Abstract
Evolution or engineering of novel metabolic pathways can endow microbes with new abilities to degrade anthropogenic pollutants or synthesize valuable chemicals. Most studies of the evolution of new pathways have focused on the origins and quality of function of the enzymes involved. However, there is an additional layer of complexity that has received less attention. Introduction of a novel pathway into an existing metabolic network can result in inhibitory cross-talk due to adventitious interactions between metabolites and macromolecules that have not previously encountered one another. Here, we report a thorough examination of inhibitory cross-talk between a novel metabolic pathway for synthesis of pyridoxal 5′-phosphate and the existing metabolic network of Escherichia coli. We demonstrate multiple problematic interactions, including (i) interference by metabolites in the novel pathway with metabolic processes in the existing network, (ii) interference by metabolites in the existing network with the function of the novel pathway, and (iii) diversion of metabolites from the novel pathway by promiscuous activities of enzymes in the existing metabolic network. Identification of the mechanisms of inhibitory cross-talk can reveal the types of adaptations that must occur to enhance the performance of a novel metabolic pathway as well as the fitness of the microbial host. These findings have important implications for evolutionary studies of the emergence of novel pathways in nature as well as genetic engineering of microbes for “green” manufacturing processes.
Keywords: molecular evolution, metabolic engineering, serendipitous pathway, synthetic biology, biodegradation
Metabolic networks typically consist of hundreds of small molecules and enzymes (1, 2). Efficient function of networks requires that proteins recognize specific small metabolites while excluding structurally similar molecules. The exquisite specificity exhibited by many proteins has evolved in response to selective pressures for metabolic efficiency as well as the flexibility to respond to environmental perturbations by directing fluxes appropriately through catabolic and anabolic pathways. Mechanisms for molecular discrimination evolve to exclude ligands (i) to which proteins are exposed in the metabolic network and (ii) that decrease fitness by interfering with a catalytical or regulatory process. Previously unencountered small molecules can cause problems in multiple ways. They can inhibit enzymes by competing with structurally similar natural substrates or by binding to allosteric sites normally occupied by natural ligands. They can serve as substrates for promiscuous enzymes, resulting in depletion of cosubstrates, generation of useless or toxic products, and interference with the flux of normal reactions. They can also interfere with regulatory processes via adventitious binding to transcriptional regulators.
Adventitious interactions between novel compounds and proteins in metabolic networks can interfere with microbial evolution of new metabolic pathways in the environment. Exposure of microbes to anthropogenic compounds such as pesticides, munitions, antibiotics, and dyes provides selective pressures that can foster evolution of new metabolic pathways that enable biodegradation of problematic environmental pollutants. For example, pathways for degradation of pesticides such as atrazine and pentachlorophenol likely arose due to selective pressure for use of a novel source of nitrogen (atrazine) or for elimination of a toxin as well as utilization of a new source of carbon (pentachlorophenol) (3). Evolution of strains capable of efficient biodegradation is likely to require not only recruitment of the enzymes necessary to degrade a novel compound but also adaptations that prevent new metabolites from interfering with the functions of preexisting enzymes and transcriptional regulators.
Adventitious interactions between novel compounds and proteins in microbial metabolic networks also pose significant problems for efforts to engineer microbes for environmentally sustainable production of fuels, chemicals, and pharmaceuticals. Often, “green” processes rely on introduction of novel metabolic pathways patched together from enzymes that have some ability to catalyze the needed transformations, even if this is not their primary activity, into suitable microbial hosts. Synthetic biologists have generated metabolic pathways that do not exist in nature for synthesis of platform chemicals such as isobutyric acid (4), specialty chemicals such as vanillin (5), and a variety of secondary metabolites that have potential uses as therapeutic agents (6, 7). Notable successes include the microbial syntheses of amorphadiene (8), a precursor of the antimalarial drug artemisinin, and taxadiene, the precursor of the potent anticancer drug Taxol (9). Assembly of a pathway that degrades the toxic insecticide paraoxon has also been reported (10).
Given the thousands of potential interactions between components of a novel pathway and an existing network, it is difficult to know a priori how many and which interactions may be particularly problematic. In this work, we report a thorough examination of inhibitory cross-talk between a new metabolic pathway and the existing metabolic network of Escherichia coli. The experimental system used for this analysis takes advantage of a recently discovered pathway that allows a strain of E. coli that lacks 4-phosphoerythronate (4PE) dehydrogenase (PdxB), an enzyme in the pathway for pyridoxal 5′-phosphate (PLP) synthesis, to synthesize PLP by diverting an intermediate from serine biosynthesis and converting it into an intermediate downstream of the block in the PLP synthesis pathway (Fig. 1). This system provides an excellent opportunity for examination of the extent and mechanisms of inhibitory cross-talk between a new pathway and an existing metabolic pathway because two of the intermediates in the previously undescribed pathway are not known to be normal metabolites in E. coli. We have uncovered multiple problematic interactions that can be grouped into three classes: (i) interference by metabolites in the new pathway with metabolic processes in the existing network, (ii) interference by metabolites in the existing network with the function of the novel pathway, and (iii) diversion of metabolites from the novel pathway by enzymes in the existing metabolic network.
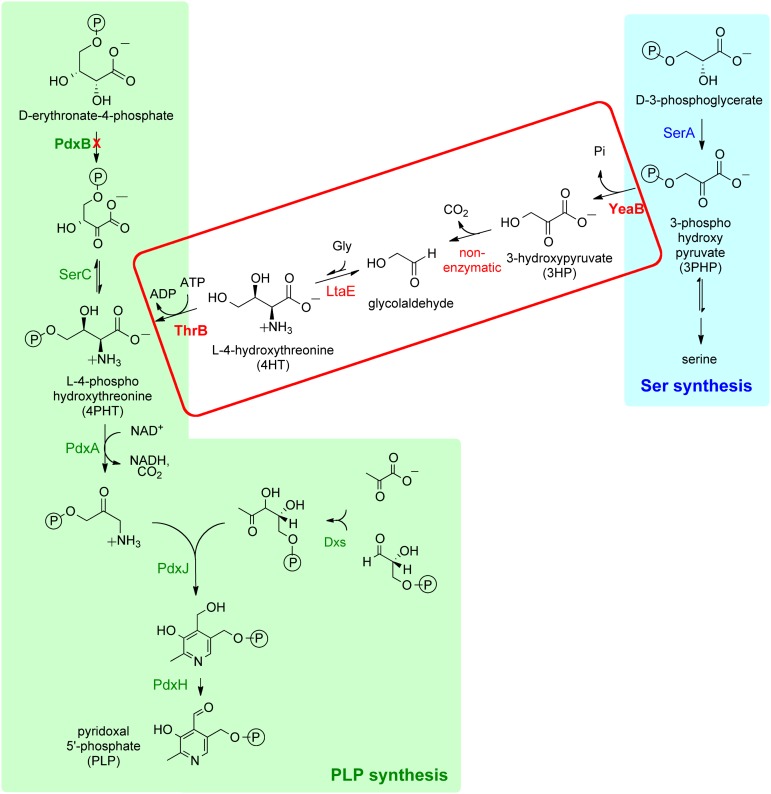
Fig. 1.
A serendipitous pathway that can restore PLP biosynthesis in a strain of E. coli that lacks pdxB when either yeaB or thrB is overexpressed.
