Cells infected with arthropod-borne flaviviruses (e.g., Dengue or Kunjin viruses) accumulate uncapped mRNAs or viral genomes, decay intermediates normally targeted by cellular 5′-to-3′ exonuclease XRN1. These viruses also generate a short noncoding RNA (sfRNA) from the viral 3′ untranslated region during infection, due to the stalling of the XRN1. This paper shows that XRN1 can be found associated with sfRNA during infection and that this results in the repression of its activity. Cellular mRNAs are also stabilized by sfRNA, likely as a result of the shutdown of the 5′-to-3′ mRNA decay pathway.
Keywords: flavivirus, mRNA decay, mRNA stability, noncoding RNA, virus–host interaction
Abstract
All arthropod-borne flaviviruses generate a short noncoding RNA (sfRNA) from the viral 3′ untranslated region during infection due to stalling of the cellular 5′-to-3′ exonuclease XRN1. We show here that formation of sfRNA also inhibits XRN1 activity. Cells infected with Dengue or Kunjin viruses accumulate uncapped mRNAs, decay intermediates normally targeted by XRN1. XRN1 repression also resulted in the increased overall stability of cellular mRNAs in flavivirus-infected cells. Importantly, a mutant Kunjin virus that cannot form sfRNA but replicates to normal levels failed to affect host mRNA stability or XRN1 activity. Expression of sfRNA in the absence of viral infection demonstrated that sfRNA formation was directly responsible for the stabilization of cellular mRNAs. Finally, numerous cellular mRNAs were differentially expressed in an sfRNA-dependent fashion in a Kunjin virus infection. We conclude that flaviviruses incapacitate XRN1 during infection and dysregulate host mRNA stability as a result of sfRNA formation.
INTRODUCTION
The cellular mRNA decay machinery plays an important role in determining both the quantity and quality of gene expression (Garneau et al. 2007). The steady-state level of a given transcript is largely determined through coordination of transcription and mRNA decay (Elkon et al. 2010; Shalem et al. 2011). In fact, recent data suggest that the processes of transcription and cytoplasmic mRNA decay may be much more tightly coordinated than previously suspected (Bregman et al. 2011; Trcek et al. 2011). Based on global analyses performed in response to various stimuli, ∼17%–50% of changes in mRNA levels are primarily a result of altered mRNA stability (Cheadle et al. 2005; Schwanhäusser et al. 2011). In addition, the cellular mRNA decay machinery rapidly and efficiently removes unwanted transcripts such as mRNAs with premature termination codons and damaged or malformed RNAs (Mühlemann and Jensen 2012). Hence, the mRNA decay machinery is clearly a major force in cellular RNA metabolism, and disrupting it can lead to a variety of biological effects (Schaljo et al. 2009; Astuti et al. 2012).
Pathways of mRNA decay are often initiated by shortening of the poly(A) tail, a process referred to as deadenylation (Chen and Shyu 2011). Decay can also be initiated through alternative mechanisms, including endonucleolytic cleavage by targeted nucleases via small regulatory miRNAs or siRNAs or other factors (Schoenberg 2011). Deadenylation-initiated decay is thought to be the primary pathway for the demise of most cellular mRNAs. These initiating events in mRNA decay feed the transcript into one of two exonucleolytic pathways. The 5′-to-3′ exonucleolytic pathway begins with a coordinated set of proteins recruiting a decapping enzyme (DCP2 or NUDT16) to remove the 5′ methylated cap structure (Li et al. 2011). This generates a transcript bearing a 5′ monophosphate that is rapidly and processively degraded by the 5′-to-3′ exoribonuclease XRN1 (Chang et al. 2011; Jinek et al. 2011). Alternatively, proteins associated with the exosome complex attack the transcript exonucleolytically from the 3′-to-5′ direction (Lykke-Andersen et al. 2011; Astuti et al. 2012). XRN1-mediated decay is considered in many current models to be the major pathway of mRNA decay in cells (Stoecklin et al. 2006), although it is clear that exosome-mediated 3′-to-5′ decay may also make a significant contribution (Murray and Schoenberg 2007). Factors associated with the 5′-to-3′ decay pathway can often be colocalized in cytoplasmic granules referred to as Processing or P-bodies, suggesting a coordination and/or coregulation of factors and steps in this pathway (Kulkarni et al. 2010).
Growing evidence indicates that when RNA viruses infect cells their transcripts use several strategies to interface successfully with and/or avoid the cellular RNA decay machinery. Some RNA viruses that encode polyadenylated transcripts appear to usurp cellular RNA-binding proteins to stabilize their transcripts (Sokoloski et al. 2010; Palusa et al. 2012). Poliovirus, interestingly, appears to cause the accelerated turnover of several proteins involved in the 5′–3′ RNA decay pathway, including XRN1 and the auxiliary decapping factor DCP1a (Dougherty et al. 2011). Poliovirus also causes the apparent degradation or inhibition of factors that initiate decay as well, including the deadenylase component PAN3 (Dougherty et al. 2011) and the endoribonuclease RNase L (Townsend et al. 2008). Disruption of the cellular RNA decay machinery can have a variety of benefits for the virus, including increased stability of viral transcripts as well as interference with the regulation of cellular gene expression that may influence the ability of the cell to mount an effective innate immune response (Dickson and Wilusz 2011).
During infection, all arthropod-borne flaviviruses tested to date generate a small RNA termed subgenomic flavivirus RNA (sfRNA) from their 3′ untranslated region (UTR) (Pijlman et al. 2008). Interestingly, sfRNA was shown not to be the result of transcription from an internal promoter. RNAi-mediated knockdowns and applications of a recombinant yeast enzyme suggest that sfRNA is generated as a viral genomic RNA decay product by the action of the XRN1 exoribonuclease (Pijlman et al. 2008). XRN1 appears to stall at a set of conserved pseudoknot-like structures at the 5′ side of the flavivirus 3′ UTR (Funk et al. 2010; Silva et al. 2010). Importantly, the ability of West Nile virus to generate sfRNA has been linked to cytopathology in infected cells and pathogenicity in mice (Pijlman et al. 2008; Funk et al. 2010; Schuessler et al. 2012). Thus, understanding the consequences of sfRNA generation in flavivirus-infected cells may provide significant insights into an underexplored area of virus–host interactions.
In this study, we have determined that stalling of the XRN1 enzyme in the process of generating sfRNA results in the repression of its activity. The repression of XRN1 by sfRNA formation is a direct result of the XRN1 protein–RNA interaction and does not require any auxiliary factors. Inhibition of XRN1 activity in flavivirus infections was demonstrated by the increased accumulation of uncapped mRNA decay intermediates and prolonged cellular mRNA half-lives in Dengue and West Nile virus (Kunjin strain) infections. Interestingly, the extension of mRNA half-lives can be directly attributed to sfRNA formation because it is not seen in cells infected with a mutant virus that cannot make sfRNA. However, cellular mRNA stabilization can be reproduced by generating sfRNA in the absence of viral infection. The inactivation of XRN1 and the associated increase in the stability of cellular transcripts coincide with an sfRNA-dependent overexpression of a variety of cellular mRNAs. Collectively, these data suggest a novel role for this small noncoding sfRNA in the dysregulation of cellular mRNA expression.
