Abstract/Synopsis
Recent technological advances in Next Generation Sequencing (NGS) methods have substantially reduced cost and operational complexity leading to the production of bench top sequencers and commercial software solutions for implementation in small research and clinical laboratories. This chapter summarizes requirements and hurdles to the successful implementation of these systems including 1) calibration, validation and optimization of the instrumentation, experimental paradigm and primary readout, 2) secure transfer, storage and secondary processing of the data, 3) implementation of software tools for targeted analysis, and 4) training of research and clinical personnel to evaluate data fidelity and interpret the molecular significance of the genomic output. In light of the commercial and technological impetus to bring NGS technology into the clinical domain, it is critical that novel tests incorporate rigid protocols with built-in calibration standards and that data transfer and processing occur under exacting security measures for interpretation by clinicians with specialized training in molecular diagnostics.
Keywords: clinical pathology, computational pathology, genomics, genomic sequencing, molecular diagnostics, next generation sequencing, tumor diagnostics
1. Introduction
The development of massively parallel sequencing, also known as Next Generation Sequencing (NGS), has provided both basic and clinical scientists with the opportunity to carry out whole genome sequencing in a manner previously restricted to genome centers performing large scale sequencing projects or developing novel sequencing technologies. NGS methods have largely replaced its predecessor, Sanger dideoxynucleotide capillary sequencing, for research purposes based on greater throughput, faster readout, decreased cost per nucleotide base identification and ease of use. Massively parallel paired-end sequencing (MPS) allows for the unprecedented global assessment of interchromosomal rearrangements while simultaneously interrogating single nucleotide substitutions (also called single nucleotide variants or SNVs), copy number variants (CNV), insertions/deletions (indels) and other structural variations (1). Discovery of novel SNVs using Next Generation Sequencing today still requires validation via Sanger methods since the trade off in generating so many parallel short templates using the polymerase chain reaction (PCR) during library construction and DNA polymerase during MPS is loss of accuracy. NGS platforms have approximately 10 fold higher error rates (1 in 1,000 bases at 20X coverage) versus Sanger sequencing (1 in 10,000 bases) (1–3). While variant call accuracy matching Sanger sequencing has been predicted at 20 fold read depth per base for NGS platforms, empirical studies indicate that average read depths exceeding 100X and potentially as high as 1000X are required for use of these platforms as an independent discovery tool of novel targets even under ideal conditions (1–3).
Next Generation platforms differ from each other predominantly in their methods of clonal amplification of short DNA fragments (50 to 400 bases) as a genomic library template and how these fragment libraries are subsequently sequenced through repetitive cycles to provide a nucleotide readout (2–4). The dominant NGS whole genome platforms are the Life Technologies SOLiD (Life Technologies Inc., Grand Island, NY), Roche 454 (Roche Diagnostics Inc., Indianapolis, IN) and Illumina systems (Illumina Inc., San Diego, CA). The SOLiD and 454 systems rely on emulsion PCR to densely decorate beads (SOLiD: 1 micron; 454: 28 microns) with monoclonal DNA templates followed by ligation sequencing or pyrosequencing, respectively, to provide a base readout. The Illumina system utilizes bridge PCR to amplify templates in discrete monoclonal clusters attached to the surface of a flow cell followed by reverse termination sequencing to define individual base incorporation. These platforms vary in performance characteristics and cost and each offers advantages for different sequencing applications such as de novo assembly versus the mapping of structural variants; but they perform comparably at saturating sequencing coverage (2–4).