The pathway shown in the red box in Fig. 1 was discovered during experiments designed to reveal multicopy suppressors that restored growth in cells in which genes encoding essential metabolic enzymes had been deleted. Remarkably, we identified seven genes that, when individually overexpressed, restored the ability of a strain lacking pdxB to grow on glucose (11). The enzymes encoded by these genes include homoserine kinase (ThrB), imidazole glycerol phosphate dehydratase/histidinol phosphate phosphatase (HisB), dehydroquinate synthase (AroB), 4-hydroxy-l-threonine phosphate dehydrogenase (PdxA), and three enzymes of unknown function (YeaB, Php, and YjbQ). Surprisingly, none of the enzymes encoded by these genes has a promiscuous activity that can replace PdxB. Rather, overexpression of these genes facilitates at least three “serendipitous” pathways patched together from promiscuous enzyme activities that generate intermediates downstream of the block in the PLP synthesis pathway.
We previously characterized the new PLP synthesis pathway that is facilitated by overexpression of yeaB or thrB (11) (Fig. 1). This pathway diverts a metabolite from serine biosynthesis and, via a series of four reactions, forms 4-phosphohydroxythreonine (4PHT). YeaB, a predicted NUDIX (nucleoside diphosphate linked to moiety X) hydrolase of unknown function, converts 3-phosphohydroxypyruvate (3PHP) to 3-hydroxypyruvate (3HP). 3HP undergoes nonenzymatic decarboxylation to produce glycolaldehyde. LtaE, a low-specificity threonine aldolase whose physiological function is also unknown, catalyzes the condensation of glycolaldehyde and glycine to a mixture of 4-hydroxythreonine (4HT) and 4-hydroxy-allo-threonine. ThrB phosphorylates 4HT, generating 4PHT, an intermediate in the normal PLP synthesis pathway. Flux through this pathway can be increased to a physiologically significant level if intermediates in the pathway are supplied in the medium or if either YeaB or ThrB is overproduced (11). Overproduction of YeaB increases flux of 3HP into the pathway by diverting 3PHP from serine biosynthesis. Overproduction of ThrB pulls material through the pathway by irreversibly phosphorylating 4HT, the product of the thermodynamically unfavorable aldol condensation of glycolaldehyde and glycine.
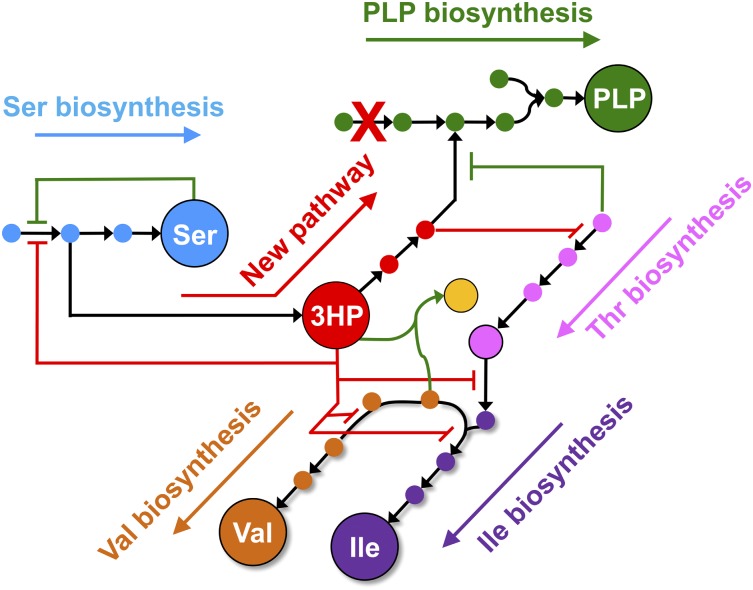
Two of the intermediates in the new PLP synthesis pathway, 3HP and 4HT, are not normal metabolites in E. coli, and thus have the potential to interfere with enzymes and/or regulatory proteins. Here, we show that 3HP inhibits production of serine and branched chain amino acids and 4HT interferes with threonine biosynthesis (Fig. 2). Conversely, components of the normal metabolic network interfere with the new PLP synthesis pathway in three ways. Serine inhibits 3-phospho-d-glycerate dehydrogenase (SerA) and prevents formation of 3PHP, the first intermediate in the new pathway. Homoserine inhibits conversion of 4HT to 4PHT in the new pathway. Finally, an isozyme of acetohydroxyacid synthase diverts 3HP from the new pathway and forms a dead-end metabolite. This work provides an important demonstration of the molecular mechanisms by which inhibitory cross-talk between a novel pathway and the existing metabolic network can impair performance of the novel pathway as well as fitness of the microbe.
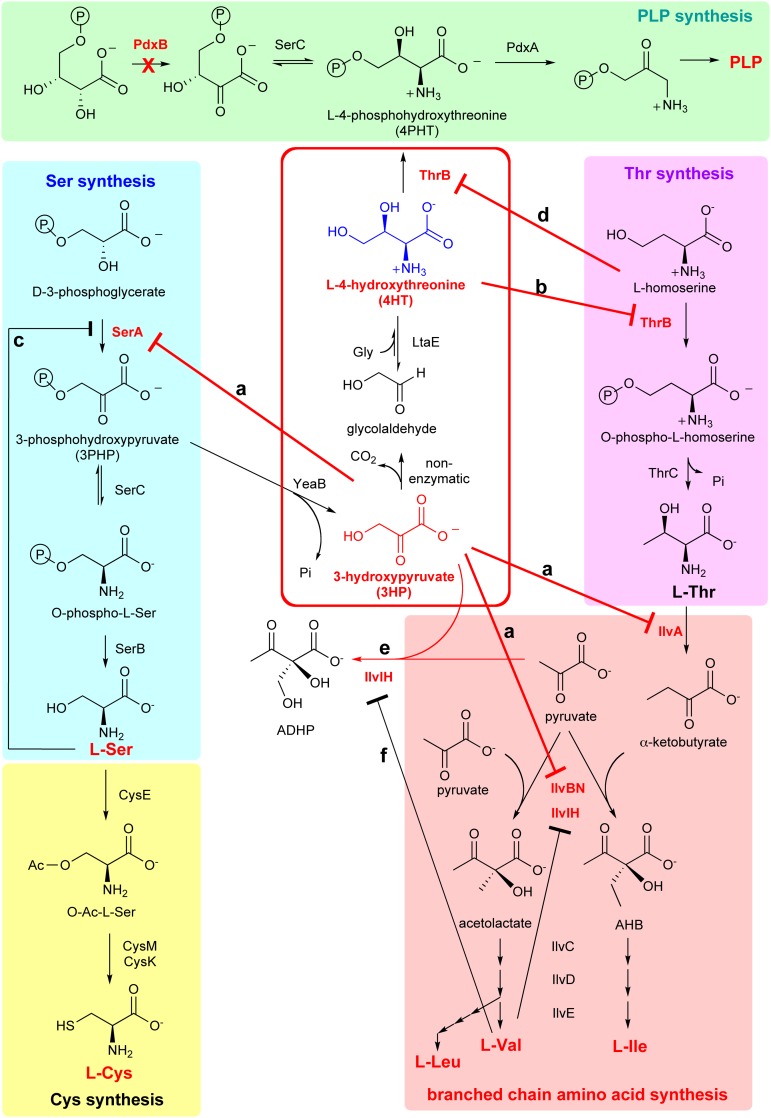
Fig. 2.
