Abstract
The host mechanisms responsible for protection against malaria remain poorly understood, with only a few protective genetic effects mapped in humans. Here, we characterize a host-specific genome-wide signature in whole-blood transcriptomes of Plasmodium falciparum-infected West African children and report a demonstration of genotype-by-infection interactions in vivo. Several associations involve transcripts sensitive to infection and implicate complement system, antigen processing and presentation, and T-cell activation (i.e., SLC39A8, C3AR1, FCGR3B, RAD21, RETN, LRRC25, SLC3A2, and TAPBP), including one association that validated a genome-wide association candidate gene (SCO1), implicating binding variation within a noncoding regulatory element. Gene expression profiles in mice infected with Plasmodium chabaudi revealed and validated similar responses and highlighted specific pathways and genes that are likely important responders in both hosts. These results suggest that host variation and its interplay with infection affect children’s ability to cope with infection and suggest a polygenic model mounted at the transcriptional level for susceptibility.
Keywords: host response, parasite load, eQTL, eSNP, genotype-by-environment interactions
Accumulating evidence has converged on the recognition that the onset of disease implicates complex biological processes. Susceptibility to infection, like any other complex trait, is multifactorial and has a significant heritable component. Genome-wide association (GWA) approaches have been extended to mapping the genetic architecture underlying susceptibility to infectious diseases (1–5), but only hemoglobin mutations and a handful of other loci conferring risk or protection to malaria have been identified (5–8). There has also been no explicit effort to characterize the effects of host regulatory variation, polygenic inheritance, and genotype-by-infection interactions on malaria phenotypes in vivo.
Host transcriptional response to malaria infection takes place in several organs. We set out to uncover the heritable and infection-response components of host immunity to malaria infection in whole blood of a sample of West African children (SI Appendix, Figs. S1 and S2). Whole blood constitutes a reservoir of circulating immune and nonimmune cells that respond to signals from the parasite while incorporating information from host genotype and play important role in controlling the course of infection. Blood is also a readily accessible system to capture these effects in regions of the world where malaria is endemic. Nonetheless, key transcriptional events in response to infection take place in other organs such as spleen, liver, and bone marrow, the signature of which may not be well preserved in blood. Also, correcting for the effects of differences of cell type proportions on differential expression can be challenging. Here, we test the hypothesis that malaria infection, host regulatory variation, and their interplay generate significant transcriptional variation that affects key immune response mechanisms. First, we uncover the magnitude at which malaria infection and parasite load impact transcript abundance and identify the immune processes influenced by these effects. Second, we identify the genetic factors that influence transcript abundance and test their dependence on infection status. Finally, we use joint analysis of genotypic and gene expression data to identify genes and mechanisms likely affecting the course of infection.
Results
Influence of Infection on Human Transcriptome.
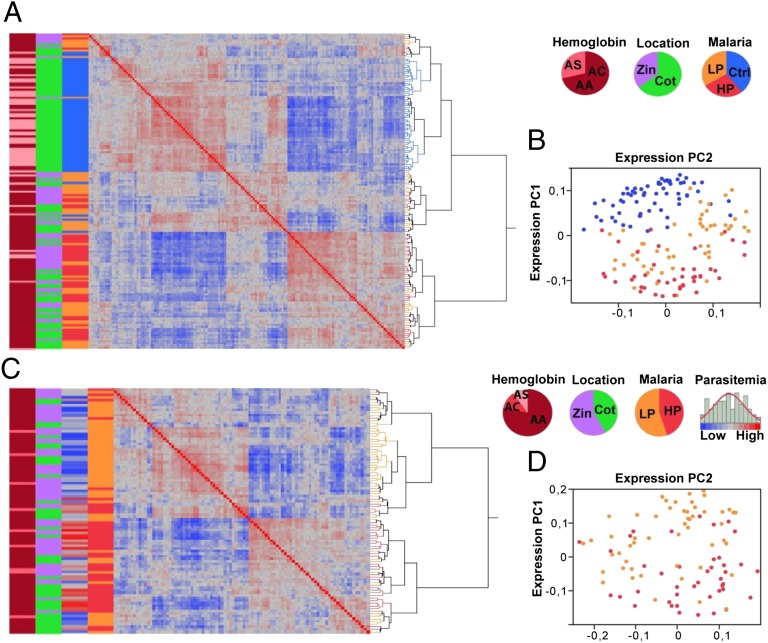
By using unbiased unsupervised statistical analysis, we first evaluated the consistency of the expression profiles between cases and controls (i.e., the combined dataset) and across the range of the parasite load within the infected sample alone (i.e., cases). Clustering of gene expression profiles based on similarity (Fig. 1 A and C), as well as principal component (PC) analysis of the genome-wide gene expression correlation matrix (Fig. 1 B and C), suggest that individuals cluster largely based on their infection status and parasite load. This analysis revealed the presence of strong correlation structure in the data such that expression PC1 (ePC1) explains 19.6% and 17.5% of total variation in the combined dataset and in the cases, respectively.
Fig. 1.
Malaria infection impacts gene expression genome-wide. Correlation structure in whole-transcriptome data for the combined dataset of 155 cases and controls (A and B) and for the 94 cases alone (C and D). (A and C) Hierarchical clustering of whole-genome gene expression correlation matrix. The colored bars from left to right indicate the following phenotypes in the proportions displayed in the pie charts: hemoglobin genotype (AA, AC, or AS), location (Cotonou and Zinvié), and malaria infections status (control and high and low parasitemia groups). Parasite load or log2 parasitemia (low to high) is shown only in C. (B and D) PC analysis of the correlation matrix. The two major expression PCs (ePC1 and ePC2) are shown and individuals are labeled to indicate their infection status (controls, blue; high parasitemia, red; low parasitemia, orange).