The instruments, dedicated servers and computational tools required to perform whole genome sequencing using NGS methodology have become progressively more affordable and available through continual technological refinements since completion of the Human Genome Project. The cost of instrumentation for DNA library preparation and sequencing of whole genomes along with the computational power for data processing, transfer, storage and analysis now fall within the price range of academic institutional core facilities ($600K to $1M). Smaller, less expensive instruments capable of whole exome and targeted resequencing have recently been developed for “research use only” (RUO) applications in individual research labs and are being avidly marketed to clinical labs (<$200K). The cost of sequencing “per base” has plummeted from the estimated 2.7 billion dollar cumulative price tag of the first genome draft sequence published in 2001 to commercial sequencing costs of $5,000 for an entire genome in 2010 (2–5). At the same time, the scope of sequencing has expanded from delineation of the prototypical whole human genome to characterization of the personal genome for individualized medicine (5, 6). In particular, genomic sequencing is expected to make a preeminent contribution to diagnosis and treatment of cancer since tumors derive from somatic DNA lesions that occur sporadically in tissues or through de novo germ line changes. Carefully designed NGS studies allow characterization of multiple modalities of genomic structural alterations in cancer while providing sufficiently deep coverage to identify single base mutations in heterogeneous specimens (7). Translation of these discoveries into the clinical domain could subsidize a new generation of diagnostic tests. For example, important challenges regarding cancer diagnostics that can be effectively addressed using whole genome sequencing are 1) identification of DNA biomarker regions for early diagnosis of various tumor classes, 2) delineation of genomic changes underlying the mechanisms of tumorigenesis, and 3) pretreatment specification of personal genomic alterations to validate tumor susceptibility to targeted molecular therapies.
2. Bench Top Instrumentation
Technical refinements in emulsion PCR methods, smaller flow cell size, faster microfluidics and reduced imaging time have improved NGS speed and throughput while enabling production of bench top size sequencing instruments. The Roche 454 GS Junior, the Illumina MiSeq and the Life Technologies Ion Torrent PGM provide throughput capabilities and cost efficiencies that support utilization of these bench top instruments in a small research laboratory environment or within a clinical diagnostic laboratory (8). Aspects of Third Generation sequencing methodology are incorporated into the Ion Torrent Personal Genome Machine (Life Technologies, Inc.), which interrogates single nucleotides as they are sequentially incorporated via DNA polymerase into a parallel strand complementary to the library template. The Ion Torrent PGM utilizes a complementary metal oxide semiconductor (CMOS) chip with individual wells that function as pH sensitive pixels to directly detect the release of a hydrogen ion upon nucleotide incorporation. This approach eliminates the need for chemilluminescent dyes, charged-coupled device (ccd) cameras, serial image acquisition and a motorized stage resulting in a faster throughput than other platforms but with shorter read lengths (8).
It is important to note that the reduction in size, cost and complexity of bench top sequencers has made them more accessible but has not improved the technical performance of their underlying NGS methodology. The Roche bench top system still relies on pyrosequencing and the Illumina and Ion Torrent systems perform sequencing by synthesis, thus error rates associated with those methodologies applied to short DNA fragments remain the same. The significant challenge of creating optimal template to bead or template to flow cell stoichiometry remains a critical determinant of successful sequencing in these platforms and is a major hurdle for users of these instruments. Bench top sequencers provide the extent and depth of coverage in a single run to create a comprehensive alignment map of a bacterial genome but require multiple runs to sequence the human exome with sufficient integrity to identify novel structural variants. However, they can readily perform high-resolution, targeted sequencing of small human genomic regions of interest in domains known to be associated with diseases such as cancer. Consequently, they may prove useful for discovery research when coupled with established methods to selectively elucidate specific genomic target regions ranging from a few hundred bases to 500 Kb utilizing primer driven amplification, hybridization based capture or restriction digest isolation (Agilent, Inc., Carlsbad, CA; Roche Nimblegen, Inc., Madison, WI; Life Technologies, Inc.) (9).
