Abstract
MHC class II-expressing thymocytes and thymic epithelial cells can mediate CD4 T-cell selection resulting in functionally distinct thymocyte-selected CD4 (T-CD4) and epithelial-selected CD4 (E-CD4) T cells, respectively. However, little is known about how T-cell receptor (TCR) signaling influences the development of these two CD4 T-cell subsets. To study TCR signaling for T-CD4 T-cell development, we used a GFP reporter system of Nur77 in which GFP intensity directly correlates with TCR signaling strength. T-CD4 T cells expressed higher levels of GFP than E-CD4 T cells, suggesting that T-CD4 T cells received stronger TCR signaling than E-CD4 T cells during selection. Elimination of Ras GTPase-activating protein enhanced E-CD4 but decreased T-CD4 T-cell selection efficiency, suggesting a shift to negative selection. Conversely, the absence of IL-2–inducible T-cell kinase that causes poor E-CD4 T-cell selection due to insufficient TCR signaling improved T-CD4 T-cell generation, consistent with rescue from negative selection. Strong TCR signaling during T-CD4 T-cell development correlates with the expression of the transcription factor promyelocytic leukemia zinc finger protein. However, although modulation of the signaling strength affected the efficiency of T-CD4 T-cell development during positive and negative selection, the signaling strength is not as important for the effector function of T-CD4 T cells. These findings indicate that innate T-CD4 T cells, together with invariant natural killer T cells and γδ T cells, receive strong TCR signals during their development and that signaling requirements for the development and the effector functions are distinct.
CD4 T cells are selected in the thymus by T-cell receptor (TCR) signaling through the interaction with MHC class II molecules (1). To ensure the responsiveness to foreign antigens and the tolerance to self-proteins, the TCR repertoire is defined by positive and negative selection (2), and the fate of thymocytes is largely determined by the magnitude of TCR signaling (3). Upon ligation, the TCR complex recruits and phosphorylates a series of protein tyrosine kinases (4). One of the Tec family kinases, IL-2–inducible T-cell kinase (Itk), is a TCR proximal signaling component vital for thymocyte positive selection (5–7). Activated by Lck, Itk activates phospholipase C-γ1, leading to calcium mobilization and Erk/MAPK activation (8). Itk deficiency has been reported to selectively impair CD4 T-cell positive selection without altering CD4/CD8 lineage decision (9). More interestingly, although the development of conventional CD8 T cells is compromised in Itk−/− mice, an innate CD8 T-cell population that is selected on hematopoietic cells is highly enriched in Itk−/− mice (10, 11).
Downstream of proximal signaling events, the Ras–Erk–MAPK cascade has long been identified as important for thymocyte positive selection (12). Ras is a small GTPase and its activity is balanced by positive and negative regulators. Ras guanine nucleotide exchange factors (GEFs) promote Ras activity by catalyzing the exchange of GDP for GTP (13, 14). One of the GEFs, Ras guanyl nucleotide releasing protein 1 (RasGRP1), is critical for mediating TCR signaling to the Ras–Erk cascade during positive selection (14). RasGRP1 deficiency completely abrogates Erk activation and thymocyte positive selection (13, 15). Ras GTPase-activating proteins (RasGAPs), on the other hand, facilitate Ras GTP hydrolysis and thereby function to inactivate Ras (16). One of the RasGAPs, RASA1 (p120 RasGAP), has been shown to function as a negative regulator of developing thymocytes and to inhibit positive selection of CD4 T cells (17).
CD4 T cells can develop by either cortical thymic epithelial cells (TEC) or hematopoietic cells, including MHC class II-expressing thymocytes (18, 19). We designated thymocyte-selected CD4 cells “T-CD4 T cells” and TEC-selected conventional CD4 T cells “E-CD4 T cells.” T-CD4 T cells are present in humans (20, 21). Using a mouse model in which MHC class II is expressed on thymocytes due to the expression of MHC class II transactivator (19), we showed that T-CD4 T cells display an innate-like phenotype, producing Th1 and Th2 cytokines simultaneously upon short stimulation in vitro and in vivo (22). Furthermore, T-CD4 T cells produce IL-4 under Th1-skewing conditions in a Stat6-independent manner (22). Interestingly, the IL-4–producing potential of T-CD4 T cells is shaped during development (22). However, the signaling events that activate IL-4 expression are not well understood. The signaling pathway mediated by signaling lymphocyte activation molecule (SLAM) is the only known pathway required for T-CD4, but not E-CD4 T-cell development (23). Currently, it is not clear whether TCR-mediated signaling regulates T-CD4 T-cell selection in the same manner as E-CD4 T-cell selection.
Similar to T-CD4 T cells, invariant natural killer T (iNKT) cells that use CD1d as TCR ligands, are selected by thymocytes and also have an innate-like phenotype, which suggests a correlation between the thymocyte-mediated selection pathway and the functional phenotype. Furthermore, the SLAM–SLAM-associated protein (SAP) pathway is essential for iNKT generation as for T-CD4 T cells (23, 24). The transcription factor promyelocytic leukemia zinc finger (PLZF) has been shown to be expressed in iNKT cells (25, 26). In the absence of PLZF, iNKT cells failed to acquire the characteristic innate T-cell effector functions and phenotype (25, 26).
In this study, we characterized TCR signaling strength during T-CD4 T-cell development in comparison with E-CD4 T cells. The data clearly demonstrated the opposing effect of TCR signaling strength on the commitment of E- and T-CD4 T-cell lineages. T-CD4 T cells developed by receiving stronger TCR signals than E-CD4 T cells, which was correlated with PLZF expression and likely to be responsible for the innate phenotype. Moreover, our data demonstrated that the signaling events responsible for T-CD4 T-cell generation versus the effector function are distinct from each other.
Results
T-CD4 T Cells Receive Stronger TCR Signals than E-CD4 T Cells During Selection.
It is known that iNKT cells and other innate T cells receive strong TCR signaling during development (27) and, therefore, we hypothesized that T-CD4 T cells likely receive stronger TCR signal than E-CD4 T cells during selection. To investigate the question of TCR signals, we used a Nur77GFP reporter system in which GFP is up-regulated by TCR stimulation and its intensity correlates with the strength of TCR signals (27).
