In this paper, the authors explore the possibility that the base-pairing strength of miRNA–mRNA target plays a role in microRNA-mediated gene regulation. The authors found a significant correlation between the physiological temperature of the organism and the average G/C content of its miRNAs. This study suggests that an organism adapts its miRNA–target free energy according to its physiological temperature, thus highlighting the importance of base-pairing strength in miRNA activity.
Keywords: base-pairing strength, G/C content, homeotherms, microRNA, poikilotherms
Abstract
MicroRNAs (miRNAs) are short, single-stranded RNAs that silence gene expression by either degrading mRNA or repressing translation. Each miRNA regulates a specific set of mRNA “targets” by binding to complementary sequences in their 3′ untranslated region. In this study, we examined the importance of the base-pairing strength of the miRNA–target duplex to repression. We hypothesized that if base-pairing strength affects the functionality of miRNA repression, organisms with higher body temperature or that live at higher temperatures will have miRNAs with higher G/C content so that the miRNA–target complex will remain stable. In the nine model organisms examined, we found a significant correlation between the average G/C content of miRNAs and physiological temperature, supporting our hypothesis. Next, for each organism examined, we compared the average G/C content of miRNAs that are conserved among distant organisms and that of miRNAs that are evolutionarily recent. We found that the average G/C content of ancient miRNAs is lower than recent miRNAs in homeotherms, whereas the trend was inversed in poikilotherms, suggesting that G/C content is associated with temperature, thus further supporting our hypothesis. In the organisms examined, the average G/C content of miRNA “seed” sequences was higher than that of mature miRNAs, which was higher than pre-miRNA loops, suggesting an association between the degree of functionality of the sequence and its average G/C content. Our analyses show a possible association between the base-pairing strength of miRNA–targets and the temperature of an organism, suggesting that base-pairing strength plays a role in repression by miRNAs.
INTRODUCTION
In recent years, post-transcriptional regulation by microRNAs (miRNAs) has emerged as an important mechanism for silencing gene expression in both plants and metazoans. miRNAs are single-stranded noncoding RNA molecules of ∼22 nt that repress the translation of genes by binding to complementary sequences in the 3′ untranslated region (3′ UTR) of messenger RNAs (mRNAs) (Lagos-Quintana et al. 2001; Lau et al. 2001). miRNAs reduce the protein level of their target genes either by inhibiting their translation or by inducing degradation of their mRNA transcripts (Reinhart et al. 2000; Slack et al. 2000; Llave et al. 2002; Krutzfeldt et al. 2005; Filipowicz et al. 2008).
The mature miRNA is produced after several modifications in the nucleus and cytoplasm. In the nucleus, the miRNA precursor molecule (pri-miRNA) undergoes an initial processing that produces an ∼70-bp hairpin structured dsRNA (pre-miRNA) (Lee et al. 2002, 2003). The pre-miRNA is then exported to the cytoplasm (Yi et al. 2003), where it is processed by Dicer, which removes the loop to leave an ∼22-bp dsRNA (Bernstein et al. 2001; Hutvagner et al. 2001; Knight and Bass 2001; Cullen 2004; Lee et al. 2004). Next, Dicer, along with transactivating response (TAR) RNA-binding protein (TRBP), separates the dsRNA into two single strands. The mature miRNA strand, called the guide, remains bound to Dicer and TRBP. Argonaute 2 (Ago2) is then recruited to induce the assembly of an active RNA-induced silencing complex (RISC) (Chendrimada et al. 2005). Finally, the miRNA directs RISC to bind to the complementary region on the 3′ UTR of its target gene and induce its down-regulation (Hammond et al. 2000, 2001; Mourelatos et al. 2002; Sontheimer and Carthew 2005).
In metazoans, miRNA–target interaction is influenced by several factors. Most importantly, the “seed” region (positions 2–8) of the miRNA base-pairs to binding sites on the 3′ UTR of the target, not allowing for any mismatches (Fig. 1A; Doench and Sharp 2004; Brennecke et al. 2005; Lewis et al. 2005; Nielsen et al. 2007). Although perfect seed base-pairing is considered a prerequisite for repression (Lai 2002; Brennecke et al. 2005; Krek et al. 2005; Lewis et al. 2005), some rare exceptions exist (Didiano and Hobert 2006). When the seed–target interaction is imperfect, the 3′ end of the miRNA can also base-pair to the target gene (Doench and Sharp 2004; Brennecke et al. 2005; Grimson et al. 2007). When both the 5′ and 3′ ends of the miRNA base-pair to the target, the mismatched nucleotides in the central region of the miRNA bulge out (Grimson et al. 2007). The miRNA binding sites on the 3′ UTR of the targets also influence miRNA–target interaction. These binding sites are usually located near either end of the target gene's 3′ UTR and reside within an A/U-rich context (Lewis et al. 2005; Ameres et al. 2007; Grimson et al. 2007; Nielsen et al. 2007). It has been suggested these A/U-rich sequences of the target form weak secondary structures (Grimson et al. 2007) that increase the efficiency of target cleavage (Ameres et al. 2007).
