Abstract
The eight metabotropic glutamate receptors (mGluRs) are key modulators of synaptic transmission and are considered promising targets for the treatment of various brain disorders. Whereas glutamate acts at a large extracellular domain, allosteric modulators have been identified that bind to the seven transmembrane domain (7TM) of these dimeric G-protein-coupled receptors (GPCRs). We show here that the dimeric organization of mGluRs is required for the modulation of active and inactive states of the 7TM by agonists, but is not necessary for G-protein activation. Monomeric mGlu2, either as an isolated 7TM or in full-length, purified and reconstituted into nanodiscs, couples to G proteins upon direct activation by a positive allosteric modulator. However, only a reconstituted full-length dimeric mGlu2 activates G protein upon glutamate binding, suggesting that dimerization is required for glutamate induced activation. These data show that, even for such well characterized GPCR dimers like mGluR2, a single 7TM is sufficient for G-protein coupling. Despite this observation, the necessity of dimeric architecture for signaling induced by the endogenous ligand glutamate confirms that the central core of signaling complex is dimeric.
Keywords: time-resolved FRET, oligomerization, SNAP-tag
Metabotropic glutamate receptors (mGluRs) play key roles in the modulation of both excitatory and inhibitory synapses in the brain. These eight G-protein-coupled receptors (GPCRs) represent major targets for pharmaceutical companies in search of new treatments for a variety of neurological and psychiatric disorders (1–3). These receptors are part of the class C GPCR family that also includes the GABAB, calcium sensing, and sweet and umami taste receptors, which are all major targets for drug development (4).
The structural complexity of class C GPCRs, compared with rhodopsin-like class A GPCRs, offers multiple possibilities in designing molecules that modulate their activity. Not only are mGluRs strict constitutive dimers (5, 6), but each protomer is composed of several domains (7, 8). Agonists bind in a bilobate venus fly-trap domain (VFT) (9), which is linked through a cysteine-rich domain (CRD) to the heptahelical transmembrane domain (7TM) that is responsible for G-protein activation (7). The 7TM is the target of a number of synthetic compounds acting either as negative or positive allosteric modulators (NAMs and PAMs, respectively). Given their ability to finely tune endogenous signaling, such compounds present exciting opportunities for drug development (10).
The functional mechanism of such a complex machine remains to be characterized, although some critical steps have been well documented. Previous studies have shown that receptor activation results from the closure of the VFT upon agonist binding (9, 11–15). This conformational change in the extracellular domain is coupled to a conformational change in the intracellular side of at least one 7TM that is responsible for G-protein coupling (16–19). The mechanism for allosteric communication between the VFT and 7TM intracellular domain remains unknown. Several data revealed that a relative movement of the two subunits within mGluR dimers is associated with receptor activation. Structural as well as mutagenesis studies demonstrated a large change in the relative position of the VFTs is associated with dimer activation (9, 14, 15, 20), but this is not consistent with all structures available (8). At the 7TM level, a movement between the two subunits has also been demonstrated using FRET studies (19, 21, 22). It remains to be determined whether such changes are only associated with the activation process, result as a consequence of other more critical steps, or represent the unique driving force for 7TM activation.
Studies of mGluRs with VFTs deleted demonstrated that the 7TM can fold independently, oscillate between active and inactive conformations, and respond to synthetic ligands (23). Indeed, it was found that PAMs that only potentiate the action of agonists on the full-length receptors display strong agonist activity on VFT truncated receptors (23). Accordingly, the VFTs are not only responsible for agonist-induced activation, but also prevent PAMs from activating the full-length receptor via a mechanism that remains to be discovered.
In the present study, we examined the role of dimerization of class C GPCRs in the coupling between agonist binding and G-protein activation. We first show that a monomeric subunit couples to G protein upon activation with a PAM, demonstrating that a dimeric 7TM of mGluRs is not required for G-protein coupling. We then demonstrated that a dimeric structure is necessary both for glutamate to activate the receptor and to prevent PAMs from activating the 7TM. Our data provide detailed insight into the molecular basis of class C GPCR activation. They further illustrate that 7TM dimers are not necessary for G-protein activation per se, but show how dimerization can provide fine control of GPCR function.
Results
Oligomerization and Function of the mGluR 7TM Domain.
We previously reported that a minimal 7TM domain of various mGluRs [receptors truncated for both of their extracellular domains (ECDs) and C-terminal intracellular domains] retains its capacity to be addressed to the cell surface and to activate G proteins upon stimulation with a PAM (23). Because dimers of mGluRs are stabilized by a disulfide bridge at the level of their VFTs (5), we assessed whether the truncated 7TM domains of mGluRs spontaneously form dimers, and whether such protein association was mandatory for G-protein coupling.