E- and T-CD4 T cells expressing Nur77GFP were generated using mixed bone marrow (BM) chimeras. To produce Nur77GFP E-CD4 T cells, Nur77GFP BM cells were cotransferred with WT BM cells into lethally irradiated B6 hosts generating [Nur77GFP+WT→B6] in which Nur77GFP and WT CD4 T cells were selected by host TEC. This group is called E-CD4 BM transplantation (E-BMT). To generate Nur77GFP T-CD4 T cells, Nur77GFP and CIITATg (Tg) BM cells were cotransferred into lethally irradiated MHC class II-deficient hosts resulting in [Nur77GFP+Tg→Aβ−/−] T-BMT (Fig. S1A). We previously showed that the expression of the CIITA transgene induces MHC class II in thymocytes and MHC class II-expressing thymocytes can select other thymocytes (19). Therefore, in T-BMT, Nur77GFP thymocytes will be selected by Tg thymocytes to become T-CD4 T cells, whereas E-CD4 development is blocked, because host TEC lack MHC class II (18, 19). Cells from the three different sources were distinguished by congenic markers. We compared GFP expression of E- and T-CD4 T cells that were originated from Nur77GFP BM cells.
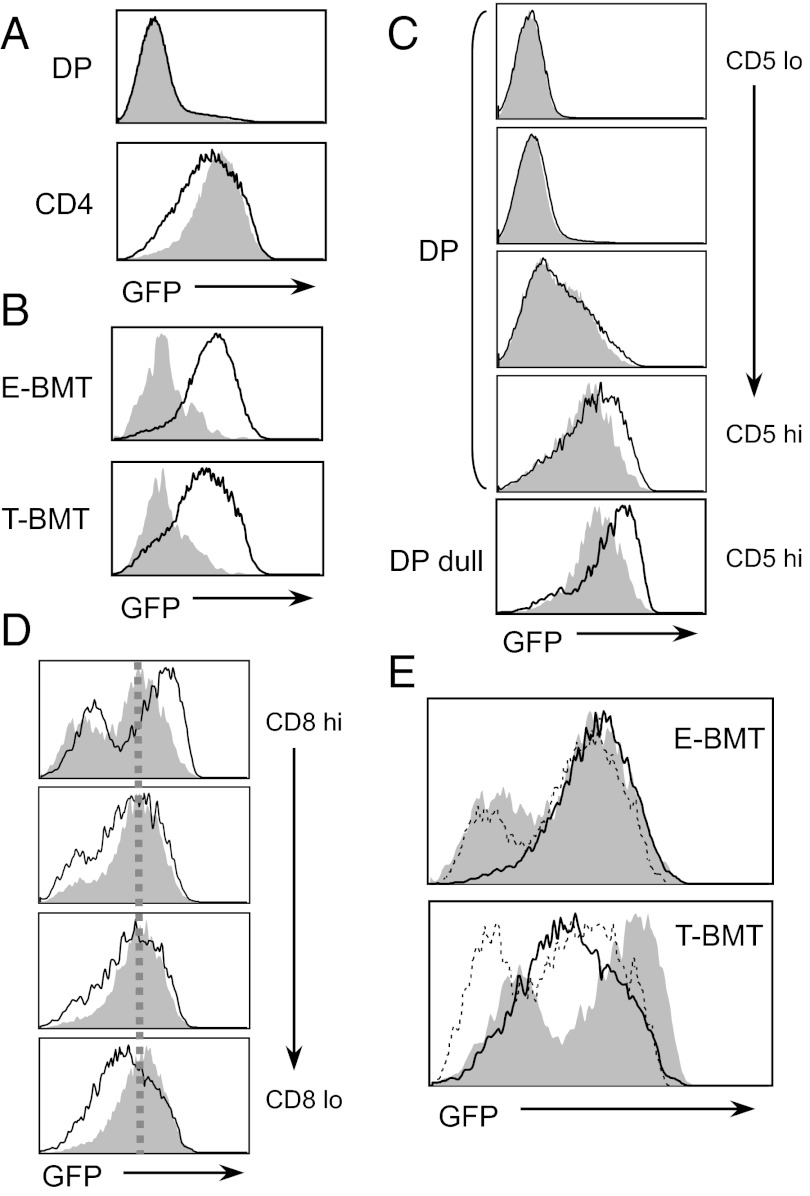
When we compared GFP expression in thymocytes from two groups of chimeras, a majority of double positive (DP) cells did not express GFP, but single positive (SP) thymocytes expressed GFP (Fig. 1A). T-CD4 T cells expressed lower levels of GFP than E-CD4 T cells (Fig. 1A). As reported (27), iNKT cells in both chimeras expressed reduced levels of GFP compared with CD4 T cells (Fig. 1B). However, the study showed that iNKT cells receive a stronger TCR signaling during early development, but down-regulate upon maturation (27). Therefore, we compared the TCR signaling strength during different phases of CD4 T-cell development. DP thymocytes were examined for the expression of CD5 because CD5 is induced upon positive selection (28). DP thymocytes were divided into four subsets on the basis of the CD5 expression levels (Fig. S1B) and GFP levels were compared. DP thymocytes from both E- and T-BMT showed the gradual increase of GFP as CD5 levels induced and notably CD5hi DP thymocytes from T-BMT cells expressed higher levels of GFP than that from E-BMT (Fig. 1C). In the CD5hi DP dull population, which is considered to be more mature DP thymocytes, GFP levels were further up-regulated with a clear difference between E- vs. T-BMT (Fig. 1C). We then examined CD5 expression levels of GFP+ DP dull populations between E- and T-BMT mice. As expected, CD5 expression level was higher in the GFP+ DP dull population from T-BMT compared with E-BMT, confirming that stronger TCR signaling was delivered to cells by thymocytes than TEC (Fig. S1C). Consistent with this observation, GFP expression was higher in T-BMT than in E-BMT thymocytes when they started to up-regulate TCR (Fig. S1D, TCRβlo), and the GFP expression level of these T-BMT cells reduced to that of the corresponding E-BMT cells (Fig. S1D, TCRβhi). This down-regulation of GFP in T-CD4 T cells continued during their maturation determined by the expression level of CD8 coreceptors (Fig. 1D) and CD24 (Fig. S1E). However, E-CD4 T cells maintained the same level of GFP. Therefore, a constant amount of TCR signaling seems to be delivered to E-CD4 T cells during and after selection, whereas T-CD4 T cells receive stronger TCR stimulation than E-CD4 T cells during their selection but the strength is reduced afterward (Fig. 1E).
Fig. 1.
Stronger TCR signaling for T-CD4 than E-CD4 T-cell development. (A) Level of GFP on DP cells and CD4 SP thymocytes from E- or T-BMT mice. Shaded and line histograms indicate cells from E- and T-BMT mice, respectively. (B) GFP levels of iNKT cells (shaded) were compared with E- or T-CD4 SP thymocytes (line). (C) Higher GFP expression on CD5hi DP cells and DP dull thymocytes from T-BMT than E-BMT mice. CD5 expression and gating of those cells are shown in Fig. S1B. Shaded and line histograms indicate cells from E- and T-BMT mice. (D) Decrease in GFP expression in T-CD4 SP thymocytes as they mature. CD4 SP thymocytes were divided based on the level of CD8 in E- and T-CD4 T cells and GFP expression of those cells was examined. Shaded and line histograms indicate cells from E- and T-BMT mice. (E) GFP levels on three different stages of E- or T-CD4 T cells were compared. Shaded, DP dull; dotted line, CD8hi CD4 SP thymocytes; solid line, CD8lo CD4 SP thymocytes. Data are representative of 6 E-BMT and 10 T-BMT mice.