FIGURE 1.
miRNA G/C content correlates to the physiological temperature of an organism. (A) A scheme of the different regions of miRNA (modified from Filipowicz et al. 2008, with permission of Nature Publishing Group, © 2008). (B) The average G/C content of miRNA sequences, their seeds, the pre-miRNA loops, and the pre-miRNA flanking regions were calculated for nine representative model organisms and plotted against the physiological temperature of each organism. (Khaki triangles) miRNAs; (light green triangles) seeds; (dark green triangles) loops; (brown shadowed squares) the control flanking regions. The regression lines and their correlation coefficients for each of the subsets of values are shown in the corresponding color. The organisms' labels are shown above the arrowheads that point to the organism's physiological temperature: (cel) Caenorhabditis elegans (nematode); (dme) Drosophila melanogaster (fly); (bmo) Bombyx mori (silkmoth); (dre) Danio rerio (zebrafish); (oan) Ornithorhynchus anatinus (platypus); (hsa) Homo sapiens (human); (mmu) Mus musculus (mouse); (bta) Bos taurus (cow); (gga) Gallus gallus (chicken). For organisms that have the same physiological temperature, we added next to the data point the first letter of the organism in a color that corresponds to the data that it represents (e.g., d in light green stands for D. melanogaster seeds, d in khaki for miRNAs, etc.). (C) For each of the organisms, we divided its miRNAs into two subsets that contain (1) miRNAs that are specific to a taxonomic group (e.g., Drosophila insect-specific miRNAs) and (2) the rest of the miRNAs that are shared by other taxonomic groups (e.g., Drosophila miRNAs that are not insect-specific). Each pair of bars shows the difference in G/C content between these two subsets for miRNAs (black) and for seeds (dark gray) ± standard error of the difference between two means. The label under each bar pair describes the organism and miRNAs classifiers (i.e., the taxonomic group according to which we sorted the organism miRNAs into two subsets). (*) p < 0.05; (**) p < 0.0005 as indicated. (D) The average G/C level of the first 10 positions of miRNAs sequence in nine model organisms: (cel) C. elegans (nematode); (dme) D. melanogaster (fly); (bmo) B. mori (silkmoth); (dre) D. rerio (zebrafish); (oan) O. anatinus (platypus); (hsa) H. sapiens (human); (mmu) M. musculus (mouse); (bta) B. taurus (cow); (gga) G. gallus (chicken).
Extensive studies on the role of miRNA–target base-pairing in repression by miRNAs has shown that miRNA–target base-pairing is crucial for miRNA specificity (Doench and Sharp 2004; Brennecke et al. 2005). While base-pairing strength is one of the key factors that affects the stability of a dsRNA duplex (Freier et al. 1986; Nicholson 1996), the contribution of the base-pairing strength to miRNA–target duplex stability is still unclear. Several recent studies have indicated that base-pairing strength is important in miRNA repression. In Arabidopsis, stress-responsive miRNAs were found to be G/C rich (Mishra et al. 2009). In mouse embryonic fibroblast cells (MEF) in which Ago2 has been knocked out, removal of the unselected miRNA strand during pre-miRNA processing is greatly influenced by the inherent thermodynamic stability of the pre-miRNA hairpin (Gu et al. 2011). Another factor that is involved in stabilizing the dsRNA duplex is the concentration of each of the ssRNAs that comprise the dsRNA (Freier et al. 1986; Nicholson 1996). Lastly, the protein components of the silencing complex are also likely to play a role in stabilizing the miRNA–target duplex.
We hypothesized that if base-pairing strength affects the functionality of miRNA repression, organisms with higher body temperature or that live in higher temperatures will have miRNAs with higher G/C content so that the miRNA–target complex will remain stable. In the nine model organisms examined, we found a significant correlation between the average G/C content of miRNAs and physiological temperature, supporting our hypothesis. Next, for each organism examined, we calculated the difference between the average G/C content of miRNAs that are conserved among distant organisms and miRNAs that are evolutionarily recent. We found that this difference is positive for homeotherms and negative for poikiloterms. This opposite directionality suggests that the G/C content is associated with temperature, which further supports our hypothesis. Thus, our results identify variability in miRNA G/C content among different organisms, which as a consequence also differs in the stability of binding association of miRNAs to their conserved targets. We show a possible association between miRNA G/C content and the temperature of an organism, suggesting that base-pairing strength plays a role in miRNA–target duplex stability.
RESULTS
The G/C content of miRNAs correlates to the organism's physiological temperature
miRNAs have been documented among diverse organisms that maintain or live in a wide range of temperatures (Bartel 2004; Sempere et al. 2006; Niwa and Slack 2007). Considering that the stability of dsRNA is affected by temperature, we examined whether base-pairing strength plays a role in miRNA–target duplex stabilization under different temperatures. For this study, we defined the term “physiological temperature” as constant body temperature for homeotherms. For poikilotherms, “physiological temperature” is the optimal temperature in which the embryo develops into a healthy adult in the shortest period of time (see Materials and Methods).
Next, using G/C content as an indicator for base-pairing strength, we calculated the average G/C content of miRNAs and of their seed sequences for nine model organisms with different physiological temperatures (five homeotherms: chicken, platypus, cow, human, and mouse; and four poikilotherms: fish, fly, silkmoth, and worm). For each organism, we plotted the average G/C content of miRNA and seed sequences against its physiological temperature (Fig. 1B). As a control, we calculated the average G/C content of the organism's pre-miRNA genomic context using the 50 nt adjacent to each of the pre-miRNA's ends (hereafter flanking regions).
