Abstract
Inflammatory Bowel Disease (IBD) is a chronic inflammatory disorder of the gastrointestinal tract for which there are few safe and effective therapeutic options for long-term treatment and disease maintenance. In this study, we applied a computational approach to discover novel drug therapies for IBD in silico using publicly available molecular data measuring gene expression in IBD samples and 164 small-molecule drug compounds. Among the top compounds predicted to be therapeutic for IBD by our approach were prednisolone, a corticosteroid known to treat IBD, and topiramate, an anticonvulsant drug not previously described to demonstrate efficacy for IBD or any related disorders of inflammation or the gastrointestinal tract. We experimentally validated our topiramate prediction in vivo using a trinitrobenzenesulfonic acid (TNBS) induced rodent model of IBD. The experimental results demonstrate that oral administration of topiramate is able to significantly reduce gross pathological signs and microscopic damage in primary affected colon tissue in a TNBS-induced rodent model of IBD. These finding suggest that topiramate might serve as a novel therapeutic option for IBD in humans, and support the use of public molecular data and computational approaches to discover novel therapeutic options for IBD.
INTRODUCTION
Inflammatory Bowel Disease (IBD), of which Crohn’s disease (CD) and Ulcerative colitis (UC) are the most prominent clinically defined manifestations, represents a group of chronic, progressive inflammatory disorders of the intestinal tract that affects more than one million individuals in North America alone(1). There is currently no known cure for IBD, and available treatment options are aimed towards controlling symptoms, promoting remission, and preventing relapse. Current treatment protocols for IBD incorporate chronic administration of corticosteroids and systemic anti-inflammatory drugs to reduce or inhibit primary inflammation, along with antibiotics treat secondary infection(2). Chronic use of corticosteroids or systemic anti-inflammatory drugs (e.g., 6-mercaptopurine) typically used to treat IBD is associated with severe side effects, and more targeted therapies such as anti-TNFα drugs incur high costs and elicit a therapeutic response in only a subset of affected patients(3). Surgical removal of affected regions of the small or large intestine is also employed as a treatment strategy, however this option is expensive and highly invasive, and the disease can often re-manifest in previously unaffected locations along the intestinal tract(2).
In this study we aimed to discover and validate novel therapeutic options for IBD using a systematic computational approach for drug repositioning that is based on integration of public gene expression signatures of drugs and diseases (Sirota M and Dudley JT et al. co-appearing in this issue). We systematically evaluated gene expression signatures of IBD derived from public microarray data against a compendium of gene expression signatures comprising 164 drug compounds to infer novel therapeutic relationships between drug-disease pairs represented in the data sets. Among the top-ranked results rendered by our approach was the corticosteroid prednisolone, a known treatment for IBD. More surprisingly, we found that the approved anti-epileptic drug topiramate, which has not previously been described to have a therapeutic association with IBD or any other intestinal disorders, was ranked higher by our method (i.e., higher predicted efficacy for IBD) than prednisolone. We evaluated the efficacy of topiramate for IBD in three independent experiments using a TNBS-induced rodent model of colitis. Concordant with our computational prediction, the experimental results demonstrate that topiramate is able to significantly ameliorate an IBD phenotype compared to untreated groups, and even exhibited greater efficacy than prednisolone, which was used as a positive control.