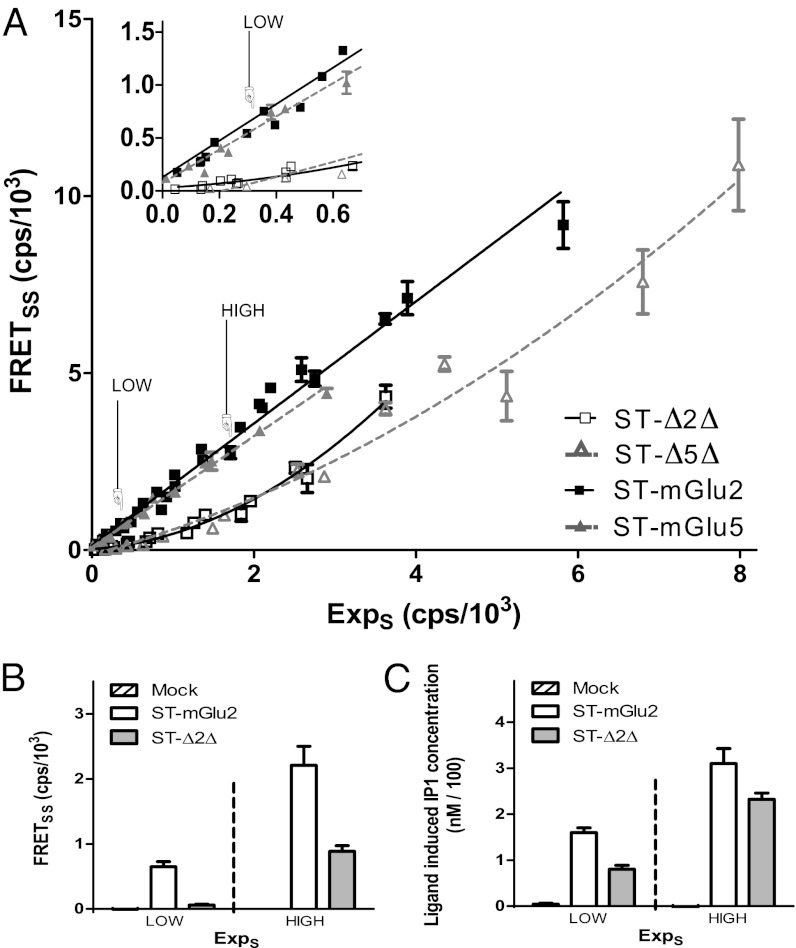
As illustrated in Fig. 1, dimers of full-length mGlu2 and mGlu5 receptors were easily detected by time-resolved FRET (tr-FRET) using N-terminal SNAP-tag fusion subunits, as reported (24). A linear dependence between FRET emission signal and the receptor expression level was observed for the full-length receptor (Fig. 1A), indicating a constant FRET efficacy over this range of receptor expression consistent with covalently stable dimers (24). In contrast, increasing the amount of SNAP-tagged truncated mGlu2 (ST-Δ2Δ) or mGlu5 (ST-Δ5Δ) receptors resulted in a nonlinear increase in FRET signals with nonsignificant FRET signals at low receptor density (Fig. 1A), suggesting an absence of interaction between two truncated protomers. Surprisingly, even under such conditions, ST-Δ2Δ retained the ability to activate G proteins when stimulated by LY487379, a mGlu2 PAM (25) (Fig. 1C). Meanwhile, we observed significant FRET for the same amount of full-length receptor (Fig. 1B). Together, these data revealed that 7TMs of mGlu receptors do not, on their own, form high-affinity stable dimers, and suggest that they exist as functional monomers at the surface of living cells.
Fig. 1.
Dimerization and functional properties of mGlu receptors at the surface of living cells. (A) Specific FRETSNAP-SNAP (FRETSS) signals were plotted as a function of SNAP-tagged (ST) receptor expression (ExpS) given by the fluorescence intensity at 620 nm. (Inset) FRETSS signals measured at low expression levels. (B) FRETSS signals of full-length mGlu2 and ∆2∆ receptors at two different expression levels. (C) Glutamate (mGlu2) and LY487379 (∆2∆)-induced IP1 production in the same batch of cells used in B. Data (mean ± SEM) are representative of five (A) or three (B and C) independent experiments performed in triplicates.
Expression, Detergent Solubilization, and Purification of mGluR 7TM.
To further study the function of monomeric mGlu 7TM domains, we developed an in vitro approach using purified mGlu 7TM reconstituted in lipid nanodiscs, an approach that proved successful for the isolation of GPCR monomers (26–28). To this end, truncated 7TM versions of both mGlu5 and mGlu2 receptors (23, 29) carrying a FLAG epitope and a β2-adrenoreceptor N-terminal domain (30) (see details in the SI Materials and Methods), were produced in insect cells using recombinant baculovirus technology (31) (Fig. S1). mGlu ∆5∆ retains its ability to bind the NAM [3H]-MPEP with a Kd of 8.8 ± 0.7 nM, similar to that reported for the full-length receptor (32) (Fig. S2A). Δ5Δ was then solubilized using a combination of the lipid-like detergent 0.12% Fos-Choline14, 0.01% (wt/vol) cholesteryl hemisuccinate (CHS) and 0.6% [3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate] (CHAPS) (Fig. S2B). These conditions were found to be the most efficient to maintain the stability of the protein and to retain its ability to bind [3H]-MPEP. The same conditions were also efficient for solubilizing the closely related receptor Δ2Δ. Solubilized mGlu 7TMs were purified by immuno-chromatography and displayed the expected molecular weight (36.5 kDa for Δ5Δ and 36.2 kDa for Δ2Δ) using SDS/PAGE (Fig. S2C). The purity of the proteins was estimated by densitometry analysis to be approximately 70%.
Isolated 7TM Reconstituted into Nanodiscs Retains Its Ability to Activate G Proteins.
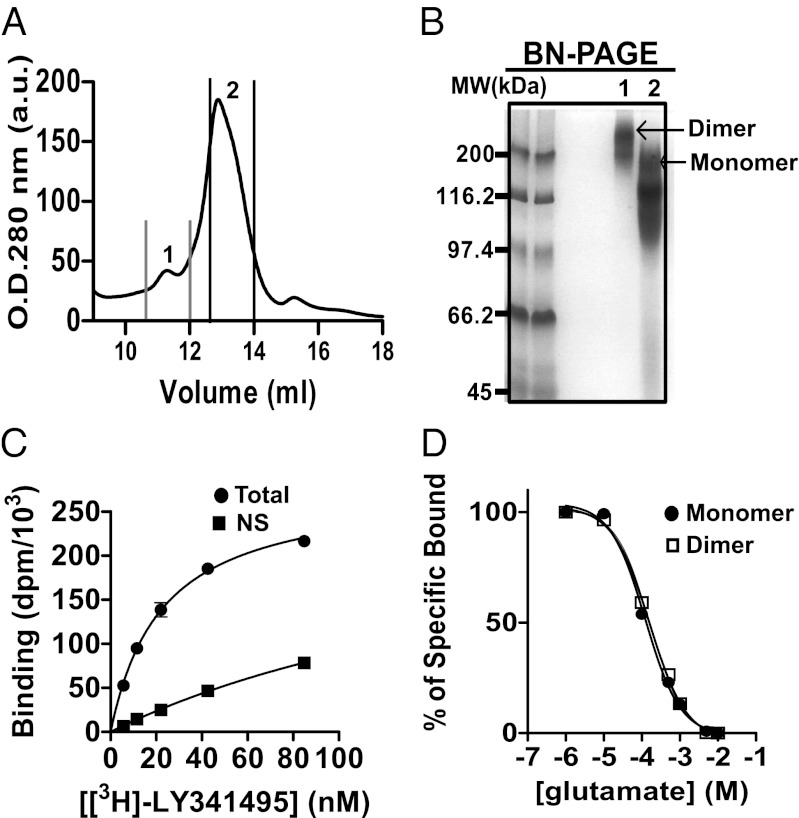
The purified ∆2∆ protein was incorporated into lipid nanodiscs following the published protocol (33). Briefly, the purified protein was mixed with a membrane scaffold protein (MSP) MSP1E3D1 together with the 1-Palmitoyl-2-oleoylphosphatidylcholine (POPC) using two different ratios. When using a POPC:MSP:Δ2Δ ratio of 150:1:1, the resulting nanodiscs preparation was largely enriched in structures containing a single monomer of ∆2∆. This enrichment is well illustrated by the size-exclusion chromatography profile (Fig. 2A) and blue native (BN)-PAGE analyses (Fig. 2B), in which the peak corresponding to the expected monomer (∼100 kDa) is predominant. In contrast, the peak corresponding to nanodiscs containing two ∆2∆ proteins (∼140 kDa) is predominant when using a 90:1:1 ratio. The use of two distinct ratios of lipids, MSP, and receptor allowed the isolation of nanodiscs containing predominantly either one or two 7TM domains of mGlu2 receptors. Electron microscopy revealed particles with diameter of 12.6 nm, which is in agreement with reported results for rhodopsin nanodiscs (26) (Fig. S3).