We found a high significant correlation between the G/C content of miRNAs and the organism's physiological temperature (R2 = 0.944, p < 1.2 × 10−5). In contrast, G/C content in pre-miRNA flanking regions and physiological temperature showed a weaker correlation (R2 = 0.67, p < 0.022). Furthermore, in some organisms (e.g., cow, platypus), the average G/C content of miRNAs is higher than that of the pre-miRNA flanking regions, whereas in other organisms (e.g., fly, worm, chicken), the average G/C content of miRNAs is lower, implying that the G/C content of miRNAs and their genomic context are only weakly associated (R2 = 0.633, p < 0.01). The correlation between miRNA G/C content and temperature suggests that an organism's physiological temperature affects its miRNA G/C content. For example, miRNAs with the highest G/C content (0.515) were found in Gallus gallus (chicken), which maintains a constant temperature of 41.5°C; and miRNAs with the lowest G/C content (0.439) were found in Bombyx mori (silkmoth), whose embryos develop optimally at 26°C (Fig. 1B; Supplemental Table 1).
The G/C content of seed sequences and physiological temperature also showed a significant correlation (R2 = 0.878, p < 1.99 × 10−4) ranging from 0.548 in chicken to 0.477 in Drosophila (Fig. 1B; Supplemental Table 1). Interestingly, the G/C content of chicken seeds is significantly 5% higher than that of Homo sapiens (human) miRNAs (p < 0.003) and 8.1% higher than that of Mus musculus (mouse; p < 1.16 × 10−5) (Fig. 1B). As in miRNAs, the G/C content of seeds and their pre-miRNA flanking regions is weakly correlated (R2 = 0.45, p < 0.04) (Fig. 1B). Furthermore, our results indicate that the average G/C content is higher in the seed sequences than in the miRNA sequences in all of the organisms examined (Fig. 1B; Supplemental Table 1). Since seed sequences play an essential role in miRNA–target interaction, a higher G/C content in seeds further supports a possible association between G/C content and miRNA repression.
Next, we examined whether the association between G/C content and physiological temperature occurs in parts of the pre-miRNA sequences that are not involved in miRNA–target interaction. For that, we calculated the average G/C content of the pre-miRNA hairpin loops that are cleaved by Dicer before repression (Zhang et al. 2004). We found a significant correlation between the pre-miRNA loops and the organism's physiological temperature (R2 = 0.693, p < 0.005), but it was lower than the correlation that was found for the miRNAs and seeds. Interestingly, the average G/C content of pre-miRNA loops is lower than that of miRNAs in all organisms examined.
Taken together, our results highlight the importance of miRNA–target base-pairing strength for miRNA activity. Based on these findings, we suggest a hierarchy in the average G/C content of an organism in which the G/C content of seeds is higher than that of miRNAs, which is higher than that of pre-miRNA loops.
A correlation between the G/C content and the organism's temperature was first suggested by Bernardi et al. (1985). They found that genomes are composed of large regions with different G/C content, which he termed “isochors” (Cuny et al. 1981). At first, it was suggested that a G/C-rich isochore structure is an adaptation to homeothermy since it was found only in mammals and birds but not in amphibians and fish. However, the discovery of similar G/C-rich structures among crocodiles and turtles led to the rejection of this hypothesis (Hughes et al. 1999). Our results here suggest that an organism's physiological temperature does influence the G/C content of its miRNAs; miRNAs differ from isochors in many characters such as size, functionality, and formation dynamic. While miRNA sequences are short and highly conserved after acquiring a sufficient number of target genes (Sempere et al. 2006; Shomron et al. 2009), isochors are large genomic regions that may change their G/C content with time (Romiguier et al. 2010). Additionally, in our study, we used the criterion of “physiological temperature,” which reflects the organism's body temperature, rather than the homeothermity or poikilothermity used by Bernardi et al. (1985).
Newly generated miRNAs have higher G/C content in homeothermic organisms and lower G/C content in poikilothermic organisms
A point mutation in an active miRNA sequence might cause two deleterious effects: It can alter the miRNA secondary structure and damage its biosynthesis, and it can change the target repertoire of the miRNA, causing a disruption in the genetic program of the organism (Shomron et al. 2009). These two effects might be the reason that a miRNA sequence is conserved almost without change once it gains a sufficient number of targets (Sempere et al. 2006; Shomron et al. 2009). Thus, considering the possible association between temperature and G/C content shown above (Fig. 1B), we predict that when an organism adapts to a new temperature, the G/C content of miRNAs generated under the new temperature regime will differ from ancestral miRNAs that were already active before the temperature change. To examine this prediction, we compared the average G/C content of “new” miRNAs, miRNAs that are specific to the organism close taxonomic group (e.g., the insect-specific miRNAs of Drosophila), and the G/C content of “old” miRNAs, miRNAs that are conserved in other taxonomic groups (e.g., the Drosophila miRNAs that are not insect-specific).
Among poikilothermic organisms, the newly generated miRNAs have lower average G/C content compared with the G/C content of “old” miRNAs (Fig. 1C). For B. mori (silkmoth), the G/C content of insect-specific miRNAs is 0.03 lower than non-insect-specific miRNAs (6.5%, p < 0.007). Similarly, for seed sequences, the difference in G/C content between insect-specific and non-insect-specific is 0.04 (8.3%, p < 0.03). The trend is the opposite in homeotherms; newly generated miRNAs have higher G/C content compared with the G/C content of “old” miRNAs. The most prominent difference in G/C content is found in the chicken (G. gallus); the average G/C content of bird-specific miRNAs is 0.54 (n = 349), 19.6% higher than the non-bird-specific ones (0.45, n = 142; p < 1.02 × 10−13). In chicken seed sequences, the difference in G/C content between bird-specific and non-bird-specific is 0.08 (16%, p < 8.07 × 10−7). Thus, the average G/C content of the chicken miRNAs and seeds (n = 491) (Fig. 1B) is mostly due to the high G/C content of miRNAs and seeds that are bird-specific.