RESULTS
Formation of sfRNA by the cellular XRN1 exoribonuclease represses enzymatic activity
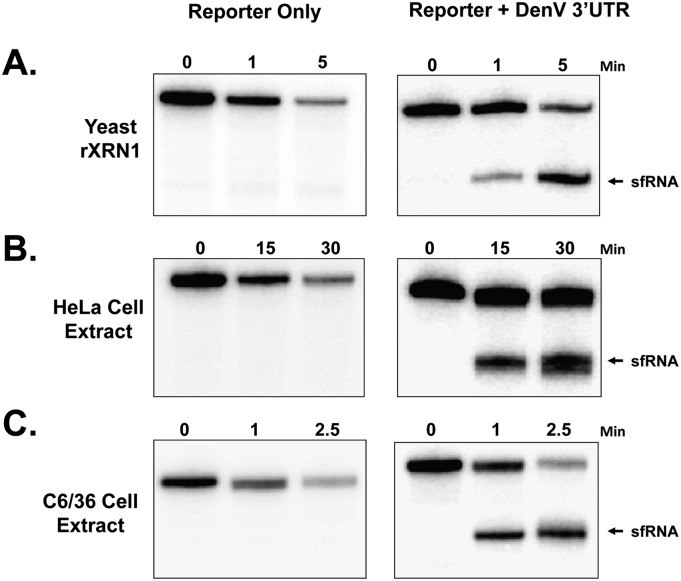
Previous work using sh/siRNA-mediated Xrn1 knockdown in infected cells or incubation with a recombinant Xrn1 protein derived from yeast have strongly implicated a role for the cellular exoribonuclease in the formation of sfRNA from West Nile virus and Yellow Fever virus 3′ UTRs (Pijlman et al. 2008; Silva et al. 2010). To extend these observations, we inserted the 3′ UTR of Dengue virus type 2 (DenV2) into a reporter construct and prepared in vitro–transcribed RNAs bearing a 5′ monophosphate so that they would be effective substrates for XRN1. As seen in Figure 1A, while the reporter RNA lacking DenV2 sequences was effectively degraded by recombinant yeast XRN1 with no apparent decay intermediates, the reporter containing the 3′ UTR efficiently generated an sfRNA-like decay product due to stalling of XRN1 at the 5′ end of the DenV2 3′ UTR. To demonstrate that XRN1-like activity from human or mosquito cells can also generate sfRNA, the same constructs were incubated in HeLa (Fig. 1B) or C6/36 mosquito cell (Fig. 1C) cytoplasmic extracts under conditions that favor almost exclusively 5′-to-3′ exonucleolytic decay of RNA substrates (Mukherjee et al. 2002). Both extracts efficiently degraded the control RNA containing a 5′ monophosphate but generated an sfRNA-like decay intermediate from RNAs containing the DenV2 3′ UTR. Thus we conclude that XRN1 from multiple organisms stalls at the proximal side of the DenV2 3′ UTR to generate an sfRNA. This is surprising given that XRN1 is a processive and highly efficient exoribonuclease on the vast majority of RNA substrates.
FIGURE 1.
XRN1 stalls at the 5′ border of the Dengue virus 3′ UTR to generate sfRNAs. Radiolabeled reporter RNAs derived from a pGem4 template (Reporter Only) or RNAs from the same plasmid containing the stem–loop 1 region of the 3′ UTR of Dengue virus type 2 (Reporter + DenV 3′ UTR) were generated with a 5′ monophosphate as substrates for the XRN1 exonuclease. The RNAs were incubated with recombinant yeast XRN1 (A), or cytoplasmic extracts from HeLa (B) or Aedes C6/36 (C) cells under conditions in which XRN1-mediated 5′–3′ decay predominates. Reaction products were analyzed on 5% polyacrylamide gels containing urea and visualized by phosphorimaging. The position of sfRNA-like reaction products is indicated on the right.
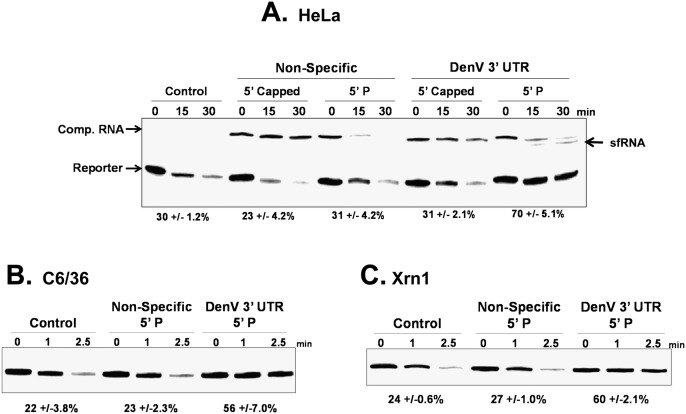
The stalling of XRN1 on RNA substrates as abundant as those generated during a viral infection could have significant impact on its ability to function effectively as an enzyme. If XRN1 was only slowly released (or not released at all) from RNA substrates that it stalls on, its ability to function as a 5′-to-3′ exonuclease would be inhibited. To test this possibility, we incubated XRN1 from a variety of sources with a reporter RNA in the presence of a 30-fold excess of a competitor RNA. The RNA competitors used in this experiment were either a transcript that contained the 3′ UTR of DenV2 capable of stalling XRN1 and forming an sfRNA-like decay intermediate or a nonspecific transcript of similar length. We also used two forms of the competitor RNAs: 5′-capped transcripts that would be naturally resistant to 5′-exonuclease digestion and transcripts containing a 5′ monophosphate that would be rapidly attacked by XRN1. In this way, we could determine whether there was a requirement for the competitor to be a substrate for XRN1 digestion in order to influence enzymatic activity or if the sequence of the RNA competitor alone was sufficient to see any effects observed. As seen in Figure 2A, the addition of nonspecific competitor RNA had no effect on XRN1-mediated decay of a radiolabeled reporter in HeLa extracts. This was true regardless of whether the 5′ end was capped or bore a monophosphate and confirmed that the XRN1 enzyme was in excess under the conditions used in the experiment. Interestingly, a competitor RNA containing the DenV2 3′ UTR repressed XRN1 activity, but only when the competitor transcript contained a 5′ monophosphate and could recruit XRN1 to the 5′ end to form sfRNA. These data indicate that RNAs containing the DenV2 3′ UTR can repress XRN1 activity, but only when they can also serve as substrates for the enzyme. The presence of the DenV2 3′-UTR sequence on a transcript with a 5′ end blocked by a 7-methyl G cap structure did not efficiently repress XRN1 activity. The DenV2 3′-UTR competitor RNA bearing a 5′ monophosphate also effectively repressed XRN1 activity in C6/36 mosquito extracts (Fig. 2B) and, notably, repressed purified recombinant XRN1 protein (Fig. 2C). These data indicate that the repression of XRN1 activity by sfRNA generation does not require auxiliary cellular factors, although cellular factors may modulate the efficiency of the repression. In summary, we conclude that XRN1 exoribonuclease from divergent eukaryotic organisms (yeast, human, and mosquito) can be inhibited as a result of stalling during formation of sfRNA.
FIGURE 2.
sfRNA formation inhibits the cellular exoribonuclease XRN1. A radiolabeled RNA containing a 5′ monophosphate (Reporter) was incubated for the time indicated in HeLa (A) or C6/36 (B) cytoplasmic extract under conditions that favor 5′-to-3′ decay or with purified recombinant XRN1 (C). The lanes labeled “Control” indicate reactions that were performed in the absence of any competitor RNA. A 30× molar excess of lightly radiolabeled nonspecific competitor RNA (“Non-Specific” lanes) or a competitor transcript containing the proximal half of the 3′ UTR of Dengue virus type 2 (“DenV 3′ UTR” lanes) was added to reactions. The 5′ end of the competitors contained either a 5′ cap or a 5′ monophosphate as indicated above the lanes. After the times indicated, reaction products were analyzed on 5% polyacrylamide gels containing urea and visualized by phosphorimaging. The numbers under the lanes represent the means ± standard deviation of the amount of starting reporter RNA remaining in three independent experimental replicates.
