10.1007/BF01309680. [DOI] [PubMed] [Google Scholar]
- 57.Bernstein HB, Tucker SP, Hunter E, Schutzbach JS, Compans RW. Human immunodeficiency virus type 1 envelope glycoprotein is modified by O-linked oligosaccharides. J Virol. 1994;68:463–468. doi: 10.1128/jvi.68.1.463-468.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doores KJ, et al. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci USA. 2010;107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Igarashi M, Ito K, Kida H, Takada A. Genetically destined potentials for N-linked glycosylation of influenza virus hemagglutinin. Virology. 2008;376:323–329. doi: 10.1016/j.virol.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 60.Das SR, Puigbo P, Hensley SE, Hurt DE, Bennink JR, Yewdell JW. Glycosylation focuses sequence variation in the influenza A virus H1 hemagglutinin globular domain. PLoS Pathog. 2010;6:e1001211. doi: 10.1371/journal.ppat.1001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 62.Wilson IA, Ladner RC, Skehel JJ, Wiley DC. The structure and role of the carbohydrate moieties of influenza virus haemagglutinin. Biochem Soc Trans. 1983;11:145–147. [PubMed] [Google Scholar]
- 63.Wei CJ, et al. Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci Transl Med. 2010;2:24ra21. doi: 10.1126/scitranslmed.3000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Jr, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Go EP, et al. Glycosylation site-specific analysis of clade C HIV-1 envelope proteins. J Proteome Res. 2009;8:4231–4242. doi: 10.1021/pr9002728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Go EP, et al. Characterization of glycosylation profiles of HIV-1 transmitted/founder envelopes by mass spectrometry. J Virol. 2011;85:8270–8284. doi: 10.1128/JVI.05053-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonomelli C, et al. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS One. 2011;6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wyatt R, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 69.Johnson WE, Desrosiers RC. Viral persistance: HIV's strategies of immune system evasion. Annu Rev Med. 2002;53:499–518. doi: 10.1146/annurev.med.53.082901.104053. [DOI] [PubMed] [Google Scholar]
- 70.Labrijn AF, et al. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol. 2003;77:10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang CC, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moore PL, et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanchez-Martinez S, Lorizate M, Katinger H, Kunert R, Nieva JL. Membrane association and epitope recognition by HIV-1 neutralizing anti-gp41 2F5 and 4E10 antibodies. AIDS Res Hum Retroviruses. 2006;22:998–1006. doi: 10.1089/aid.2006.22.998. [DOI] [PubMed] [Google Scholar]
- 74.Moore PL, et al. The c3-v4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J Virol. 2008;82:1860–1869. doi: 10.1128/JVI.02187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Willey S, Aasa-Chapman MM. Humoral immunity to HIV-1: neutralisation and antibody effector functions. Trends Microbiol. 2008;16:596–604. doi: 10.1016/j.tim.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 76.Conley AJ, et al. The consequence of passive administration of an anti-human immunodeficiency virus type 1 neutralizing monoclonal antibody before challenge of chimpanzees with a primary virus isolate. J Virol. 1996;70:6751–6758. doi: 10.1128/jvi.70.10.6751-6758.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mascola JR. Defining the protective antibody response for HIV-1. Curr Mol Med. 2003;3:209–216. doi: 10.2174/1566524033479799. [DOI] [PubMed] [Google Scholar]
- 78.Hessell AJ, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hessell AJ, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hessell AJ, et al. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J Virol. 2009;84:1302–1313. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doria-Rose NA, et al. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol. 2009;83:188–199. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sather DN, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009;83:757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simek MD, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 85.Barbas CF, 3rd, et al. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc Natl Acad Sci U S A. 1992;89:9339–9343. doi: 10.1073/pnas.89.19.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barbas CF, 3rd, et al. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J Mol Biol. 1993;230:812–823. doi: 10.1006/jmbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- 87.Buchacher A, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 88.Calarese DA, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 89.Ofek G, et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78:10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cardoso RM, et al. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22:163–173. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 91.Zhou T, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Julien JP, Bryson S, Nieva JL, Pai EF. Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site. J Mol Biol. 2008;384:377–392. doi: 10.1016/j.jmb.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 93.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu X, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mouquet H, et al. Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses. PLoS One. 2011;6:e24078. doi: 10.1371/journal.pone.0024078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kwong PD, Mascola JR, Nabel GJ. Mining the B cell repertoire for broadly neutralizing monoclonal antibodies to HIV-1. Cell Host Microbe. 2009;6:292–294. doi: 10.1016/j.chom.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Clapham PR, Lu S. Vaccinology: precisely tuned antibodies nab HIV. Nature. 2011;477:416–417. doi: 10.1038/477416a. [DOI] [PubMed] [Google Scholar]
- 100.Overbaugh J, Morris L. The Antibody Response against HIV-1. Cold Spring Harb Perspect Med. 2012;2:a007039. doi: 10.1101/cshperspect.a007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huse WD, et al. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science. 1989;246:1275–1281. doi: 10.1126/science.2531466. [DOI] [PubMed] [Google Scholar]
- 103.Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ryu SE, Hendrickson WA. Structure and design of broadly-neutralizing antibodies against HIV. Mol Cells. 2012 doi: 10.1007/s10059-012-0104-4. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen L, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Diskin R, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nelson JD, et al. An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10. J Virol. 2007;81:4033–4043. doi: 10.1128/JVI.02588-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Montero M, van Houten NE, Wang X, Scott JK. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol Mol Biol Rev. 2008;72:54–84. doi: 10.1128/MMBR.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gach JS, Leaman DP, Zwick MB. Targeting HIV-1 gp41 in close proximity to the membrane using antibody and other molecules. Curr Top Med Chem. 2011;11:2997–3021. doi: 10.2174/156802611798808505. [DOI] [PubMed] [Google Scholar]
- 111.Bryson S, Julien JP, Hynes RC, Pai EF. Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications. J Virol. 2009;83:11862–11875. doi: 10.1128/JVI.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Salzwedel K, West JT, Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J Virol. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bellamy-McIntyre AK, et al. Functional links between the fusion peptide-proximal polar segment and membrane-proximal region of human immunodeficiency virus gp41 in distinct phases of membrane fusion. J Biol Chem. 2007;282:23104–23116. doi: 10.1074/jbc.M703485200. [DOI] [PubMed] [Google Scholar]
- 114.Poumbourios P, el Ahmar W, McPhee DA, Kemp BE. Determinants of human immunodeficiency virus type 1 envelope glycoprotein oligomeric structure. J Virol. 1995;69:1209–1218. doi: 10.1128/jvi.69.2.1209-1218.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Suarez T, Gallaher WR, Agirre A, Goni FM, Nieva JL. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J Virol. 2000;74:8038–8047. doi: 10.1128/jvi.74.17.8038-8047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Suarez T, Nir S, Goni FM, Saez-Cirion A, Nieva JL. The pre-transmembrane region of the human immunodeficiency virus type-1 glycoprotein: a novel fusogenic sequence. FEBS Lett. 2000;477:145–149. doi: 10.1016/s0014-5793(00)01785-3. [DOI] [PubMed] [Google Scholar]
- 117.Saez-Cirion A, et al. Sphingomyelin and cholesterol promote HIV-1 gp41 pretransmembrane sequence surface aggregation and membrane restructuring. J Biol Chem. 2002;277:21776–21785. doi: 10.1074/jbc.M202255200. [DOI] [PubMed] [Google Scholar]
- 118.Alfsen A, Iniguez P, Bouguyon E, Bomsel M. Secretory IgA specific for a conserved epitope on gp41 envelope glycoprotein inhibits epithelial transcytosis of HIV-1. J Immunol. 2001;166:6257–6265. doi: 10.4049/jimmunol.166.10.6257. [DOI] [PubMed] [Google Scholar]
- 119.Alfsen A, Bomsel M. HIV-1 gp41 envelope residues 650–685 exposed on native virus act as a lectin to bind epithelial cell galactosyl ceramide. J Biol Chem. 2002;277:25649–25659. doi: 10.1074/jbc.M200554200. [DOI] [PubMed] [Google Scholar]