In a previous study, Shomron et al. (2009) found a correlation between the antiquity of a miRNA and its number of predicted targets in Tetrapoda. They also reported that miRNAs with the highest number of predicted targets have a high A/U content while those with the lowest number of predicted targets have a low A/U content, implying that the age of a miRNA inversely correlates with its G/C content. Shomron et al. (2009) suggested that the mechanism behind this correlation is the biased occurrence of G-to-A, C-to-T substitutions that take place more frequently than the reverse direction. Our results show that among homeothermic organisms, the G/C content of “old” miRNAs is lower than that of the “new” miRNAs, whereas, among poikilotherms, the “old” miRNA G/C content is higher compared with that of the “new” miRNAs. Thus, biased substitutions of G to A, C to T, which is also the directionality found in Drosophila and Caenorhabditis elegans, cannot explain the higher G/C content among the conserved miRNAs found in poikilothermic organisms (Vicario et al. 2007; Denver et al. 2009; Keightley et al. 2009). Moreover, because miRNAs are highly conserved after gaining a critical number of functional targets (Sempere et al. 2006; Shomron et al. 2009), it is suggested that random changes in the miRNA sequences, which are caused by neutral mutations, can take place in a narrow time window in which miRNAs have few targets. A change in the miRNA sequence later will likely be selected against because it might disrupt the genetic network of the organism.
miRNA sequences have different G/C level signatures among the examined organisms
We showed that the extent of average G/C content of miRNAs and seed sequences is different among the examined organisms (Fig. 1B). Next, we examined whether this difference in G/C content is a result of distinct G/C level in specific positions of the miRNA sequence or is uniformly divided along the sequence of the miRNA. For that, we calculated the average G/C level of the first 10 positions in the 5′ ends of the miRNAs (Fig. 1D). We focused on the 5′ end because it contains the seed region, which is well defined (Lewis et al. 2005). We found a lower G/C level in positions 1 and 9 among all of the organisms examined, in accordance with previous studies (Lau et al. 2001; Lewis et al. 2005; Gorodkin et al. 2006; Zhang et al. 2009). These positions, which define the borders of the seed region, do not contribute to targeting in miRNA–target base-pairing (Nielsen et al. 2007). In positions 1 to 2 of the miRNA, the G/C level increases for all organisms, with the highest level observed in the chicken (0.55) and the lowest in fish (0.37). The increase in G/C level continues to position 3 of fish and also of platypus and chicken, which maintain a high G/C level (0.55) unlike the rest of the organisms. Among insects, positions 4–6 have a unique pattern; both in B. mori and D. melanogaster, the G/C level decreases from positions 4 to 5 and increases from positions 5 to 6. Taken together, we found different patterns of G/C level in positions 1–6 of miRNA sequences, which also suggests the existence of the complementary pattern in the miRNA binding sites. Our findings might explain in part the variance in miRNA average G/C content among the different organisms demonstrated above (Fig. 1B).
The free energy of miRNA-predicted targets duplex inversely correlates with the organism's physiological temperature
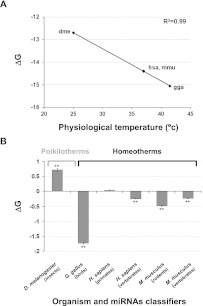
Although G/C content of miRNAs is an indicator for the strength of miRNA–target hybridization, the actual base-pairing is determined by the sequences of both miRNA and its 3′-UTR target binding site. Hence, the free energy (ΔG) value reflects overall the miRNA–target base-pairing strength. To examine the importance of Watson–Crick base-pairing strength in miRNA activity, we tested if organisms with different temperatures have different ΔG of the miRNA–target base-pairing. Since TarBase, the database that contains the proven in vivo miRNA–target interactions, is partial and not representative (Sturm et al. 2010), we used putative miRNA–target couples that were predicted in silico by TargetSpy, a recent algorithm that showed good predictability when compared with experimental data (Sturm et al. 2010). Unlike other predictions, TargetSpy does not rely on species conservation, thus, the targets for each species are predicted independently. We used the miRNA–target prediction of four model organisms available in TargetSpy: human, mouse, chicken, and fly. For each model organism, we have calculated the average ΔG for the miRNA–target base-pairing and plotted these values against the organism's physiological temperature (Fig. 2A). We found a highly significant inverse correlation (R2 = 0.99, p < 8.8 × 10−5) between the temperature and the average ΔG of miRNA–target base-pairing, which is in agreement with the correlation between the organism's physiological temperature and miRNA average G/C content demonstrated above (Fig. 1B). The ΔG of the chicken was the lowest (−15.05 kcal/mol), and that of fly the highest (−12.69 kcal/mol). This correlation between the temperature and the average ΔG of miRNA–target base-pairing suggests that the free energy of the miRNA–target dsRNA is affected by the physiological temperature of the organism, which further highlights the importance of base-pairing strength to miRNA activity.
FIGURE 2.