The falling cost of genomic sequencing has driven rapid growth of NGS utilization in RUO applications involving the sequencing of DNA from fresh-frozen and formalin-fixed paraffin embedded (FFPE) specimens. While there are peculiarities specific to FFPE preparations, studies have revealed critical common sources of errors involved with implementation of whole genome sequencing platforms, which should be considered in the use of bench top versions of these instruments. However, the short reads and depth of coverage associated with NGS systems yield high fidelity single base interrogation of FFPE samples compared to microarray and PCR assays. Extrinsic or pre-analytic variables that can affect sequence accuracy and integrity at the level of sample acquisition and processing include specimen cellular heterogeneity, DNA extraction method, reagent batch effects, protocol drift, nucleotide or barcode cross contamination, personnel training and study site. Many of these factors can be balanced by randomizing the order of sample processing, procuring large reagent lots, altering the position of DNA samples within plates and including inter-batch positive and negative sequencing controls. The sequencing platforms and instrumentation are themselves an intrinsic source of variability affecting technical reproducibility, accuracy, error rates and the specificity and sensitivity to detect genomic structural changes down to the level of mutant alleles. The performance of NGS methods and instruments should be routinely validated against a laboratory DNA standard such as a Hap Map cell line without somatic variants and a tumor cell line with stable, structural changes and mutations. An average depth of sequence coverage ≥ 50X is adequate for validation of these homogeneous samples. However, for clinical samples in which tumor cells are contained within an admixture of heterogeneous cells (stroma, infiltrating immune cells, capillary endothelial cells, etc.) a much deeper coverage (100X to 1,000X) is generally required. Despite precise error control and calibration standards, concern persists as to whether NGS platforms independently provide the confidence levels required for using previously uncharacterized novel individual variant calls in clinical samples for patient diagnostic applications.
The value that NGS technology could bring to clinical laboratory diagnostic services is accentuated by several recent genetic developments. First, Next Generation genomic sequencing was critical to the characterization of genetic disorders over the past decade with nearly 3,000 single gene Mendelian disorders identified by the year 2011 (10). Second, increased numbers of small molecule targeted cancer therapies have been introduced over the past decade that require a sequence based companion diagnostic test, e.g., the drug, PLX-4032, targets papillary thyroid cancers and metastatic malignant melanoma that feature the V600E mutation of the BRAF gene (11). As the number of these therapeutic products increases, the demand for sequencing solid tumors and hematologic malignancies will commensurately increase. Third, multi-gene cancer biomarker panels have emerged that provide diagnostic information in demand by both patients and physicians. For example, OncotypeDX, PAM50 and Mammaprint® are separate gene expression assays that supply information on the risk of breast cancer recurrence and help inform therapy choices through evaluation of 21, 50 and 70 genes, respectively (12,13). The role of the clinical diagnostic lab in generating genomic information associated with these developments is not clear. However, it is certain that increased demand for detailed patient genomic information for diagnosis and treatment cannot be met by scaling up traditional Sanger sequencing, pyrosequencing or PCR methods while NGS can acutely meet the challenge.
In order for bench top sequencing platforms to be effective in hospital clinical laboratories for diagnostic purposes, they must provide rapid sample throughput and turnaround times (minutes to hours), have very low technical error rates (e.g. 0.001 to 0.0001), employ standardized protocols including positive and negative technical controls, and obtain reimbursement within current acceptable guidelines (hundreds to thousands of dollars). The turnaround time marketed for bench top sequencing instruments attempts to satisfy these clinical lab requirements, particularly with the use of DNA bar-coding adapters that allow multiple samples to be evaluated simultaneously. The significant labor and costs of capturing, amplifying and preparing templates for targeted sequencing and the time and resources required to perform the data analysis are currently within the timeframe and price-point for clinical labs using the fastest bench top NGS systems. Protocols for NGS are marketed as semi-automated and easy to use. In our experience, commercial sequencing protocols are still undergoing routine revision, lack important QA/QC checkpoints, and are subject to version “drift”. Furthermore, the highest acuity for identification of critical genomic alterations in tumor samples is by direct comparison to DNA obtained from a “normal” sample, preferably blood. The addition of a matched “normal” reference doubles the cost, required reagents and work effort. Consequently, the development, validation, and accreditation of sequencing tests to supplant current accredited “stand-alone” assays are unlikely. A more probable scenario for clinical laboratory sequencing is implementation of new diagnostic tests revealed through NGS on whole genome platforms but translated into targeted assays for bench top instruments. These tests will evaluate larger genomic domains at high resolution to provide the physician with knowledge that is currently unattainable through Sanger sequencing or qPCR, e.g., the presence of “unexpected” sequence structural abnormalities, somatic base pair and indels, balanced and unbalanced somatic rearrangements, and gene copy number information including homozygous and heterozygous deletions associated with specific diseases.