- 120.Yu H, Alfsen A, Tudor D, Bomsel M. The binding of HIV-1 gp41 membrane proximal domain to its mucosal receptor, galactosyl ceramide, is structure-dependent. Cell Calcium. 2008;43:73–82. doi: 10.1016/j.ceca.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 121.Schibli DJ, Montelaro RC, Vogel HJ. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry. 2001;40:9570–9578. doi: 10.1021/bi010640u. [DOI] [PubMed] [Google Scholar]
- 122.Sun ZY, et al. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28:52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 123.Pejchal R, et al. A conformational switch in human immunodeficiency virus gp41 revealed by the structures of overlapping epitopes recognized by neutralizing antibodies. J Virol. 2009;83:8451–8462. doi: 10.1128/JVI.00685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Song L, et al. Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc Natl Acad Sci USA. 2009;106:9057–9062. doi: 10.1073/pnas.0901474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim M, et al. Antibody mechanics on a membrane-bound HIV segment essential for GP41-targeted viral neutralization. Nat Struct Mol Biol. 2011;18:1235–1243. doi: 10.1038/nsmb.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Walker LM, et al. Rapid development of glycan-specific, broad, and potent anti-HIV-1 gp120 neutralizing antibodies in an R5 SIV/HIV chimeric virus infected macaque. Proc Natl Acad Sci USA. 2011;108:20125–20129. doi: 10.1073/pnas.1117531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lavine CL, Lao S, Montefiori DC, Haynes BF, Sodroski JG, Yang X. High-mannose glycan-dependent epitopes are frequently targeted in broad neutralizing antibody responses during human immunodeficiency virus type 1 infection. J Virol. 2012;86:2153–2164. doi: 10.1128/JVI.06201-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- 129.Eggink D, et al. Lack of complex N-glycans on HIV-1 envelope glycoproteins preserves protein conformation and entry function. Virology. 2010;401:236–247. doi: 10.1016/j.virol.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Scanlan CN, et al. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Walker BD, et al. Inhibition of human immunodeficiency virus syncytium formation and virus replication by castanospermine. Proc Natl Acad Sci U S A. 1987;84:8120–8124. doi: 10.1073/pnas.84.22.8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Geijtenbeek TB, et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 133.Bonsignori M, et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J Virol. 2012;86:4688–4692. doi: 10.1128/JVI.07163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Walker LM, Bowley DR, Burton DR. Efficient recovery of high-affinity antibodies from a single-chain Fab yeast display library. J Mol Biol. 2009;389:365–375. doi: 10.1016/j.jmb.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pejchal R, et al. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci USA. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pancera M, et al. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J Virol. 2010;84:8098–8110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 139.Whittle JR, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci USA. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ekiert DC, et al. Neutralization of influenza A viruses by insertion of a single antibody loop into the receptor binding site. Nature. 2012 doi: 10.1038/nature11414. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liao HX, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ekiert DC, Wilson IA. Broadly neutralizing antibodies against influenza virus and prospects for universal therapies. Curr Opin Virol. 2012;2:134–141. doi: 10.1016/j.coviro.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kashyap AK, et al. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci USA. 2008;105:5986–5991. doi: 10.1073/pnas.0801367105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Corti D, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wei CJ, et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science. 2010;329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 150.Ekiert DC, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 152.Dreyfus C, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012 doi: 10.1126/science.1222908. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kwong PD, Wilson IA. HIV-1 and influenza antibodies: seeing antigens in new ways. Nat Immunol. 2009;10:573–578. doi: 10.1038/ni.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Barbey-Martin C, et al. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology. 2002;294:70–74. doi: 10.1006/viro.2001.1320. [DOI] [PubMed] [Google Scholar]