Formation of sfRNA impairs XRN1 activity in flavivirus-infected cells
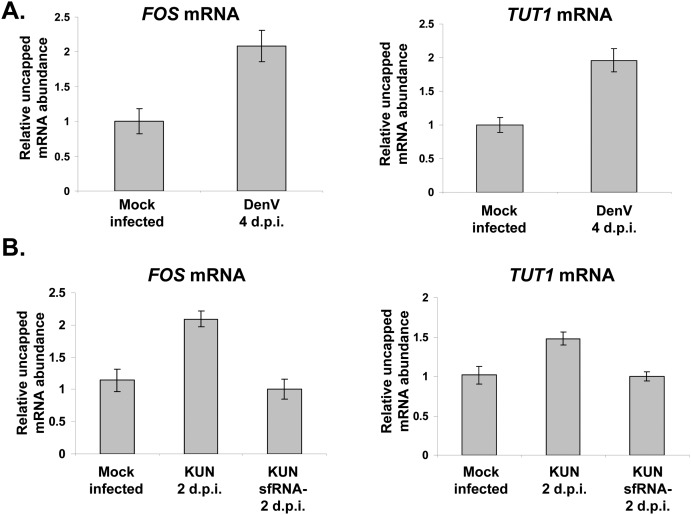
Since sfRNA formation effectively blocked XRN1 activity in cell-free assays, the next important question to address was whether XRN1 activity is also repressed in flavivirus-infected cells. A major pathway of cellular mRNA decay involves decapping and subsequent rapid degradation of the resulting transcripts that contain a 5′ monophosphate by the XRN1 exonuclease (Garneau et al. 2007). Therefore, an increased abundance of uncapped mRNA decay intermediates would be a strong indication of XRN1 inhibition in flavivirus-infected cells. Human 293T cells were infected with DenV2, and total RNA was obtained at 4 d post-infection when levels of sfRNA in cells were high. Total RNA samples were immunoprecipitated with an excess amount of an antibody specific to the methylated cap structure, and the supernatant fraction was analyzed for uncapped FOS or TUT1 mRNAs by qRT-PCR. These mRNAs were chosen to represent transcripts that were relatively short-lived (FOS) or have an average half-life (TUT1). The 7SL RNA, which naturally lacks a 5′ cap, was used as a reference. In addition, values were normalized to the relative amount of each mRNA present in the cap-containing precipitated fraction. As seen in Figure 3A, the amount of uncapped FOS or TUT1 mRNAs was reproducibly almost twofold higher in DenV2-infected cells as compared with mock-infected controls. The relative amount of uncapped RNAs was ∼4% in mock-infected cells and ∼8% of the total mRNA in DenV2-infected cells. To generalize this observation to other arthropod-borne flaviviruses that generate an sfRNA, the experiment was repeated with the Kunjin subtype of West Nile virus recovered from the infectious cDNA clone FLSDX (Khromykh et al. 1998). As seen in Figure 3B, the amounts of uncapped FOS or TUT1 mRNAs were substantially higher in Kunjin virus–infected cells. The relative amount of uncapped RNAs was ∼4% in mock-infected cells and ∼6%–8% of the total mRNA in Kunjin virus–infected cells. Finally, we performed the experiment using a mutant Kunjin virus (FL-IRAdCS3) that replicated to similar levels as wild-type Kunjin virus in 293T cells as measured by qRT-PCR (Supplemental Fig. 1A) but failed to produce sfRNA (see Fig. 7; Pijlman et al. 2008). Importantly, the sfRNA-deficient Kunjin virus strain failed to cause the accumulation of uncapped FOS or TUT1 mRNAs in infected cells (Fig. 3B). Similar differences were evident when uncapped RNA levels were normalized to total mRNA levels (data not shown). These data strongly suggest that XRN1 activity is specifically repressed by the generation of sfRNA during flavivirus infections.
FIGURE 3.
Uncapped mRNAs accumulate in flavivirus-infected cells due to XRN1 inhibition by sfRNA formation. 293T cells were either mock-treated or infected with Dengue virus (DenV) (A), Kunjin virus (KUN) (B), or a Kunjin virus mutant that cannot make sfRNA (KUN sfRNA-) (B) for the times indicated. Total RNA was isolated and immunoprecipitated using a cap-specific antibody. The presence of FOS and TUT1 mRNAs was measured in the input, capped, and uncapped fractions by qRT-PCR. The relative amount of uncapped RNAs was normalized to 7SL RNA and to the total amount of capped RNA present in the sample and expressed in the graphs relative to the level of uncapped mRNAs in mock-infected samples. Error bars represent the standard error of the mean in the indicated samples.
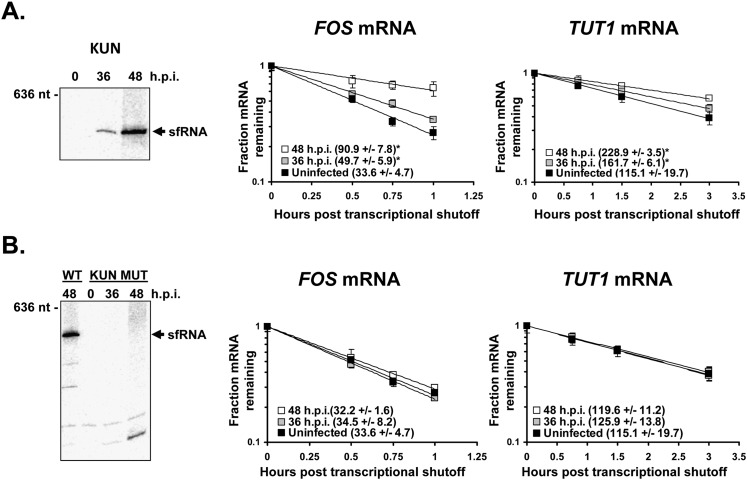
FIGURE 7.
Cellular mRNAs are stabilized in an sfRNA-dependent fashion in Kunjin virus infection. Human 293T cells were infected with either wild-type Kunjin virus (A) or an sfRNA-deficient mutant Kunjin virus that fails to produce sfRNA due to structural mutations in the 3′ UTR (B). Northern blot analysis showed the presence or absence of sfRNA accumulation in infected cells (panels at left). Transcriptional shutoff by actinomycin D treatment was done at 48 hpi, and the half-lives of FOS and TUT1 mRNAs were determined by qRT-PCR. Half-lives were calculated from two independent experiments and are expressed as ±standard deviations. (*) P-values <0.05.
XRN1 is preferentially associated with the flavivirus 3′ UTR/sfRNA during infection
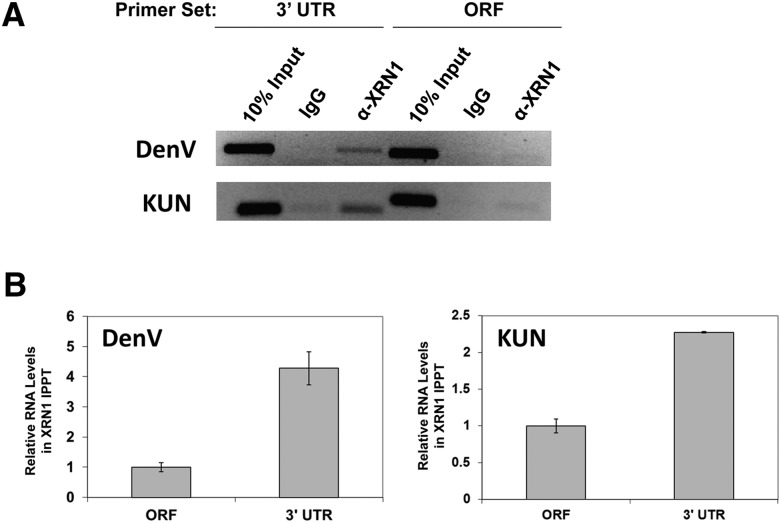
Given the results in Figure 2C showing that purified recombinant XRN1 can be inhibited by sfRNA formation, the most likely mechanism for the repression of XRN1 activity by sfRNA formation is that the enzyme remains bound to the RNA. To demonstrate this in flavivirus-infected cells, RNA–protein complexes in human 293T cells infected with Kunjin virus or Dengue virus were cross-linked with formaldehyde at 2 or 4 d post-infection (respectively), and XRN1 antibodies were used to immunoprecipitate XRN1 and any associated RNAs from cell lysates. Precipitated RNAs were isolated and RT-PCR was performed using primers to a region of the NS3 portion of the Kunjin virus open reading frame (ORF) or the NS5 ORF of DenV2 just upstream of the 3′ UTR or using primers to the viral 3′ UTR that would be present in sfRNA species. As seen in Figure 4A, XRN1 antibodies preferentially precipitated viral RNAs that contained the 3′-UTR sequences as compared with the upstream NS3 or NS5 sequences. To confirm these data, quantitative RT-PCR was performed using the same primers on independently immunoprecipitated samples. As seen in Figure 4B, the 3′ UTR was significantly enriched over the NS3 or NS5 regions, respectively, of the Kunjin or Dengue viral ORFs in the XRN1 coprecipitated fractions. The twofold higher relative levels of 3′-UTR-containing RNAs associated with XRN1 in the DenV2 infections is likely a factor of the longer time point (4 d post-infection) compared with the 2-d post-infection point used for the Kunjin virus–infected samples. The small overall increase in the amount of viral 3′ UTR relative to full-length genomic RNA due to the generation of sfRNA is not sufficient to account for the enrichment observed in the immunoprecipitated fractions. In addition, the efficiencies of the primers used in each set of qPCR reactions were within 2% of each other, indicating that the differences observed were not due to primer sensitivities. Therefore, we conclude that XRN1 was preferentially associated with viral transcripts that contained the 3′ UTR but lacked the upstream ORF (i.e., sfRNA).