Supervised multiple regression and variance component analyses accounting for sex, hemoglobin genotype, location, total blood cell counts, and ancestry confirmed the strong effect exerted by malaria infection and parasite load on the transcriptome. The majority of variation captured by the first ePCs is explained largely by malaria infection status (74% of total variation in the combined dataset; P < 10−5) and by parasitemia class (47% of total variation within the cases; P < 10−4) when modeled as a function of sex, hemoglobin genotype, location, total blood cell counts, and ancestry (SI Appendix, Fig. S3). To estimate the effect of parasite load independently of the hemoglobin genotype, we rerun PC analysis on 73 infected individuals who are AA homozygotes for the hemoglobin locus. The expression profiles again strongly correlate with parasitemia class explaining 39% (P < 10−4) of the variance of ePC1–3.
Next, we evaluated the magnitude and significance of differential expression of individual transcripts first between cases and controls, and second between the controls, the high and low parasitemia groups. ANOVA (accounting for location, sex, hemoglobin genotype, and infection status) and analysis of covariance (ANCOVA; accounting also for total blood cell counts and ancestry) revealed a strong effect of infection status on whole-blood transcriptome. A statistical significance threshold at 1% false discovery rate (FDR; per Benjamini and Hochberg) was applied to all tests of differential expression. A total of 3,334 transcripts (23%) were differentially expressed between cases and controls, whereas 3,177 and 3,154 of these transcripts remained differentially expressed even after accounting for total blood cell counts and ancestry, respectively (Table 1). Breaking down the ANOVA into pair-wise comparisons, we observed that the effect of malaria infection on differential expression of individual transcripts is highest when comparing controls vs. the high parasitemia group (4,085 transcripts), and less so when comparing controls vs. the low parasitemia group (2,377 transcripts), with evidence for a within malaria-infected sample differentiation (2,078 transcripts; Table 1, Fig. 2 A and B, and Dataset S1).
Table 1.
Number of transcripts differentially expressed
| Effect | ANOVA | ANCOVA I | ANCOVA II |
| Malaria | |||
| Parasite load | 2,971 | 3,014 | 1,990 |
| Cases:controls | 3,334 | 3,177 | 3,154 |
| High parasitemia:control | 4,852 | 4,402 | 4,085 |
| Low parasitemia:control | 2,493 | 2,438 | 2,377 |
| High parasitemia:low parasitemia | 2,772 | 2,601 | 2,078 |
| Three-way comparison | 6,178 | 5,856 | 5,180 |
| Location | |||
| Village:city | 1,089 | 310 | 30 |
| Sex | |||
| Female:male | 40 | 48 | 43 |
All contrasts shown in this table are from analyses performed on the cases and controls combined dataset (155 individuals), except the parasite load effect, which was estimated by analyzing the 94 cases alone. ANOVA accounts for the infection status effect, sex, location, hemoglobin genotype and pair-wise interactions. ANCOVA I and ANCOVA II additionally account for total blood cell counts and significant gPCs (gPC1-3; Tracy–Widom statistic < 0.01), respectively. The FDR was evaluated by using the Benjamini and Hochberg method.
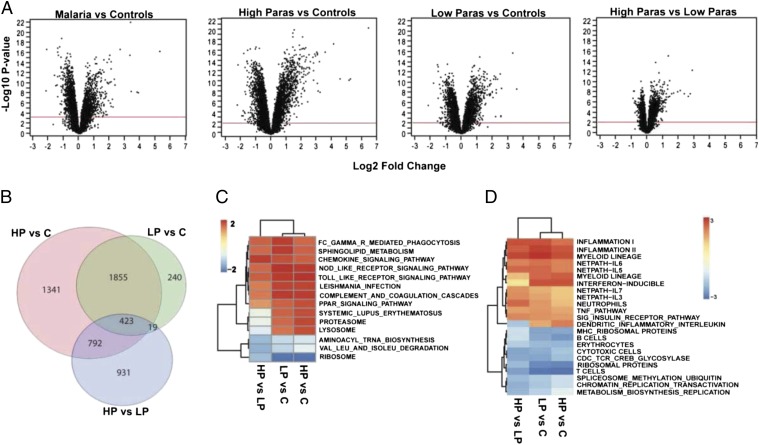
Fig. 2.
Differential expression in whole-blood transcriptome. (A) Volcano plots of statistical significance vs. magnitude of differential expression for the two-way contrasts between the controls (marked as “C”) and high parasitemia (HP) and low parasitemia (LP) groups. For each transcript, significance is shown as the –log10 P value on the y axis, and the log2 of magnitude of mean expression difference is on the x-axis. The red horizontal line indicates the 1% FDR threshold. (B) Venn diagram shows numbers of differentially expressed transcripts for each comparison and the overlaps between them. For each contrast, GSEA was performed for KEGG pathways (C) and the C2, C3, and C5 collections of the Molecular Signatures Database (D) as previously described (9, 16). Only pathways and modules significantly enriched (Bonferroni-adjusted P < 0.05) from at least one contrast are shown. Colors in the heat map indicate the enrichment score from the GSEA analysis.
Gene Set Enrichment Analysis.
Pathway analysis (9) of the differentially expressed genes implicates divergence in core immune processes. We particularly note a strong signature of induced innate immunity (up-regulation of IFN-inducible genes, neutrophil-associated modules, and markers of FcGR-mediated phagocytosis) and suppression of several adaptive immune processes (down-regulation of MHC genes, T cells, B cells, and cytotoxic T cell signaling pathways) in the cases relative to controls (Fig. 2 C and D). Few studies that report whole blood or peripheral blood mononuclear cell (PBMC) transcriptional signatures associated with malaria infection in African populations have been carried out (10–12). Among these studies, Griffiths et al. (10) detected two main signatures in whole blood related to neutrophil and erythroid activity differentiating acutely ill and convalescent Kenyan children. The authors reported a list of genes implicated in these two processes as being differentially regulated between the two groups. We highlight the replication of the expression patterns of the following loci: C1QB (Hochberg and Benjamini q-value = 8.72 × 10−19; fold change, 11.15), MMP9 (q-value = 1.12 × 10−12; fold change, 11), C3AR1 (q-value = 5.8 × 10−7; fold change, 1.33), IL18R (q-value = 7.96 × 10−7; fold change, 2.83), and HMOX1 (q-value = 1.1 × 10−8; fold change, 2.08). These genes seem a promising target for focused evaluation as circulating biomarkers of malaria infection. Several other genes that paralleled the intensity of the infection in our dataset have been reported by others (13, 14), but a systematic comparison with these reports is difficult given differences in study design and the different in vitro cell populations profiled.