The free energy (ΔG) of the base-pairings between miRNAs and their predicted targets inversely correlates with the organism's physiological temperature. We used the ΔG values for the base-pairing of miRNAs and their targets predicted from TargetSpy (Sturm et al. 2010). (A) Shown is the average ΔG for the miRNA–target base-pairing of four model organisms available in TargetSpy plotted against the organism's physiological temperature. The regression line and the correlation coefficient are shown. (dme) D. melanogaster (fly); (hsa) H. sapiens (human); (mmu) M. musculus (mouse); (gga) G. gallus (chicken). (B) For each of the organisms, we divided the miRNAs into two subsets that contain (1) miRNAs that are specific to a taxonomic group and (2) the rest of the miRNAs that are shared with other taxonomic groups. Each bar shows the difference between the average ΔG values of the two subsets for the predicted miRNA–target binding sites ± standard error of the difference between two means. The label under each bar describes the organism and miRNA classifiers (i.e., the taxonomic group according to which we sorted the organism miRNAs into two subsets). (**) p < 0.0005 as indicated.
Newly generated miRNAs have lower miRNA–target free energy (ΔG) in homeothermic organisms and higher in poikilothermic organisms
Since we found a difference in the average G/C content of miRNAs that are specific to the species' close taxonomic group compared with miRNAs that are conserved in other taxonomic groups (Fig. 1C), we tested if that is also the case for the ΔG of the miRNA-predicted targets' base-pairing. For each of the four organisms examined (Fig. 2A), we divided the miRNAs into two subsets; the first contains miRNAs that are unique to a taxonomic group (“new”), and the second subset contains the rest of the miRNAs that are conserved in other taxonomic groups (“old”). We then calculated the difference in the average ΔG of miRNA-predicted targets' base-pairing between the two subsets (Fig. 2B). We found that in homeotherms the ΔG of “old” miRNA-predicted targets' base-pairing is higher compared with that of the “new” miRNAs in accordance with the lower G/C content of “old” miRNAs demonstrated above (Fig. 1C). In D. melanogaster, the only poikilothermic organism that was examined, the trend was inversed; the ΔG of insect-specific miRNA-predicted targets is 0.71 kcal/mol, 5.43% higher than the ΔG of non-insect-specific miRNAs (p < 5 × 10−37). Among the homoeothermic organisms, chicken miRNAs had the most pronounced significant difference, 11% (−1.73 kcal/mol, p < 1 × 10−100), between the ΔG of bird-specific miRNA-predicted targets' base-pairing and the non-bird-specific subset. For humans, the average ΔG of miRNA-predicted targets that are primate-specific was the same as the non-primate-specific. Interestingly, the average ΔG of miRNA-predicted targets that are vertebrate-specific is 0.24 kcal/mol lower than the ΔG of miRNA-predicted targets with human miRNAs that are conserved in groups other than vertebrates (p < 5 × 10−36). In mouse, both rodent-specific and vertebrate-specific miRNAs have lower ΔG in their predicted binding sites compared with their non-rodent-specific and non-vertebrate-specific miRNAs, respectively (−0.48 kcal/mol, p < 2 × 10−39; −0.23 kcal/mol, p < 1.9 × 10−23, respectively).
Thus, we show a difference in the free energy of miRNA-predicted target interaction between newly generated miRNAs and miRNAs that are conserved among distant organisms. The inverse directionality of the difference between the homeotherms and poikilotherms further supports an existing association between temperature and base-pairing strength.
DISCUSSION
The base-pairing of miRNA to its target genes is a fundamental step in repression by miRNA. Thus far it has been shown that miRNA–target base-pair matching is important for miRNA activity. In this study, we examined the importance of base-pairing strength and its contribution to the stability of the repression complex. We hypothesized that if miRNA–target base-pairing strength plays a role in repression by miRNA, then the G/C content of the miRNAs of an organism would correlate to its physiological temperature so that the stability of the miRNA–target duplex is not affected by temperature. We show a significant correlation between the physiological temperature of an organism and the average G/C content of its miRNA and seed sequences; furthermore, we found a high inverse correlation between temperature and the average free energy of miRNA-predicted target base-pairing. Together, these correlations suggest that an organism adapts its miRNA–target free energy according to its physiological temperature, thus highlighting the importance of base-pairing strength in miRNA activity. Among the organisms tested, the chicken, which has the highest constant body temperature (41.5°C), has the highest average G/C content and the lowest miRNA-predicted target base-pairing free energy. Interestingly, this result is derived from the high G/C content that is found among miRNAs that are specific to birds. Finally, in the nine organisms examined, we found that the average G/C content of seed sequences was higher than that of mature miRNAs, which is higher than that of pre-miRNA loops, suggesting an association between the degree of functionality of the sequence and its average G/C content.
The correlation between the miRNA–target free energy and the body temperature of the organism suggests that miRNA–target base-pairing strength contributes to miRNA activity. In other cell processes that involve base-pairing, associations between base-pairing strength and reaction efficacy have not been detected. For example, the speed of translation is not affected by the strength of codon anti-codon base-pairing in an in vitro system derived from Escherichia coli (Andersson et al. 1984). Another process in the cell that involves base-pairing is pre-mRNA splicing. A study that compared splice site motifs in different organisms found no difference between mammals and chicken (Abril et al. 2005). However, the splice site motifs of human are significantly different from those of fish and fly. The 5′ splice site motifs of fish and fly have a reduced level of G/C in position −1, and the fish also has a reduced G/C level in position +5. However, we cannot conclude that there is an association between base-pairing strength and the physiological temperature of the examined organisms in splicing. Thus, the suggested association between base-pairing strength and miRNA activity shown in our study is unique and requires further attention.