3. Enterprise Sequencing
The advent of massively parallel sequencing has enabled an intense effort at the “enterprise” level to characterize various normal and tumor genomes by drawing on the infrastructure and expertise developed during the Human Genome Project. Multiple large-scale sequencing projects are underway in an effort to accumulate genomic data from cancer patients in centralized databases and concurrently develop analytic tools for interrogating these data. Examples in cancer genomics include The Cancer Genome Atlas (NCI and NHGRI), the Cancer Genome Project (Wellcome Trust Sanger Institute) and the International Cancer Genome Consortium (Ontario Institute for Cancer Research) (14, 15). Individual institutions such as the Genome Institute at Washington University (Saint Louis, MO) have developed independent programs such as the Pediatric Cancer Genome Project building on expertise developed during the Human Genome Project. Commercial targeted and whole genome services are also rapidly proliferating both as service providers and as data repositories (Table 1). These programs will no doubt continue to multiply under the assumption that accumulation of DNA sequence along with detailed clinical data will achieve a critical mass when the appropriate analysis of a comprehensive sequence repository will answer pertinent scientific and clinical questions, e.g., identification of driver mutations as therapeutic targets and predicting patient response to therapy.
Table 1.
Select vendors of commercial genomics and data storage services as of June 2012. The list is not exhaustive and is intended for initial guidance only
| Service | Vendor | Website | Remarks |
|---|---|---|---|
| Whole Genome Sequencing (WGS) |
|
Using various NGS technologies they provide WGS services within several weeks to months of sample submission. The data is provided in a raw format without any interpretation of the variants. | |
| Whole Exome Sequencing (WES) |
|
The focus of WES services is to sequence the protein (± microRNA) coding part of the genome, including their splice sites. The whole exome is “captured” by using specially designed bait probes. | |
| Genomic data storage providers |
|
Commercial storage and computing power in the “cloud”. Prices are highly competitive with enormous computing power at one’s fingertips. However, issues related to patient privacy and HIPAA compliance remain. Amazon has taken initial steps to ensure compliance with HIPAA. Data transfer of the huge WES and WGS data files over the Internet is a significant problem. | |
| Genome/ exome interpretation software/ providers |
|
The goal of commercial companies in the “data inter-pretation” space is to interpret the raw sequence data for a fee. The data may be generated in-house or from an external source. Most of the bioinformatics tools are developed on a proprietary basis. Information related to pathway analysis, sequence variants and data querying services are provided. Most of these companies are in the incubator stage. |
4. NGS Data Analysis
Most commercial NGS platforms generate light (fluorescence) as the underlying raw signal output when the genomic template is interrogated with serial images accumulating until they are converted to a base readout. Once the base coding is obtained for the templates, it requires a computationally intensive process to map these sequence fragments in register with an established reference sequence as opposed to the complex task of de novo assembly. The present standard is the latest build of the human genome provided by the Genome Reference Consortium (15).