Summary of interactions between a new metabolic pathway for synthesis of PLP (in red box) and the existing metabolic network of E. coli K12. 3HP inhibits enzymes in serine and branched chain amino acid biosynthesis (a); 4HT inhibits threonine biosynthesis (b); serine inhibits SerA and prevents formation of 3PHP, the precursor for the serendipitous pathway (c); homoserine inhibits the serendipitous pathway for PLP synthesis (d); 3HP is diverted from the serendipitous pathway by promiscuous activity of IlvIH (e); and valine inhibits IlvIH and prevents diversion of 3HP from the serendipitous pathway (f).
Results
3HP Is Profoundly Toxic to E. coli.
3HP, an intermediate in the novel pathway for PLP synthesis, is not a common metabolite in E. coli. It is formed from tartronate semialdehyde as a nonproductive side product during degradation of glycolate (Fig. S1). The glycolate degradation operon contains a gene encoding hydroxypyruvate isomerase, which likely facilitates return of 3HP to the pathway for conversion to 3-phospho-d-glycerate. However, in the absence of glycolate, 3HP is not known to be formed or used by any metabolic enzyme.
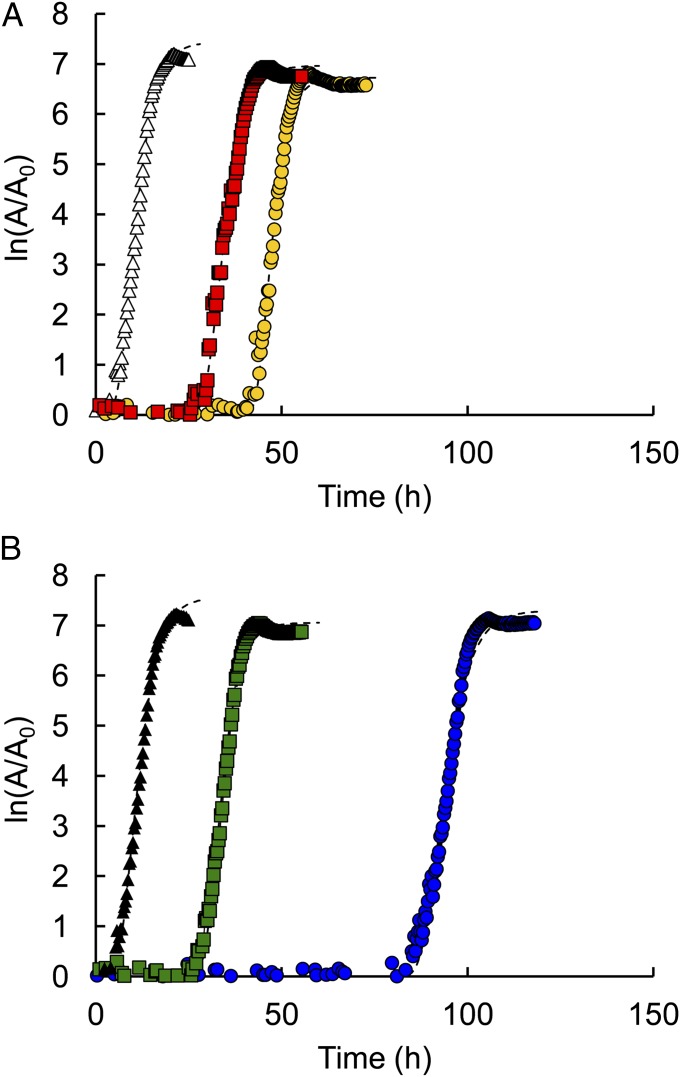
We previously showed that addition of 3HP to the medium allows ΔpdxB E. coli to synthesize PLP via the serendipitous pathway shown in Fig. 1, and thus to grow on glucose (11). However, growth under these conditions is peculiar (Fig. 3A). At least 0.5 mM 3HP is required for growth, but the cells cannot grow with more than 1 mM 3HP. Further, there is a surprisingly long lag phase, particularly in the presence of 1 mM 3HP. Even though the lag phase was longer in the presence of 1 mM 3HP, the overall growth yield was substantially higher. These results suggest that 3HP exerts both stimulatory and inhibitory effects on growth of the ΔpdxB strain. The stimulatory effect can be attributed to the synthesis of PLP (11); the basis for the inhibitory effect is unknown.
Fig. 3.
3HP inhibits the growth of E. coli K12 on M9/glucose. (A) Growth of the ΔpdxB strain with 10 μM pyridoxine (●), 0.5 mM 3HP (red △), or 1.0 mM 3HP (○). (B) Growth of WT E. coli with no supplement (●) or with 0.2 mM 3HP (yellow □), 0.5 mM 3HP (red △), or 1.0 mM 3HP (○). (C) Growth of replicate cultures of WT E. coli K12 on M9/glucose. 3HP (final concentration of 1 mM) was added to one culture (○) at the time indicated by the first arrow. After 90 min (second arrow), supplements were added to give the following final concentrations: 1 mM Ser and 40 μM Ile (yellow ○), 1 mM Ser (□), or 40 μM Ile (blue □). (D) Growth of WT E. coli K12 on M9/glucose with no supplement (●); with 1.0 mM 3HP (○); or with 1.0 mM 3HP, 1 mM Ser, and 40 μM Ile (yellow ○). A = OD600.
We also examined the effect of 3HP on the growth of WT E. coli K12 BW25113. The presence of 3HP caused a substantial lag in the growth of WT E. coli, and the duration of the lag was proportional to the concentration of 3HP in the medium (Fig. 3B). However, neither the eventual growth rate nor the yield was affected by the concentration of 3HP. In a separate experiment, we found that addition of 3HP to a growing culture of E. coli caused nearly immediate cessation of growth (Fig. 3C). Taken together, these data suggest that 3HP is so toxic that growth cannot occur until 3HP is converted to something else.
3HP is converted to 4HT in E. coli by means of the reactions shown in Fig. 1 (11). 4HT has previously been shown to be toxic to E. coli (12). We confirmed that formation of 4HT from 3HP contributes to the toxic effects of 3HP by comparing the growth of WT and ΔltaE strains of E. coli K12 on glucose in the presence of 3HP. (LtaE is required to form 4HT from glycolaldehyde and glycine.) Loss of LtaE does not affect growth of WT E. coli on glucose. However, the lag phase in the presence of 3HP is much shorter for the ΔltaE strain than for the WT strain, demonstrating that LtaE is partially responsible for the toxic effects of 3HP (Fig. 4). We attribute this effect to its ability to generate 4HT. Thus, some of the toxicity of 3HP is due to production of a downstream metabolite.
Fig. 4.
Addition of homoserine or deletion of ltaE diminishes the toxic effects of 3HP. (A) Growth of ΔltaE E. coli K12 on M9/glucose (△), on M9/glucose containing 1 mM 3HP (yellow ○), and on M9/glucose containing 1 mM 3HP and 0.1 mM homoserine (red □). (B) Growth of E. coli K12 on M9/glucose (▲), M9/glucose containing 1 mM 3HP (blue ○), and M9/glucose containing 1 mM 3HP and 0.1 mM homoserine (green □). A = OD600.
Toxicity Caused by 3HP Can Be Relieved by Addition of Specific Amino Acids.