RESULTS
Prediction of known and novel drugs for IBD
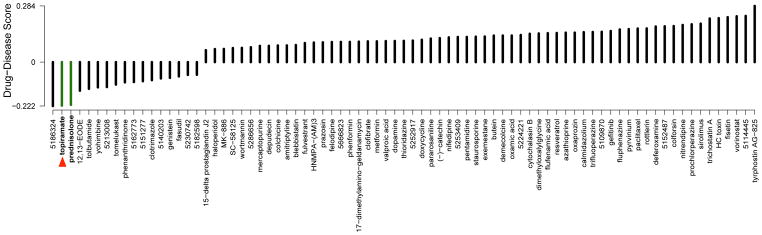
To discover novel therapeutic relationships for IBD, we compared the gene expression profiles from a compendium of 164 drug compounds(4) to a gene expression signature of IBD derived from publicly available experiments obtained from NCBI Gene Expression Omnibus (GEO) (5). We derived therapeutic predictions for drug-disease pairs based on the hypothesis that if a drug has a gene expression signature that is opposite of a disease signature, that drug could potentially be used as a treatment for that disease. Our method produces a negative score when the drug signature is oppositional to the disease signature, and a positive score when they are concordant (see Methods). Among the strongest therapeutic predictions for Crohn’s disease is the corticosteroid prednisolone (score=−0.216), a well-known treatment for these conditions (6) (Fig 1). Interestingly, we observed that topiramate, an anticonvulsant drug currently used to treat epilepsy, was determined to have a stronger therapeutic score for Crohn’s (−0.220) (Fig. 1; red arrow) than the established therapeutic prednisolone. Topiramate is also one of the strongest predicted therapies for ulcerative colitis based on our analysis, with a score of −0.219 (data not shown). Although another compound, 5186324, was scored higher than topiramate by our method, we chose to focus on topiramate for experimental validation because it is an FDA-approved compound known to be generally safe in humans and is readily available for clinical use.
Figure 1.
Significant drug-disease scores for Crohn’s disease. The names of the drugs are placed along the bottom axis, and the vertical bars above the drug name indicate the computationally predicted therapeutic score for the drug based on comparison of the gene expression signature of the drug with the gene expression signature of Crohn’s disease. A positive score indicates that the drug exhibits an expression pattern that is synergistic with the disease, while a negative score indicates that the drug exhibits an expression pattern that is oppositional to the disease. Drugs are sorted from left-to-right starting with those predicted to be most efficacious for the disease. Green bars indicate drugs that are discussed in the text. The red triangle points towards the anticonvulsant drug topiramate, which was selected for experimental validation.
Experimental validation of topiramate as an indication for IBD
To determine whether our in silico drug indication predictions would translate into therapeutic efficacy in vivo, we tested whether topiramate would show efficacy for IBD using a trinitrobenzenesulfonic acid (TNBS) induced rat model of IBD. We performed an initial pilot validation experiment followed by two independent replication experiments using male Sprague-Dawley rats administered TNBS (5% w/v total of 100 mg/kg) intrarectally to induce colitis. The initial experiment consisted of 18 rats that were divided into three groups of equal size (n=6 per group). One group was not induced with TNBS and treated with vehicle, another group was induced with TNBS and treated with vehicle (TNBS+vehicle), and the final group was induced with TNBS and treated with topiramate (80 mg/kg/day) by oral gavage (TNBS+topiramate). The independent replicate experiments used 60 total animals each (n=12 animals per group) and added a fourth group that was induced with TNBS and treated with prednisolone (3 mg/kg/day) as a positive control (TNBS+prednisolone). After the initial TNBS induction, animals were treated for seven consecutive days, and the induced IBD phenotype was assessed in in vivo by video endoscopy at days 3 and 7. Rats were euthanized at the end of treatment and whole colons were excised for pathological assessment between groups.
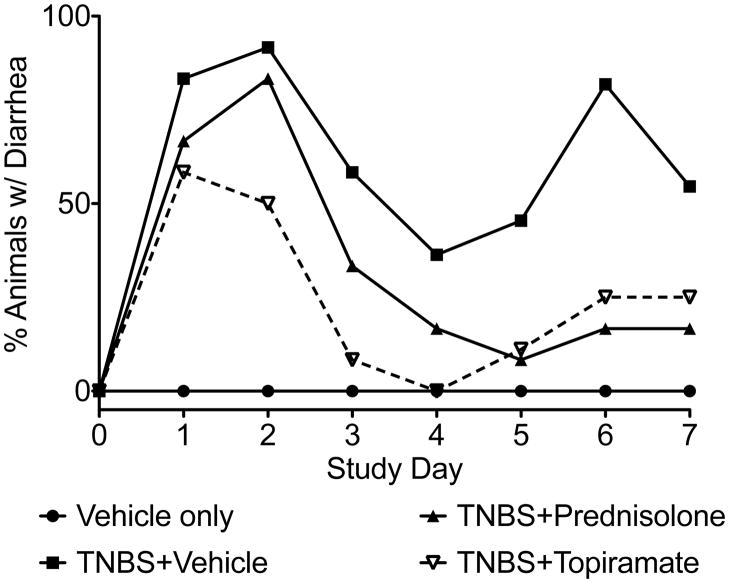
Disease severity was assessed over the course of treatment by observation of clinical signs and by gross inspection of affected colon tissues after treatment termination. Animals treated with both prednisolone (TNBS+prednisolone) and topiramate (TNBS+topiramate) exhibited reduced incidence of diarrhea over the course of treatment compared to affected animals administered vehicle alone (TNBS+vehicle) (Fig. 2). Assessment of colitis by visual inspection of endoscopy video captured on day 7 of treatment demonstrated significantly reduced gross pathological inflammation and ulceration in the topiramate and predinisolone treated groups relative to the untreated (TNBS+vehicle) group (Fi3. 4A; Supplementary video S1–4). Quantitative assessment of the gross pathological characteristics revealed that animals in the topiramate-treated group (TNBS+topiramate) exhibited significantly reduced swelling, ulceration and other gross pathological characteristics compared to animals in the untreated (TNBS+vehicle) group (P< 0.0001; Mann-Whitney U test; n=12 per group) (Fig. 3B).