Fig. 2.
Formation and biochemical characterization of ∆2∆ containing nanodiscs. (A) Size-exclusion chromatography profile of Δ2Δ nanodisc preparations. (B) BN-PAGE analyses of peaks 1 and 2 from A with 5 μg of total proteins. Δ2Δ monomer nanodisc corresponds to the ∼100 kDa [2MSP (2× 30 kDa) + 1Δ2Δ (36.2 kDa) + lipids] band, whereas the nanodisc containing two Δ2Δ corresponds to the ∼140 kDa (2× 30 + 2× 36.2 kDa + lipids) band. (C) Binding properties of the Δ2Δ monomeric nanodiscs. Nanodiscs (20 μM) were incubated for 15 min at room temperature with increasing amount of MNI-136. Tryptophan fluorescence values (λexc = 300 nm, λem = 345 nm) were plotted against the ligand over receptor concentration ratio (L/R).
We evaluated the ability of the 7TM monomers reconstituted in nanodiscs to bind ligands and couple to G proteins. As revealed by changes in intrinsic tryptophan fluorescence, the monomeric fraction could still bind the mGlu2 NAM, MNI-136 (34), with an apparent affinity of 770 ± 60 nM and a maximal effect of 36% change in the total fluorescence intensity (SI Results and Fig. S4A). In an attempt to estimate the binding stoichiometry, we determined the break in the equilibrium titration carried out at high nanodisc concentrations (20 μM). Here, ligand binding, monitored by a change in fluorescence, followed a linear increase as a function of ligand concentration and is saturated when all available binding sites are occupied. We observed that a ligand:receptor ratio of 0.8–0.9 (Fig. 2C) results in a saturation of binding sites, suggesting that, in our preparation, 80–90% of the receptors are correctly folded and competent to bind ligands.
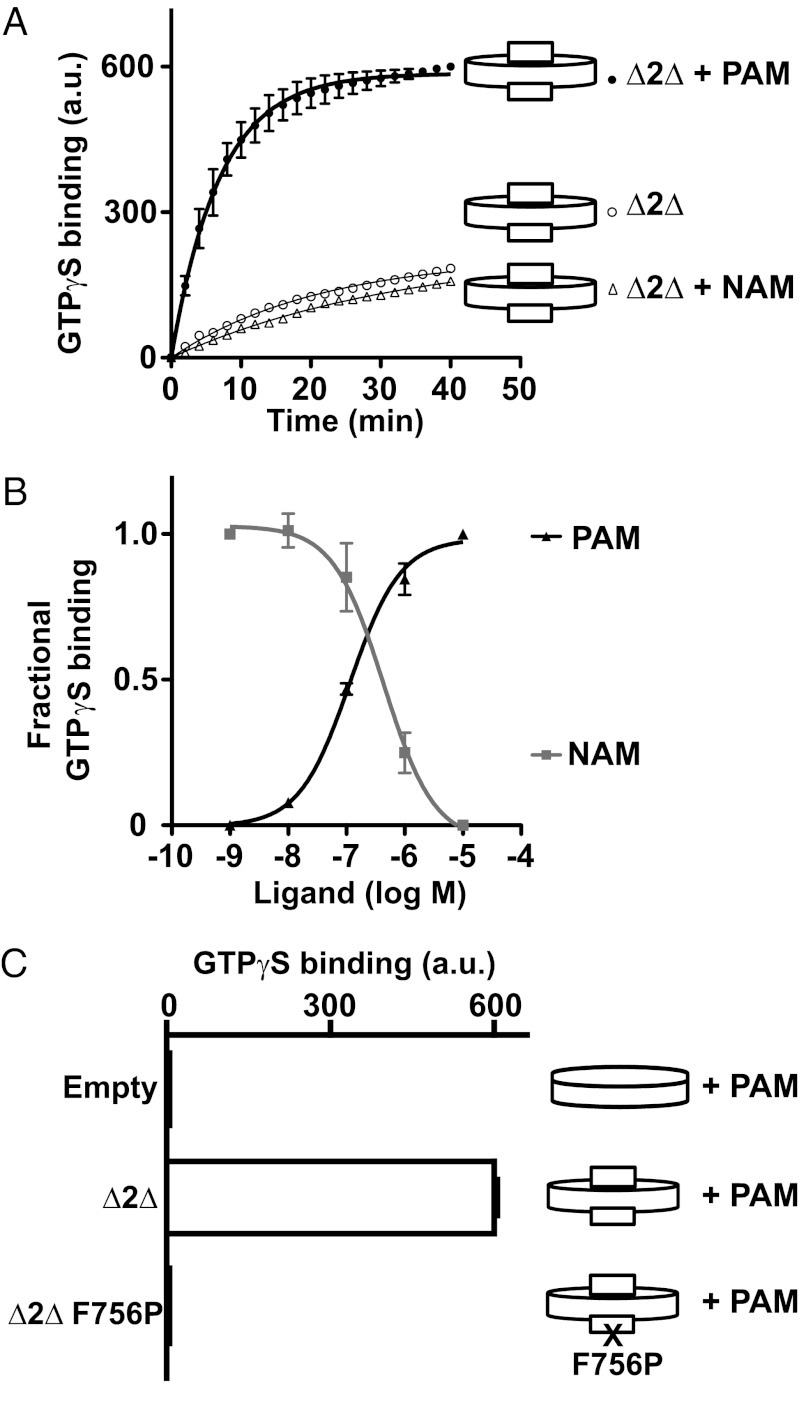
Reconstituted monomeric ∆2∆ could also activate purified Gαi2 proteins efficiently, as demonstrated by the well characterized nucleotide exchange assay described by Hamm and colleagues and based on tryptophan fluorescence measurements (35). Whereas the PAM LY487379 largely increased GTPγS exchange, the NAM MNI-136 significantly decreased the basal rate measured in the presence of monomer ∆2∆ nanodiscs (Fig. 3A), consistent with a slight constitutive activity of ∆2∆, also observed in transfected cells (Fig. S5). When comparing the values at t = 0 and t = 40 min, we routinely observed a three- to fourfold increase of the basal values after PAM treatment, with a basal exchange rate (k value) of 0.05 ± 0.01 min−1 and a PAM-treated k value of 0.15 ± 0.02 min−1. These values are in agreement with those reported for basal and rhodopsin-catalyzed GTPγS exchange in Gαi (35, 36). Moreover, the PAM and NAM potencies (EC50 and IC50, respectively) were 115 ± 19 nM and 430 ± 60 nM, respectively (Fig. 3B), consistent with data obtained in living cells (29). No such PAM- or NAM-induced GTPγS exchange was observed with empty nanodiscs or nanodiscs containing a mutant ∆2∆ impaired in its ability to activate G protein (Phe756Pro) (37) (Fig. 3C), even though this mutant perfectly binds the PAM (SI Results and Fig. S5B).
Fig. 3.