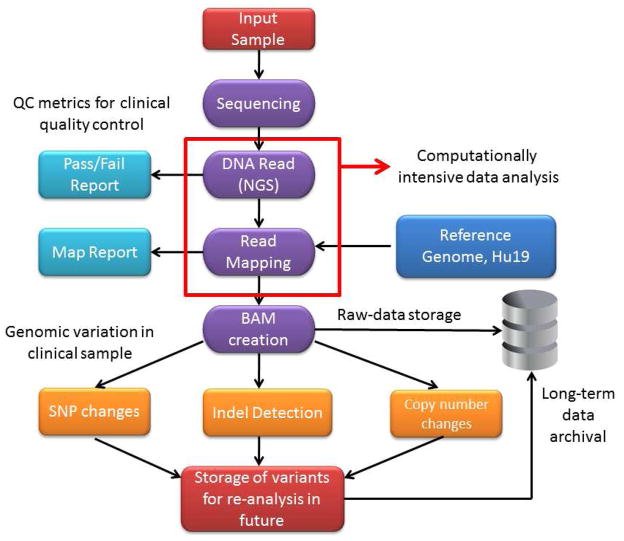
A typical supercomputing DNA alignment solution utilizes multiple parallel processing nodes to assemble different genomic components of the data. These data are subsequently aggregated by the head node to provide final, mapped genomic sequence. The computer processing time required for mapping depends on the extent of the mapped genome (genome, exome or target region) and the redundancy of coverage. A single base of the human haploid genome occupies roughly 2.5 bytes in the FASTQ format. Mapping the 3.1 Gb whole human genome at 30X coverage generates 2.5 × 3.1 × 30X or ~ 230 GB of raw base calls requiring hours of parallel processing. The alignment process is complicated by the fact that the fragment data comprises inherently self-similar FASTQ text lines. It is currently possible to implement NGS technology in a medium sized facility (e.g., an academic medical center) as computer capabilities have increased in speed and decreased in cost. A potential schematic of the NGS workflow is shown in Figure 1.
Figure 1. Prototypical workflow in a clinical next generation sequencing laboratory.
The entire workflow process occurs under the auspices of a CLIA-certified laboratory for clinical diagnostic application. An important distinction of the workflow process in the clinical laboratory relative to a research environment is enforcement of strict process and quality metrics. At the present time, a national standard for quality assurance in a NGS laboratory remains to be defined.
The goal of clinical NGS for cancer diagnostics is the identification of pertinent point mutations and larger structural variations such as translocations, rearrangements, inversions, deletions and amplifications in tumour samples compared to the normal genome. At present an array of free and commercially available software are available for NGS data analysis. The workflow includes three major steps.
Step 1: Alignment and Assembly
There are multiple free mapping software tools available including MAQ (16), BWA (17), Bowtie (18), SOAP (19), ZOOM (20), SHRiMP (21) and Novoalign (22). Illumina and SOLiD provide their own alignment software as well. Commercial, third party software vendors such as CLC Genomics also provide mapping programs. Disadvantages of free, open source software are the lack of documentation and a reliance on Unix and its command line environment. However, open source software based on the Burrows-Wheeler transformation (BWT) algorithm remains significantly faster than commercial solutions for mapping and alignment. Software based on the BWT algorithm can map a human genome in hours instead of days required by other software tools such as MAQ (16) and Novoalign (22). Commercial vendors provide access to proprietary mapping algorithms but at a substantial cost for mid-level academic institutions. Software for de novo assembly of cancer genomes is a powerful tool for detection of unique rearrangements and chromosomal breakpoints in a tumor sample albeit by a slower method than mapping against the reference genome. These include Velvet (23, 24), EULER-SR (25), EDENA (26), QSR (27), and AbYSS (28).
Step 2: Variant detection
Once alignment is completed, downstream bioinformatics analysis is performed to detect structural genomic alterations relevant to the clinical diagnosis.
Single nucleotide polymorphisms (SNPs) and point mutations – Molecular diagnostic assays for cancer have focused on discovery of mutations in tumor-related genes or small panels of these genes. For example, certain mutations in the epidermal growth factor receptor (EGFR) gene are associated with favorable responses in lung cancers treated with gefitinib compared to lung cancers with wild type EGFR (29). An impediment to finding these somatic mutations in cancer is specimen cellular heterogeneity. Recent studies indicate that there is a 5% probability of detecting a mutation in 25% of tumor cells sequenced at 30- to 40-fold coverage (30). Laser capture microdissection to obtain DNA from a population highly enriched for cancer cells can reduce the cellular variability of the specimen. There is a variety of software tools for detection of single nucleotide variants based on different statistical models of base calling including SNVmix (31), VarScan (32) and SomaticSniper (33). Open source tools such as SAMtools (34), use Bayesian detection to identify somatic SNP variants.