Figure 2.
Gross clinical evaluation of IBD severity between treatment groups by percent of animals with diarrhea within each group. Both the topiramate treated group and the prednisolone treated group show reduced proportions of animals with diarrhea relative to the disease-induced group receiving only vehicle. (n=12 animals per group)
Figure 3.
Pathological assessment of IBD severity between treatment groups. A) Clinical endoscopy captured from live animals on day 7 of the study. B) Gross pathology score. C) Pictomicrographs of H&E stained colon tissues showing microscopic damage to the mucosal and epithelial layers of the colon wall between treatment groups. D) Macroscopic damage score assessed from light microscopy of fixed colon tissues. Data graphs represent the mean and SEM estimated from three independent experiments. (n=12 rats per group) (* = P<0.05, ****=P<0.00005; Mann Whitney U test).
Microscopic damage was assessed by histopathology analysis of fixed colon tissues sections. Visual inspection of fixed colon tissues revealed extensive destruction of the colon mucosal layer in the untreated colitis-induced group (TNBS+vehicle), which was substantially ameliorated in colitis-induced animals receiving topiramate (TNBS+topiramate) (Fig. 3C). Quantitative evaluation of microscopic damage between groups demonstrated significantly reduced macroscopic damage in animals treated with topiramate compared to animals in the untreated (TNBS+vehicle) group (P< 0.05; Mann-Whitney U test; n=12 per group) (Fig. 3D). Taken together, these data provide evidence that topiramate exhibits efficacy against IBD in the TNBS model, as predicted by the computational method.
Expression signature evaluation
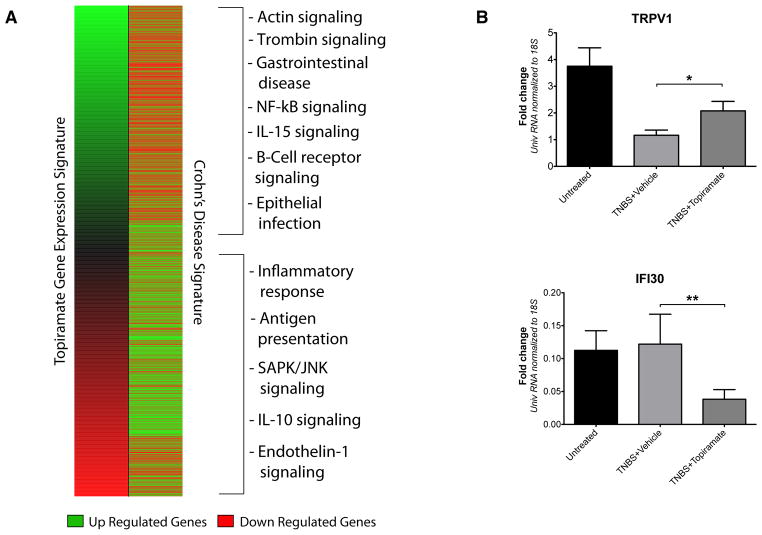
The predicted efficacious relationship between IBD and topiramate was inferred from public data suggesting that particular sets of genes would exhibit oppositional expression between drug and disease. Comparative visual inspection of the Crohn’s and topiramate expression signatures revealed the expected antithetical expression patterns between the drug and disease, and functional enrichment analysis indicated that genes involved with gastrointestinal disease, inflammatory response, and other immune-related functions were divergently expressed between the drug-affected and disease-affected conditions (Fig. 2A)
We performed qPCR analysis spot checks of post-mortem colon tissues to evaluate if the expected expression patterns of genes reflected in the public gene expression data driving our prediction were observed in the animal validation study. We randomly selected eight genes for qPCR analysis from a set of genes exhibiting opposing expression patterns between drug and disease expression signatures, and for which commercial primers were available (see Methods). qPCR analysis revealed that two of these genes, TRPV1 and IFI30 were differentially expressed between treatment groups in the direction expected from comparison of the public molecular data (Fig. 4B). TRPV1 was significantly up-regulated in the topiramate treated group (TNBS+topiramate) relative to the untreated disease-induced group (TNBS+Vehicle) (P<0.05; Mann Whitney U test; n=12 per group), and that IFI30 was significantly down-regulated in the topiramate treated group relative to the untreated disease-affected group (TNBS+Vehicle) (P<0.005; Mann Whitney U test; n=12 per group). Both of these findings corroborate the expected oppositional relationships between these genes reflected in the expression signatures that were used to computationally predict an efficacious relationship between topiramate and IBD.
Figure 4.