- 155.Klein JS, Gnanapragasam PN, Galimidi RP, Foglesong CP, West AP, Jr, Bjorkman PJ. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc Natl Acad Sci USA. 2009;106:7385–7390. doi: 10.1073/pnas.0811427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mouquet H, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ward CW, Gleeson PA, Dopheide TA. Carbohydrate composition of the oligosaccharide units of the haemagglutinin from the Hong Kong influenza virus A/Memphis/102/72. Biochem J. 1980;189:649–652. doi: 10.1042/bj1890649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mir-Shekari SY, Ashford DA, Harvey DJ, Dwek RA, Schulze IT. The glycosylation of the influenza A virus hemagglutinin by mammalian cells. A site-specific study. J Biol Chem. 1997;272:4027–4036. doi: 10.1074/jbc.272.7.4027. [DOI] [PubMed] [Google Scholar]
- 159.Phogat S, Wyatt R. Rational modifications of HIV-1 envelope glycoproteins for immunogen design. Curr Pharm Des. 2007;13:213–227. doi: 10.2174/138161207779313632. [DOI] [PubMed] [Google Scholar]
- 160.Walker LM, Burton DR. Rational antibody-based HIV-1 vaccine design: current approaches and future directions. Curr Opin Immunol. 2010;22:358–366. doi: 10.1016/j.coi.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dey B, et al. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 2009;5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bhattacharyya S, et al. Design of a non-glycosylated outer domain-derived HIV-1 gp120 immunogen that binds to CD4 and induces neutralizing antibodies. J Biol Chem. 2010;285:27100–27110. doi: 10.1074/jbc.M110.152272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Azoitei ML, et al. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science. 2011;334:373–376. doi: 10.1126/science.1209368. [DOI] [PubMed] [Google Scholar]
- 164.Kumar R, Tuen M, Li H, Tse DB, Hioe CE. Improving immunogenicity of HIV-1 envelope gp120 by glycan removal and immune complex formation. Vaccine. 2011;29:9064–9074. doi: 10.1016/j.vaccine.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ahmed FK, Clark BE, Burton DR, Pantophlet R. An engineered mutant of HIV-1 gp120 formulated with adjuvant Quil A promotes elicitation of antibody responses overlapping the CD4-binding site. Vaccine. 2012;30:922–930. doi: 10.1016/j.vaccine.2011.11.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Depetris RS, et al. Partial Enzymatic Deglycosylation Preserves the Structure of Cleaved Recombinant HIV-1 Envelope Glycoprotein Trimers. J Biol Chem. 2012;287:24239–24254. doi: 10.1074/jbc.M112.371898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ofek G, et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci USA. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stanfield RL, Julien JP, Pejchal R, Gach JS, Zwick MB, Wilson IA. Structure-based design of a protein immunogen that displays an HIV-1 gp41 neutralizing epitope. J Mol Biol. 2011;414:460–476. doi: 10.1016/j.jmb.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Azoitei ML, et al. Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope. J Mol Biol. 2012;415:175–192. doi: 10.1016/j.jmb.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Alam SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ofek G, et al. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J Virol. 2010;84:2955–2962. doi: 10.1128/JVI.02257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Xu H, et al. Interactions between lipids and human anti-HIV antibody 4E10 can be reduced without ablating neutralizing activity. J Virol. 2010;84:1076–1088. doi: 10.1128/JVI.02113-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Julien JP, et al. Ablation of the complementarity-determining region H3 apex of the anti-HIV-1 broadly neutralizing antibody 2F5 abrogates neutralizing capacity without affecting core epitope binding. J Virol. 2010;84:4136–4147. doi: 10.1128/JVI.02357-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Guenaga J, Wyatt RT. Structure-guided Alterations of the gp41-directed HIV-1 Broadly Neutralizing Antibody 2F5 Reveal New Properties Regarding its Neutralizing Function. PLoS Pathog. 2012;8:e1002806. doi: 10.1371/journal.ppat.1002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Luallen RJ, et al. An engineered Saccharomyces cerevisiae strain binds the broadly neutralizing human immunodeficiency virus type 1 antibody 2G12 and elicits mannose-specific gp120-binding antibodies. J Virol. 2008;82:6447–6457. doi: 10.1128/JVI.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Astronomo RD, et al. A glycoconjugate antigen based on the recognition motif of a broadly neutralizing human immunodeficiency virus antibody, 2G12, is immunogenic but elicits antibodies unable to bind to the self glycans of gp120. J Virol. 2008;82:6359–6368. doi: 10.1128/JVI.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dunlop DC, et al. Antigenic mimicry of the HIV envelope by AIDS-associated pathogens. AIDS. 2008;22:2214–2217. doi: 10.1097/QAD.0b013e328314b5df. [DOI] [PubMed] [Google Scholar]