Structural changes in the cancer genome – Cancer genomes are highly unstable including diverse chromosomal abnormalities such as large genomic insertions and deletions. While karyotyping is the standard method to identify chromosomal abnormalities, it cannot identify structural abnormalities smaller than ~5 megabases. SNP and oligonucleotide microarrays have revolutionized the field of cytogenetics providing high resolution (~1Kb) to identify copy number variants and copy neutral loss of heterozygosity. NGS technologies also identify structural variations in the genome, although typical alignment tools cannot identify more than a few nucleotide mismatches. Specialized software for analyzing indels from paired-end reads such as Pindel (35) identify structural variants by defining the flanking regions of the read data while the GATK indel genotyper (36) employs heuristic cut offs for indel calling. Nevertheless, delineation of large amplifications and deletions in cancer chromosomes remains a formidable challenge. Algorithms to identify large variations include the circular binary segmentation algorithm of arrays (37) and the SegSeq algorithm which uses a merging procedure to join localized SNP changes with whole chromosome changes to compare tumor to normal samples (38). Several programs are available to identify large scale structural variations in the genome, such as BreakDancer (39). While NGS technology has revealed variations in lung cancer, melanomas and breast cancer at the single nucleotide level (40–43), significant hurdles remain to address changes at the chromosomal level.
Step 3: Beyond genome sequencing
NGS platforms offer the versatility to perform transcriptomic profiling, chromatin immunoprecipitation, small RNA sequencing and epigenomics studies. Transcriptomics via NGS can probe alternate splicing, the process by which multiple RNA isoforms arise from a single gene. These isoforms contribute to cell type specificity and may play a role in specification of cancerous cells. Identification of novel splicing variants is important for understanding biological specificity in the context of normal and abnormal cellular function. Software tools such as TOPHAT (44) facilitate de novo discovery of splicing variants.
RNA discovery
The role of small RNAs (18–35 bp) in the regulation of gene expression and translation of mRNAs has been recently recognized. NGS methods can perform deep sequencing of small RNA species for discovery and analysis. There is a specific advantage to platforms such as Illumina and SOLiD in small RNA discovery due to the short reads generated by those technologies. There are many small RNA databases and bioinformatics tools e.g. MirCat (45) and mirDeep (46, 47), that can facilitate identification and discovery of small RNAs.
Epigenomic discovery
Epigenomics refers to chemical modifications (e.g., methylation) of DNA and RNA and its impact on gene expression. Traditional methods of assessing gene methylation rely on bisulfite conversion of unmethylated cytosines to uracil for identification using sequencing methods or restriction endonuclease analysis. One pitfall associated with this approach is the labor intensive methodology required to identify epigenetic changes on an individual gene basis. In contrast, NGS technologies can interrogate broad changes in DNA methylation patterns across the entire genome, simultaneously capturing epigenetic information from multiple genes while providing information regarding normal or tumor tissue methylation status.
5. Scientific Challenges for the Implementation of NGS
The acuity of cancer related sequencing studies is enhanced by differential comparison of patient tumor genomic sequences to matched normal reference sequences e.g. a paired blood sample. However, routine assembly and comparison of each of these paired samples is dependent on the existence of an accurate representation of the reference human normal genome, including its intrinsic variability encompassing benign structural modifications and polymorphisms. A database comprising normal whole genome sequences is being compiled through concomitant large parallel sequencing projects, including the 1000 Genomes Project (NHGRI: an extension of the International HapMap Project), the Genome Reference Consortium (Wellcome Trust, Genome Institute at Washington University, EMBL, NCBI) and the Personal Genome Project (Harvard University) (48–50). The initial reference genome was constructed de novo with DNA comprising a small number of anonymous subjects with the bulk of the clones (~60%) from a single male donor. The current iteration of the reference genome (Genome Reference Consortium 37 or HG Build19) is estimated to be 99.99% accurate containing 2.95 billion bases and 210 gaps (49). Sequencing of personal genomes has established diversity as high as 3% among individuals and personal sequences can differ from the reference genome in hundreds of thousands of bases (51, 52). These estimates will likely change drastically with accumulating numbers of personal genome sequences. Thus the current reference genome represents only a small sampling of human genetic variation and contains thousands of both common and rare risk alleles that remain to be defined.