Evaluation and comparison of gene expression signatures for the drug compound topiramate and Crohn’s disease. A) Comparison of the Crohn’s disease signature (right) to the topiramate gene expression profile (left) showing a generally anti-correlated pattern, with genes at the top- and bottom-most ends of the expression pattern showing generally oppositional expression patterns. Genes that are up regulated are shown in green. Genes that are down regulated are shown in red. The pathways and functional groups significantly enriched (enrichment P-value < 0.05) in the top- and bottom 25% of genes are shown on the right hand side. B) qPCR results showing that gene transcripts for TRPV1 and IFI30 have oppositional gene expression levels in the disease condition (TNBS+vehicle) relative to the drug treated condition (TNBS+topiramate) in colon tissues collected from the animal validation study, which corroborates their expected oppositional relationship from initial comparison of the topiramate and Crohn’s expression signatures. (n=12 rats per group) (*=P<0.05, **=P<0.005; Mann Whitney U test; Error bars=SEM)
DISCUSSION
Using an in silico approach based on the integration of publicly available gene expression data, we inferred that the anticonvulsant topiramate could serve as a novel therapeutic for IBD, and performed an experimental validation which demonstrated topiramate’s efficacy in ameliorating a TNBS-induced rodent model of IBD. The precise mechanism of action for topiramate is unknown, but it is known to enhance GABA-activated chloride channels, shows activity on kainate and AMPA receptors, and inhibits the activity of some carbonic anhydrase enzymes (7). Topiramate is administrated orally and is often used to treat seizures and migraines, and has some antidepressant activity. Although topiramate has not previously been suggested as a therapy for IBD, it has been investigated for off-label use in treating obesity and type II diabetes (8, 9), and was recently shown to have efficacy against multiple sclerosis (10). Although elucidation of the precise mechanism of action by which topiramate acts to ameliorate the induced IBD phenotype in our study requires follow-up investigations that exceed the scope of this pilot investigation, functional enrichment analysis of reveals that sets of genes related to NF-kB signaling, inflammatory response, antigen presentation, and other functional processes relevant to the pathophysiology of IBD are oppositionally expressed between the disease and drug expression profiles (Fig. 4).
Using a TNBS-induced rodent model of IBD, we demonstrated that induced animals treated with topiramate (TNBS+topiramate) showed improvements in both gross and microscopic measures of disease pathology relative to the relevant vehicle treated group (TNBS+vehicle), with endpoints exceeding even the prednisolone treated positive control group. Both topiramate and predinisolone reduced the severity of diharrhea over the course of treatment relative to the vehicle treated group (TNBS+vehicle) (Fig. 2), however only the topiramate treated group exhibited significantly reduced gross pathology and microscopic damage scores (Fig. 3B and 3D). Although prednisolone is an established treatment for IBD in humans and was correctly identified as such by our method, previous studies have reported limited efficacy of prednisolone in chemically induced rodent models of IBD (11). Prednisolone does prevent the complete loss of the mucosal layer observed in the induced vehicle treated animals (TNBS+vehicle); however, it’s likely that the potent immunosuppressive effects or prednisolone are such that it substantially slows healing from the initial chemical insult, and possibly renders damaged tissues more susceptible to secondary bacterial infections.