Gi coupling properties of ∆2∆ reconstituted into nanodiscs. (A) Kinetics of GTPγS binding to Gi in Δ2Δ nanodiscs treated with 10 μM LY487379 (PAM) or 10 μM MNI-136 (NAM). Data were analyzed as described in SI Materials and Methods. Data points represent the mean ± SEM of triplicate determinations from a representative experiment out of three. (B) Dose–response of LY487379 and MNI-136 on Δ2Δ nanodiscs. Values correspond to the fractional GTPγS binding given by (Δ − Δref)/(Δmax − Δref), where Δref is the It40 − It0 value in the absence of ligand for the PAM curve or in the presence of 10 μM PAM for the NAM curve. (C) GTPγS binding (It40 − It0 value) measured with empty nanodiscs, nanodiscs ∆2∆ WT, or ∆2∆ F756P in the presence of 10 μM PAM. Data points in B and C represent the mean ± SEM of three independent experiments.
We then examined the possibility that the observed activity of the monomeric preparation could be due to a small amount of nanodiscs containing Δ2Δ dimer. If such a possibility was true, the nanodisc preparation containing two Δ2Δ species should give a signal higher than the monomeric preparation. This is clearly not the case (SI Results and Fig. S6), because a similar response was obtained with both preparations. This finding was not due to a saturation of the assay, because larger signals could be obtained with nanodiscs containing the BLT1 receptor (SI Results and Fig. S6). Taken together, these data demonstrate that a single 7TM of mGlu receptors can activate G proteins in a monomeric form.
Coupling Properties of Full-Length mGlu2 Monomers and Dimers.
The data obtained with the truncated 7TM domains raised the question of whether mGluR dimerization is indeed necessary for G-protein coupling. To purify both mGlu2 monomers and dimers, we produced full-length mGlu2 receptors carrying the Cys121Ala mutation to prevent covalent linkage between the two subunits. This protein was produced in Sf9 cells as for the truncated receptors and was solubilized in 0.2% Cholate, 0.5% DDM, and 0.03% CHS, in the presence of 500 mM NaCl. We found this condition to result in higher yield, as assessed by immunoblotting (Fig. S7A). The receptor was purified by immuno-chromatography using M1 FLAG antibody, as for the truncated receptors, yielding 1.5 mg of protein per liter of Sf9 cell culture. Protein purity was checked by SDS/PAGE (Fig. S7B) revealing immunoreactive species at the expected molecular weight, as well as after Coomassie blue staining (Fig. S7C).
The purified full-length mGlu2-C121A receptor was reconstituted into nanodiscs using a shorter version of MSP (MSP1D1), because it was expected to favor the isolation of nanodiscs containing a monomer and because of the better stability of such complexes (33). The reconstitution was essentially as described for the truncated receptors, but using an optimized ratio of POPC:MSP1D1:mGlu2-C121A of 60:1:0.1. Under these conditions, two main populations of nanodiscs could be separated by size-exclusion chromatography (Fig. 4A) and analyzed by BN-PAGE (Fig. 4B), corresponding to nanodiscs containing either one or two mGlu2 proteins per nanodisc (160 versus 260 kDa, respectively).
Fig. 4.
Formation and biochemical characterization of mGlu2-C121A nanodisc preparations. (A) Size-exclusion chromatography profile of mGlu2-C121A nanodisc preparations. (B) BN-PAGE analyses of peaks 1 and 2 from A with 5 μg of total proteins. (C) Total and nonspecific (NS) binding of [3H]-LY341495 on monomeric full-length mGlu2-C121A reconstituted into nanodiscs (10 nM). l-glutamate (1 mM) was used to define nonspecific binding. A representative curve (from a total of three) is shown (each point corresponds to the mean value of triplicates). (D) Displacement with glutamate of [3H]-LY341495 binding to nanodiscs containing monomeric or dimeric full-length mGlu2-C121A.