Opposing Effect of Ras–MAPK Signaling Modulation on E- vs. T-CD4 T-Cell Development.
Having observed that T-CD4 T cells receive stronger TCR signal than E-CD4 T cells during selection, we asked whether changing TCR-induced signaling strength will affect T-CD4 T-cell selection. To test the hypothesis, we first examined Ras signaling because signaling through the Ras–MAPK pathway is known to be important for positive selection of E-CD4 T cells (12). Recently, we reported improved selection of E-CD4 T cells in the absence of a RasGTPase-activating protein RASA1 further supporting that increased Ras–MAPK signaling enhances E-CD4 T-cell development (17). To determine whether T-CD4 T cells require the same signaling principle as E-CD4 T cells, we evaluated T-CD4 T-cell development in the absence of RASA1 (RASA1−/−). RASA1−/− E- and T-CD4 T cells were generated using the same strategy as for Nur77GFP cells.
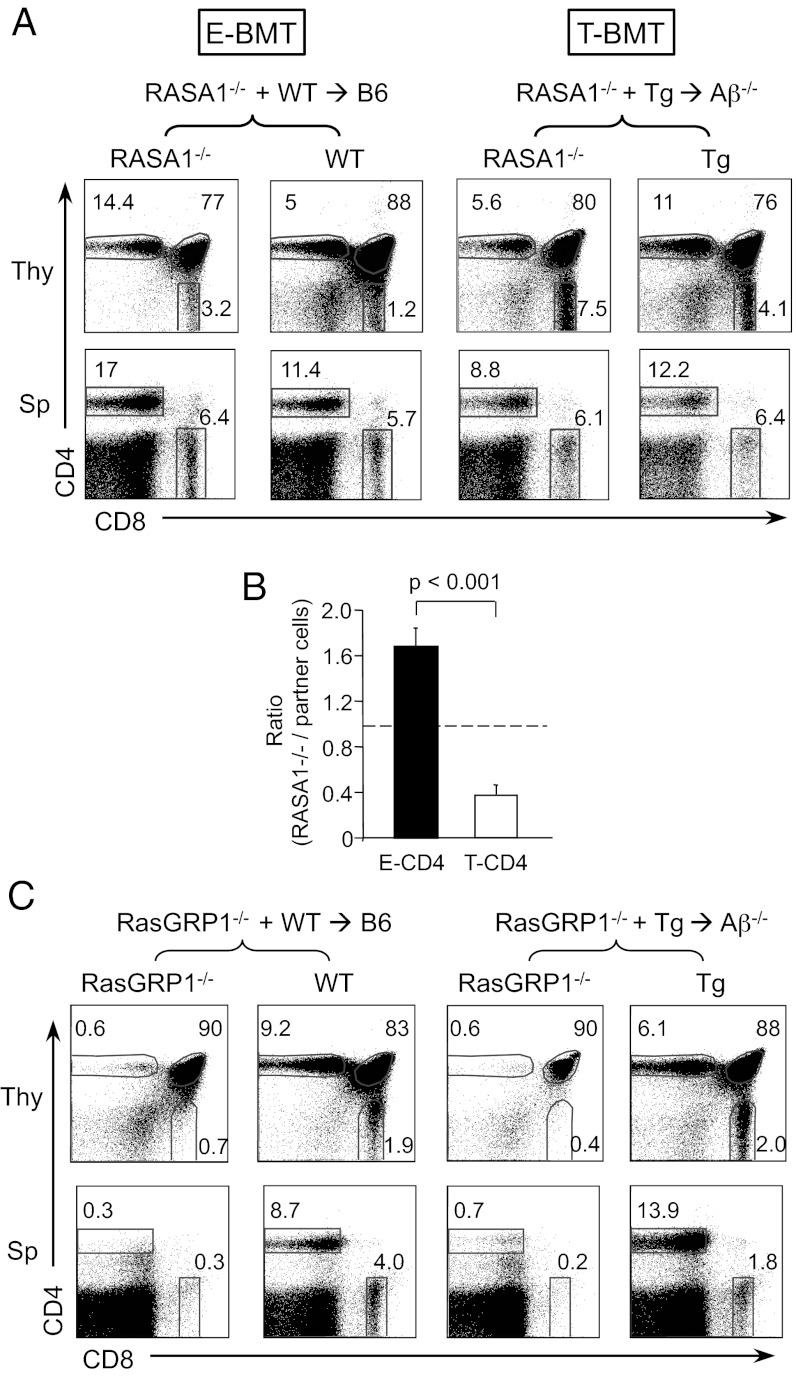
Consistent with our previous study (17), RASA1−/− BM cells generated CD4 SP thymocytes more efficiently than WT partner cells when they were selected by TEC (Fig. 2A, Upper Left). In contrast, the percentages of RASA1−/− CD4 SP were lower than the partner in T-BMT (Fig. 2A, Upper Right). To quantify the selection efficiency from several BM chimeras, the ratio of RASA1−/− CD4 SP to DP thymocytes was calculated. Previously, we have shown that WT and Tg CD4 T cells were generated with comparable efficiency with a ratio of 1 in [WT+Tg→Aβ−/−] chimeras (23). Therefore, Tg or WT partner cells in the same chimeric mouse serve as an internal control. As shown in Fig. 2B, the selection efficiency of RASA1−/− CD4 SP thymocytes was greater than that of WT partner cells in the E-BMT group, whereas, in the T-BMT group, the efficiency was reduced. Therefore, RASA1 deficiency compromised thymocyte-mediated CD4 T-cell selection. To determine whether the selection efficiency is due to the enhanced TCR signaling strength in the absence of RASA1, we assessed CD69 expression in DP dull populations because CD69 expression is up-regulated by TCR signaling (29). Indeed, more CD69hi cells were present in RASA1−/− cells than the partner in both E- and T-CD4 BMT (Fig. S2A). Therefore, TCR signaling is enhanced at a similar level in RASA1−/− E- and T-CD4 DP dull thymocytes but its effect on selection is opposite between E- vs. T-CD4 T cells. In the periphery, the proportion of RASA1−/− E- or T-CD4 T cells was not significantly different from their partner cells (Fig. 2A, Lower and Fig. S2B). The CD8 T-cell compartments were comparable among the different groups (Fig. S2B). In addition, peripheral T-CD4 T cells expressed high levels of CD44, but reduced levels of CD62L regardless of RASA1 status, suggesting that RASA1 plays a role in CD4 T-cell development, but has little impact in the effector phenotype (Fig. S2C).