A fraction of the expression differences detected for the parasite load effect after accounting for total cell counts is likely caused by average differences in the proportions of subtypes of PBMCs (15). To infer these effects in our sample, we used the genomic signature of flow cytometry-sorted immune cell types (16) in which cell type-specific modules are constructed based on transcript abundance of each gene relative to each other cell type in the PBMC mixture. These expression signatures are constructed from healthy individuals and therefore can be used as a reference panel. We computed Pearson correlation between parasite load and average transcript abundance of each module across all 94 infected individuals (SI Appendix, Fig. S4). This analysis shows a significant effect of parasite load on the six cell type-specific expression profiles investigated (B cells, T cells, myeloid dendritic cells, plasmacytoid cells, natural killer cells, and monocytes; P < 10−7) that can result from modulation of cell type-specific transcription, a shift of cell type mixture in the bloodstream, or a combination of both. Particularly, we note that parasite load is positively correlated with average transcript abundance of myeloid antigen-processing cells and negatively correlated with average transcript abundance of B and T cells, along with the other innate immunity cell types (SI Appendix, Fig. S4).
Contrasting Host Whole-Blood Response in Humans and Mice.
Animal models represent a valuable companion to the study of human clinical material for understanding host–parasite interactions in malaria (17). In particular, mouse models allow detailed characterization of pathogenesis and host response in an experimental framework in which the genetic contribution of the host and environmental factors (including parasite type and infectious doses) are carefully controlled. To test the role for some of the genes and pathways uncovered in our human study in host response to malaria, we infected mice (C57BL/6J) with Plasmodium chabaudi AS (106 parasitized erythrocytes, i.v.), and blood from infected mice was collected 4 d (3.6 ± 0.9% parasitemia) and 6 d (32.8 ± 2% parasitemia) postinfection. Globin-depleted total RNA was prepared, and gene expression profiles were generated by hybridization to microarrays (MouseWG-6 BeadChips; Illumina).
ANOVA revealed 1,783 transcripts differentially expressed (1% FDR) in at least one of the pair-wise contrasts, with the effect of infection being highest in the uninfected mice vs. high parasitemia comparison (1,575 transcripts; Dataset S2 and SI Appendix, Fig. S5A). Gene set enrichment analysis (GSEA; 5% FDR) revealed a strong induction of IFN response, antigen processing and presentation, and the proteasome modules, along with a suppression of the B-cell module, which were all consistent with the human signature (SI Appendix, Fig. S5B). Next, we compared the gene lists derived from the ANCOVA II and ANOVA analyses of the human and mouse datasets, respectively. This contrast was limited to genes significantly regulated (1% FDR) in both hosts, with 47 genes showing fold change greater than two in the human dataset. Thirteen genes were significantly regulated when specifically comparing the high parasitemia group vs. controls (SI Appendix, Fig. S6A). Of these genes, 11 show the same pattern of response in both hosts, notably for three Fc receptors (FCER2, FCGR3B and FCRLA), indicating the importance of FcGR-mediated phagocytosis in host whole-blood response to malaria infection (SI Appendix, Fig. S6B).
Uncovering the Genetic Basis for Gene Regulation in Children Infected with Malaria.
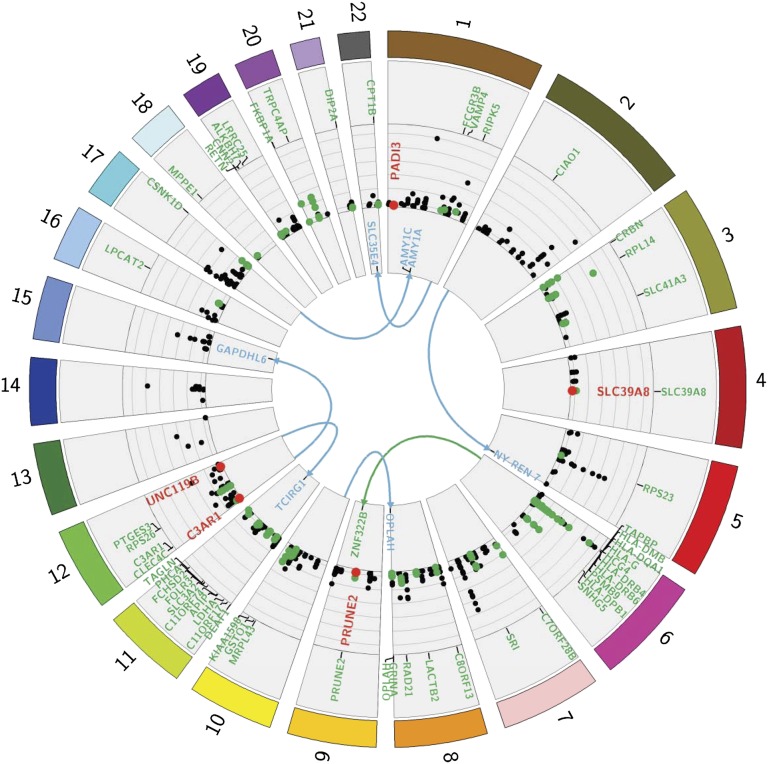
Next, we uncovered the genetic basis of gene expression variation in malaria by performing a GWA test of transcript abundance in the human host. We applied Bonferroni correction for all associations performed in this study. Each of 544,672 SNPs was tested for association with each of the 18,876 expressed transcripts, and a genome-wide Bonferroni correction for multiple testing accounting for the number of SNPs and loci was applied. This analysis gave rise to (i) a genome-wide Bonferroni threshold of 4.86 × 10−12 [0.05/(18,876 × 544,672)] for distal associations, which is likely to be conservative given the linkage disequilibrium structure across the genome; and (ii) a genome-wide Bonferroni threshold for local associations considering the number of SNPs within the region spanning from 100 kb upstream to 100 kb downstream of the transcript (including the transcript itself) and accounting for the number of loci tested. This analysis revealed 263 peak local SNP-probe associations at P < 1.3 × 10−8 and five peak distal SNP–probe associations at P < 4.86 × 10−12 in the combined dataset (Fig. 3, SI Appendix, Fig. S7A, and Dataset S3). The threshold P = 1.3 × 10−8 is the most conservative threshold for local associations and corresponds to a test against 196 markers [P = 1.3 × 10−8, or (0.05/(18,876 × 196)]. The effect sizes of regulatory variation in our dataset are more than an order of magnitude larger than typical SNP–disease associations (SI Appendix, Fig. S7C), thus providing sufficient power to uncover these associations at genome-wide significance. Applying the same global association test of gene expression to the cases alone revealed 149 and six peak local and distal associations, respectively (SI Appendix, Fig. S7B and Dataset S4). In total, both analyses revealed 265 local and eight distal peak SNP–gene associations.