The toxicity of 3HP can be relieved by addition of the 20 proteinogenic amino acids (Fig. S2). By adding individual amino acids to cultures growing on glucose in the presence of 3HP, we found that several amino acids were able to restore growth partially. Serine, cysteine, and branched chain amino acids showed the most dramatic effects (Fig. S3A). Lesser effects were observed with methionine, lysine, arginine, tryptophan, glutamate, a mixture of aspartate and asparagine, and proline (Fig. S3B). Notably, a combination of serine and isoleucine nearly completely prevented the toxic effects of 3HP, whether they were present in the initial medium (Fig. 3D) or were added after cessation of growth due to addition of 3HP during growth (Fig. 3C). These findings suggest that 3HP interferes, either directly or indirectly, with the synthesis of several amino acids and has particularly potent effects on synthesis of serine and isoleucine.
3HP Interferes with Serine Biosynthesis by Binding to an Allosteric Regulatory Site on SerA.
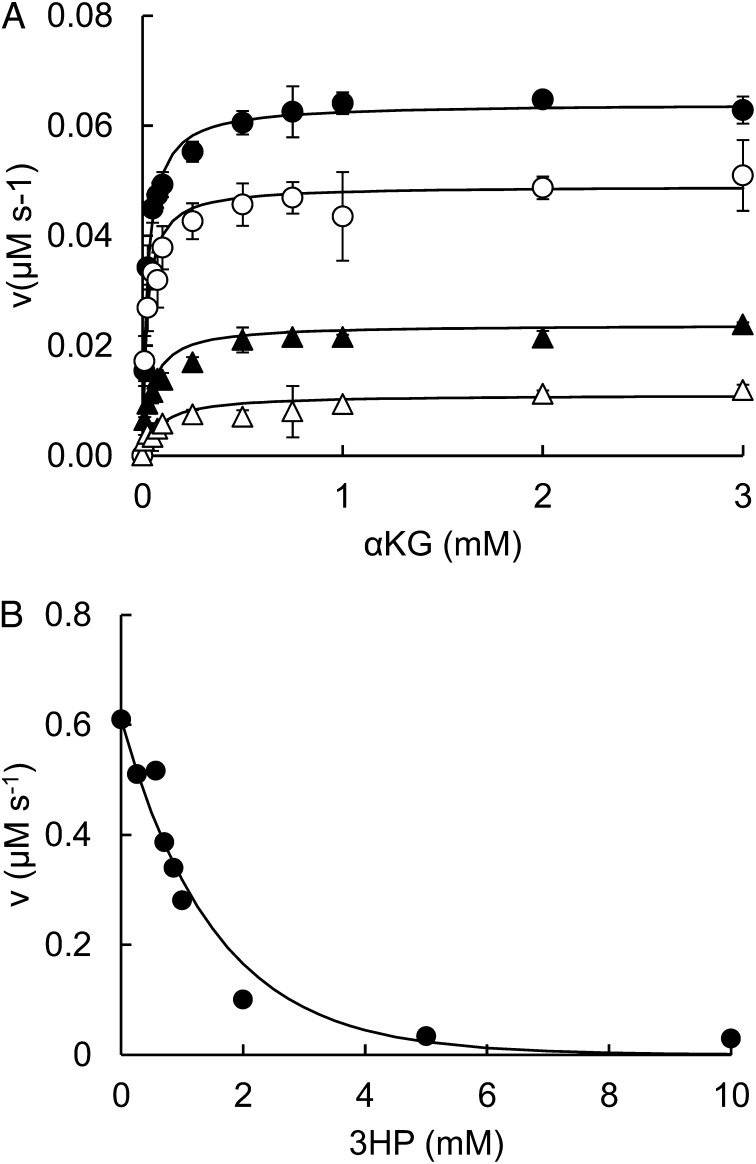
We purified all the enzymes involved in serine and cysteine biosynthesis and assayed their activities in the presence of 3HP. Inhibition was observed only in the case of the first enzyme in serine biosynthesis, SerA, which is inhibited by 3HP with an I0.5 (inhibitor concentration at half the maximal inhibition) of 0.27 ± 0.02 mM and a Hill coefficient of 1.9 ± 0.2 (Fig. 5A and Fig. S4). (SerA has 3-phospho-d-glycerate dehydrogenase and α-ketoglutarate reductase activity. We measured the effect of 3HP on the latter activity.) 3HP is a noncompetitive inhibitor that decreases Vmax while leaving KM unaffected. Feedback inhibition of SerA by serine mediated by the ACT (Aspartate kinase, Chorismate mutase, and TyrA) regulatory domain is also noncompetitive; the I0.5 for serine is 2 μM (13). The structural similarity between 3HP and serine and the observation of noncompetitive inhibition suggest that 3HP might also bind to the ACT regulatory domain of SerA. To test this hypothesis, we generated a serine-insensitive version of SerA by site-directed mutagenesis of Asn364 to alanine (14). The side chain of Asn364 forms a hydrogen bond with the amino group of serine. A change from Asn to Ala at this position abolishes inhibition of SerA by serine (14). Inhibition of N364A SerA by 3HP was weaker than inhibition of the WT enzyme; the I0.5 was 0.42 ± 0.02 mM as opposed to 0.27 ± 0.02 mM. The Hill coefficient remained unchanged (1.9 ± 0.2) (Fig. S4). (Note that Asn364 would be expected to be less critical for binding of 3HP than for binding of serine because 3HP has a carbonyl in place of the amino group of serine.) In sum, the noncompetitive inhibition and the effect of amino acid substitution in the allosteric site are consistent with the hypothesis that 3HP inhibits SerA by binding at the allosteric site. This adventitious interaction has the effect of inappropriately shutting down serine biosynthesis.
Fig. 5.
3HP inhibits SerA (A) and IlvA (B). (A) Inhibition of the α-ketoglutarate (αKG) reductase activity of SerA by 3HP. Assays were carried out using variable concentrations of α-ketoglutarate and no 3HP (●), 0.125 mM 3HP (○), 0.25 mM 3HP (▲), or 0.5 mM 3HP (△). (B) Inhibition of the threonine dehydratase activity of IlvA by 3HP. Assays were performed with 10 mM l-threonine and variable amounts of 3HP. v, velocity.
Inhibition of serine biosynthesis will compromise the synthesis of other metabolites because serine is the precursor of glycine, cysteine, tryptophan, and phospholipids. The effects will not necessarily be equivalent, however, because fluxes toward these different products will be affected by the Km,Ser of the enzymes that use serine, as well as by compensatory regulatory responses that may affect fluxes toward different products to different degrees.
3HP Interferes with Branched Chain Amino Acid Synthesis by Competing with the Substrates for Threonine Synthase and Acetohydroxyacid Synthases.
The initial step in the synthesis of branched chain amino acids involves the condensation of two molecules of pyruvate to make acetolactate, the precursor of valine and leucine, and the condensation of pyruvate and α-ketobutyrate to make acetohydroxybutyrate, the precursor of isoleucine (Fig. 2). These reactions are catalyzed by two isozymes of acetohydroxyacid synthase (IlvIH and IlvBN). (A third isozyme, IlvGM, is not active in E. coli K12 due to a frame shift mutation.) The mechanism of the reaction involves the initial formation of an adduct between pyruvate and thiamin pyrophosphate (TPP) at the active site, followed by reaction with either a second molecule of pyruvate or with α-ketobutyrate. IlvBN and IlvIH are subject to feedback regulation by valine, isoleucine, and leucine, which is mediated by the regulatory subunit.