To characterize the binding properties of mGlu2-C121A monomeric and dimeric nanodiscs, we took advantage of the availability of a radioligand [3H]-LY341495. This ligand is an antagonist that binds specifically the VFTs of mGlu2 (38). Both monomeric and dimeric nanodiscs bound the radioligand with an affinity (Kd = 15.7 nM and 17.3 nM for monomers and dimers, respectively) close to that measured on the recombinant receptor expressed in cell lines (38) (Fig. 4C and Fig. S8). Displacement with glutamate revealed a Ki of 38.6 μM and 56.2 μM for monomer and dimer preparations, respectively (Fig. 4D). These values are three- and fourfold higher than those reported on mGlu2 expressed in RGT cells (38). This small difference may be explained by the fact that purified mGluRs are reconstituted in a different lipidic environment than that of mammalian cells or may result from the difference in the glycosylation status between proteins expressed in Sf9 and mammalian cells.
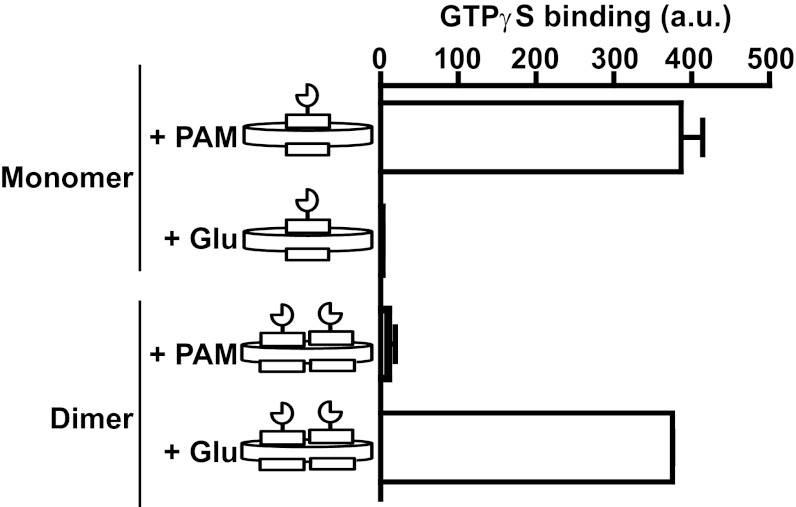
The G-protein-coupling properties of both fractions of nanodiscs containing either one or two mGlu2-C121A proteins were examined, as described above with the truncated receptors. Glutamate was unable to stimulate GTPγS exchange when the monomer fraction was used, even though a large response could be measured when the PAM LY487379 was applied (Fig. 5). In contrast, when the fraction enriched in the dimeric form was used, glutamate was fully active, and no significant response could be detected with LY487379. The absence of glutamate-induced response in the monomer fraction supports the presence of a very low proportion of nanodiscs containing two mGlu2 receptors per disk. Additionally, the absence of PAM agonist activity in the dimer fraction is consistent with a very low contamination with nanodiscs containing a single mGlu2 protomer. These data provide clear evidence that the dimeric form of mGlu2 is required (i) for agonist activation of the receptor and (ii) to prevent PAMs from activating the receptor in the absence of agonist.
Fig. 5.
Comparison of Gi-coupling properties of mGlu2-C121A nanodiscs. GTPγS binding (It40 − It0 value) induced by 10 μM LY487379 (PAM) or 1 mM l-glutamate (Glu) on nanodiscs containing monomeric or dimeric full-length mGlu2-C121A. Data points represent the mean ± SEM of three independent experiments.
To clarify whether the reconstitution of full-length mGlu2-C121A with MSP1D1 may affect Gi coupling properties, we reconstituted the full-length dimer with either MSP1D1 or the larger MSP1E3D1. As illustrated in Fig. S9, we get the same result with both MSPs, indicating that the receptor coupling properties observed with MSP1D1-containing discs do not result from specific constraints resulting from a smaller size of the discs.
Discussion
Although class C GPCRs, and especially mGluRs, are recognized as constitutive dimers, the functional importance of dimer formation remains unclear. Our present data show that the dimeric organization of the 7TM is not required for G-protein coupling per se, but that the dimer is required for agonist activation and for limiting the agonist activity of positive allosteric modulators.
The large extracellular domain of mGluRs is known to play an important role in their dimeric organization because their VFTs assemble as stable dimers through a large hydrophobic dimerization interface and an intersubunit disulfide bond (5, 9). Whether the 7TM domain could also spontaneously form dimers, and whether such a dimeric assembly is required for G-protein activation, was not known. Our data demonstrate that the 7TMs do not form high-affinity stable dimers, as revealed by the very low FRET efficacy measured at low receptor density, in contrast to the full-length receptor, demonstrating the importance of the ECD in the stabilization of mGlu dimers. They also show that a 7TM monomer can activate G proteins. Indeed, nanodiscs containing the purified monomeric 7TM domain of mGlu2 receptors (Δ2Δ) are able to activate the Gi proteins upon binding of a PAM, with kinetics and efficacy compatible with what was previously observed with class A GPCRs (35). Interestingly, the reconstituted receptor displays a slight constitutive activity that can be reversed by a NAM, as observed with 7TM domain of mGlu2 or mGlu5 expressed in cellular systems (Fig. S5) (23), suggesting that the isolated monomer recapitulates some of the features of the signaling repertoire observed in living cells.
Such a finding is consistent with the observation that, within the dimeric class C GPCRs, a single 7TM is directly responsible for G-protein activation. This proposal is best illustrated in the heterodimeric GABAB and taste receptors (39, 40), as well as in the homodimeric mGlu receptors where an asymmetrical activation of the dimeric 7TM is responsible for G-protein activation (17–19). This finding is also consistent with the asymmetric functioning reported for many class A GPCR dimers (41–43). However, these data did not exclude the involvement of the second 7TM associated with the one coupled to the G protein. Indeed, for the GABAB receptor, agonist binding to GABAB1 increases G-protein coupling efficacy of the GABAB2 subunit, and this partly involves the GABAB1 7TMs (39, 44, 45). Although a similar coupling efficacy could be measured with nanodiscs containing one or two 7TMs, more work will be necessary before such a comparison can be done. Indeed, it is not known whether a dimeric organization similar to that found in the full-length receptor dimer can be achieved in the nanodiscs containing two 7TM domains. Moreover, the proportion of nanodiscs in which both 7TMs are similarly oriented in the bilayer is unknown. Solving these issues will be necessary to clarify the roles of each 7TM in the signaling properties of mGluRs.