- 178.Luallen RJ, et al. A yeast glycoprotein shows high-affinity binding to the broadly neutralizing human immunodeficiency virus antibody 2G12 and inhibits gp120 interactions with 2G12 and DC-SIGN. J Virol. 2009;83:4861–4870. doi: 10.1128/JVI.02537-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dunlop DC, et al. Polysaccharide mimicry of the epitope of the broadly neutralizing anti-HIV antibody, 2G12, induces enhanced antibody responses to self oligomannose glycans. Glycobiology. 2010;20:812–823. doi: 10.1093/glycob/cwq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Doores KJ, et al. A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity. Proc Natl Acad Sci USA. 2010;107:17107–17112. doi: 10.1073/pnas.1002717107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Huber M, et al. Very few substitutions in a germ line antibody are required to initiate significant domain exchange. J Virol. 2010;84:10700–10707. doi: 10.1128/JVI.01111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chen W, Streaker ED, Russ DE, Feng Y, Prabakaran P, Dimitrov DS. Characterization of germline antibody libraries from human umbilical cord blood and selection of monoclonal antibodies to viral envelope glycoproteins: Implications for mechanisms of immune evasion and design of vaccine immunogens. Biochem Biophys Res Commun. 2012;417:1164–1169. doi: 10.1016/j.bbrc.2011.12.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.West AP, Jr, Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci USA. 2012;109:E2083–E2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang TT, et al. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci USA. 2010;107:18979–18984. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Steel J, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio. 2010;1:e000018–e000010. doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pica N, et al. Hemagglutinin stalk antibodies elicited by the 2009 pandemic influenza virus as a mechanism for the extinction of seasonal H1N1 viruses. Proc Natl Acad Sci USA. 2012;109:2573–2578. doi: 10.1073/pnas.1200039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hai R, et al. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J Virol. 2012;86:5774–5781. doi: 10.1128/JVI.00137-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fleishman SJ, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Whitehead TA, et al. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol. 2012;30:543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE., Jr A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol. 2011;85:10905–10908. doi: 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Krause JC, et al. Epitope-specific human influenza antibody repertoires diversify by B cell intraclonal sequence divergence and interclonal convergence. J Immunol. 2011;187:3704–3711. doi: 10.4049/jimmunol.1101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Krause JC, et al. Human monoclonal antibodies to pandemic 1957 H2N2 and pandemic 1968 H3N2 influenza viruses. J Virol. 2012;86:6334–6340. doi: 10.1128/JVI.07158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Okada J, et al. Monoclonal antibodies in man that neutralized H3N2 influenza viruses were classified into three groups with distinct strain specificity: 1968–1973, 1977–1993 and 1997–2003. Virology. 2010;397:322–330. doi: 10.1016/j.virol.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 195.Okada J, et al. Localization of epitopes recognized by monoclonal antibodies that neutralized the H3N2 influenza viruses in man. J Gen Virol. 2011;92:326–335. doi: 10.1099/vir.0.026419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ohshima N, Iba Y, Kubota-Koketsu R, Asano Y, Okuno Y, Kurosawa Y. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol. 2011;85:11048–11057. doi: 10.1128/JVI.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yoshida R, et al. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 2009;5:e1000350. doi: 10.1371/journal.ppat.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 199.Sheriff S, Hendrickson WA, Smith JL. Structure of myohemerythrin in the azidomet state at 1.7/1.3 Å resolution. J Mol Biol. 1987;197:273–296. doi: 10.1016/0022-2836(87)90124-0. [DOI] [PubMed] [Google Scholar]