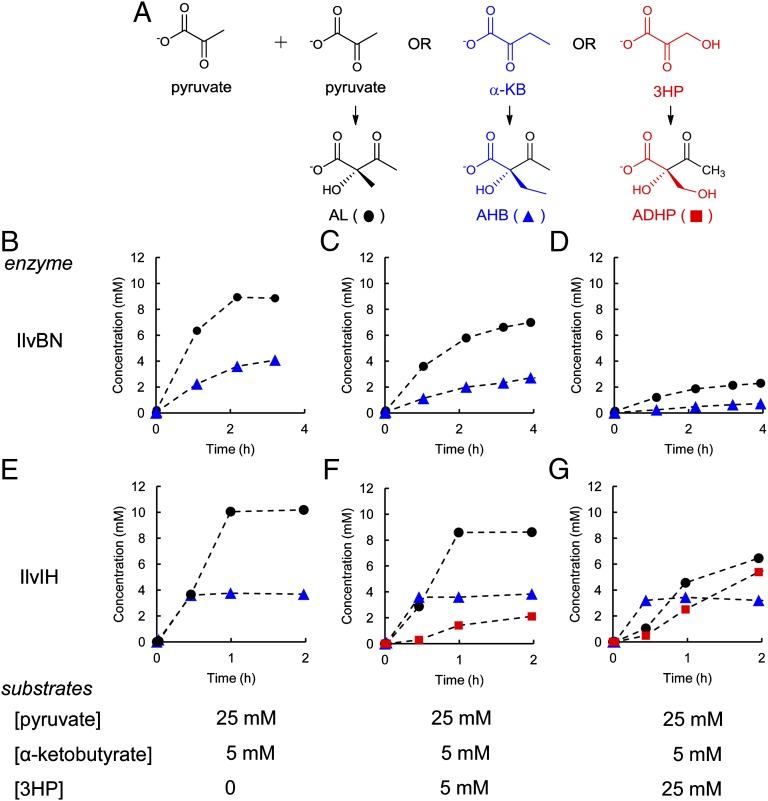
3HP interferes with product formation by IlvBN and IlvIH, but in remarkably different ways (Fig. 6). Addition of 3HP decreases both the initial rate of formation of acetolactate and acetohydroxybutyrate as well as the total amount of product that is formed by IlvBN. These findings are consistent with the observation of suicide inactivation of IlvGM (15) and other TPP-containing enzymes by 3HP and analogs such as 3-fluoropyruvate and 3-bromopyruvate (16–18). The proposed mechanism for this effect (17) is shown in Fig. S5. When 3HP reacts with TPP, the dihydroxyethyl-TPP intermediate formed can eliminate H2O to form an intermediate that tautomerizes to acetyl-TPP. If hydrolysis of the acetyl-TPP is slow, the enzyme can be tied up in a nonproductive form for a considerable period, leading to decreased flux into the branched chain amino acid pathways.
Fig. 6.
Products formed from pyruvate, α-ketobutyrate, and 3HP by acetohydroxyacid synthases IlvBN and IlvIH. (A) Structures of possible products. Formation of acetolactate (AL) and acetohydroxybutyrate (AHB) by IlvBN (B–D); formation of AL, AHB, and ADHP by IlvIH (E–G). Initial concentrations of substrates are given below each time profile.
3HP has a remarkably different effect on IlvIH (Fig. 6). In this case, the enzyme uses 3HP as the second substrate in the reaction, forming 2-aceto-2,3-dihydroxypropanoate (ADHP). α-Ketobutyrate is clearly the preferred substrate, because ADHP is not formed until α-ketobutyrate levels are depleted (Fig. 6). In contrast to the situation with IlvBN, the total amount of product formed over the course of the experiment shown in Fig. 6 is not decreased even in the presence of 25 mM 3HP, although the distribution of products formed is altered. (In Fig. 6 E–G, the total amounts of all products are 13.9 mM, 14.6 mM, and 15 mM, respectively.) The lack of substrate inactivation by 3HP, coupled with the observation that 3HP can be used to form ADHP, suggests that IlvIH is more specific than IlvBN for pyruvate as the first substrate but can use 3HP as the second substrate. The net effect in this case is also diminished flux into the branched chain amino acid pathway due to diversion of pyruvate to nonproductive formation of ADHP.
Because addition of isoleucine and serine to the medium alleviates the toxicity of 3HP, we also examined the effect of 3HP on an enzyme that is required only for synthesis of isoleucine. Threonine deaminase (IlvA) catalyzes the synthesis of α-ketobutyrate, which is required for the first step of isoleucine biosynthesis. The activity of IlvA was inhibited by 3HP (Fig. 5B). IlvA retained approximately half of its activity in the presence of 1 mM 3HP. The practical constraints of the assay make it difficult to determine the type of inhibition caused by 3HP. However, the structural resemblance between 3HP and the product formed by threonine deaminase, α-ketobutyrate, suggests that competitive inhibition by binding of 3HP to the active site is most likely.
These results demonstrate that 3HP interferes with synthesis of all branched chain amino acids by inhibiting IlvBN and IlvIH; in addition, it interferes specifically with synthesis of isoleucine by inhibiting IlvA.
4HT Inhibits ThrB in Vivo.
We demonstrated above that 4HT formed from 3HP contributes to the toxicity of 3HP. A mechanism for the toxic effect of 4HT was suggested by our previous demonstration that ThrB can use 4HT as a substrate (11); this reaction is part of the serendipitous pathway for synthesis of PLP shown in Fig. 1. When a single enzyme is used for catalysis of two different reactions, each substrate acts as a competitive inhibitor of the other. Thus, 4HT should be a competitive inhibitor of homoserine phosphorylation, which is required for synthesis of threonine. It should be possible to lessen this inhibitory effect by supplying homoserine in the medium. Indeed, the duration of the lag phase observed when WT E. coli K12 is grown on glucose in the presence of 3HP is substantially reduced when homoserine is added (Fig. 4B). (Note that we would not expect the lag phase to be completely eliminated because of the inhibitory effect of 3HP on branched chain amino acid synthesis.) This effect cannot be entirely attributed to relief of the inhibition of ThrB, however, because addition of homoserine also decreases the lag phase, but to a lesser degree, for the ΔltaE strain in the presence of 3HP. (The ΔltaE strain does not make 4HT; thus, inhibition of ThrB should not contribute to the overall toxicity due to 3HP in this strain.) Adding homoserine should increase the ability of the cells to make threonine. Because threonine is a precursor of isoleucine, addition of homoserine alleviates both the inhibition of isoleucine synthesis caused by 3HP and the inhibition of threonine synthesis caused by 4HT.
Inhibition of Amino Acid Synthesis by 3HP and 4HT Triggers the Stringent Response.
The data discussed above indicate that 3HP interferes with synthesis of serine, and thus its downstream metabolites, and branched chain amino acids. In addition, the downstream metabolite 4HT interferes with synthesis of threonine. We hypothesize that the immediate cessation of growth when 3HP is added to growing cultures of E. coli (Fig. 3C) is attributable to activation of the stringent response due to inadequate levels of certain amino acids. During the stringent response, expression of about 500 genes is altered, growth ceases, and resources are directed toward survival (19). For example, genes involved in glycolysis and the TCA cycle are down-regulated. Consistent with our hypothesis that the stringent response was activated in response to 3HP, the level of the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase in cell extracts was diminished by 98% by 1 h after addition of 3HP (Table S2). Notably, the loss of glyceraldehyde 3-phosphate dehydrogenase was even more severe when cells were treated with 3HP than when they were treated with serine hydroxamate, which is often used to trigger the stringent response (19). This observation makes sense because serine hydroxamate interferes only with synthesis of serine, whereas 3HP interferes with synthesis of serine and branched chain amino acids as well.