Fig. 3.
Genome-wide eSNP map in malaria-infected children. Circos plot displaying all genome-wide significant associations detected in the combined dataset of cases and controls and in the cases alone. Each chromosome is shown in a different color. Distal associations are displayed in the center of the plot, with the links indicating target transcripts. Circularized Manhattan plot displays local associations and their respective significance (−log10 P value). Associations significant for the genotype-by-infection effect are shown in red, and those implicating genes differentially expressed at 1% FDR in at least one of the two-way contrasts among control and high and low parasitemia groups (Table 1) are shown in green.
We observed significant overlap between these associations and those reported in 13 published expression quantitative trait locus (eQTL) studies of various tissues, including peripheral blood and its derivatives at nominal P values >10−7 and 10−12 for local and distal associations, respectively. A total of 147 of 272 genes (54%) are replicated, including one distal association with AMY1A. Approximately half of these associations (76 of 147) are exact, namely implicating the same SNP–gene pair and most of the remaining report a SNP in the same linkage group. The other associations in our dataset are novel, of weaker strength in the 13 eQTL studies, or might have been reported in other studies.
Joint Action of Host Genotype and Infection on Gene Expression.
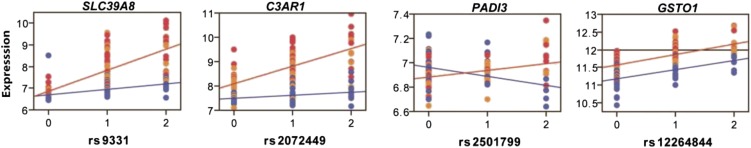
To test for genotype-by-infection interactions, we ran a model that accounts for SNP, infection, SNP × malaria status, sex, location, RBCs, and WBCs. This analysis identified five peak local genotype-by-infection interactions at Bonferroni significance: PRUNE2 (P = 4.17 × 10−9), SLC39A8 (P = 8.37 × 10−7), C3AR1 (P = 1.07 × 10−6), PADI3 (P = 1.61 × 10−6), and UNC119B (P = 2.15 × 10−6; Fig. 3 and SI Appendix, Table S1). The associations implicating C3AR1, PADI3, and SLC39A8 are shown in Fig. 4, and the remaining associations are shown in SI Appendix, Fig. S8. These findings demonstrated the existence of genome-wide significant interactions in malaria, and our data also suggest that interaction effects are pronounced for several associations beneath genome-wide significance.
Fig. 4.
Transcriptional additive and multiplicative effects in malaria. Examples of transcriptional interaction effects implicating the genes SLC39A8, C3AR1, and PADI3. The case of GSTO1 illustrates the scenario of an interaction effect for a disease phenotype in the absence of a transcriptional interaction. This example illustrates how the effect of the gene is conditional on genotype with only the minor allele homozygote individuals shifting to the resistance zone (transcript abundance >12.0 indicated by horizontal line) when infected, giving rise to an interaction effect for the disease phenotype. Genotypes on the x-axis are labeled to indicate the number of minor alleles and individuals are labeled to indicate their infection status (controls, blue; high parasitemia, red; low parasitemia, orange). The y axis shows normalized expression values.
Our survey of the sources of gene expression variation revealed dozens of genes under statistically significant joint effects of malaria infection and host genotype. The genes for which the infection effect is highly dependent on host genotype translate into statistically significant interactions. These genes show a substantial expressed SNP (eSNP) effect in the infected group or the control group but not in both, or show the effect in opposite directions in the two different groups. Other genes subject to interaction effects beneath genome-wide significance show different magnitudes of eSNP effects between the two groups and likely have important roles in modulating the course of infection, and several of them have previously been associated with malaria (i.e., FCGR3B, PSMB9, and GSTO1) (18–20). In addition, we discovered several associations implicating key immune processes, particularly antigen processing and presentation, plasmacytoid dendritic cell activation, and T-cell activation and expansion (i.e., RAD21, LRRC25, CLEC4C SLC3A2, and TAPBP) (21–25). The genes that are associated with an eSNP and that are differentially regulated by the infection are shown in green in Fig. 3 (Datasets S3 and S4 provide further details). We also note that expression of five genetically regulated HLA (HLA) class II loci is negatively correlated (r2 = 0.31) with parasite load and with key immune effectors such as IL18R1, TLR4, TLR5, IFNGR1, and IFNGR2 (P < 10−4), indicating an impairment of antigen processing and likely of subsequent priming of host immune response.