A recent study in Arabidopsis found that stress-responsive miRNAs that were expressed in response to high salinity, drought, and low temperature are G/C-enriched. Mishra et al. (2009) explain that this G/C enrichment increases the miRNA–target duplex stability, which accelerates the stress response. The association between miRNA–target base-pairing strength and miRNA activity we proposed here supports this idea. Following the same logic, we propose that the G/C content of miRNAs that function as “noise-buffers” and are constantly coexpressed with their targets is relatively low since high G/C content might cause an increased repression of targets, thus disrupting the maintenance of their targets at a constant level.
The association between temperature and G/C content may further suggest that miRNA–target interaction hinders the adaptation of an organism to a new temperature. A temperature change may impair miRNA activity by altering the stability of miRNA–target duplex, thus disrupting the genetic program of the organism. Sempere et al. (2006) and Shomron et al. (2009) suggested that the sequence of a miRNA is almost unchanged during evolution once it accumulates a critical number of targets. We propose that in a scenario of a temperature change (either intrinsic in homeotherms or environmental in poikilotherms), the expression level of the currently existing miRNAs will be modified so that the miRNA repression level would be unaffected. New miRNAs that are generated under the new temperature regime will have a different G/C content that allows them to obtain a stabilized miRNA–target duplex. Such a scenario may explain the difference that we found in the average G/C content and also in the average free energy of miRNA-predicted target base-pairing between “new” miRNAs, miRNAs that are unique to the organism close taxonomic group, and “old” miRNAs, miRNAs that are also conserved in other taxonomic groups (Figs. 1C, 2B). For example, in homeothermic organisms, we found that the average G/C content of “new” miRNAs is higher than that of “old” ones. This suggests that the current physiological temperature of the homeothermic organisms is higher compared with their past temperature. More specifically, in chicken, the difference between bird-specific miRNAs and non-bird-specific miRNAs implies a high temperature difference between birds and the common ancestor of birds and mammals. Furthermore, additional analysis will allow us to evaluate when the change in G/C content occurred during evolution by calculation of specific/nonspecific difference using recent and previous taxonomic groups as classifiers. For example, comparison of (1) the difference in G/C content between vertebrate-specific and non-vertebrate-specific chicken miRNAs to (2) the difference between bird-specific and non-bird-specific, can indicate whether the change in G/C content occurred before or after the emergence of birds.
Because our study used few model organisms, we cannot rule out the possibility that the G/C content of miRNAs is generated by nonselective mutations rather than adaptation to the organism's physiological temperature. Since miRNAs are conserved almost without change once they accumulate a sufficient number of targets, it is possible under a nonselective scenario that miRNAs “snapshot” the G/C content of their loci close to the time in which they were generated (Sempere et al. 2006). Under each of the scenarios, the G/C content of miRNAs is highly informative either as an indicator of the organism's past and present physiological temperature or as evidence of the past G/C content of the genomic region in which the miRNA was located.
MATERIALS AND METHODS
Physiological temperature
We defined “physiological temperature” as follows: In homeotherms, it is the constant body temperature of the species—33°C for Ornithorhynchus anatinus (platypus), 37°C for H. sapiens (human) and M. musculus (mouse), 38.9°C for Bos taurus (cow), and 41.5°C for G. gallus (chicken) (Grant and Dawson 1978; Bitman et al. 1984; Whittow 1986). In poikilotherms, “physiological temperature” is defined by the optimal temperature at which the embryo develops to a healthy adult in the shortest period of time, because the expression levels of miRNA targets are higher in embryos than in later life stages, which indicates the important role of miRNAs for embryo development (Yu et al. 2007). “Physiological temperature” is 25°C in D. melanogaster (Ashbruner 1989) and C. elegans (Byerly et al. 1976), 26°C for B. mori (Muniraju et al. 2004), and 28.5°C for Danio rerio (Kimmel et al. 1995).
miRNA data set
We used miRBase v18 (http://www.mirbase.org).
Seed sequence
The seed sequence is a 7 to 8 nt sequence in the 5′ side that base-pairs to its target gene contiguously and perfectly in most miRNA–target interactions (Filipowicz et al. 2008). Based on Brennecke et al. (2005), we defined the seed as a 7-mer from position 2 to 8 of miRNA, because this sequence is conserved in both “canonical” and “seed only” miRNA–target binding sites.
G/C content calculation
The related sequences for each of the organisms were extracted from miRBase, and the average G/C content was calculated as follows:
miRNAs and seeds
We used nonredundant miRNAs that were documented by deep sequencing and/or miRNAs that were categorized as “experimental” in miRBase.
pre-miRNA loops
We used pre-miRNAs that were documented by deep sequencing and/or were categorized as “experimental” in at least one of their miRNAs' strands. We defined “loops” as (1) in pre-miRNAs with two documented miRNAs on each of the strands (“5p” and “3p”) as the sequence between the miRNAs strands. To reduce base-pairing constraints of loop sequences, we trimmed 3 nt from each end because miRBase predicts these positions to base-pair in many pre-miRNA secondary structures. (2) For the cases in which pre-miRNAs have one documented miRNA, loops are 8-mers starting from 3 nt downstream from the 3′ end of the miRNA if the miRNA is on the 5′ strand of the pre-miRNA hairpin (“5p”); and are 8-mers starting 3 nt upstream of the miRNA 5′ end if the miRNA is located in the 3′ strand of pre-miRNA (“3p”). We chose to begin the loop 3 nt downstream from or upstream of the 5′ or 3′ end of the miRNAs to keep constancy and reduce base-pairing constraint.