The suggestion that 3HP triggers the stringent response, and thereby shuts down glycolysis, provides an explanation for the observation that addition of several amino acids can partially alleviate the long lag phase caused by 3HP (Fig. S3). Glutamate and proline can be converted to α-ketobutyrate, which would allow synthesis of isoleucine even when IlvA is inhibited by 3HP. Tryptophan is degraded to pyruvate, a direct precursor of branched chain amino acids as well as a precursor for gluconeogenesis, which would generate the intermediate needed for synthesis of serine. Degradation of arginine forms succinate, which can also provide pyruvate and support gluconeogenesis. It is less obvious why addition of methionine and lysine decreases the lag phase caused by 3HP, because these amino acids cannot be degraded by E. coli K12. However, their availability should decrease the amount of carbon that must be drained from the TCA cycle for their synthesis, and thus increase the amount available for synthesis of other amino acids.
Homoserine, an Intermediate in the Normal Metabolic Network, Interferes with the Serendipitous Pathway for Synthesis of PLP.
ThrB plays two important roles during growth of the ΔpdxB strain on glucose. Its normal activity is required for synthesis of threonine. Its promiscuous activity (phosphorylation of 4HT) is required for synthesis of PLP. As mentioned above, each of two alternative substrates for the same enzyme acts as a competitive inhibitor of the reaction involving the other substrate. Thus, l-homoserine would be expected to interfere with synthesis of PLP by the serendipitous pathway. Indeed, addition of l-homoserine significantly impairs growth of the ΔpdxB strain using the serendipitous pathway shown in Fig. 1 when YeaB, the first enzyme in the serendipitous pathway, is overproduced (Table 1). Only very mild inhibition occurs when ThrB is overproduced, suggesting that sufficient flux through both pathways can be achieved in the presence of homoserine if extra ThrB is available.
Table 1.
Effect of homoserine on the growth of the ΔpdxB strain when PLP is synthesized via the normal pathway (when pdxB is overexpressed) or via the serendipitous pathway (when yeaB or thrB is overexpressed)
| Overexpressed gene |
||||
| Supplement | None* | pdxB | yeaB | thrB |
| None | — | ++++†,‡ | +++ | ++++ |
| l-homoserine (100 μM) | — | ++++ | — | +++ |
*Empty vector (pTrc histidinol phosphate phosphatase) was used for the negative control.
†ΔpdxB strain grows on plates but not in liquid medium when YeaB or ThrB is overproduced for reasons that are not understood (11).
‡Colonies reached 1 mm in diameter in 1–2 d (++++), 3–5 d (+++), or 6–8 d (++).
IlvIH Diverts 3HP from the Serendipitous Pathway for PLP Synthesis.
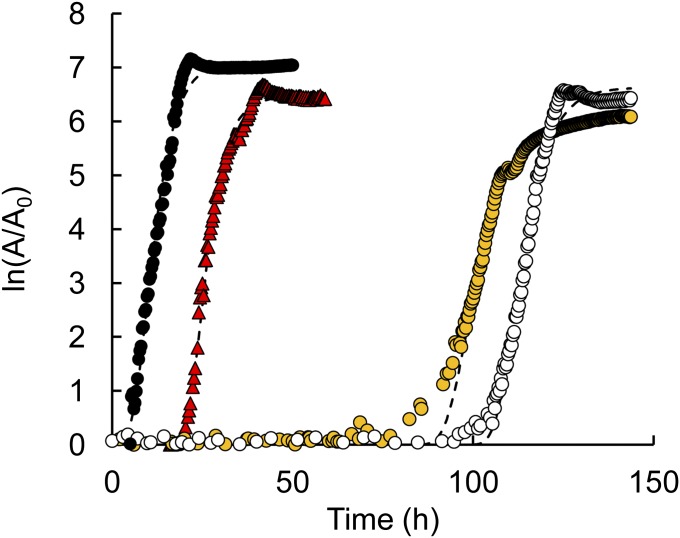
The ability of IlvIH to use 3HP as a substrate could diminish flux through the serendipitous pathway for PLP synthesis by diverting 3HP from the pathway. 3HP is a poor substrate for the enzyme, and is only used when levels of α-ketobutyrate are low (Fig. 6). Because 3HP inhibits the synthesis of α-ketobutyrate by IlvA, this situation may well be obtained in vivo. Evidence that 3HP is indeed diverted from the novel pathway for PLP synthesis is demonstrated by the effects of amino acids on the growth of ΔpdxB E. coli K12 in the presence of 3HP. Addition of serine and isoleucine overcomes the toxic effects of 3HP on WT E. coli K12 (Fig. 3); the mechanisms of these effects have been discussed in the previous sections. However, addition of valine as well as serine and isoleucine is required to shorten the lag phase for the ΔpdxB strain dramatically (Fig. 7). The beneficial effect of valine in this strain is likely due to its action as a feedback inhibitor of IlvIH (Fig. 2). By inhibiting IlvIH, valine should prevent the diversion of 3HP toward the unnecessary metabolite ADHP, and thereby increase flux through the novel pathway for synthesis of PLP. This will also prevent unproductive consumption of pyruvate and generation of a useless metabolite that may or may not be toxic itself.
Fig. 7.
Addition of valine in addition to serine and isoleucine is required to alleviate the toxicity of 3HP to ΔpdxB E. coli K12. Growth of the ΔpdxB mutant on M9/glucose containing 10 μM pyridoxine (●); 1.0 mM 3HP (○); and 1.0 mM 3HP, 1 mM Ser, and 40 μM Ile (yellow ○) and on M9/glucose containing 1 mM 3HP, 1 mM Ser, 40 μM Ile, and 60 μM Val (red △). A = OD600.
Discussion
The emergence of a novel pathway, either by elevation of the flux through a serendipitous pathway or as a result of genetic engineering, can cause unexpected interactions between metabolites and enzymes in the novel pathway and the existing metabolic network. Toxicity due to previously unencountered metabolites is a common phenomenon when heterologous pathways are engineered. For example, a heterologous pathway that produces amorphadiene, a precursor of the antimalarial drug artemisinin, in E. coli results in accumulation of 3-hydroxy-3-methyl-glutaryl–CoA and severe growth inhibition (20). Indole, an intermediate in an engineered pathway for synthesis of taxadiene (9), and vanillin, formed via a heterologous pathway in yeast, both cause substantial inhibition of growth (5). The targets of toxicity were not identified in any of these cases, although considerable effort was invested in the case of the amorphadiene synthesis pathway. In the absence of information about the targets of toxicity, synthetic biologists have primarily dealt with toxic intermediates by elevating levels of the enzymes that convert such intermediates to less toxic products.