Fig. 2.
Opposing effects of Ras signaling on E- and T-CD4 T-cell development. (A) BM chimeras were constructed as in Fig S1. Representative FACS profile of the thymocytes (Thy) and splenocytes (Sp) derived from indicated BM donors. Numbers in the dot plots indicate the percentages of gated cells. Data are representative of six pairs of mice. (B) Ratio of RASA1−/− CD4 T-cell selection efficiency to the partner cell selection efficiency in the same host, using the formula [% CD4 SP of RASA1−/−/% DP of RASA1−/−]/[% CD4 SP of the partner/% DP of the partner]. Dotted line indicates the ratio of 1. (C) CD4 and CD8 profile of thymocytes and splenocytes from RasGRP1−/− E- and T-BMT. Data are representative of five pairs of mice. RasGRP1−/− BM and the partner BM cells were mixed at a ratio of 3:1.
Having observed that enhancing Ras activity by RASA1 deficiency increased the selection efficiency of E-CD4 T cells, but reduced that of T-CD4 T cells, we asked whether repressing Ras activity would improve T-CD4 selection. For this purpose, we used mice deficient in RasGRP1, an activator of Ras, because E-CD4 T-cell development has been shown to be greatly reduced in the absence of RasGRP1 (14). As expected, CD4 SP thymocytes and CD4 splenic cells were severely diminished in E-CD4 BMT hosts (Fig. 2C, Left group). Similar to E-CD4 T cells, we also observed the disappearance of T-CD4 T cells in the absence of RasGRP1 (Fig. 2C, Right group). Therefore, RasGRP1 plays an indispensable role for both E- and T- CD4 T-cell development. In addition, both groups of chimera showed reduced CD8 T-cell generation as well (Fig. 2C).
Because the deficiency in both RASA1- and RasGRP1-impaired T-CD4 T-cell development, we examined the role of these signaling molecules for iNKT cell development (Fig. S3). As for T-CD4 T cells, RASA1−/− BM-derived iNKT cells were greatly diminished compared with WT iNKT cells. Similarly, iNKT cells were barely detectable with RasGRP1 deficiency as reported (30). Therefore, even though both T-CD4 and iNKT cell development is compromised when too much signaling in Ras pathway is available, generation of these cell types requires Ras-mediated signaling.
Enhanced T-CD4 T-Cell Development in the Absence of Itk.
The data showing that the increase in Ras activation is detrimental to T-CD4 T-cell selection prompted us to consider the possibility that dampening TCR-induced Ras activation could enhance T-CD4 T-cell selection. To test this, we used an Itk-deficient model in which Ras activation is defective and, as a consequence, E-CD4 generation is known to be compromised (5, 6).
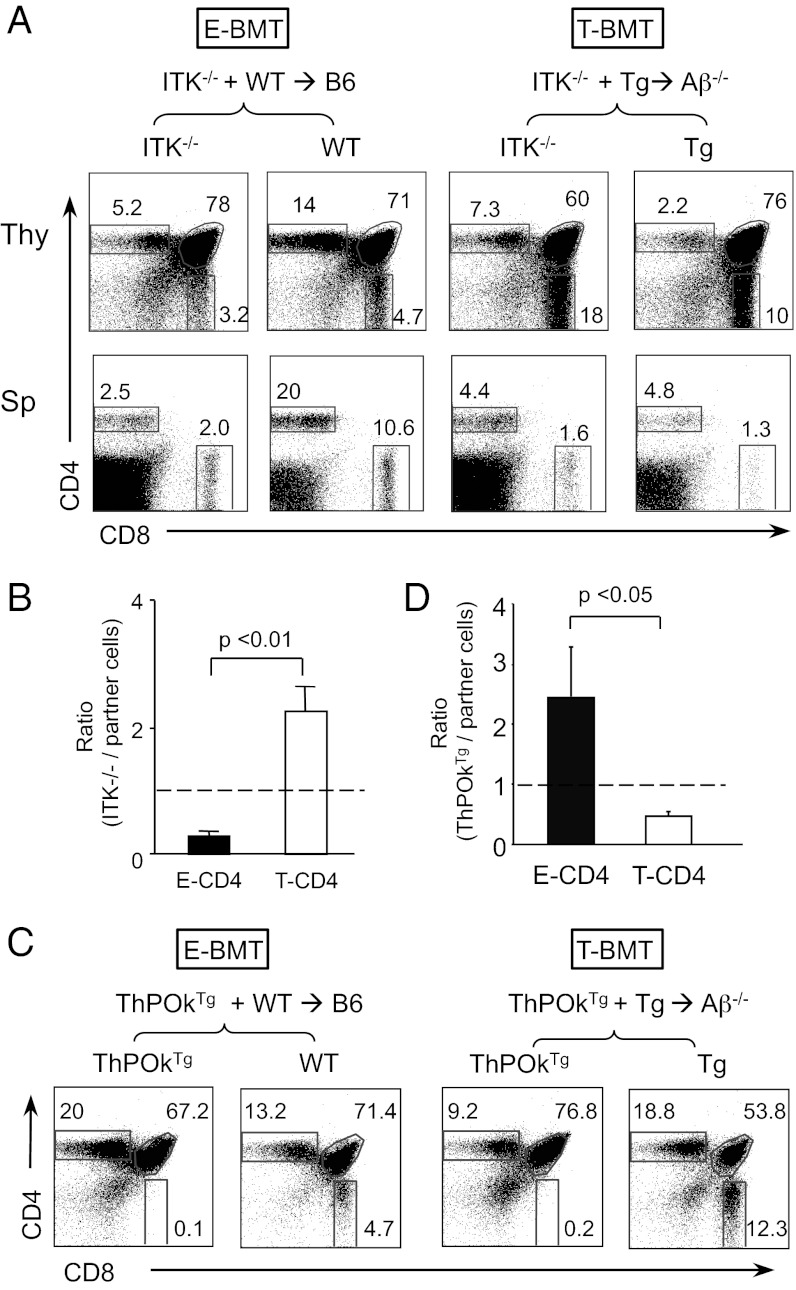
When Itk−/− CD4 T cells were selected by TEC, the CD4 SP compartment was reduced compared with the WT partner cells (Fig. 3A, Upper Left), which is consistent with the published observations that the loss of Itk substantially impairs but does not totally abolish E-CD4 T-cell development (6). By contrast, in T-BMTs, the percentage of Itk−/− CD4 SP thymocytes increased compared with Tg BM-driven partners (Fig. 3A, Upper Right), showing that the absence of Itk enhances T-CD4 T-cell generation. The selection efficiencies further confirmed the opposing effect of Itk deficiency on E-and T-CD4 T-cell development (Fig. 3B). DP dull thymocytes from E- and T-BMT showed the decrease in CD69hi cells, although cells from T-BMT were affected more severely than those of E-BMT, suggesting that TCR signaling strength was weakened in the absence of ITK (Fig. S4A). Therefore, reduced TCR signaling is beneficial and detrimental for T- and E-CD4 T-cell selection, respectively. In the periphery, E-BMT showed the reduction in Itk−/− CD4 and CD8 T cells compared with Itk+/+ partner cells (Fig. S4B) as previously reported (5). However, this reduction was smaller in T-BMT than E-BMT (Fig. S4B). With regard to the cell phenotype, Itk−/− CD4 T cells resembled that of Itk-sufficient cells in both E- and T-BMT hosts (Fig. S4C), suggesting that Itk contributes to the selection process, but not the phenotype of the resulting cells.