Although the experimental results from the rodent model of IBD corroborate our computational predictions derived from gene expression measurements of human disease, there are several caveats that could potentially limit the interpretation of the results. Foremost, we have only demonstrated the efficacy of topiramate for IBD in a TNBS-induced animal model, which we acknowledge may not be entirely representative of IBD pathology in humans. Secondly, the nature of the model is such that it is more representative an acute IBD flare-up, and therefore additional studies are needed to determine if topiramate could serve as an effective long-term therapeutic solution for chronic IBD in humans. We also note that only two of the eight genes chosen for PCR validation showed statistical significance between the induced untreated (TNBS+Vehicle) and topiramate treated groups (TNBS+topiramate), however this may be explained by species differences in gene expression between human intestinal tissue, which was used to select the genes, and rat intestinal tissue. Also, the experiment reflected an acute treatment scenario, and it’s possible that several of these genes may only reach distinguishing expression levels after a longer period of treatment. Finally, we note that our study evaluated only a single, albeit relatively conservative dose of topiramate, and therefore additional studies are needed to evaluate the possible optimal dose ranges for use of topirmate in treating both acute and chronic IBD in humans.
In this study, we demonstrate that computational approaches leveraging public gene expression microarray data can be used to infer novel drug therapies for IBD, and offer experimental evidence that the anticonvulsant topiramate is capable of ameliorating disease pathophysiology in a TNBS-induced rodent model of IBD. Because topiramate is already established as a safe and effective drug for treating neurological diseases in humans, and the side effect profile is generally more favorable than most drugs typically used to treat IBD(2, 12), these results support the need for additional clinical investigation into the use of topiramate for treating IBD in human subjects. Additionally, these findings support and motivate the need for future studies by which computational approaches for leveraging publicly available molecular data for drug repurposing are further developed and applied towards additional diseases contributing significantly to the morbidity and mortality of modern human populations.
METHODS
Computational prediction and assessment of novel IBD therapies
Computational prediction and assessment of novel IBD therapies was performed as described in a co-appearing paper. In brief, publicly available gene expression measurements of Crohn’s disease and Ulcerative colitis were obtained from NCBI GEO(5) using data from a previously published experiment measuring Crohn’s disease and Ulcertive colitis in human intestinal tissue obtained by biopsy (GDS2642). Array probes were mapped to NCBI Entrez gene identifiers using the AILUN system (13). In cases where multiple microarray probes mapped to the same NCBI gene identifier, we averaged across individual probe expression values and assigned the averaged value to the gene. We created a disease gene expression signature (i.e., disease signature) by deriving the set of differentially expressed genes between the disease affected and healthy control samples using the Significance Analysis of Microarrays (SAM) software (14).
We systematically compared to gene expression profiles of drugs obtained from the Connectivity Map(4) to derive a therapeutic score. Describe score. A randomization approach was used to determine the statistical significance, and drugs with predicted therapeutic scores below the significance threshold are ranked in reverse order according to their score. Drugs with significant negative scores have gene expression patterns that are anti-correlated, or oppositional to the disease gene expression pattern, and therefore represent putative novel therapeutic indications.
Functional analysis of IBD and Topiramate expression signatures
Functional analysis of the up- and down-regulated genes in the Crohn’s and topiramate signatures (Fig. 4) was performed through the use of IPA (Ingenuity® Systems, www.ingenuity.com). This functional analysis identified the biological functions and/or diseases that were most significant to the data set. Genes in the top and bottom 25% of rank change vs. normal in both drug affected and disease conditions and were associated with biological functions and/or diseases in the Ingenuity Knowledge Base were considered for the analysis. Right-tailed Fisher’s exact test was used to calculate a p-value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone.