FIGURE 4.
XRN1 protein preferentially binds to sfRNA in Kunjin or Dengue virus infections. 293T cells were infected with Kunjin virus for 2 d or Dengue virus for 4 d. RNA–protein complexes were stabilized with formaldehyde and immunoprecipitated with XRN1 antibody (α-XRN1 lanes) or matched amounts of a control IgG (IgG lanes). Coprecipitating RNAs were detected by RT-PCR (A) or qRT-PCR (B) using the primers indicated.
The formation of sfRNA and depletion of XRN1 activity are associated with the stabilization of cellular mRNAs
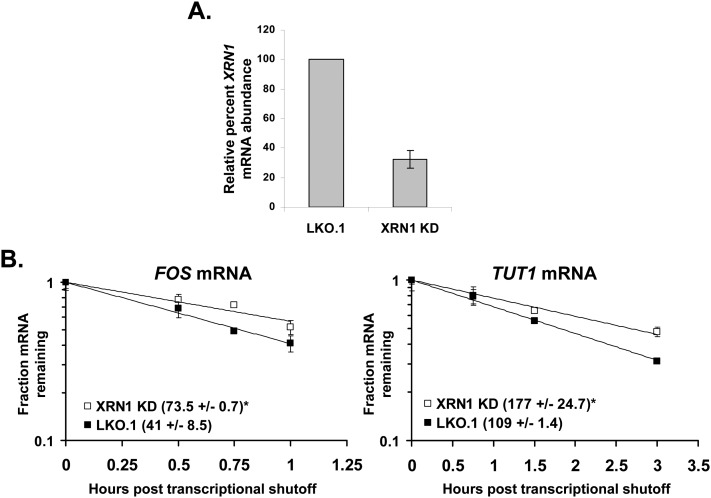
Since XRN1 is a key component in the major 5′-to-3′ pathway of mRNA decay in cells, we hypothesized that repression of its function could result in a significant stabilization of cellular mRNAs. While the accumulation of uncapped cellular mRNA is increased in flavivirus-infected cells (Fig. 3), the absolute amount of uncapped species likely remains very small compared with the overall amount of capped mRNA species due to the only ∼2× increase observed in this experiment. It is possible that the inhibition of XRN1 by sfRNA formation could feed back and generally inhibit the 5′-to-3′ decay pathway. In support of this idea, P-bodies representing subcellular sites where factors involved in 5′-to-3′ mRNA decay can be concentrated are dispersed in flavivirus infections (Emara and Brinton 2007). This observation could suggest a general disruption of the 5′-to-3′ decay pathway. As seen in Figure 5, knockdown of XRN1 to ∼32% of normal levels resulted in a significant increase in the half-lives of the two endogenous mRNAs sampled. This demonstrates that as predicted, XRN1 depletion resulted in an increase in the stability of cellular mRNAs. Therefore, we set out to assess relative mRNA half-lives in mock- and flavivirus-infected cells.
FIGURE 5.
Cellular mRNAs are stabilized upon XRN1 depletion. (A) XRN1 was depleted in 293T cells using specific XRN1 KD, or cells were treated with control (LKO.1) shRNA vectors. The bar graph illustrates the efficiency of XRN1 knockdown as measured by qRT-PCR. (B) TUT1 and FOS mRNA half-lives were determined after actinomycin D treatment by qRT-PCR in the respective graphs. Average half-lives are reported ±standard deviations from two independent experiments. (*) P-values <0.05.
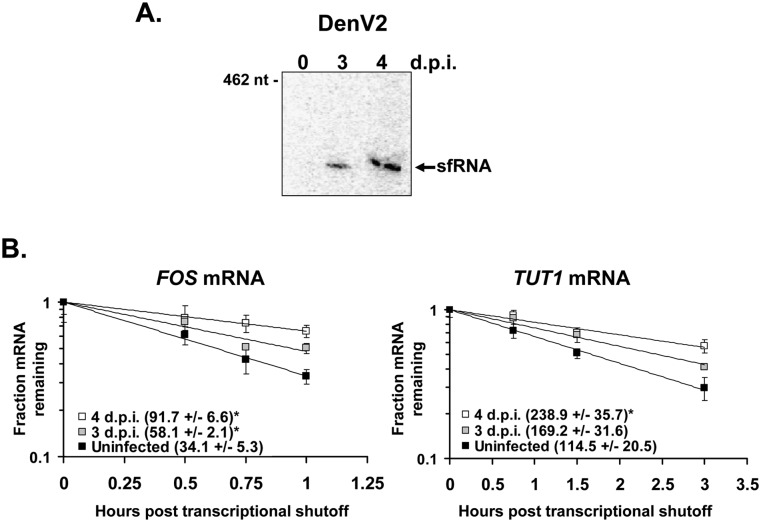
Mock- or virus-infected cells were treated with actinomycin D for 30 min to shut off cellular transcription, and RNA samples were isolated at the indicated times post–drug treatment. Levels of the FOS and TUT1 mRNAs at each time point were assessed by qRT-PCR and compared to generate mRNA decay curves and determine transcript half-lives. Infection with either DenV2 (Fig. 6) or Kunjin virus (Fig. 7A) resulted in a significant stabilization of both the FOS and TUT1 mRNAs compared with uninfected cells. Notably, the degree of stabilization as seen in the decay curves or calculated mRNA half-lives was strongly associated with the amount of sfRNA present in the infected cells (cf. the graphs to the Northern blots in Figs. 6 and 7A). Similar data were obtained in flavivirus infections of mosquito cells (data not shown). Importantly, infection with a mutant Kunjin virus that cannot produce sfRNA (Fig. 7B) also failed to cause significant stabilization of cellular mRNAs. This Kunjin virus mutant replicated to similar levels as the parent wild-type Kunjin virus in these 293T cells (Supplemental Fig. 1A). Collectively, these data indicate that cellular mRNAs are stabilized in infections with independent flaviviruses that are capable of producing sfRNA.
FIGURE 6.
Cellular mRNAs are stabilized by sfRNA formation in Dengue virus infection. (A) 293T cells were infected with Dengue virus type 2 for the times indicated (days post-infection, d.p.i.) and the levels of sfRNA were determined by Northern blotting. (B) The half-lives of the TUT1 and FOS mRNAs were determined after actinomycin D treatment by qRT-PCR. Average mRNA half-lives from two independent infections are reported ±standard deviation in parentheses. (*) P-values <0.05.