A number of studies surveyed transcriptional genotype-by-environment interactions in humans and reported dozens of response eQTLs in vitro under a variety of environmental challenges such as radiation (26) and treatment with various agents (27–29). The number of interacting loci in response to malaria infection in our in vivo study is lower than the number of response eQTLs reported in these studies despite the fact that similar sample sizes were used. This is likely because of a combination of factors, notably the strong induction of transcriptional response in vitro, the homogeneity of the cell population investigated, and the differences in the experimental design and statistical thresholds applied. Nonetheless, our results are consistent with the concept that transcriptional genotype-by-environment interactions are pervasive in human populations and can be detected in vivo.
Other eSNP associations deserved attention, but the case of SCO1, which encodes an inner mitochondrial membrane metallochaperone, stands out. This gene was implicated in the second top GWA hit by Jallow et al. (5) (rs6503319; P = 7.2 × 10−7; 10 kb from the TSS of SCO1), and, here, we detected two genome-wide significant local eSNP associations for this locus. The strongest eSNP we detected (rs201621; P = 8.91 × 10−14) is located 4 kb upstream of the SCO1 transcription start site in a strong enhancer (30, 31) (SI Appendix, Fig. S9). This finding implicates allelic variation of rs201621 in the effect captured by the malaria GWA study likely through contribution of differential expression of SCO1 to detoxification pathways of reactive oxygen species (32).
Discussion
Joint analysis of gene expression and genotypic data demonstrated that malaria infection and host genotype alters immune gene expression genome-wide in additive and multiplicative manners. The interactions we report here show the existence of robust interactions in vivo in an infectious disease. One of these associations implicates the SLC39A8 locus, which encodes a zinc transporter protein highly up-regulated in response to primary T-cell activation, especially in the presence of low concentrations of zinc (33). Several studies and initiatives have proposed zinc supplementation as a strategy to help reduce the risk of malaria episodes (34, 35), and our data implicate a gene whose action is potentiated by zinc but also clearly and robustly conditioned by host regulatory variation. The interaction implicating SLC39A8 illustrates a robust in vivo genotype-by-infection effect that is directly linked to the key process of T-cell development.
Our data also suggest the scenario of the presence of interactions for higher-level malaria phenotypes in the absence of robust genotype-by-infection interactions for transcription (36). The case of GSTO1, which encodes a protein involved in the metabolism of a broad range of xenobiotics, illustrates this scenario (Fig. 4). Supposing only individuals with a transcript abundance of >12.0, indicated by the horizontal line (Fig. 4), have an efficient detoxification capacity, certain individuals will have a greater capacity for parasite clearance and subsequently show resistance to malaria. Although hypothetical, the example of GSTO1 illustrates how such effects can be conditional on genotype, with only the minor allele homozygote individuals shifting to the resistance zone when infected, giving rise to an interaction effect for the disease phenotype. A corollary of these interactions might mask associations of genotype with disease if the exposure increases disease risk in one genotype group and decreases it in another to yield an overall null effect.
In summary, we have provided a genome-wide picture of host in vivo regulatory variation events in malaria-infected whole-blood transcriptome and highlighted the implication of regulatory variation and interactions in modulating host immune response. The underlying genetic variation of such effects would predispose to how children mount an effective immune response to infection and likely to immunization. We also demonstrate that a systems genetics approach interrogating whole blood as one of the disease tissues can facilitate mapping of susceptibility genes and pinpoint causal mechanisms. Although challenging, it is equally important to extend this approach to investigate the key in vivo transcriptional events in malaria control that take place in other organs such as spleen, liver, and bone marrow. Last, we believe this approach is promising to uncover the genetic basis of response to infection and to immunization in vivo, particularly in African populations in which GWA studies are typically underpowered.
Materials and Methods
Study Population.
The human study was approved by the Ethical Review Committee of Sainte-Justine Research Center and by the Faculté des Sciences de la Santé of the University of Abomey-Calavi in Benin. A total of 94 malaria-infected children under the age of 10 y (median age, 3.7 y) and 61 age-matched control subjects were sampled under informed consent (Dataset S5). Cases were children admitted to a secondary level hospital in Cotonou, the cosmopolitan city of the Republic of Benin, and in a rural primary level health care center in the village of Zinvié, located 36 km from Cotonou. Cases were sampled within a period of 10 wk in spring 2010.
After an initial assessment by a pediatrician, children with fever and who were diagnosed as having uncomplicated acute malaria were considered for the study. Children whose malaria infection status was confirmed by using the Parascreen P. falciparum malaria rapid diagnosis test and standard thick blood smear analysis were enrolled. Children presenting symptoms for other diseases or with known history of HIV were not included. Following blood sampling, all cases received antimalarial treatment and had an uneventful course of the disease, except for two children who underwent transfusion at D+1 and D+2 for worsening anemia. Age-matched controls were from the city of Cotonou and were siblings of a large cohort of children with sickle-cell disease registered at the health clinic of the National Center of Sickle Cell Disease in Cotonou. Hemoglobin testing was done by thin-layer agarose isoelectric focusing (Pharmacia LKB Biotechnology) on dried blood collected on Guthrie paper, and S-hemoglobin genotypes were confirmed by genotyping the sickle cell mutation (rs334) using the Sequenom assay. None of the control subjects have sickle-cell disease, and only those without clinical signs of malaria and who tested negative on both malaria detection tests were retained. All children recruited in our study were of a similar age and sampled within similar geographic and hence environmental settings.
Sampling and Genomic Profiling.