Fig. 3.
Itk deficiency enhances T-CD4 T-cell development. (A) BM chimeras were constructed as in Fig. S1 using 3:1 ratio of ITK−/− to partner cells. Representative FACS profile of the thymocytes (Thy) and splenocytes (Sp) derived from indicated BM donors. (B) Selection efficiency of Itk−/− CD4 T-cell to the partner cell calculated as in Fig. 2B. n = 4 pairs. (C) Impaired T-CD4 T-cell development by constitutive expression of ThPOK. ThPOKTg E- and T-CD4 T cells were generated using BMT chimeras constructed using the same strategy as described in Fig. S1. Representative FACS profile of the thymocytes from indicated BM donors. (D) Ratio of ThPOkTg CD4 T-cell selection efficiency to the partner cell selection efficiency in the same host. Dotted line indicates the ratio of 1. Data are representative of four pairs of mice.
We next tested the role of ITK in T-CD4 T-cell selection using ThPOKTg mice. It is reported that ThPOK functions downstream of ITK, and its expression correlates with T-cell signaling strength, as a part of the mechanisms of CD4 vs. CD8 T-cell lineage commitment (31). Therefore, overexpression of ThPOK would create a similar environment that mimics high ITK activity. Indeed, constitutive expression of ThPOK resulted in poor development of T-CD4 T cells (Fig. 3 C and D). Together, T-CD4 cells selected by strong TCR signals survived less than E-CD4 T cells upon the increased strength of signaling.
Strong Signal Induces PLZF Expression in Positively Selected DP Thymocytes.
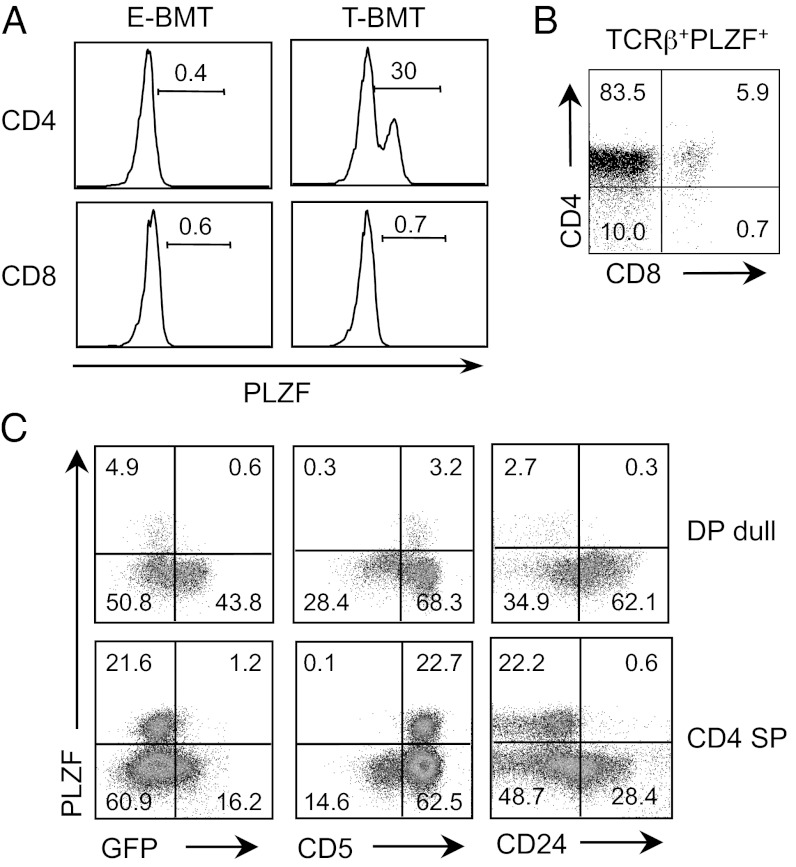
In addition to T-CD4 T cells, iNKT and γδ T cells receive strong TCR signals during selection and these three cell types express the transcription factor PLZF (25, 26, 32, 33). We hypothesized that PLZF expression in T-CD4 T cells is induced by receiving strong TCR signals given by MHC class II+ thymocytes. To test this, we first examined PLZF expression in E- and T-CD4 T cells from WT→B6 (E-BMT) and Tg→Aβ−/− (T-BMT) BM chimeric mice. We observed a PLZF+ non-iNKT cell population in CD4 SP thymocytes from T-BMT (Fig. 4A).
Fig. 4.
PLZF is expressed upon receiving strong TCR signaling. (A) PLZF expression of CD1d-tetramer negative CD4 SP (Upper) and CD8 SP (Lower) thymocytes from E-BMT (Left, WT→WT) and T-BMT hosts (Right, Tg→Aβ−/−). Data are representatives of four E-BMTs and three T-BMTs. (B) CD4 and CD8 profile of TCRβ+ PLZF+ cells from total thymocytes of [Nur77GFP+Tg → Aβ−/−] chimeras. (C) Expression of PLZF vs. Nur77GFP (Left), PLZF vs. CD5 (Center) and PLZF vs. CD24 (Right) of DP dull and CD4 SP thymocytes from Nur77GFP donor of the chimeric mice shown in Fig. 1.
Next, we examined PLZF expression together with Nur77GFP expression using the same [Nur77GFP+Tg → Aβ−/−] chimeras shown in Fig. 1. To do this, total thymocytes were stained for PLZF, and TCRβ+ PLZF+ cells were gated to assess CD4 and CD8 expression. Two observations were made. First, CD4 SP thymocytes comprised most of the PLZF+ population. In addition, a small fraction of DP thymocytes also expressed PLZF (Fig. 4B), suggesting that PLZF expression likely started at the late DP stage. A significant fraction of PLZF+ cells were CD4CD8 double negative (DN) thymocytes. These cells were not immature cells because they expressed TCRβ (Fig. 4B). Second, PLZF+ cells expressed low levels of Nur77 with high CD5 and low CD24, indicating that the PLZF+ cells arose from DP cells that had received a TCR stimulus (Fig. 4C). Together, the data show that DP thymocytes express PLZF by strong signal but subsequent signaling strength is down-regulated, as evidenced by decreased GFP.