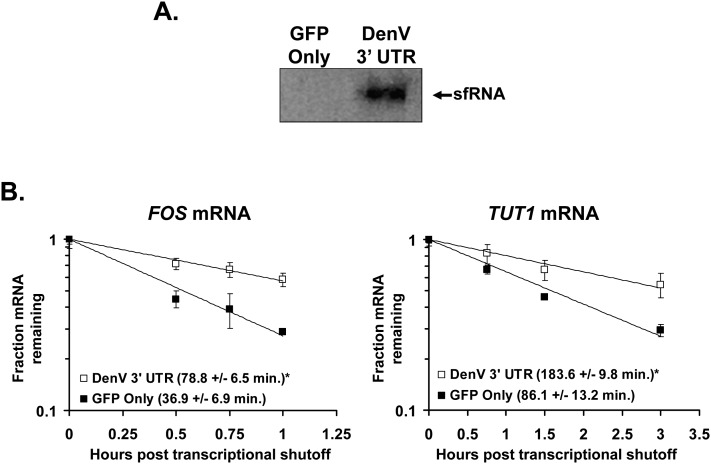
To demonstrate that sfRNA production is sufficient for the stabilization of mRNAs, we transfected 293T cells with either a control GFP-expressing reporter construct or with a GFP reporter containing the DenV2 3′-UTR sequence downstream from GFP and thus producing sfRNA. As seen in Figure 8A, sfRNA was efficiently detected only from the reporter construct containing the DenV2 3′-UTR sequences. Interestingly, the sfRNA-generating transfected reporter construct also caused a significant stabilization of cellular mRNAs compared with the GFP-only reporter. Collectively, these data indicate that a reporter RNA that possesses the DenV2 3′ UTR (and generates sfRNA) causes stabilization of cellular mRNAs in the absence of a virus infection. This demonstrates that sfRNA generation on its own is necessary and sufficient to cause the stabilization of cellular mRNAs.
FIGURE 8.
The formation of sfRNA in the absence of flaviviral infection induces cellular mRNA stabilization. GFP expression plasmids either expressing GFP only (“GFP Only” lane) or a GFP mRNA with the sfRNA-producing Dengue virus type 2 3′ UTR in its 3′ UTR (“DenV 3′ UTR” lane) were transfected into 293T cells. (A) Northern blot probed to detect the Dengue virus 3′ UTR present in sfRNA-like fragments. (B) Transfected cells were treated with actinomycin D, and RNA samples were taken at the indicated times post-transcriptional shutoff. The levels of the indicated TUT1 or FOS mRNAs were assessed by qRT-PCR, and the means of three experiments are presented graphically. The half-lives shown in the graph insets are means of at least two independent experiments ± standard deviations. (*) P-values <0.05.
Cellular gene expression is significantly dysregulated by an sfRNA-producing flavivirus
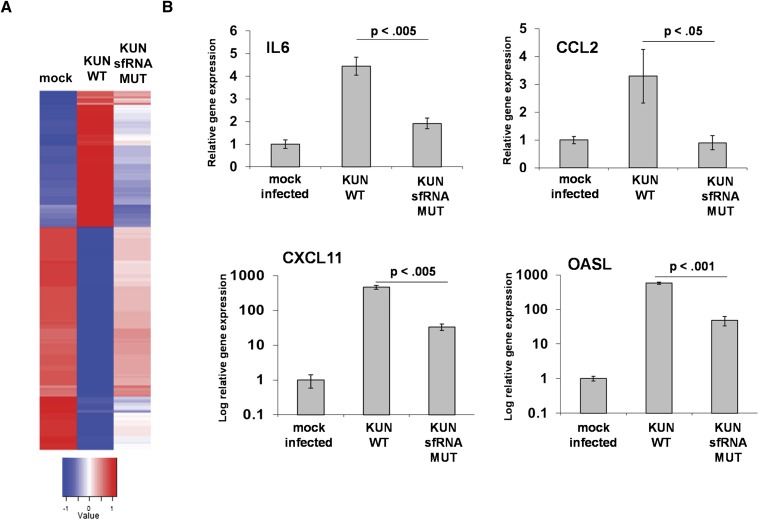
Finally, we wished to assess the scale of the global effects on cellular gene expression mediated by sfRNA production/XRN1 repression in a flavivirus infection. Interferon α/β receptor-negative mouse embryo fibroblasts (MEFs) that are highly sensitive to flavivirus infection were mock-infected, infected with wild-type Kunjin virus, or infected with an sfRNA mutant Kunjin virus for 48 h. Both wild-type and mutant viruses replicated to the same level in these cells as measured by qRT-PCR for Kunjin RNA (Supplemental Fig. 1B). Total RNA samples were obtained and relative expression level of mRNAs was determined by microarray analysis (Supplemental Fig. 2). The heat map shown in Figure 9A illustrates two interesting points from this analysis. First, a set of 416 cellular mRNAs was found to be significantly (3× or more) overexpressed in a wild-type Kunjin virus–infected cell compared with mock control. Interestingly, infection with a Kunjin virus mutant incapable of producing sfRNA failed to increase expression of most (378) of these mRNAs to the levels seen in wild-type Kunjin virus–infected cells. Analysis of Gene Ontogeny (GO) terms associated with this up-regulated set of transcripts indicates that mRNAs associated with the innate immune, inflammation, cell adhesion, stress responses, and cell proliferation are significantly (p ≤ 0.01) over-represented. The relative overexpression of four of these mRNAs in wild-type as compared with sfRNA mutant Kunjin virus infection was confirmed and generalized in an independent cell line by qRT-PCR analyses of total RNA samples obtained from infected 293T cells (Fig. 9B). Collectively, these data indicate that a significant amount of up-regulation of cellular gene expression is associated with sfRNA production during a flavivirus infection. Since the amount of uncapped mRNA that accumulated during a Kunjin virus infection likely remains relatively small compared with the total amount of capped mRNA (Fig. 3), this increase in mRNA levels noted in the array analysis may largely represent capped, functional mRNA. The shutdown of XRN1 by the generation of sfRNA could be responsible for the up-regulation of many of these transcripts.
FIGURE 9.
Numerous cellular mRNAs are up-regulated in Kunjin virus-infected cells due to sfRNA formation. (A) Interferon α/β receptor–deficient mouse embryo fibroblasts were mock-infected or infected with either wild-type (wt) or sfRNA− Kunjin virus for 48 h. Total RNA samples were isolated at 48 hpi, and cellular gene expression levels were assessed globally by microarray analysis. Robust multiarray analysis (RMA) values were calculated, and hierarchical clustering was performed for genes having threefold or more difference in RMA values after transforming the values to have mean 0 and standard deviation 1. The color key represents the transformed values. (B) Levels of the four indicated mRNAs were assessed by qRT-PCR of total RNA isolated from independent infections of 293T cells using wild type or an sfRNA-deficient Kunjin virus at 48 hpi. The results shown are the mean of three independent infections, and error bars represent the standard error of the means. Significance was determined using a one-tailed t-test.
Curiously, another set of mRNAs was significantly down-regulated in wild-type Kunjin infection whose expression was only mildly reduced in infections with an sfRNA mutant Kunjin virus (Fig. 9A, bottom portion of heat map). Analysis of GO terms associated with this down-regulated set of transcripts indicates that mRNAs associated with the cellular response to virus infections, stress, and anti-apoptosis are significantly (p ≤ 0.01) over-represented. The down-regulation of these transcripts associated with sfRNA-producing Kunjin virus infections may be a direct or indirect effect of sfRNA formation. In summary, the production of sfRNA during a Kunjin virus infection is associated with major global changes in cellular gene expression.
DISCUSSION
This study describes the characterization of a novel virus-derived RNA inhibitor of the major cellular RNA decay factor XRN1. While poly(G) tracts and related structural elements have been used in the past to stall the exoribonuclease in yeast and trypanosomes (Muhlrad et al. 1994; Li et al. 2006), these types of structures have not consistently worked in mammalian cells. The observation that the pseudoknot structures in sfRNA cannot only stall XRN1 from a variety of sources but also inhibit the enzymatic activity was, therefore, rather unexpected. Pseudoknots and related structures have been shown in the past to hinder the action of other enzymatic complexes that move directionally along RNA. They can serve as roadblocks for ribosomes (Tholstrup et al. 2012) and influence frameshifting rates (Namy et al. 2006).