The same collection protocol was followed for all samples to minimize heterogeneity for technical reasons. Peripheral blood samples were collected between 9:00 AM and 2:00 PM and stored at −30 °C until shipping to Montreal at −20 °C. Approximately 4 mL of blood was collected: 3 mL for RNA work collected in Tempus Blood RNA Tubes (Life Technologies) in which blood cells are immediately lysed after collection and total RNA stabilized, 0.5 mL stored in EDTA tubes for DNA work, and the remaining blood for thick smear analysis and total cell counts work with the use of an automated KX-21 blood cell analyzer (Sysmex). Total RNA was extracted by using a Tempus Spin RNA Isolation kit (Life Technologies) followed by globin mRNA depletion by using a GLOBINclear-Human kit (Life Technologies). Total RNA samples were quantified and quality-checked with the RNA 6000 Nano LabChip kit and the 2100 Bioanalyzer (Agilent). Only samples of high RNA quality (Agilent RNA Integrity Number >7.5) were retained for expression profiling. HumanHT-12 BeadChips (48k probes; Illumina) were used to generate expression profiles following the manufacturer’s recommended protocols. To minimize chip and batch effects, the order in which the samples were processed was randomized across all fixed effects in the sample at the extraction, cDNA synthesis, and hybridization steps.
Hybridization was performed on two different dates, and five samples from the first batch were rehybridized with the second batch. Clustering of these technical replicates with themselves indicated negligible batch effects in our data. This was confirmed by testing for batch effect in the probe-by-probe ANOVA. Only well annotated probes (RefSeq) were retained for the analysis. Furthermore, 472 probes aligning to more than one location in the African reference genome or overlying SNPs reported in dbSNP Build 135 and with minor allele frequency (MAF) >5% in the Yoruba sample were removed. Expression intensities were log2-transformed and quantile-normalized by using JMP Genomics version 5.0 (SAS) after an outlier filtering procedure (37) was applied to provide further quality control. The distribution of the probe-level expression data was assessed for normality by using a Levene test, and those that showed deviation from normality (P < 0.01) were removed from the analysis. The probes with expression greater than background levels averaged across all of the arrays were retained for further analyses as previously described (38). These probes correspond to 23,826 and 27,546 features in the combined dataset of cases and controls and in the cases alone, respectively.
For the mouse experiment, ten 9-wk-old female C57BL/6J mice were injected i.v. with 106 P. chabaudi AS parasites to model blood-stage malaria infection. Animal research has been approved by McGill University review board and all mice were maintained at the Animal Care Facility according to the guidelines of the Canadian Council on Animal Care. Parasitemia was monitored by microscopy of Hemacolor (Harleco)-stained thin blood smears, and mice were euthanized by CO2 inhalation followed by cardiac puncture to exsanguinate at day 4 (low parasitemia, n = 5) and day 6 (high parasitemia, n = 5). Blood was also collected from age- and sex-matched uninfected controls. For each condition, blood was pooled in Tempus tubes (Life Technologies). Total RNA was extracted by using a Tempus Spin RNA Isolation kit (Life Technologies) followed by globin mRNA depletion by using a GLOBINclear-Mouse kit (Life Technologies). RNA samples were quantified and quality-checked with the RNA 6000 Nano LabChip kit and the 2100 Bioanalyzer (Agilent). MouseWG-6 v2 BeadChips (Illumina) were used to generate expression profiles by using three technical replicates for each condition. The replicates started at the stage of the RNA sample at which equal quantities of input RNA from the original stock were subject to the entire procedure. Expression intensities were log2-transformed and quantile-normalized.
Genome-wide genotyping data were generated by using OmniExpress arrays (733k SNPs) and extracted with the Genotyping Module in BeadStudio software (Illumina). Only samples with call rates >99% were retained, and all SNPs that had a cluster separation value below 0.3 or call frequency below 99% were removed. The process of quality-control checks resulted in retention of 544,672 SNPs (MAF >10%) in 151 individuals for the population structure analysis and eSNP analysis. Global genotypic variation and ancestry was inferred by using Eigenstrat (39), retaining the first three eigenvectors [genotypic PCs (gPCs) 1–3) according to the Tracy–Widom test statistic (P < 0.01). gPC1–3 scores are used to account for ancestry in the analysis detailed later.
Statistical Analysis of Gene Expression Data.
All statistical analyses on the gene expression data were performed by using JMP Genomics version 5.0 and SAS 9.3 (SAS). Two datasets were subject to the analyses described later: (i) the combined dataset (94 cases and 61 controls for the gene expression data-alone analysis, or 92 cases and 59 controls for the joint genotypic and gene expression data analysis), and (ii) the cases alone (94 cases for the gene expression data-alone analysis, or 92 for the joint genotypic and gene expression data analysis). The malaria effect was considered in three different ways: (i) cases vs. controls, (ii) log2-scale transformed parasitemia counts as a quantitative measure of infection severity, and (iii) high vs. low parasitemia groups using the median value of the log2 parasitemia counts as a cutoff. PC analysis, PC variance analysis, and multiple regression analyses were performed such that the first three ePC are modeled simultaneously or individually as a function of various effects in the data: malaria infection status, log2 parasitemia, location, hemoglobin genotype, sex, pair-wise combination of fixed effects, total cell counts (RBCs and WBCs), and ancestry (gPC1–3).
SAS GLM was used to evaluate the magnitude and significance of differential expression of individual expressed probes. Variance was partitioned among the malaria effect, sex, location, hemoglobin, pairwise contrasts, total cell counts, and ancestry as covariates. Batch effect, age, and pair-wise contrasts (i.e., malaria × location, malaria × sex and sex × location) were evaluated and found to be insignificant. Results from the following full ANCOVA model (ANCOVA II in Table 1) for each malaria effect contrast were used for GSEA and for the contrast with genotypic effects:
The malaria effect was considered in the ways indicated in Table 1 and the error ε was assumed to be normally distributed with a mean of zero. For the mouse dataset, ANOVA was used to evaluate the magnitude and significance of differential expression among controls and high and low parasitemia groups. Orthology was inferred by using the Ensembl Biomart tool. A statistical significance threshold at 1% Benjamini and Hochberg FDR was applied to each term in all tests of differential expression.