Minimal Role of Signaling Molecules for T-CD4 T-Cell Effector Function.
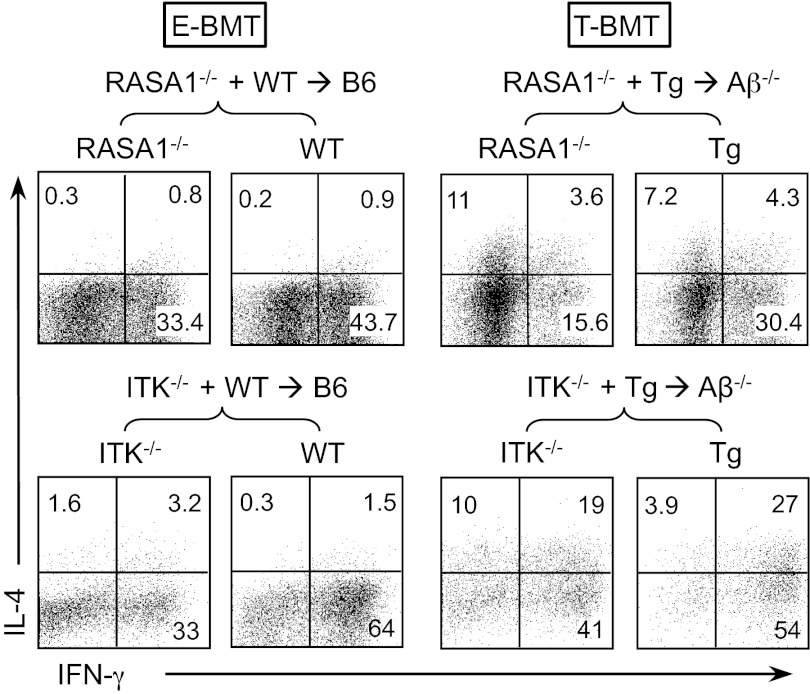
Having observed that T-CD4 T-cell development is affected by signaling strength, we asked whether a change in signaling strength influences the function of T-CD4 T cells. The key signature defining T-CD4 T-cell function is IL-4 expression under Th1-skewing conditions. To test this, splenic CD4 T cells from RASA1−/− and Itk−/− chimeras were differentiated under Th1 conditions. E-CD4 T cells showed a typical Th1 cytokine profile, defined by IFN-γ+ but not IL-4+ cells regardless of genetic modifications (Left groups in Fig. 5 and Fig. S5). In contrast, all T-CD4 T cells, including RASA1−/− or Itk−/− cells, expressed IL-4 in addition to IFN-γ, suggesting that RASA1 and Itk are not critical for the effector function despite their important roles in the developmental process of T-CD4 T cells (Right groups in Fig. 5 and Fig. S5). However, we observed that RASA1 or Itk deficiency resulted in reduced IFN-γ production in both E- and T-CD4 T cells (Fig. 5 and Fig. S5).
Fig. 5.
Effector function of T-CD4 T cells is not affected by the strength of TCR signaling. Splenic CD4 T cells from indicated mice were enriched and cultured under the Th1 differentiating condition followed by intracellular staining of IFN-γ and IL-4. Shown is the representative staining profiles of the indicated chimeras using RASA1−/− (Upper) or Itk−/− (Lower) BM. RASA1−/−, six pairs of mice; Itk−/−, three pairs of mice.
Discussion
T-CD4 T cells, innate CD8 T cells, iNKT cells, and γδ T cells have been recognized as innate-like T cells because of their ability to act as effector cells immediately after activation (34). Interestingly, except γδ T cells, these cells are selected by hematopoietic cells. Both iNKT and γδ T cells have been demonstrated or suggested to perceive stronger TCR stimulus than conventional T cells during development (27, 32). Here with T-CD4 T cells, our data clearly showed that thymocytes receive stronger TCR signaling upon interaction with thymocytes than with TEC. Thymocyte–thymocyte (T–T) interaction changes the intracellular environment of thymocytes, induces new transcriptional programming of genes including IL-4 and PLZF, and consequently generates effector T-CD4 T cells with distinct functions. This is consistent with other innate-like T cells such as iNKT and γδ T cells. Although modulation of signal strength resulted in altered T-CD4 T-cell development, it did not prevent T-CD4 T cells from producing IL-4 under Th1-skewing conditions (Fig. 5), suggesting the separation of the signaling events for development and function.
A recent study reported that persistent high expression of Egr2, an early transcription factor induced by TCR signaling, correlated with the expression of PLZF, which specifies the iNKT cell lineage (35). Moreover, iNKT cells also receive stronger TCR signaling measured by Nur77 expression (27). Therefore, both T-CD4 and iNKT cells receive stronger TCR signaling than E-CD4 T cells during selection and then the two cell types lose GFP signals subsequently. It is not clear what triggers the quick loss of the GFP signals on T-CD4 T cells but not E-CD4 T cells. However, the loss occurs in the thymus immediately after selection and hence the lack of tonic TCR signals is not likely responsible for the reduction of GFP levels on T-CD4 T cells. Furthermore, peripheral T-CD4 T cells maintain the GFP levels similar to that of thymic T-CD4 T cells, indicating that T-CD4 T cells continue to receive the same levels of TCR signals as they emigrate to the periphery.
An opposing role of RASA1 and Itk in E- vs. T-CD4 T-cell selection suggests that the two CD4 T-cell types are selected by different spectrums of TCR signaling strength. CD69 expression profiles of RASA1−/− and ITK−/− cells further support the increase and decrease in signaling strength, respectively. In the absence of RASA1, E-CD4 T cells with low-to-moderate TCR affinity receive enhanced Ras–MAPK signaling, which would help the survival of E-CD4 T cells. In contrast, the RASA1 deficiency would make high affinity TCR bearing T-CD4 T cells reach the signaling strength beyond the tolerance level, placing them on negative selection. As a result, more T-CD4 T cells die tipping the balance favorable to E-CD4 T cells. iNKT cell development was also compromised when Ras signaling is enhanced by RASA1 deficiency (Fig. S3), which further supported the notion that both T-CD4 and iNKT cells receive strong TCR signaling during their selection.