Based on previous work (Pijlman et al. 2008; Funk et al. 2010; Silva et al. 2010) and our observations for Dengue and West Nile viruses reported here, the ability to generate sfRNA and repress XRN1 is likely to be conserved throughout the arthropod-borne flaviviruses. Since the formation of sfRNA inactivates XRN1 in both mosquito and human cell extracts (Figs. 1, 2), other non-arthropod-borne members of the Flaviviridae such as hepatitis C virus and Bovine Viral Diarrhea virus may interface with the cellular RNA decay machinery by a related mechanism. It is clear that naturally uncapped viral mRNAs from HCV have defined mechanisms to protect viral mRNAs from 5′ exonucleases like XRN1 (Shimakami et al. 2012). Human enteroviruses/rhinoviruses also have developed ways to protect their transcripts from XRN1 (Murray et al. 2001; Kempf and Barton 2008a,b). Other positive-strand RNA viruses have well-described structural elements that might be able to stall and/or inhibit XRN1. These include the large stem–loop structures present in the ambisense RNA segments of arenaviruses (Pinschewer et al. 2005) and pseudoknot/double hairpin structures of coronaviruses (Stammler et al. 2011). Several positive-sense plant viruses also generate some apparent subgenomic stable decay products that may arise by a mechanism similar to the flavivirus sfRNA (Iwakawa et al. 2008; Peltier et al. 2012). We are currently assessing if these viruses may interact with XRN1 through mechanisms related to those we describe here for Dengue and West Nile viruses.
The repression of XRN1 by flaviviruses could have several important consequences during an infection. While the level of uncapped mRNAs is increased (Fig. 3), the relative amount of uncapped mRNAs is still rather small compared with the large amount of capped versions of individual mRNAs in infected cells. Therefore, the increase in the relative level of uncapped mRNA in infected cells is almost certainly not sufficient to account for the increase in cellular mRNA half-lives that we detect in infected cells (Figs. 6–8). Uncapped mRNA levels, however, could serve to send a signal to the cell, perhaps through novel pattern recognition receptors. Shutting down the major 5′-to-3′ decay pathway in cells is very likely to have significant ramifications for the cell. Factors of this pathway demonstrate a great deal of cross-communication (Garneau et al. 2007; Kulkarni et al. 2010). Thus the repression of XRN1 activity is likely to feed back and disrupt other components of the pathway. P-bodies, for example, which are known to contain concentrations of interacting 5′-to-3′ decay pathway components (Beckham and Parker 2008), are disrupted in West Nile and Dengue infections by an as-yet-unknown mechanism (Emara and Brinton 2007). Perhaps this is related to sfRNA formation and is a consequence of the direct feedback of XRN1 repression that disrupts the entire pathway. Disrupting 5′–3′ mRNA decay should result in a significant interference of cellular RNA regulation—as evidenced by the changes in mRNA half-lives (Figs. 6–8) and inferred from the dramatic changes in mRNA expression (Fig. 9) observed in this study.
Alterations on cellular mRNA stability by flavivirus sfRNA are likely to have a significant mechanistic role in viral-induced pathogenesis. Based on models of the kinetics of mRNA accumulation (Elkon et al. 2010), the alterations in mRNA half-lives that we observe during flavivirus infections are likely to have a much greater influence on the expression of naturally short-lived mRNAs as compared with longer-lived transcripts. Interestingly, many cytokines and factors involved in aspects of innate immunity are often encoded by short-lived transcripts (Schott and Stoecklin 2010). Thus flaviviruses may use this strategy to disturb the normal regulation of these factors and escape cellular surveillance mechanisms that would normally limit their replication and perhaps induce immunopathogenesis. As a timely example of this, West Nile virus sfRNA has been recently shown to interfere with an interferon response (Schuessler et al. 2012). In addition, cell cycle genes and several major transcriptional regulators in mammalian cells are also encoded by short-lived mRNAs. Thus, disrupting these families of mRNAs may contribute to cytopathology during viral infections. Previous work has indeed demonstrated that West Nile virus variants that are incapable of generating sfRNA replicate less efficiently in some cell types and cause a significantly reduced cytopathology compared with the wild-type virus (Pijlman et al. 2008). We are currently assessing global mRNA half-life changes in flavivirus-infected cells to obtain a clear picture of the effect of sfRNA production on a variety of cellular pathways and RNA regulons.
In summary, flavivirus sfRNA demonstrates a novel way that an RNA virus interfaces with the cellular mRNA decay machinery. This may assist in the stabilization of viral transcripts as well as cause a disruption of the regulation of host cell gene expression. The data described here, along with other recently reported examples of viral RNA interactions with the cellular RNA decay machinery (Sokoloski et al. 2010; Dougherty et al. 2011; Palusa et al. 2012), demonstrate interesting aspects of viral RNA–host interactions that have potentially broad biological implications. These findings also present a potential target for the development of broad-spectrum antivirals as well as discovery tools for novel insights into mechanisms and regulation of cellular mRNA stability.
MATERIALS AND METHODS
Cell lines and virus propagation
C6/36, LLC-MK2, and BHK-21 cells were grown in MEM (Mediatech, Inc.) with 10% fetal bovine serum (FBS). 293T and Vero cells were maintained in DMEM (Mediatech, Inc.) with 10% FBS. HeLa cells were grown in MEM supplemented with 10% horse serum. Mouse embryonic fibroblasts were generated at Osaka University as previously described. Mouse protocols were approved by the Osaka University Animal Care and Use Committee (H19-16-0 and H20-03-0). All experiments were in accordance with the specifications of the Association for Assessment and Accreditation of Laboratory Animal Care.
Dengue virus type 2 (Jamaica 1409) was propagated on C6/36 cells and titers determined by plaque titration on LLC-MK2 cells. Kunjin viruses (FLSDX and FL-IRAdCS3) were recovered from corresponding infectious cDNA clones (Pijlman et al. 2008), propagated on Vero cells and titers determined using plaque titration on BHK-21 cells. 293T cells were infected with Dengue virus or Kunjin virus at a multiplicity of infection of 1 PFU/cell. Virus was allowed to adsorb for 2 h, cells were rinsed twice with maintenance medium, and experiments were performed at the indicated times after adsorption.
Plasmids and transfections
The Dengue virus type 2 3′ untranslated region was amplified from total infected cellular RNA using RT-PCR with the following primers: 5′-GCGGCCGCAAGGCAAAACTAACATGAAC, 5′-GCGGCCGCAGAACCTGTTGATTCAACAG and cloned into the NotI site in the 3′ untranslated region of eGFP in peGFP-N1 to create the DenV 3′-UTR-containing reporter plasmid. Purified plasmid DNA treated with MiraClean endotoxin reagent was transfected into 293T cells using Lipofectamine 2000 (Invitrogen).
Cell-free XRN1 RNA decay assays
The reporter RNA for XRN1 assays was derived from pGem4 digested with Ear I. To prepare the Reporter-DenV 3′-UTR RNA, the stem–loop 1 region of the 3′ UTR of Dengue virus type 2 that contains the first pseudoknot-type structure previously implicated in sfRNA formation (Funk et al. 2010; Silva et al. 2010) (5′-AAGAAGUCAGGCCAUCACAAAUGCCACAGCUUGAGUAAACUGUGCAGCCUGUAGCUCCACC) was PCR-amplified and cloned into pGem4 at the XbaI and HindIII sites. Plasmid DNA was linearized with EarI. The nonspecific competitor RNA was derived from pGem4 templates digested with SmlI. To prepare the DenV 3′-UTR competitor RNA, the proximal half of the 3′ UTR of Dengue virus type 2 that contains both of the pseudoknot-type structures previously implicated in sfRNA formation (Funk et al. 2010; Silva et al. 2010) (5′-AAGGCAAAACUAACAUGAAACAAGGCUAAAAGUCAGGUCGGAUCAAGCCAUAGUACGGAAAAAACUAUGCUACCUGUGAGCCCCGUCCAAGGACGUUAAAAGAAGUCAGGCCAUCACAAAUGCCACAGCUUGAGUAAACUGUGCAGCCUGUAGCUCCACCUGAGAAGGUGUAAAAAAUCUGGGAGGCCACAAACCAUGGAAGCUGUACGCAUGGCGUAG) was PCR-amplified and cloned into pGem4 at the XbaI and HindIII sites. Plasmid DNA was linearized with EarI. RNAs were made by in vitro transcription using SP6 polymerase in the presence of a 10× molar excess of cap analog or 5′ GMP to GTP in the reaction in order to obtain transcripts with a 5′ cap or 5′ monophosphate, respectively. Approximately 30 fm of radioactive RNAs was incubated in HeLa or C6/36 cytoplasmic S100 extracts prepared as described previously (Ford et al. 1999; Sokoloski et al. 2008) under conditions that strongly favor 5′-to-3′ exonucleolytic decay (Mukherjee et al. 2002) or with recombinant yeast XRN1 protein (New England Biolabs) for the times indicated. Reaction products were analyzed on 5% polyacrylamide gels containing 7 M urea and visualized by phosphorimaging.