GSEA.
Enrichment analysis for each contrast (high parasitemia vs. controls, low parasitemia vs. controls, and high vs. low parasitemia) was performed by using GSEA (9). The analysis was performed on the C2, C3, and C5 collections of MsigDB database (http://www.broad.mit.edu/gsea/msigdb). Appended to C2 canonical pathways are curated signaling pathways from NetPath (40), molecular signature gene sets of sorted PBMC cell types (16), and gene sets collected from transcriptional analyses of PBMC samples (41). The resulting P values from the GSEA were adjusted for multiple testing by using a Bonferroni correction (P < 0.05). Pearson correlations were computed between parasitemia and average transcript abundance of each module of genes from six PBMC cell type subsets obtained from Nakaya et al. (16) across all 94 infected individuals.
GWA of Gene Expression.
Marker properties and association tests were performed by using JMP Genomics version 5.0 and SAS 9.3 (SAS). Regression tests for association of gene expression levels with each numeric genotype (coded as 0, 1, or 2, with each number representing the number of copies of the minor allele) were performed. Only autosomal SNPs with an MAF >10%, with missing data <1%, and in Hardy–Weinberg equilibrium (P < 0.01) were retained for the GWA tests. Tests of association were carried out with two models for each dataset (the combined dataset and cases only) separately. We distinguished between local and distal associations based on the location of the genotype and the associated transcript. We applied Bonferroni correction for all associations performed in this study. Each of 544,672 SNPs was tested for association with each of the 18,876 expressed transcripts. This analysis gave rise to (i) a genome-wide Bonferroni threshold of 4.86 × 10−12 [0.05 / (18,876 × 544,672); (−log10[P] > 11.3) for distal associations and (ii) to a genome-wide Bonferroni threshold of 2.65 × 10−6 to 1.3 × 10−8 for local association [−log10(P) > 5.57–7.88], considering the number of SNPs within the region spanning 100 kb upstream and 100 kb downstream of the transcript. Only linkage disequilibrium block tagging SNPs (based on D′ > 0.90) were used in the full model testing for the interaction effects. The analysis on the infected sample was performed by using 535,838 SNPs (with no more than one missing genotype per parasitemia group) and 18,974 probes.
First, a model in which m is the mean measure of transcript abundance, and the error ε is assumed to be normally distributed with a mean of zero was used (model 1):
The results from this model provided a list of significant associations that we compared with the associations reported in 13 published eQTL studies of peripheral blood or its derivatives at nominal P values > 10−7 and 10−12 for local and distal associations, respectively. These published associations were accessed by using the eQTL Browser (http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/), and we also included the results of our own eQTL study of the leukocyte transcriptome in the Moroccan population (36).
To test for genotype-by-infection interactions, we ran a model on the combined dataset (544,672 SNPs and 18,876 expressed transcripts) that accounts for SNP, malaria status, SNP × malaria status, sex, location, RBCs, and WBCs, where m is the mean measure of transcript abundance, and the error ε is assumed to be normally distributed with a mean of zero (model 2):
Because testing for multiplicative SNP effects between the control and the infected group might be sensitive to differences in the representation of each group within each genotype class, we applied an additional filter to the list of SNPs in model 2 and excluded all SNPs not in Hardy–Weinberg equilibrium and with a MAF <10% within each of the subgroups tested. ENCODE data (30, 31) retrieved from the University of California (Santa Cruz), browser was used to facilitate the interpretation of the detected eSNP signal for the SCO1 gene.
Supplementary Material
Acknowledgments
We thank all the study participants in Cotonou and Zinvié, Republic of Benin, as well as numerous individuals who facilitated sample collection, in particular the staff of the National Center of Sickle Cell Disease in Cotonou and the staff of Hopital de la Croix and Congrégation Filles de Sainte Camille in Zinvié. We thank Greg Gibson, Julie Hussin, Ferran Casals, Martine Zilversmit, and Alan Hodgkinson for helpful discussions and suggestions. This work was funded by Human Frontiers in Science Program Grant RGP0054/2006-C and Canadian Institute of Health Research Grants 11284 and 200183 (to P.A.), and Canadian Institute of Health Research Grant MOP-119342 (to P.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.C.K. is a guest editor invited by the Editorial Board.
Data deposition: The gene expression data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE34404).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204945109/-/DCSupplemental.