Our data revealed that a monomeric full-length mGlu2 protein can bind glutamate and can couple to G protein upon activation by a PAM but cannot be activated by glutamate. This finding demonstrates that dimerization is required for the signaling induced by agonist binding to be transferred from the extracellular domain to the 7TM domain. In addition, our data show that the dimeric organization of the receptor is also required to prevent PAMs from acting as agonists, because such PAMs act as full agonists not only on truncated mGluRs deleted of their extracellular domain, but also on a full-length mGlu monomer. Accordingly, it is not the extracellular domain per se that prevents agonist action of the PAM, but rather the dimeric arrangement of either the VFTs or the 7TMs.
How do the dimeric VFTs control the activity of the 7TM? It has been shown that a relative movement at the level of the VFTs, the CRDs (9, 20, 46), and the 7TMs of the two subunits (19, 21, 22) occurs during activation of class C GPCRs. More recently, we documented that the change in conformation of one of the 7TM domains within the dimer occurs after the relative movement between the protomers (19), consistent with a concerted activation mechanism that results from dimer rearrangement. One can then propose that the relative position of the 7TM domains is critical in controlling their activity. As such, our data are consistent with a model in which the specific orientation of the two VFTs tightly controls the association mode (relative position) of the two 7TM domains, leading to the activation of at least one of these (18, 19). However, we cannot exclude at the present time a direct control of the 7TM conformation by the dimer of ECDs when in the active orientation.
In conclusion, we found that a 7TM monomer of an obligatory dimeric receptor can signal through G proteins illustrating that it is not because a monomer can activate G proteins that dimers/oligomers cannot constitute the central element of the signaling repertoire of GPCRs. In such dimeric complexes, even if a single 7TM is responsible for G-protein activation, the associated 7TMs can be involved in other functions, which may include coupling efficacy (39, 44), controlling G-protein-independent signaling pathways (47), or recruiting any number of intracellular associated proteins to be part of the larger signaling complexes (48, 49). Although dimers are likely important for GPCR biology (50, 51), our study also indicates that understanding and studying structural changes that occur in the context of individual 7TM monomers could be relevant and useful for the development of class C allosteric modulators as new therapeutic tools.
Materials and Methods
Fluorescence, Time-Resolved FRET, and Myo-Inositol 1 Phosphate Measurement in Living Cells.
Reagents, plasmids, transfection, and labeling protocols are described in SI Materials and Methods. Fluorescence and FRET measurements are described in SI Materials and Methods. Measurements of IP accumulation were performed using the IP-One HTRF Assay (Cisbio Bioassays) as described in SI Materials and Methods.
Expression and Purification of mGluRs.
Construction and characterization of the truncated mGlu and mGlu2-C121A receptors are described in SI Materials and Methods. Sf9 insect cells were grown at 28 °C in suspension cultures in Insect-Xpress medium (Lonza) or Ex-Cell 420 (Sigma) medium. Recombinant baculoviruses were generated in Sf9 cells using the Bac-to-Bac Baculovirus Expression System (Invitrogen). For receptor purification, Sf9 cell cultures were infected at a density of ∼3 × 106 cells per mL with appropriate viruses and harvested after 60 h by centrifugation (10 min at 5,000 × g). The cell pellets were kept at −80 °C until used for purification. Membrane preparations and binding experiments protocols are given in SI Materials and Methods. Receptor purification was performed on a M1 Flag antibody affinity resin (Sigma) as described (31, 52).
Preparation of Nanodiscs.
The membrane scaffold protein MSP1E3D1 and MSP1D1 were purified as described (53). The appropriate amount of MSP1E3D1 or MSP1D1 and 1-palmitoyl-2-oleoyl- phosphatidylcholine (POPC) (Avanti Polar Lipids) were incubated in the absence (empty discs) or with purified receptors for 1 h at 4 °C. The self-assembly process was initiated by detergent removal using 0.5 g of BioBeads SM-2 (Biorad) for 1 mL of the mixture and allowed to process overnight at 4 °C. The Biobeads were then removed by centrifugation and the recovered supernatant was concentrated to 500 μL and centrifuged 5 min 20,000 × g before being loaded on a homemade column (10 mm in diameter and 400 mm in length) prepared with Superdex S200 resin (GE Healthcare). The size-exclusion chromatography was performed at room temperature in a buffer containing 20 mM Hepes (pH 7.5) and 50 mM NaCl. The peaks of interest were pooled and analyzed by BN-PAGE electrophoresis as described by the group of Schägger (54) and by negative stain transmission electron microscopy (as described in SI Materials and Methods). Nanodisc concentration was estimated by UV absorbance considering a molar absorption coefficient ɛ of 30,000 M−1cm−1 for MSP1E3D1, 21,400 M−1cm−1 for MSP1D1, 36,000 M−1cm−1 for Δ2Δ, and 100,000 M−1cm−1 for mGlu2-C121A.
In Vitro Gi Activation Assays,
The nucleotide-exchange assay using the purified Gαi2 subunit was carried out as described by Hamm and colleagues (35). Gαi2 was prepared as described (36). For measuring activation of the G protein, the basal rate of GTPγS binding was determined by monitoring every minute the relative increase in the intrinsic fluorescence (λexc = 300 nm, λem = 345 nm) of Gαi2 (500 nM of purified Gαi2) in the presence of purified Gβ1γ2 subunits (500 nM) and of nanodiscs (100 nM) in buffer containing 20 mM Hepes (pH 7.5), 130 mM NaCl, and 2 mM MgCl2 for 40 min at 15 °C after the addition of 10 μM GTPγS. The receptor-catalyzed rate was measured under the same conditions in the presence of 10 μM of the PAM LY487379. The β1γ2 subunits of the G protein were prepared as described (42). For comparing the different experimental conditions and for the dose–response experiments we only measured the fluorescence at t = 0 and t = 40 min.