XRN1 knockdown was achieved by lentivirus-mediated transduction of a Mission shRNA vector (Sigma-Aldrich) specific to XRN1 (TRCN0000049677) or the empty LKO.1 vector lacking a specific shRNA sequence. Two days after lentivirus transduction, 293T cell pools were selected for shRNA expression with puromycin (5 μg/mL). XRN1 expression was determined by qRT-PCR (forward primer: 5′-GCGGCACCTGCCATCTTCTGA; and reverse primer: 5′-GAACTGTCAGCTTGCTGTGCTCAA).
mRNA half-life analysis
Maintenance medium was replaced with fresh medium containing the transcriptional inhibitor actinomycin D (5 μg/mL) and RNA collections begun after 30 min of drug treatment to ensure complete shutoff of cellular transcription. Total RNA was isolated using TRIzol (Invitrogen) and treated with DNase I (Fermentas). Reverse transcription was done using 1 μg of total RNA with Improm-II Reverse Transcriptase (Promega) and random hexamer primers. The abundance of GAPDH, FOS, and TUT1 mRNA was determined by quantitative real-time PCR using the CFX96 real-time PCR detection system and Bio-Rad SYBR-Green supermix. The primers used were as follows: GAPDH forward: 5′-TCTTTTGCGTCGCCAGCCGA; GAPDH reverse: 5′-ACCAGGCGCCCAATACGACC; TUT1 forward: 5′-AGGCAGCCCAAGAATGGTC; TUT1 reverse: 5′-GCCACCACTCAGTCCTTTCA; FOS forward: 5′-GTGGGAATGAAGTTGGCACT; FOS reverse: 5′-CTACCACTCACCCGCAGACT. The ΔΔCt method was used to analyze data using GAPDH as a reference gene. The data shown represent the mean values from at least two independent experiments and error bars and ±calculations represent the standard deviation of each measurement. P-values were determined by a Student's t-test.
Northern blotting
One microgram of total RNA was run on a 5% denaturing polyacrylamide gel and transferred to a nylon membrane (Amersham Hybond-XL). Internally radiolabeled probes containing the antisense 3′ untranslated regions of Dengue virus or Kunjin virus were hybridized to the membrane, and RNAs were visualized by phosphorimaging.
Immunoprecipitation of capped mRNAs
Total RNA from cells infected with Dengue virus or Kunjin virus at the indicated times post-infection was fractionated into capped and uncapped portions using a mouse monoclonal antibody to m3G-cap/m7G-cap (Synaptic Systems) following the manufacturer's protocol. RNA was purified from 10% of the input sample, the supernatant (unbound) fraction, and the immunoprecipitated fraction and reverse-transcribed using Improm II reverse transcriptase (Promega). The abundance of 7SL, FOS, and TUT1 mRNA was determined by qRT-PCR using the Bio-Rad CFX96 real-time PCR detection system and Bio-Rad SYBR-Green supermix using the TUT1 and FOS primers described above and the following: 7SL forward: 5′-GGAGTTCTGGGCTGTAGTGC; 7SL reverse: 5′-ATCAGCACGGGAGTTTTGAC. The relative abundance of FOS and TUT1 mRNA in the uncapped fractions was determined relative to the immunoprecipitated, capped fractions using the ΔΔCt method with 7SL as the reference gene. Total relative amount of uncapped FOS and TUT1 mRNAs in each sample was determined by comparing with the abundance in the 10% input sample.
Coimmunoprecipitation of XRN1 and viral RNAs
293T cells infected with Dengue virus (at 4 d post-infection) or Kunjin virus (at 2 d post-infection) were washed with PBS, resuspended in 1% formaldehyde, and rocked 15 min at room temperature. The reaction was stopped after 15 min with 0.25 M glycine. Cells were rinsed with PBS and resuspended in cold RIPA buffer (50 mM Tris at pH 7.5, 1% NP-40 [v/v], 0.5% sodium deoxycholate, 0.05% SDS [w/v], 1 mM EDTA, 150 mM sodium chloride) plus 1 μL of RNase inhibitor (Fermentas). Lysates were sonicated and insoluble particles removed by centrifugation. Lysates plus 2 μL of rabbit monoclonal XRN1 antibody (the kind gift of Dr. J. Lykke-Andersen) or 2 μL of normal rabbit IgG and 1 μL of RNase inhibitor (Fermentas) were rotated for 1 h at 4°C. Pansorbin Staph A-positive cells were added to the lysates and incubated for 15 min at 4°C with rotation. Pellets were resuspended in RIPA containing 1 M urea and vortexed vigorously, and washed five times with RIPA with 1 M urea. All samples were heated to 70°C with shaking for 45 min to reverse formaldehyde cross-links. Supernatants were collected following centrifugation to remove insoluble particles and RNA extracted using TRIzol (Invitrogen) following the manufacturer's instructions. RNA was treated with DNase I (Fermentas) and reverse-transcribed with random hexamer primers. Viral 3′ UTR, NS3, or NS5 was amplified using Taq polymerase (GoTaq Flexi; Promega) with the following primers: Kunjin 3′ UTR forward: 5′-GGAGACCCCGTGCCGCAAAA and reverse: 5′-CACTGTGCCGTGTGGCTGGT; Kunjin NS3 forward: 5′-TGGACGGGGAATACCGACTTAGAGG and reverse: 5′-ACCCCAGCTGCTGCCACCTT; Dengue 3′ UTR forward: 5′-AGGCCATCACAAATGCCACAGC and reverse: 5′-ACGCCATGCGTACAGCTTCCA; Dengue NS5 forward: 5′-GTGCGTGTGCAAAGACCAACACC and reverse: 5′-AAGGCCATAGGTGCCGACTTGC. PCR products were run on a 2% agarose gel and stained with ethidium bromide, and band intensities were quantified using Quantity One software.
Microarray analysis
IFNAR-deficient MEF cells were infected with wild-type Kunjin virus or the sfRNA-deficient IRAdCS3 mutant virus at an MOI of 1 PFU/cell. At 48 h post-infection, total RNA was isolated using TRIzol (Invitrogen), further purified with the RNeasy Mini kit (QIAGEN), and then subjected to cDNA library preparation with the WT-Ovation RNA Amplification System and Encore Biotin Module (Nugen) according to the manufacturers' instructions. The resulting cDNA was hybridized to Mouse 430 2.0 Arrays (Affymetrix). The arrays were washed, stained, and scanned according to the manufacturer's recommendations. Robust multiarray analysis (RMA) values were calculated and hierarchical clustering was performed for genes having threefold or more differences in RMA values after transforming the values to have mean 0 and standard deviation 1. Genes were placed into six clusters according to Akaike's information criterion. All calculations were done using R and Bioconductor. Relative increases in expression were confirmed for the four indicated cellular mRNAs by qRT-PCR analysis of samples obtained from three independent infections using an MOI of 5 PFU/cell. The following primers were used in the analysis: CXCL11: 5′-CATGAGTGTGAAGGGCATGGCT and 5′-CCAGGGCCTATGCAAAGACAGCG; CCL2: 5′-TGAAGCTCGCACTCTCGCCT and 5′-TGAGCGAGCCCTTGGGGAATGA; OASL: 5′-TCCTTCAGCGAGCTGCAGAGAAAT and 5′-CCGGATCCAGGATGATGGGCCT; and IL6: 5′-TCGAGCCCACCGGGAACGAA and 5′-GCAACTGGACCGAAGGCGCT.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Jens Lykke-Andersen (UCSD) for the kind gift of XRN1 antibody and members of the Wilusz laboratory for helpful comments. These studies were supported by NIH grant AI063434, an NIAID award through the Rocky Mountain Regional Center of Excellence (U54 AI-065357) to J.W., and an NH&MRC grant 631365 to A.A.K. S.L.M. received support from a USDA NIFA NNF training grant (2010-38420-20367).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.034330.112.