A majority of E-CD4 T cells die in the absence of Itk due to insufficient TCR signaling (9). In contrast, Itk−/− T-CD4 T cells can survive better, perhaps because they are rescued from negative selection that would normally be mediated by their high affinity TCR, likely due to their ability to exert intracellular signaling delivered by high affinity TCR. We further tested the role of ITK in T-CD4 T-cell selection using ThPOKTg mice. Together, T-CD4 cells selected by strong TCR signals are more susceptible to negative selection than E-CD4 T cells upon the increased strength of signaling.
Although development of T-CD4 T cells improved in the absence of Itk, Itk deficiency compromises iNKT cell generation (36). One explanation for this difference could be the diversity of TCR repertoires between T-CD4 and iNKT cells. T-CD4 T cells are polyclonal, allowing a wide window of survival, whereas iNKT cells with invariant TCR would have a very limited range of tolerance, making them more susceptible to a change in the strength of signaling. In this regard, it would be informative to compare the signaling requirements to select CD4 T cells by thymocytes vs. TEC using the same monoclonal TCR such as the DO11.10 or AND TCR transgenic mice. Unfortunately, thymocytes expressing the DO11.10 or AND TCR were poorly selected by MHC class II expressed by other thymocytes, and residual CD4 T cells did not express transgenic TCR (19). Although underlying mechanisms for this observation are not clear, two different selection pathways seem to generate CD4 T-cell populations composed of nonoverlapping TCR repertoires and that the TCR specificity may instruct the development of E- and T-CD4 T cells by TEC and thymocytes, respectively. If this were the case, thymocytes expressing a TCR cloned from a T-CD4 T cell would be selected preferentially by thymocytes. In addition to the difference in the TCR repertoire between T-CD4 T and iNKT cells, IL-15R and T-bet are critical for iNKT but not for T-CD4 T-cell survival and maturation (23, 37, 38), which indicates that although thymocytes deliver similar TCR signaling regardless of TCR recognition with MHC class II or CD1d during selection, the signaling recipients, T-CD4 and iNKT cells, respectively, take different courses and requirements for maturation afterward. Therefore, T–T interactions generate both types of innate T cells, but the two undergo distinct signaling events forming functional cells in the end, which may be mediated by other receptors expressed in a cell-type–specific manner. Furthermore, the maintenance of mature T-CD4 and iNKT cells in the periphery seems to be distinct. As we showed here, T-CD4 T cells appear to receive TCR signaling continuously, as evidenced by Nur77GFP expression, albeit at a reduced level, whereas iNKT cells are reported to receive very weak TCR signals at the steady state (27).
PLZF was detected in DP thymocytes from T-BMT that express high CD5, low CD24, and low TCRβ, which indicates that PLZF expression starts at the late stage of DP that have undergone selection processes by receiving strong TCR signals. Because DP thymocytes can receive signaling via both MHC classes I and II, the MHC class I signal might have contributed to Nur77 up-regulation and PLZF expression. However, this is unlikely the case on the basis of the following two reasons. First, the same Nur77GFP BM cells were introduced to WT or Aβ−/− hosts in which MHC class I expression is not different. Therefore, DP thymocytes in both E- and T-BMT should have received the same MHC class I-mediated signals during development. Second, we compared the Nur77GFP levels of postselected DP (TCRβ+ CD69hi) thymocytes developed in Nur77GFP/WT and Nur77GFP/Aβ−/− mice. As shown in Fig. S6, TCRβ+ CD69hi DP thymocytes from Aβ−/− mice expressed lower levels of GFP than WT, indicating that postselected DP thymocytes received weaker signaling by MHC class I than MHC class I together with class II. Therefore, the elevated Nur77GFP levels on CD5hi DP thymocytes in T-BMT are likely contributed by TCR–MHC class II interactions.
Some PLZF+ cells expressed TCRβ but neither CD4 nor CD8, suggesting that these PLZF+ DN cells are mature T cells. Identification of these mature DN cells is not clear. A recent study showed the presence of TCRαβ+ DN cells in the thymus that survive negative selection and become CD8αα+ intraepithelial lymphocytes (39). Therefore, it is tempting to speculate that PLZF+ DN cells might have a similar fate. We also observed that PLZF is not uniformly expressed in T-CD4 SP thymocytes, although all were selected by thymocytes (Fig. 4A). Perhaps, PLZF expression might be an indicator of two subpopulations of T-CD4 T cells. Along this line, a similar observation has been made in γδ T cells and it has been proposed that PLZF+ and PLZF− γδ T cells represent two distinct lineages (32). Human T-CD4 T cells also show PLZF+ and PLZF− T-CD4 T cells (20) and further characterization of these two populations of T-CD4 T cells is warranted.
In conclusion, the current study shows differential requirements of TCR signaling strength for E- vs. T-CD4 T-cell development. Receiving stronger signaling from thymocytes contributes to T-CD4 PLZF expression, which makes them become innate-like effector cells immediately after selection. Their functional phenotype seems to be intrinsic to the strength of TCR signals they received, but the signaling events responsible for the generation and the effector function of the resulting CD4 T cells are not identical.
Materials and Methods
Mice.
CIITATg mice carrying CD45.1 and CD45.2 congenic markers were described previously (19). CD45.1+ B6.SJL mice and CD45.1+ Aβ−/− mice were purchased from Taconic. Itk−/− (5), RASA1−/− (17), RasGRP1−/− (14), ThPOKTg (40), and Nur77GFP (27) mice were described elsewhere. All mice were housed in the animal facility at the University of Michigan Medical School under specific pathogen-free conditions and were used at 6–12 wk of age. All animal experiments were performed under protocols approved by the University Committee on Use and Care of Animals.
Reagents.
TCRβ (H57), CD4 (L3T4), CD5 (53-7.3), CD8 (53-6.7), CD24 (M1/69), CD45.1 (A20), CD45.2 (104), CD62L (MEL-14), CD44 (IM7), NK1.1 (NK1.1), anti–IL-4 (11B11), and anti–IFN-γ (XMG1.2) were purchased from BD PharMingen or eBioscience. PBS 57-loaded CD1d-tetramer was provided by the National Institutes of Health tetramer core facility. Antimouse PLZF mAb (Mags.21F7) was previously described (32).
Full methods are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Terry Geiger for critical reading of the manuscript. This research was supported in part by National Institutes of Health (NIH) Grants T32 CA009138 (to G.L.S.), CA153260 (to D.J.K.); HL096498 (to P.D.K); AI083988 and AI059739 and Robert Wood Johnson Foundation Grant 67038 to the Child Health Institute (to D.B.S.); AI073677 (to C.-H.C.); and by intramural funds from the National Human Genome Research Institute (to P.L.S.). CD1d tetramers were provided by the NIH tetramer facility.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207528109/-/DCSupplemental.