Supplementary Material
Acknowledgments
We thank Juan Jose Fung, Aashish Manglik (Stanford University), Cathy Royer (Centre de Biologie Structurale, Montpellier), and Laëtitia Comps-Agrar (Institut de Génomique Fonctionnelle, University of Montpellier) for their critical readings of the manuscript; Gilles Tamagnan (Institute for Neurodegenerative Disorders, New Haven) for providing us with the mGlu2-negative allosteric modulator; and Thierry Durroux (Institut de Génomique Fonctionnelle, University of Montpellier) for constructive discussions. This work was supported by the CNRS, INSERM, “Agence Nationale pour la Recherche” Grant ANR-09-PIRI-0011, and Fondation Bettencourt Schueller.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205838109/-/DCSupplemental.
References
- 1.Nicoletti F, et al. Metabotropic glutamate receptors: from the workbench to the bedside. Neuropharmacology. 2011;60:1017–1041. doi: 10.1016/j.neuropharm.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niswender CM, Conn PJ. Metabotropic glutamate receptors: Physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 2010;50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swanson CJ, et al. Metabotropic glutamate receptors as novel targets for anxiety and stress disorders. Nat Rev Drug Discov. 2005;4:131–144. doi: 10.1038/nrd1630. [DOI] [PubMed] [Google Scholar]
- 4.Bräuner-Osborne H, Wellendorph P, Jensen AA. Structure, pharmacology and therapeutic prospects of family C G-protein coupled receptors. Curr Drug Targets. 2007;8:169–184. doi: 10.2174/138945007779315614. [DOI] [PubMed] [Google Scholar]
- 5.Romano C, Yang WL, O’Malley KL. Metabotropic glutamate receptor 5 is a disulfide-linked dimer. J Biol Chem. 1996;271:28612–28616. doi: 10.1074/jbc.271.45.28612. [DOI] [PubMed] [Google Scholar]
- 6.Doumazane E, et al. A new approach to analyze cell surface protein complexes reveals specific heterodimeric metabotropic glutamate receptors. FASEB J. 2011;25:66–77. doi: 10.1096/fj.10-163147. [DOI] [PubMed] [Google Scholar]
- 7.Kniazeff J, Prézeau L, Rondard P, Pin J-P, Goudet C. Dimers and beyond: The functional puzzles of class C GPCRs. Pharmacol Ther. 2011;130:9–25. doi: 10.1016/j.pharmthera.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Rondard P, Goudet C, Kniazeff J, Pin J-P, Prézeau L. The complexity of their activation mechanism opens new possibilities for the modulation of mGlu and GABAB class C G protein-coupled receptors. Neuropharmacology. 2011;60:82–92. doi: 10.1016/j.neuropharm.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Kunishima N, et al. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 10.Urwyler S. Allosteric modulation of family C G-protein-coupled receptors: From molecular insights to therapeutic perspectives. Pharmacol Rev. 2011;63:59–126. doi: 10.1124/pr.109.002501. [DOI] [PubMed] [Google Scholar]
- 11.Bessis A-S, et al. Closure of the Venus flytrap module of mGlu8 receptor and the activation process: Insights from mutations converting antagonists into agonists. Proc Natl Acad Sci USA. 2002;99:11097–11102. doi: 10.1073/pnas.162138699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kniazeff J, et al. Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nat Struct Mol Biol. 2004;11:706–713. doi: 10.1038/nsmb794. [DOI] [PubMed] [Google Scholar]
- 13.Kniazeff J, et al. Locking the dimeric GABA(B) G-protein-coupled receptor in its active state. J Neurosci. 2004;24:370–377. doi: 10.1523/JNEUROSCI.3141-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muto T, Tsuchiya D, Morikawa K, Jingami H. Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc Natl Acad Sci USA. 2007;104:3759–3764. doi: 10.1073/pnas.0611577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuchiya D, Kunishima N, Kamiya N, Jingami H, Morikawa K. Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+ Proc Natl Acad Sci USA. 2002;99:2660–2665. doi: 10.1073/pnas.052708599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binet V, et al. Common structural requirements for heptahelical domain function in class A and class C G protein-coupled receptors. J Biol Chem. 2007;282:12154–12163. doi: 10.1074/jbc.M611071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goudet C, et al. Asymmetric functioning of dimeric metabotropic glutamate receptors disclosed by positive allosteric modulators. J Biol Chem. 2005;280:24380–24385. doi: 10.1074/jbc.M502642200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hlavackova V, et al. Evidence for a single heptahelical domain being turned on upon activation of a dimeric GPCR. EMBO J. 2005;24:499–509. doi: 10.1038/sj.emboj.7600557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hlavackova V, et al. Sequential inter- and intra-subunit rearrangements during asymmetric activation of dimeric metabotropic glutamate receptor 1. Sci Signal. 2012 doi: 10.1126/scisignal.2002720. 10.1126/scisignal.2002720. [DOI] [PubMed] [Google Scholar]
- 20.Rondard P, et al. Functioning of the dimeric GABA(B) receptor extracellular domain revealed by glycan wedge scanning. EMBO J. 2008;27:1321–1332. doi: 10.1038/emboj.2008.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcaggi P, Mutoh H, Dimitrov D, Beato M, Knöpfel T. Optical measurement of mGluR1 conformational changes reveals fast activation, slow deactivation, and sensitization. Proc Natl Acad Sci USA. 2009;106:11388–11393. doi: 10.1073/pnas.0901290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tateyama M, Abe H, Nakata H, Saito O, Kubo Y. Ligand-induced rearrangement of the dimeric metabotropic glutamate receptor 1alpha. Nat Struct Mol Biol. 2004;11:637–642. doi: 10.1038/nsmb770. [DOI] [PubMed] [Google Scholar]
- 23.Goudet C, et al. Heptahelical domain of metabotropic glutamate receptor 5 behaves like rhodopsin-like receptors. Proc Natl Acad Sci USA. 2004;101:378–383. doi: 10.1073/pnas.0304699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maurel D, et al. Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: Application to GPCR oligomerization. Nat Methods. 2008;5:561–567. doi: 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson MP, et al. Discovery of allosteric potentiators for the metabotropic glutamate 2 receptor: Synthesis and subtype selectivity of N-(4-(2-methoxyphenoxy)phenyl)-N-(2,2,2- trifluoroethylsulfonyl)pyrid-3-ylmethylamine. J Med Chem. 2003;46:3189–3192. doi: 10.1021/jm034015u. [DOI] [PubMed] [Google Scholar]