REFERENCES
- Astuti D, Morris MR, Cooper WN, Staals RH, Wake NC, Fews GA, Gill H, Gentle D, Shuib S, Ricketts CJ, et al. 2012. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet 44: 277–284 [DOI] [PubMed] [Google Scholar]
- Beckham CJ, Parker R 2008. P-bodies, stress granules, and viral life cycles. Cell Host Microbe 3: 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman A, Avraham-Kelbert M, Barkai O, Duek L, Guterman A, Choder M 2011. Promoter elements regulate cytoplasmic mRNA decay. Cell 147: 1473–1483 [DOI] [PubMed] [Google Scholar]
- Chang JH, Xiang S, Xiang K, Manley JL, Tong L 2011. Structural and biochemical studies of the 5′→3′ exoribonuclease Xrn1. Nat Struct Mol Biol 18: 270–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, Gorospe M, Becker KG 2005. Control of gene expression during T cell activation: Alternate regulation of mRNA transcription and mRNA stability. BMC Genomics 6: 75 doi: 10.1186/1471-2164-6-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Shyu AB 2011. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA 2: 167–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson AM, Wilusz J 2011. Strategies for viral RNA stability: Live long and prosper. Trends Genet 27: 286–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD, White JP, Lloyd RE 2011. Poliovirus-mediated disruption of cytoplasmic processing bodies. J Virol 85: 64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Zlotorynski E, Zeller KI, Agami R 2010. Major role for mRNA stability in shaping the kinetics of gene induction. BMC Genomics 11: 259 doi: 10.1186/1471-2164-11-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Brinton MA 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci 104: 9041–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford LP, Watson J, Keene JD, Wilusz J 1999. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev 13: 188–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori Melian E, Dong H, Shi PY, Khromykh AA 2010. RNA structures required for production of subgenomic flavivirus RNA. J Virol 84: 11407–11417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ 2007. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126 [DOI] [PubMed] [Google Scholar]
- Iwakawa HO, Mizumoto H, Nagano H, Imoto Y, Takigawa K, Sarawaneeyaruk S, Kaido M, Mise K, Okuno T 2008. A viral noncoding RNA generated by cis-element-mediated protection against 5′ → 3′ RNA decay represses both cap-independent and cap-dependent translation. J Virol 82: 10162–10174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Coyle SM, Doudna JA 2011. Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol Cell 41: 600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf BJ, Barton DJ 2008a. Poliovirus 2APro increases viral mRNA and polysome stability coordinately in time with cleavage of eIF4G. J Virol 82: 5847–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf BJ, Barton DJ 2008b. Poly(rC) binding proteins and the 5′ cloverleaf of uncapped poliovirus mRNA function during de novo assembly of polysomes. J Virol 82: 5835–5846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Kenney MT, Westaway EG 1998. Trans-complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J Virol 72: 7270–7279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni M, Ozgur S, Stoecklin G 2010. On track with P-bodies. Biochem Soc Trans 38: 242–251 [DOI] [PubMed] [Google Scholar]
- Li CH, Irmer H, Gudjonsdottir-Planck D, Freese S, Salm H, Haile S, Estevez AM, Clayton C 2006. Roles of a Trypanosoma brucei 5′ → 3′ exoribonuclease homolog in mRNA degradation. RNA 12: 2171–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Song M, Kiledjian M 2011. Differential utilization of decapping enzymes in mammalian mRNA decay pathways. RNA 17: 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Tomecki R, Jensen TH, Dziembowski A 2011. The eukaryotic RNA exosome: Same scaffold but variable catalytic subunits. RNA Biol 8: 61–66 [DOI] [PubMed] [Google Scholar]
- Mühlemann O, Jensen TH 2012. mRNP quality control goes regulatory. Trends Genet 28: 70–77 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ → 3′ digestion of the transcript. Genes Dev 8: 855–866 [DOI] [PubMed] [Google Scholar]
- Mukherjee D, Gao M, O'Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J 21: 165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EL, Schoenberg DR 2007. A+U-rich instability elements differentially activate 5′–3′ and 3′–5′ mRNA decay. Mol Cell Biol 27: 2791–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray KE, Roberts AW, Barton DJ 2001. Poly(rC) binding proteins mediate poliovirus mRNA stability. RNA 8: 1126–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I 2006. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441: 244–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palusa S, Ndaluka C, Bowen RA, Wilusz CJ, Wilusz J 2012. The 3′ untranslated region of the rabies virus glycoprotein mRNA specifically interacts with the cellular PCBP2 protein and promotes transcript stability. PLoS ONE 7: e33561 doi: 10.1371/journal.pone.0033561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier C, Klein E, Hleibieh K, D'Alonzo M, Hammann P, Bouzoubaa S, Ratti C, Gilmer D 2012. Beet necrotic yellow vein virus subgenomic RNA3 is a cleavage product leading to stable non-coding RNA required for long distance movement. J Gen Virol 93: 1093–1102 [DOI] [PubMed] [Google Scholar]
- Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, et al. 2008. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4: 579–591 [DOI] [PubMed] [Google Scholar]
- Pinschewer DD, Perez M, de la Torre JC 2005. Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation. J Virol 79: 4519–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaljo B, Kratochvill F, Gratz N, Sadzak I, Sauer I, Hammer M, Vogl C, Strobl B, Muller M, Blackshear PJ, et al. 2009. Tristetraprolin is required for full anti-inflammatory response of murine macrophages to IL-10. J Immunol 183: 1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR 2011. Mechanisms of endonuclease-mediated mRNA decay. Wiley Interdiscip Rev RNA 2: 582–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott J, Stoecklin G 2010. Networks controlling mRNA decay in the immune system. Wiley Interdiscip Rev RNA 1: 432–456 [DOI] [PubMed] [Google Scholar]
- Schuessler A, Funk A, Lazear HM, Cooper DA, Torres S, Daffis S, Jha BK, Kumagai Y, Takeuchi O, Hertzog P, et al. 2012. West Nile virus non-coding subgenomic RNA contributes to viral evasion of type 1 interferon-mediated antiviral response. J Virol 86: 5708–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Shalem O, Groisman B, Choder M, Dahan O, Pilpel Y 2011. Transcriptome kinetics is governed by a genome-wide coupling of mRNA production and degradation: A role for RNA Pol II. PLoS Genet 7: e1002273 doi: 10.1371/journal.pgen.1002273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM 2012. Stabilization of hepatitis C virus RNA by an Ago2–miR-122 complex. Proc Natl Acad Sci 109: 941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva PA, Pereira CF, Dalebout TJ, Spaan WJ, Bredenbeek PJ 2010. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J Virol 84: 11395–11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloski KJ, Wilusz J, Wilusz CJ 2008. The preparation and applications of cytoplasmic extracts from mammalian cells for studying aspects of mRNA decay. Methods Enzymol 448: 139–163 [DOI] [PubMed] [Google Scholar]
- Sokoloski KJ, Dickson AM, Chaskey EL, Garneau NL, Wilusz CJ, Wilusz J 2010. Sindbis virus usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells. Cell Host Microbe 8: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stammler SN, Cao S, Chen SJ, Giedroc DP 2011. A conserved RNA pseudoknot in a putative molecular switch domain of the 3′-untranslated region of coronaviruses is only marginally stable. RNA 17: 1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Mayo T, Anderson P 2006. ARE-mRNA degradation requires the 5′–3′ decay pathway. EMBO Rep 7: 72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholstrup J, Oddershede LB, Sørensen MA 2012. mRNA pseudoknot structures can act as ribosomal roadblocks. Nucleic Acids Res 40: 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend HL, Jha BK, Han JQ, Maluf NK, Silverman RH, Barton DJ 2008. A viral RNA competitively inhibits the antiviral endoribonuclease domain of RNase L. RNA 14: 1026–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Larson DR, Moldón A, Query CC, Singer RH 2011. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell 147: 1484–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]