G/C content of pre-miRNA flanking regions
For each organism examined, we calculated the G/C content of the 50 nt adjacent to the pre-miRNA 5′ and 3′ ends using the UCSC Galaxy suite (Giardine et al. 2005; Blankenberg et al. 2010; Goecks et al. 2010). First, we uploaded into Galaxy the genomic positions of all of the pre-miRNAs described in “gff” files in the miRBase v18 ftp site (ftp://mirbase.org/pub/mirbase/18/genomes/). Next, we used the Galaxy “Get flanks” application to obtain the coordinates of 50 nt from each end of the pre-miRNA. We downloaded the coordinates of the flanking regions, corrected the chromosome names, and uploaded them again into Galaxy to get their sequences with the “Extract genomic DNA” application. Using the “geecee” application (Blankenberg et al. 2007), we calculated the G/C level of the flanking sequences, converted white spaces into “tabs” (with “Convert” application), and averaged the results using Galaxy “Summary statistics.”
Position analysis
We used miRNAs that were documented by deep sequencing and/or miRNAs that were categorized as “experimental” in miRBase and calculated the percent of G/C in each position (1–10) of the 5′ end of miRNAs that contain the seed region (Filipowicz et al. 2008).
Generating lists of miRNAs that are specific or nonspecific to a taxonomic group
We used miRBase v18 to obtain lists of miRNAs for each of the organisms examined. Next, we divided each of the lists into two subgroups: (1) miRNAs that are specific to a taxonomic group (e.g., insect-specific for D. melanogaster) and (2) miRNAs that are found in organisms that are not included in the specific taxonomic group (e.g., non-insect-specific for D. melanogaster).
To compile a list of miRNAs that are, for example, insect-specific and a list of miRNAs that are not insect-specific, we applied the following algorithm using a Perl script:
(A) Extract from miRBase a list of all of the miRNAs found in insects, remove the organism prefix and suffix from the miRNA name (e.g., dme-miR-7a-1 and bmo-miR-7b transform into miR-7), and filter out redundant miRNAs, because miRNAs from different species that share the same name are homologous (Griffiths-Jones et al. 2006).
(B) Prepare another list of miRNAs from organisms that are not insects in a similar manner as in step A above.
(C) Subtract from the list of insect miRNAs generated in step A miRNAs that were also found in non-insect organisms (generated in step B) to obtain an insect-specific miRNAs list.
- (D) Divide Drosophila miRNAs into two groups:
- (1) “Drosophila insect-specific miRNAs” that appear in the list of insect-specific miRNAs (generated in step C) and
- (2) “non-insect-specific Drosophila miRNAs” that comprise the remaining miRNAs.
Statistical analyses
Significance of the difference between two G/C content subsets was tested by using a t-test with an equal variance assumption. We used the Pearson correlation coefficient to measure the linear relationship between G/C content or ΔG and physiological temperature. For the standard error of the difference between two means, we used a formula that is suitable for large sample sizes:
 |
where si is standard deviation and ni is sample size (Sokal and Rohlf 1995).
TargetSpy
To examine the average ΔG of the miRNA–target duplex for several model organisms, we used the ΔG values that are provided by TargetSpy for miRNA-predicted target base-pairing (http://www.targetspy.org/). TargetSpy is a recent, highly reliable miRNA targets prediction program that does not rely on species conservation (Sturm et al. 2010). It uses miRBase v12 to predict miRNA binding sites for a few model organisms. We have downloaded the target prediction for human, mouse, chicken, and fly with a low false-positive rate of 1% (labeled “spec” by the authors). For each organism, we have downloaded two data sets: (1) with seed hybridization (“seed”), and (2) without seed hybridization (“no seed”). Redundant predicted binding sites were excluded. The “seed” data sets analysis is shown in Figure 2, and the “no seed” analysis is shown in Supplemental Figure 1.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Uyen Tram for critical proofreading of this manuscript. This work is supported by a United States–Israel Binational Science Foundation (BSF, 2009270) grant.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.032185.111.