- 26.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 27.Whorton MR, et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci USA. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whorton MR, et al. Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer. J Biol Chem. 2008;283:4387–4394. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rondard P, et al. Coupling of agonist binding to effector domain activation in metabotropic glutamate-like receptors. J Biol Chem. 2006;281:24653–24661. doi: 10.1074/jbc.M602277200. [DOI] [PubMed] [Google Scholar]
- 30.Kobilka BK. Amino and carboxyl terminal modifications to facilitate the production and purification of a G protein-coupled receptor. Anal Biochem. 1995;231:269–271. doi: 10.1006/abio.1995.1533. [DOI] [PubMed] [Google Scholar]
- 31.Granier S, et al. Structure and conformational changes in the C-terminal domain of the beta2-adrenoceptor: insights from fluorescence resonance energy transfer studies. J Biol Chem. 2007;282:13895–13905. doi: 10.1074/jbc.M611904200. [DOI] [PubMed] [Google Scholar]
- 32.Malherbe P, et al. Mutational analysis and molecular modeling of the binding pocket of the metabotropic glutamate 5 receptor negative modulator 2-methyl-6-(phenylethynyl)-pyridine. Mol Pharmacol. 2003;64:823–832. doi: 10.1124/mol.64.4.823. [DOI] [PubMed] [Google Scholar]
- 33.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 34.Hemstapat K, et al. A novel family of potent negative allosteric modulators of group II metabotropic glutamate receptors. J Pharmacol Exp Ther. 2007;322:254–264. doi: 10.1124/jpet.106.117093. [DOI] [PubMed] [Google Scholar]
- 35.Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nat Struct Mol Biol. 2006;13:772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- 36.Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Mapping allosteric connections from the receptor to the nucleotide-binding pocket of heterotrimeric G proteins. Proc Natl Acad Sci USA. 2007;104:7927–7932. doi: 10.1073/pnas.0702623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Francesconi A, Duvoisin RM. Role of the second and third intracellular loops of metabotropic glutamate receptors in mediating dual signal transduction activation. J Biol Chem. 1998;273:5615–5624. doi: 10.1074/jbc.273.10.5615. [DOI] [PubMed] [Google Scholar]
- 38.Johnson BG, et al. [3H]-LY341495 as a novel antagonist radioligand for group II metabotropic glutamate (mGlu) receptors: Characterization of binding to membranes of mGlu receptor subtype expressing cells. Neuropharmacology. 1999;38:1519–1529. doi: 10.1016/s0028-3908(99)00053-2. [DOI] [PubMed] [Google Scholar]
- 39.Galvez T, et al. Allosteric interactions between GB1 and GB2 subunits are required for optimal GABA(B) receptor function. EMBO J. 2001;20:2152–2159. doi: 10.1093/emboj/20.9.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu H, et al. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc Natl Acad Sci USA. 2004;101:14258–14263. doi: 10.1073/pnas.0404384101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Albizu L, et al. Time-resolved FRET between ligands bound on asymmetric G protein-coupled receptor reveals oligomers in native tissues. Nat Chem Biol. 2010;6:587–594. doi: 10.1038/nchembio.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Damian M, Martin A, Mesnier D, Pin JP, Banères JL. Asymmetric conformational changes in a GPCR dimer controlled by G-proteins. EMBO J. 2006;25:5693–5702. doi: 10.1038/sj.emboj.7601449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han Y, Moreira IS, Urizar E, Weinstein H, Javitch JA. Allosteric communication between protomers of dopamine class A GPCR dimers modulates activation. Nat Chem Biol. 2009;5:688–695. doi: 10.1038/nchembio.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monnier C, et al. Trans-activation between 7TM domains: implication in heterodimeric GABAB receptor activation. EMBO J. 2011;30:32–42. doi: 10.1038/emboj.2010.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duthey B, et al. A single subunit (GB2) is required for G-protein activation by the heterodimeric GABA(B) receptor. J Biol Chem. 2002;277:3236–3241. doi: 10.1074/jbc.M108900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang S, et al. Interdomain movements in metabotropic glutamate receptor activation. Proc Natl Acad Sci USA. 2011;108:15480–15485. doi: 10.1073/pnas.1107775108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 48.Bockaert J, Perroy J, Bécamel C, Marin P, Fagni L. GPCR interacting proteins (GIPs) in the nervous system: Roles in physiology and pathologies. Annu Rev Pharmacol Toxicol. 2010;50:89–109. doi: 10.1146/annurev.pharmtox.010909.105705. [DOI] [PubMed] [Google Scholar]
- 49.Maurice P, et al. Molecular organization and dynamics of the melatonin MT₁ receptor/RGS20/G(i) protein complex reveal asymmetry of receptor dimers for RGS and G(i) coupling. EMBO J. 2010;29:3646–3659. doi: 10.1038/emboj.2010.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bouvier M. Oligomerization of G-protein-coupled transmitter receptors. Nat Rev Neurosci. 2001;2:274–286. doi: 10.1038/35067575. [DOI] [PubMed] [Google Scholar]
- 51.Lohse MJ. Dimerization in GPCR mobility and signaling. Curr Opin Pharmacol. 2010;10:53–58. doi: 10.1016/j.coph.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 52.Granier S, Kim S, Fung JJ, Bokoch MP, Parnot C. FRET-based measurement of GPCR conformational changes. Methods Mol Biol. 2009;552:253–268. doi: 10.1007/978-1-60327-317-6_18. [DOI] [PubMed] [Google Scholar]
- 53.Bayburt TH, Grinkova YV, Sligar SG. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002;2:853–856. [Google Scholar]
- 54.Wittig I, Braun HP, Schägger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.