Experimental evaluation of topiramate using a rodent TNBS model of IBD
Experimental validation of treatment efficacy was performed using a TNBS (trinitrobenzenesulfonic acid, also known as picryl sulfonic acid) induced rodent model of IBD(15, 16). Male Sprague Dawley rats weighing ~ 300–350 g were used in the study. Rats were fasted over night prior to initialization of the study. TNBS 100mg/kg of body weight (BW) diluted in 50% ethyl alcohol was administered via a polyethylene catheter (PE-90) 4–5 cm from the anus as a rectal enema to rats under 2% isofluorane inhalation anesthesia. Control group of rats got equal volume of the vehicle (50 % ethyl alcohol in water). Topiramate (80mg/kg BW) dissolved in 1M sodium hydroxide (2.5% V/V) or vehicle used for topiramate was administered thru a stainless steel feeding needle as oral gavage daily for 7 days starting a day after TNBS administration. Rats were monitored daily for signs of colitis (diarrhea). On day 8 all the rats were sacrificed and tissues (distal segment of the colon) harvested for macroscopic damage scores & histology. The initial pilot study used 8 animals per treatment group. Two independent replication studies used 12 animals per treatment group. The initial animal study was performed at Stanford University School of Medicine. The independent replicate studies were performed by Biomodels LLC (Watertown, MA, USA) and MD Biosciences, Inc. (St. Paul, MN, USA).
qRT-PCR Analysis
Extraction of RNA
RNA was extracted from tissue using a standard protocol for TRIzol® (Invitrogen, Life Technologies, Carlsbad, CA)/Chloroform. In brief, frozen colon tissues were transferred into 1mL Trizol and homogenized using a rotor stator homogenizer (Polytron PT 1200 E Kinematica AG, Lucerne, Switzerland). After homogenization and addition of Chloroform, tissue suspensions were centrifuged at 4°C for 15 min. resulting in the formation of three phases. Total RNA, located in the upper, aqueous phase was subsequently precipitated using Isopropanol (100%). After 2 wash steps in Ethanol 75%, RNA was pelleted and diluted in TE-Buffer (Ambion, Life Technologies, Foster City, CA). Concentration of RNA was assessed by NanoDrop (NanoDrop, Thermo Fisher Scientific Inc., Wilmington, DE). Standard animal care procedures were followed in each experimental situation.
Reverse Transcription
For cDNA synthesis, 2 μg of total RNA was reversed transcribed using Superscript III (Invitrogen, Carlsbad, CA) according to the manufacture’s protocol.
Gene Expression Analyses
Transcriptional levels for 9 genes, including 18S as an endogenous control gene, were assessed in 36 samples (12 controls, 12 vehicle treated, 12 drug treated rats) in duplicates using qRT-PCR. All reagents were purchased from Applied Biosystems and reactions were run on a TaqMan HT7900 machine (Applied Biosystems, Life Technologies, Foster City, CA) for a total of 40 cycles. Gene expression assays had the following accession numbers: 18S (Hs99999901_s1); ALOX5 (Rn00563172_m1); ALOX5AP (Rn00568506_m1); TRPV1 (Rn00583117_m1); CHRM3 (Rn00560986_s1); IFI30 (Rn01420317_m1); MAPK3 (Rn00820922_g1); IL-6 (Rn01410330_m1); IL-10 (Rn00563409_m1). Gene Expression levels were calculated using the ΔΔCt method.
Macroscopic damage assessment
Macroscopic Damage Score (MDS) was assessed in a blinded fashion by examination of excised colons harvested at study termination. After removal of the distal colon segment (~5–7cm), tissue specimens were cut longitudinally, flushed gently with normal saline to clear the fecal matter and photographed. They were scored by a blinded observer for tissue damage using the following criteria: 0 – no inflammation, 1 – swelling or redness, 2 – swelling and redness, 3 – 1 or 2 ulcers, 4 – more than 2 ulcers or 1 large ulcer, 5 – necrosis.
Histopathology and microscopic damage assessment
A piece of the colon tissue was placed in 10 % buffered formalin and fixed over night. The next day they were paraffin embedded and cut into 5 um sections. The sections were stained with hematoxylin and eosin (H&E) and examined under light microscopy. The slides were scored according to a grading system established previously (17).