REFERENCES
- Abril JF, Castelo R, Guigo R 2005. Comparison of splice sites in mammals and chicken. Genome Res 15: 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Martinez J, Schroeder R 2007. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130: 101–112 [DOI] [PubMed] [Google Scholar]
- Andersson SG, Buckingham RH, Kurland CG 1984. Does codon composition influence ribosome function? EMBO J 3: 91–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. 1989. Drosophila: A laboratory handbook, pp. 181–200. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bernardi G, Olofsson B, Filipski J, Zerial M, Salinas J, Cuny G, Meunier-Rotival M, Rodier F 1985. The mosaic genome of warm-blooded vertebrates. Science 228: 953–958 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Bitman J, Lefcourt A, Wood DL, Stroud B 1984. Circadian and ultradian temperature rhythms of lactating dairy cows. J Dairy Sci 67: 1014–1023 [DOI] [PubMed] [Google Scholar]
- Blankenberg D, Taylor J, Schenck I, He J, Zhang Y, Ghent M, Veeraraghavan N, Albert I, Miller W, Makova KD, et al. 2007. A framework for collaborative analysis of ENCODE data: Making large-scale analyses biologist-friendly. Genome Res 17: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J 2010. Galaxy: A web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol 89: 19.10.1–19.10.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM 2005. Principles of microRNA–target recognition. PLoS Biol 3: e85 doi: 10.1371/journal.pbio.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byerly L, Cassada RC, Russell RL 1976. The life cycle of the nematode Caenorhabditis elegans. I. Wild-type growth and reproduction. Dev Biol 51: 23–33 [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR 2004. Transcription and processing of human microRNA precursors. Mol Cell 16: 861–865 [DOI] [PubMed] [Google Scholar]
- Cuny G, Soriano P, Macaya G, Bernardi G 1981. The major components of the mouse and human genomes. 1. Preparation, basic properties and compositional heterogeneity. Eur J Biochem 115: 227–233 [DOI] [PubMed] [Google Scholar]
- Denver DR, Dolan PC, Wilhelm LJ, Sung W, Lucas-Lledo JI, Howe DK, Lewis SC, Okamoto K, Thomas WK, Lynch M, et al. 2009. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc Natl Acad Sci 106: 16310–16314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D, Hobert O 2006. Perfect seed pairing is not a generally reliable predictor for miRNA–target interactions. Nat Struct Mol Biol 13: 849–851 [DOI] [PubMed] [Google Scholar]
- Doench JG, Sharp PA 2004. Specificity of microRNA target selection in translational repression. Genes Dev 18: 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N 2008. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH 1986. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci 83: 9373–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. 2005. Galaxy: A platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J 2010. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86 doi: 10.1186/gb-2010-11-8-r86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodkin J, Havgaard JH, Enstero M, Sawera M, Jensen P, Ohman M, Fredholm M 2006. MicroRNA sequence motifs reveal asymmetry between the stem arms. Comput Biol Chem 30: 249–254 [DOI] [PubMed] [Google Scholar]
- Grant T, Dawson T 1978. Temperature regulation in the platypus, Ornithorhynchus anatinus: Maintenance of body temperature in air and water. Physiol Zool 51: 1–6 [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP 2007. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang F, Huang Y, Grimm D, Rossi JJ, Kay MA 2011. Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes. Proc Natl Acad Sci 108: 9208–9213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150 [DOI] [PubMed] [Google Scholar]
- Hughes S, Zelus D, Mouchiroud D 1999. Warm-blooded isochore structure in Nile crocodile and turtle. Mol Biol Evol 16: 1521–1527 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Keightley PD, Trivedi U, Thomson M, Oliver F, Kumar S, Blaxter ML 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res 19: 1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310 [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. 2005. Combinatorial microRNA target predictions. Nat Genet 37: 495–500 [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438: 685–689 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- Lai EC 2002. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 30: 363–364 [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN 2002. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J 21: 4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056 [DOI] [PubMed] [Google Scholar]
- Mishra AK, Agarwal S, Jain CK, Rani V 2009. High GC content: Critical parameter for predicting stress regulated miRNAs in Arabidopsis thaliana. Bioinformation 4: 151–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G 2002. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev 16: 720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniraju E, Sekharappa BM, Raghuraman R 2004. Food intake and utilization efficiency in silkworm Bombyx mori L. (Pure Mysore) reared at different temperature combinations. Int J Trop Insect Sci 24: 135–142 [Google Scholar]
- Nicholson AW 1996. Structure, reactivity, and biology of double-stranded RNA. Prog Nucleic Acid Res Mol Biol 52: 1–65 [DOI] [PubMed] [Google Scholar]
- Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB 2007. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA 13: 1894–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R, Slack FJ 2007. The evolution of animal microRNA function. Curr Opin Genet Dev 17: 145–150 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906 [DOI] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Douzery EJ, Galtier N 2010. Contrasting GC-content dynamics across 33 mammalian genomes: Relationship with life-history traits and chromosome sizes. Genome Res 20: 1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Cole CN, McPeek MA, Peterson KJ 2006. The phylogenetic distribution of metazoan microRNAs: Insights into evolutionary complexity and constraint. J Exp Zoolog B Mol Dev Evol 306: 575–588 [DOI] [PubMed] [Google Scholar]
- Shomron N, Golan D, Hornstein E 2009. An evolutionary perspective of animal microRNAs and their targets. J Biomed Biotechnol 2009: 594738 doi: 10.1155/2009/594738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 5: 659–669 [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ 1995. Biometry: The principles and practice of statistics in biological research, p. 836. Freeman, New York [Google Scholar]
- Sontheimer EJ, Carthew RW 2005. Silence from within: Endogenous siRNAs and miRNAs. Cell 122: 9–12 [DOI] [PubMed] [Google Scholar]
- Sturm M, Hackenberg M, Langenberger D, Frishman D 2010. TargetSpy: A supervised machine learning approach for microRNA target prediction. BMC Bioinformatics 11: 292 doi: 10.1186/1471-2105-11-292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario S, Moriyama EN, Powell JR 2007. Codon usage in twelve species of Drosophila. BMC Evol Biol 7: 226 doi: 10.1186/1471-2148-7-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittow GC. 1986. Regulation of body temperature. In Avian physiology (ed. PD Sturkie), pp. 221–252. Springer-Verlag, New York. [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Jian Z, Shen SH, Purisima E, Wang E 2007. Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos. Nucleic Acids Res 35: 152–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57–68 [DOI] [PubMed] [Google Scholar]
- Zhang B, Stellwag EJ, Pan X 2009. Large-scale genome analysis reveals unique features of microRNAs. Gene 443: 100–109 [DOI] [PubMed] [Google Scholar]