References
- 1.Thye T, et al. African TB Genetics Consortium Wellcome Trust Case Control Consortium Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet. 2010;42:739–741. doi: 10.1038/ng.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davila S, et al. International Meningococcal Genetics Consortium Genome-wide association study identifies variants in the CFH region associated with host susceptibility to meningococcal disease. Nat Genet. 2010;42:772–776. doi: 10.1038/ng.640. [DOI] [PubMed] [Google Scholar]
- 3.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang FR, et al. Genomewide association study of leprosy. N Engl J Med. 2009;361:2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 5.Jallow M, et al. Wellcome Trust Case Control Consortium Malaria Genomic Epidemiology Network Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat Genet. 2009;41:657–665. doi: 10.1038/ng.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verra F, Mangano VD, Modiano D. Genetics of susceptibility to Plasmodium falciparum: From classical malaria resistance genes towards genome-wide association studies. Parasite Immunol. 2009;31:234–253. doi: 10.1111/j.1365-3024.2009.01106.x. [DOI] [PubMed] [Google Scholar]
- 7.Ayodo G, et al. Combining evidence of natural selection with association analysis increases power to detect malaria-resistance variants. Am J Hum Genet. 2007;81:234–242. doi: 10.1086/519221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths MJ, et al. Genomewide analysis of the host response to malaria in Kenyan children. J Infect Dis. 2005;191:1599–1611. doi: 10.1086/429297. [DOI] [PubMed] [Google Scholar]
- 11.Ockenhouse CF, et al. Common and divergent immune response signaling pathways discovered in peripheral blood mononuclear cell gene expression patterns in presymptomatic and clinically apparent malaria. Infect Immun. 2006;74:5561–5573. doi: 10.1128/IAI.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torcia MG, et al. Functional deficit of T regulatory cells in Fulani, an ethnic group with low susceptibility to Plasmodium falciparum malaria. Proc Natl Acad Sci USA. 2008;105:646–651. doi: 10.1073/pnas.0709969105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevenson MM, Riley EM. Innate immunity to malaria. Nat Rev Immunol. 2004;4:169–180. doi: 10.1038/nri1311. [DOI] [PubMed] [Google Scholar]
- 14.Oakley MS, Gerald N, McCutchan TF, Aravind L, Kumar S. Clinical and molecular aspects of malaria fever. Trends Parasitol. 2011;27:442–449. doi: 10.1016/j.pt.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Shen-Orr SS, et al. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010;7:287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Longley R, et al. Host resistance to malaria: Using mouse models to explore the host response. Mamm Genome. 2011;22:32–42. doi: 10.1007/s00335-010-9302-6. [DOI] [PubMed] [Google Scholar]
- 18.Omi K, et al. Fcgamma receptor IIA and IIIB polymorphisms are associated with susceptibility to cerebral malaria. Parasitol Int. 2002;51:361–366. doi: 10.1016/s1383-5769(02)00040-5. [DOI] [PubMed] [Google Scholar]
- 19.Niesporek S, Meyer CG, Kremsner PG, May J. Polymorphisms of transporter associated with antigen processing type 1 (TAP1), proteasome subunit beta type 9 (PSMB9) and their common promoter in African children with different manifestations of malaria. Int J Immunogenet. 2005;32:7–11. doi: 10.1111/j.1744-313X.2005.00484.x. [DOI] [PubMed] [Google Scholar]
- 20.Oakley MS, et al. Molecular correlates of experimental cerebral malaria detectable in whole blood. Infect Immun. 2011;79:1244–1253. doi: 10.1128/IAI.00964-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seitan VC, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rissoan MC, et al. Subtractive hybridization reveals the expression of immunoglobulin-like transcript 7, Eph-B1, granzyme B, and 3 novel transcripts in human plasmacytoid dendritic cells. Blood. 2002;100:3295–3303. doi: 10.1182/blood-2002-02-0638. [DOI] [PubMed] [Google Scholar]
- 23.Cao W, et al. BDCA2/Fc epsilon RI gamma complex signals through a novel BCR-like pathway in human plasmacytoid dendritic cells. PLoS Biol. 2007;5:e248. doi: 10.1371/journal.pbio.0050248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cantor J, Slepak M, Ege N, Chang JT, Ginsberg MH. Loss of T cell CD98 H chain specifically ablates T cell clonal expansion and protects from autoimmunity. J Immunol. 2011;187:851–860. doi: 10.4049/jimmunol.1100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Praveen PV, Yaneva R, Kalbacher H, Springer S. Tapasin edits peptides on MHC class I molecules by accelerating peptide exchange. Eur J Immunol. 2010;40:214–224. doi: 10.1002/eji.200939342. [DOI] [PubMed] [Google Scholar]
- 26.Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG. Genetic analysis of radiation-induced changes in human gene expression. Nature. 2009;459:587–591. doi: 10.1038/nature07940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romanoski CE, et al. Systems genetics analysis of gene-by-environment interactions in human cells. Am J Hum Genet. 2010;86:399–410. doi: 10.1016/j.ajhg.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maranville JC, et al. Interactions between glucocorticoid treatment and cis-regulatory polymorphisms contribute to cellular response phenotypes. PLoS Genet. 2011;7:e1002162. doi: 10.1371/journal.pgen.1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barreiro LB, et al. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc Natl Acad Sci USA. 2012;109:1204–1209. doi: 10.1073/pnas.1115761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leary SC, Sasarman F, Nishimura T, Shoubridge EA. Human SCO2 is required for the synthesis of CO II and as a thiol-disulphide oxidoreductase for SCO1. Hum Mol Genet. 2009;18:2230–2240. doi: 10.1093/hmg/ddp158. [DOI] [PubMed] [Google Scholar]
- 33.Aydemir TB, Liuzzi JP, McClellan S, Cousins RJ. Zinc transporter ZIP8 (SLC39A8) and zinc influence IFN-gamma expression in activated human T cells. J Leukoc Biol. 2009;86:337–348. doi: 10.1189/jlb.1208759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shankar AH. Nutritional modulation of malaria morbidity and mortality. J Infect Dis. 2000;182(Suppl 1):S37–S53. doi: 10.1086/315906. [DOI] [PubMed] [Google Scholar]
- 35.Zeba AN, et al. Major reduction of malaria morbidity with combined vitamin A and zinc supplementation in young children in Burkina Faso: A randomized double blind trial. Nutr J. 2008;7:7. doi: 10.1186/1475-2891-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Idaghdour Y, et al. Geographical genomics of human leukocyte gene expression variation in southern Morocco. Nat Genet. 2010;42:62–67. doi: 10.1038/ng.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chu TM, Weir B, Wolfinger R. A systematic statistical linear modeling approach to oligonucleotide array experiments. Math Biosci. 2002;176:35–51. doi: 10.1016/s0025-5564(01)00107-9. [DOI] [PubMed] [Google Scholar]
- 38.Idaghdour Y, Storey JD, Jadallah SJ, Gibson G. A genome-wide gene expression signature of environmental geography in leukocytes of Moroccan Amazighs. PLoS Genet. 2008;4:e1000052. doi: 10.1371/journal.pgen.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 40.Kandasamy K, et al. NetPath: A public resource of curated signal transduction pathways. Genome Biol. 2010;11:R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaussabel D, et al. A modular analysis framework for blood genomics studies: Application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.