Endoscopy
Endoscopy was performed in a blinded fashion using a small animal endoscope (Karl Storz Endoskope, Germany). To evaluate colitis severity, animals were anesthetized with isoflurane and subjected to video endoscopy of the lower colon.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of General Medical Sciences (R01 GM079719), Howard Hughes Medical Institute, Pharmaceutical Research and Manufacturers of America Foundation, Lucile Packard Foundation for Children’s Health, the Hewlett Packard Foundation, US National Library of Medicine (T15 LM007033). We thank Alex Skrenchuk and Boris Oskotsky from Stanford University for computer support, and Eugene Davydov, Chuong Do, Samuel Gross and Marc Schaub from Stanford University for constructive discussion.
References
- 1.Loftus EV., Jr Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology. 2004 May;126:1504. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 2.Baumgart DC, Sandborn WJ. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007 May 12;369:1641. doi: 10.1016/S0140-6736(07)60751-X. [DOI] [PubMed] [Google Scholar]
- 3.Sands BE, et al. Infliximab maintenance therapy for fistulizing Crohn’s disease. N Engl J Med. 2004 Feb 26;350:876. doi: 10.1056/NEJMoa030815. [DOI] [PubMed] [Google Scholar]
- 4.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006 Sep 29;313:1929. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 5.Barrett T, et al. NCBI GEO: mining millions of expression profiles--database and tools. Nucleic Acids Res. 2005 Jan 1;33:D562. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irving PM, Gearry RB, Sparrow MP, Gibson PR. Review article: appropriate use of corticosteroids in Crohn’s disease. Aliment Pharmacol Ther. 2007 Aug 1;26:313. doi: 10.1111/j.1365-2036.2007.03379.x. [DOI] [PubMed] [Google Scholar]
- 7.Thiry A, Dogne JM, Supuran CT, Masereel B. Anticonvulsant sulfonamides/sulfamates/sulfamides with carbonic anhydrase inhibitory activity: drug design and mechanism of action. Curr Pharm Des. 2008;14:661. doi: 10.2174/138161208783877956. [DOI] [PubMed] [Google Scholar]
- 8.Khanna V, Arumugam S, Roy S, Mittra S, Bansal VS. Topiramate and type 2 diabetes: an old wine in a new bottle. Expert Opin Ther Targets. 2008 Jan;12:81. doi: 10.1517/14728222.12.1.81. [DOI] [PubMed] [Google Scholar]
- 9.Stenlof K, et al. Topiramate in the treatment of obese subjects with drug-naive type 2 diabetes. Diabetes Obes Metab. 2007 May;9:360. doi: 10.1111/j.1463-1326.2006.00618.x. [DOI] [PubMed] [Google Scholar]
- 10.Bhat R, et al. Inhibitory role for GABA in autoimmune inflammation. Proc Natl Acad Sci U S A. 2010 Feb 9;107:2580. doi: 10.1073/pnas.0915139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodruff TM, et al. A Potent Human C5a Receptor Antagonist Protects against Disease Pathology in a Rat Model of Inflammatory Bowel Disease. The Journal of Immunology. 2003;171:5514. doi: 10.4049/jimmunol.171.10.5514. [DOI] [PubMed] [Google Scholar]
- 12.Roy Chengappa KN, et al. Adjunctive topiramate therapy in patients receiving a mood stabilizer for bipolar I disorder: a randomized, placebo-controlled trial. J Clin Psychiatry. 2006 Nov;67:1698. doi: 10.4088/jcp.v67n1105. [DOI] [PubMed] [Google Scholar]
- 13.Chen R, Li L, Butte AJ. AILUN: reannotating gene expression data automatically. Nat Methods. 2007 Nov;4:879. doi: 10.1038/nmeth1107-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001 Apr 24;98:5116. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jurjus AR, Khoury NN, Reimund JM. Animal models of inflammatory bowel disease. Journal of Pharmacological and Toxicological Methods. 2004;50:81. doi: 10.1016/j.vascn.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Wirtz S, Neufert C, Weigmann B, Neurath MF. Chemically induced mouse models of intestinal inflammation. Nat Protocols. 2007;2:541. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
- 17.Cooper HS, Murthy SN, Shah RS, Sedergran DJ. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 1993 Aug;69:238. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.