The pathway for synthesis of PLP shown in Fig. 1 offers an excellent model system for examining the inhibitory cross-talk that can occur between a novel pathway and an existing metabolic network. Two intermediates in the novel pathway perturb normal metabolic pathways by inhibiting enzymes due to their resemblance to either substrates or allosteric regulators. 3HP inhibits SerA, IlvA, and IlvBN by different mechanisms; it appears to act as a noncompetitive inhibitor of SerA, a competitive inhibitor of IlvA, and a suicide inactivator of IlvBN. It also acts as an alternative substrate for IlvIH. 4HT competes with homoserine for the active site of ThrB. The normal metabolic network interferes with PLP synthesis by the novel pathway in three ways. First, in a mirroring of the effect of 4HT on threonine synthesis, homoserine competes with 4HT for the active site of ThrB. Second, 3HP is diverted from the novel pathway by the adventitious reaction with pyruvate catalyzed by IlvIH. Finally, we have previously shown that the presence of serine in the medium prevents operation of the novel pathway due to inhibition of SerA (11), which produces the 3PHP that is diverted toward PLP synthesis by the novel pathway.
There is no doubt that further evolutionary processes or genetic engineering could resolve the conflicts between the serendipitous pathway for PLP synthesis and the normal metabolic network. Alteration of the active sites or regulatory sites that bind 3HP could lead to proteins that are able to exclude 3HP from these sensitive sites. Duplication of the gene encoding ThrB, followed by divergence of one copy to provide a specific 4HT kinase and divergence of the other to provide a protein that discriminates against 4HT, would prevent the competition between homoserine and 4HT. Alternatively, or perhaps in addition, “balancing” of the activities that form and use problematic metabolites could minimize their concentrations, and thus their ability to interfere with enzymes in the metabolic network.
Notably, the serendipitous pathway for synthesis of PLP involves three intermediates, only one of which is normally encountered in E. coli. Glycolaldehyde is formed during synthesis of folate. It is not used in any known pathway, and it is likely lost from cells by diffusion through the membrane. The other two intermediates, 3HP and 4HT, are charged, and therefore will be retained in the cytoplasm. Both of these intermediates cause toxicity by at least one mechanism, and in the case of 3HP, by at least three mechanisms. These findings support the notion that introduction of novel pathways is more likely than not to engender inhibitory cross-talk because proteins do not preemptively evolve mechanisms for exclusion of unusual molecules from sensitive binding sites.
This work has important implications for both synthetic biology and evolutionary biology. It suggests that multiple interactions between a newly introduced pathway and regulatory and metabolic networks will occur when synthetic biologists build novel pathways for synthesis of desirable products or degradation of toxins. From an evolutionary standpoint, it suggests that emergence of a new metabolic pathway may require a variety of adaptations in the metabolic and regulatory networks, as well as in enzymes in the pathway itself. Our observations also suggest that the severity of these interactions will be environment-dependent; 3HP does not cause toxicity in rich medium when serine and branched chain amino acids are available. Thus, inhibitory cross-talk between an engineered pathway and the existing metabolic network may be ameliorated under certain environmental conditions. Further, the emergence of serendipitous pathways in nature may only occur under environmental conditions in which inhibitory cross-talk is minimized.
Materials and Methods
Strains.
E. coli K12 BW25113 and strains in which pdxB, ilvI, or ilvB is replaced with a kanamycin resistance gene were obtained from the Keio collection (21). A mutant strain in which ltaE is replaced with a chloramphenicol resistance gene was constructed using the method of Datsenko and Wanner as described previously (11, 22). An ASKA (A complete Set of E. coli K-12 ORF Archive) clone expressing ilvA was obtained from the Nara Institute of Science and Technology (23). Construction of a clone expressing serC was described previously (24). Construction of other overexpression strains, primers for PCR amplification of genes (Table S1), and bacterial growth conditions are described in SI Materials and Methods.
Preparation of Enzymes.
SerA, N364A SerA, SerB, CysE, CysM, and CysK were overproduced from pET46 in E. coli BL21(DE3). IlvA was overproduced from pCA24N in E. coli K12 AG1. IlvIH was overproduced from the ΔilvB::kan strain, and IlvBN was overproduced from the ΔilvI::kan strain to prevent contamination by the other isozyme in each case. Preparation of SerC was described previously (24). All enzymes were prepared with N-terminal His6-tags and purified by nickel affinity chromatography. Additional details are available in SI Materials and Methods.
Assay of SerA Activity.
The α-ketoglutarate reductase activity of SerA was measured by following the oxidation of NADH at 340 nm in reaction mixtures containing 0.25 mM NADH, variable amounts of α-ketoglutarate, and 0.5 μM SerA or N364A SerA in 100 mM potassium phosphate (pH 7.5) at 25 °C (14). Inhibition of activity by 3HP was measured using variable amounts of 3HP and 5 mM α-ketoglutarate. The Hill equation was used to calculate I0.5, as described by Grant et al. (14).
Assay of SerB Activity.
The p-nitrophenyl phosphate phosphatase activity of SerB was measured by following release of p-nitrophenol from p-nitrophenyl phosphate (10 mM) at 405 nm in 50 mM Tris⋅HCl (pH 7.5) containing 0.25 M KCl, 2.5 mM MgCl2, 3HP (up to 5 mM), and SerB (2 µM) at 25 °C (25).
Assay of SerC Activity.
The transaminase activity of SerC was followed using a coupled assay with glutamate dehydrogenase (26). Activity was measured by following the formation of NADH at 340 nm in mixtures containing 1 mM NAD, 0.1 mM α-ketoglutarate, 2 μM SerC, 0.25 mg/mL glutamate dehydrogenase, and variable amounts of O-phospho-l-serine in 0.1 M Tris⋅HCl (pH 8.5). Inhibition by 3HP was measured in the presence of 20 and 200 μM O-phospho-l-serine and variable concentration of 3HP up to 5 mM.
Assay of CysE Activity.
The serine acetyltransferase activity of CysE was measured by following the release of CoA from acetyl CoA. CoA was quantified based on its reaction with 5,5′-dithiobis(2-nitrobenzoic acid), which yields 2-nitro-5-thiobenzoic acid (ε412 = 13.6 mM−1 cm−1) (27). Assays were carried out at 25 °C in 50 mM Tris⋅HCl (pH 7.5) containing 100 mM NaCl, 0.1 mM EDTA, 0.8 mM acetyl CoA, varying amounts of serine, and 20 nM CysE. Inhibition by 3HP was measured in the presence of 0.5 mM l-serine and variable concentrations of 3HP up to 5 mM.
Assay of CysK and CysM Activity.
The cysteine synthase activity of CysK and CysM was measured by following the formation of cysteine from O-acetyl-l-serine at 25 °C using a ninhydrin assay (28). Assay mixtures contained O-acetyl-l-serine (4 mM for CysK and 1 mM for CysM), 2 mM sodium sulfide, variable amounts of 3HP (up to 5 mM), and CysK or CysM (1 nM) in 50 mM Tris⋅HCl (pH 7.5).
Assay of IlvA Activity.
Formation of α-ketobutyrate from threonine catalyzed by IlvA was followed by 1H-NMR in mixtures containing 0.5 μg IlvA, 20 μM PLP, and 10 mM l-threonine in 700 μL of 50 mM potassium phosphate (pH 7.5). D2O [10% (vol/vol)] was included to provide a lock signal, and 1 mM dioxane was added as an internal standard. Reaction mixtures were incubated at 25 °C, and 1H-NMR spectra were collected at intervals on a Varian Inova 500-MHz NMR spectrometer. The residual H2O signal was suppressed using the WET (Water suppression Enhanced through T1 effects) solvent suppression technique (29). Initial rates were measured by assessing concentrations of threonine and α-ketobutyrate between 20 and 90 min. Consumption of threonine was followed by integrating peaks from protons on the C4-methyl group at δ 1.27. Formation of α-ketobutyrate was followed by integrating peaks from protons on the C4-methyl group at δ 1.01.