References
- 1.Davis MM, et al. Ligand recognition by alpha beta T cell receptors. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 2.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 3.Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. 2009;9:833–844. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 4.Guy CS, Vignali DA. Organization of proximal signal initiation at the TCR:CD3 complex. Immunol Rev. 2009;232:7–21. doi: 10.1111/j.1600-065X.2009.00843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao XC, Littman DR. Altered T cell receptor signaling and disrupted T cell development in mice lacking Itk. Immunity. 1995;3:757–769. doi: 10.1016/1074-7613(95)90065-9. [DOI] [PubMed] [Google Scholar]
- 6.Schaeffer EM, et al. Tec family kinases modulate thresholds for thymocyte development and selection. J Exp Med. 2000;192:987–1000. doi: 10.1084/jem.192.7.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaeffer EM, et al. Requirement for Tec kinases Rlk and Itk in T cell receptor signaling and immunity. Science. 1999;284:638–641. doi: 10.1126/science.284.5414.638. [DOI] [PubMed] [Google Scholar]
- 8.Berg LJ, Finkelstein LD, Lucas JA, Schwartzberg PL. Tec family kinases in T lymphocyte development and function. Annu Rev Immunol. 2005;23:549–600. doi: 10.1146/annurev.immunol.22.012703.104743. [DOI] [PubMed] [Google Scholar]
- 9.Lucas JA, Atherly LO, Berg LJ. The absence of Itk inhibits positive selection without changing lineage commitment. J Immunol. 2002;168:6142–6151. doi: 10.4049/jimmunol.168.12.6142. [DOI] [PubMed] [Google Scholar]
- 10.Broussard C, et al. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 11.Horai R, et al. Requirements for selection of conventional and innate T lymphocyte lineages. Immunity. 2007;27:775–785. doi: 10.1016/j.immuni.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alberola-Ila J, Hernández-Hoyos G. The Ras/MAPK cascade and the control of positive selection. Immunol Rev. 2003;191:79–96. doi: 10.1034/j.1600-065x.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- 13.Priatel JJ, Teh SJ, Dower NA, Stone JC, Teh HS. RasGRP1 transduces low-grade TCR signals which are critical for T cell development, homeostasis, and differentiation. Immunity. 2002;17:617–627. doi: 10.1016/s1074-7613(02)00451-x. [DOI] [PubMed] [Google Scholar]
- 14.Dower NA, et al. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat Immunol. 2000;1:317–321. doi: 10.1038/79766. [DOI] [PubMed] [Google Scholar]
- 15.Priatel JJ, Chen X, Dhanji S, Abraham N, Teh HS. RasGRP1 transmits prodifferentiation TCR signaling that is crucial for CD4 T cell development. J Immunol. 2006;177:1470–1480. doi: 10.4049/jimmunol.177.3.1470. [DOI] [PubMed] [Google Scholar]
- 16.Sprang SR. G proteins, effectors and GAPs: Structure and mechanism. Curr Opin Struct Biol. 1997;7:849–856. doi: 10.1016/s0959-440x(97)80157-1. [DOI] [PubMed] [Google Scholar]
- 17.Lapinski PE, Qiao Y, Chang CH, King PD. A role for p120 RasGAP in thymocyte positive selection and survival of naive T cells. J Immunol. 2011;187:151–163. doi: 10.4049/jimmunol.1100178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi EY, et al. Thymocyte-thymocyte interaction for efficient positive selection and maturation of CD4 T cells. Immunity. 2005;23:387–396. doi: 10.1016/j.immuni.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Li W, et al. An alternate pathway for CD4 T cell development: Thymocyte-expressed MHC class II selects a distinct T cell population. Immunity. 2005;23:375–386. doi: 10.1016/j.immuni.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Lee YJ, et al. 2010 Generation of PLZF+ CD4+ T cells via MHC class II-dependent thymocyte-thymocyte interaction is a physiological process in humans. J Exp Med 207(1):237–246, S231–S237. [Google Scholar]
- 21.Min HS, et al. MHC class II-restricted interaction between thymocytes plays an essential role in the production of innate CD8+ T cells. J Immunol. 2011;186:5749–5757. doi: 10.4049/jimmunol.1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, et al. Thymic selection pathway regulates the effector function of CD4 T cells. J Exp Med. 2007;204:2145–2157. doi: 10.1084/jem.20070321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W, et al. The SLAM-associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity. 2007;27:763–774. doi: 10.1016/j.immuni.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasquier B, et al. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J Exp Med. 2005;201:695–701. doi: 10.1084/jem.20042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Azzam HS, et al. Fine tuning of TCR signaling by CD5. J Immunol. 2001;166:5464–5472. doi: 10.4049/jimmunol.166.9.5464. [DOI] [PubMed] [Google Scholar]
- 29.Swat W, Dessing M, von Boehmer H, Kisielow P. CD69 expression during selection and maturation of CD4+8+ thymocytes. Eur J Immunol. 1993;23:739–746. doi: 10.1002/eji.1830230326. [DOI] [PubMed] [Google Scholar]
- 30.Shen S, et al. Critical roles of RasGRP1 for invariant NKT cell development. J Immunol. 2011;187:4467–4473. doi: 10.4049/jimmunol.1003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu J, Qi Q, August A. Itk derived signals regulate the expression of Th-POK and controls the development of CD4 T cells. PLoS ONE. 2010;5:e8891. doi: 10.1371/journal.pone.0008891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alonzo ES, et al. Development of promyelocytic zinc finger and ThPOK-expressing innate gamma delta T cells is controlled by strength of TCR signaling and Id3. J Immunol. 2010;184:1268–1279. doi: 10.4049/jimmunol.0903218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreslavsky T, et al. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci USA. 2009;106:12453–12458. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alonzo ES, Sant’Angelo DB. Development of PLZF-expressing innate T cells. Curr Opin Immunol. 2011;23:220–227. doi: 10.1016/j.coi.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seiler MP, et al. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol. 2012;13:264–271. doi: 10.1038/ni.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felices M, Berg LJ. The Tec kinases Itk and Rlk regulate NKT cell maturation, cytokine production, and survival. J Immunol. 2008;180:3007–3018. doi: 10.4049/jimmunol.180.5.3007. [DOI] [PubMed] [Google Scholar]
- 37.Chang CL, Lai YG, Hou MS, Huang PL, Liao NS. IL-15Rα of radiation-resistant cells is necessary and sufficient for thymic invariant NKT cell survival and functional maturation. J Immunol. 2011;187:1235–1242. doi: 10.4049/jimmunol.1100270. [DOI] [PubMed] [Google Scholar]
- 38.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 39.Pobezinsky LA, et al. Clonal deletion and the fate of autoreactive thymocytes that survive negative selection. Nat Immunol. 2012;13:569–578. doi: 10.1038/ni.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.