Identification of Reaction Products Formed by Acetohydroxyacid Synthases IlvIH and IlvBN.
The formation of acetohydroxyacids by acetohydroxyacid synthases was followed by 1H-NMR in mixtures containing 25 mM pyruvate, 5 mM α-ketobutyrate, 5 mM 3HP, 75 μM FAD, 0.1 mM TPP, 10 mM MgCl2, 10% (vol/vol) D2O, and 5 μM IlvIH or 1 μM IlvBN in 700 μL of 50 mM potassium phosphate (pH 7.6). Dioxane (1 mM) was added as an internal standard. Mixtures were prepared in NMR tubes and incubated at 37 °C. Disappearance of substrates and appearance of products (acetolactate, acetohydroxybutyrate, and ADHP) over a period of 2–6 h were monitored by integration of peaks in the 1H-NMR spectrum from protons on the following carbons: C3 of pyruvate hydrate, δ 2.21; C3 of α-ketobutyrate, δ 2.70; C3 of 3HP, δ 3.61; C3 of acetolactate, δ 1.41; C3 of acetohydroxybutyrate, δ 1.97; and C5 of ADHP, δ 2.23.
Supplementary Material
Acknowledgments
This work was supported by National Institute of General Medical Sciences Grant GM083285 (to S.D.C.).
Footnotes
The authors declare no conflict of interest.
See Author Summary on page 16766 (volume 109, number 42).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208509109/-/DCSupplemental.
References
- 1.Nobeli I, Ponstingl H, Krissinel EB, Thornton JM. A structure-based anatomy of the E. coli metabolome. J Mol Biol. 2003;334:697–719. doi: 10.1016/j.jmb.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabási AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 3.Copley SD. Evolution of efficient pathways for degradation of anthropogenic chemicals. Nat Chem Biol. 2009;5:559–566. doi: 10.1038/nchembio.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang K, Woodruff AP, Xiong M, Zhou J, Dhande YK. A synthetic metabolic pathway for production of the platform chemical isobutyric acid. ChemSusChem. 2011;4:1068–1070. doi: 10.1002/cssc.201100045. [DOI] [PubMed] [Google Scholar]
- 5.Hansen EH, et al. De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker’s yeast (Saccharomyces cerevisiae) Appl Environ Microbiol. 2009;75:2765–2774. doi: 10.1128/AEM.02681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siddiqui MS, Thodey K, Trenchard I, Smolke CD. Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res. 2012;12:144–170. doi: 10.1111/j.1567-1364.2011.00774.x. [DOI] [PubMed] [Google Scholar]
- 7.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 8.Westfall PJ, et al. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc Natl Acad Sci USA. 2012;109:E111–E118. doi: 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boghigian BA, Salas D, Ajikumar PK, Stephanopoulos G, Pfeifer BA. Analysis of heterologous taxadiene production in K- and B-derived Escherichia coli. Appl Microbiol Biotechnol. 2012;93:1651–1661. doi: 10.1007/s00253-011-3528-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Peña Mattozzi M, Tehara SK, Hong T, Keasling JD. Mineralization of paraoxon and its use as a sole C and P source by a rationally designed catabolic pathway in Pseudomonas putida. Appl Environ Microbiol. 2006;72:6699–6706. doi: 10.1128/AEM.00907-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Kershner JP, Novikov Y, Shoemaker RK, Copley SD. Three serendipitous pathways in E. coli can bypass a block in pyridoxal-5′-phosphate synthesis. Mol Syst Biol. 2010;6:436. doi: 10.1038/msb.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drewke C, et al. Growth response to 4-hydroxy-L-threonine of Escherichia coli mutants blocked in vitamin B6 biosynthesis. FEBS Lett. 1993;318(2):125–128. doi: 10.1016/0014-5793(93)80005-f. [DOI] [PubMed] [Google Scholar]
- 13.Thompson JR, Bell JK, Bratt J, Grant GA, Banaszak LJ. Vmax regulation through domain and subunit changes. The active form of phosphoglycerate dehydrogenase. Biochemistry. 2005;44:5763–5773. doi: 10.1021/bi047944b. [DOI] [PubMed] [Google Scholar]
- 14.Grant GA, Hu Z, Xu XL. Identification of amino acid residues contributing to the mechanism of cooperativity in Escherichia coli D-3-phosphoglycerate dehydrogenase. Biochemistry. 2005;44:16844–16852. doi: 10.1021/bi051681j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duggleby RG. Suicide inhibition of acetohydroxyacid synthase by hydroxypyruvate. J Enzyme Inhib Med Chem. 2005;20(1):1–4. doi: 10.1080/14756360400020553. [DOI] [PubMed] [Google Scholar]
- 16.Thomas G, Diefenbach R, Duggleby RG. Inactivation of pyruvate decarboxylase by 3-hydroxypyruvate. Biochem J. 1990;266:305–308. doi: 10.1042/bj2660305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams KP, Leadlay PF, Lowe PN. Inhibition of pyruvate:ferredoxin oxidoreductase from Trichomonas vaginalis by pyruvate and its analogues. Comparison with the pyruvate decarboxylase component of the pyruvate dehydrogenase complex. Biochem J. 1990;268(1):69–75. doi: 10.1042/bj2680069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gish G, Smyth T, Kluger R. Thiamin diphosphate catalysis. Mechanistic divergence as a probe of substrate activation of pyruvate decarboxylase. J Am Chem Soc. 1988;110:6230–6234. doi: 10.1021/ja00226a044. [DOI] [PubMed] [Google Scholar]
- 19.Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitera DJ, Paddon CJ, Newman JD, Keasling JD. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab Eng. 2007;9(2):193–207. doi: 10.1016/j.ymben.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Baba T, et al. 2006. Construction of Escherichia coli K12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol 2:2006.0008.
- 22.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 24.Rudolph J, Kim J, Copley SD. Multiple turnovers of the nicotino-enzyme PdxB require α-keto acids as cosubstrates. Biochemistry. 2010;49:9249–9255. doi: 10.1021/bi101291d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuznetsova E, et al. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J Biol Chem. 2006;281:36149–36161. doi: 10.1074/jbc.M605449200. [DOI] [PubMed] [Google Scholar]
- 26.Drewke C, et al. 4-O-phosphoryl-L-threonine, a substrate of the pdxC(serC) gene product involved in vitamin B6 biosynthesis. FEBS Lett. 1996;390(2):179–182. doi: 10.1016/0014-5793(96)00652-7. [DOI] [PubMed] [Google Scholar]
- 27.Kredich NM, Tomkins GM. The enzymic synthesis of L-cysteine in Escherichia coli and Salmonella typhimurium. J Biol Chem. 1966;241:4955–4965. [PubMed] [Google Scholar]
- 28.Gaitonde MK. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem J. 1967;104:627–633. doi: 10.1042/bj1040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smallcombe SH, Patt SL, Keifer PA. WET solvent suppression and its applications to LC NMR and high-resolution NMR spectroscopy. J Magn Reson A. 1995;117:295–303. [Google Scholar]