It is also important to consider relevant limitations learned from previous public and private enterprise computing efforts regarding population based genomics. For example, the Hap Map Project and deCODE Genetics were initiated over a decade ago as public and private enterprises with the goal of exhaustively mapping population based genetic diversity on a worldwide basis or within the restricted population of the Icelandic Health Sector (53). The hypothesis underlying these efforts was that disease and treatment related variants would emerge as these databases and analytic tools succeeded in precisely characterizing normal specimens and delineating differences specific to diseased samples. While significant discoveries continue to be made from these efforts, several important issues have emerged pertinent to cancer initiatives. First, the classification of “normal” specimens is challenging since they may originate from either truly “disease free” subjects or derive from “asymptomatic” patients harboring undiagnosed disease. Second, classification of a tumor specimen may vary. Some tumors are difficult to classify. Other definitive tumor types have undergone subsequent phenotype reclassification further confounding their annotation and interrogation due to the persistence of legacy classifications. Third, multi-site genomic data generation produces differences in data fidelity, variability, precision and accuracy associated with the use of different methodologies, instruments, reagent lots, experimental batches and personnel. Because of the increased noise to signal ratio at the consortium level, many frustrated investigators have created their own “in house” reference databases to obviate issues encountered at the enterprise level. This experience from the previous decade is equally relevant today as genomic sequencing databases are being generated.
6. Institutional Challenges for the Implementation of NGS
Bioinformatics is currently the single largest bottleneck to implementation of next generation sequencing in clinical practice. A general guideline is that each dollar spent on sequencing hardware will require an equal investment in informatics (54). Smaller labs cannot absorb these costs, even with the availability of open-source software (55). There are several critical considerations in developing a NGS bioinformatics facility. NGS hardware implementation requires substantial investment in infrastructure. Alternatively, this task can be outsourced to a commercial third party provider. However, “in-house” sequencing and analysis enables important control of sample substrate, library creation, sequence generation and data processing. This is critical for clinical diagnostic sequencing, where process and quality control are of utmost priority. As NGS technology is applied to clinical problems, it is critical to standardize quality metrics for acquisition of data. These include standards for calibration, validation and comparison among platforms, data reliability, robustness and reproducibility, and quality of assemblers. It will be necessary to develop guidelines for standardization of NGS protocols such as occurred in the microarray quality control (MAQC) project (56) and the sequencing quality control (SEQC, also called MAQC-III) project (http://www.fda.gov/MicroArrayQC/) particularly as lab developed tests (LDT) emerge outside the federal regulatory domain.
Most academic centers have existing centralized computing resources which can be leveraged for in-house NGS analysis by upgrading to high performance computer clusters. However, the need for advanced network infrastructure is a formidable barrier to implementation of NGS in a research or clinical setting. The typical academic network architecture comprises 100 megabyte shared ethernet services or lower bandwidth wide area network (WAN) infrastructure. Since NGS data sets are hundreds of gigabytes per run, efficient data transfer requires 10 gigabit network connectivity with gigabit cabling between locations including high speed network switching and network cards on devices that serve the network. Consequently, most academic and many commercial service providers transport their data via portable hard drives and other mobile transfer solutions including transfer from sequencer to server. Advantages of an “in house” network for data transfer and analysis include scheduled maintenance, professional back-up facilities, direct security oversight and dedicated and/or shared nodes for research. Figure 1 depicts a routine bioinformatics workflow required for analysis of NGS data.
The amount of data generated by the sequencing of a single genome comprises hundreds of gigabytes of base calls and quality scores. Multiple runs rapidly accumulate in the terabyte range and a clinical NGS center could produce terabytes to a petabyte of data in a year. Data management on this scale requires well-defined policies and standards although few exist. Furthermore, there is no industry wide standardization for data output from NGS platforms with the most commonly accepted forms comprising SAM (Sequence Alignment/Map) and BAM (Binary Alignment Map) formats (34). BAM files store the data in a compressed, indexed, binary data file format (binary text based format) for efficient storage. NGS data can be stored either a) locally on the instrument, b) at an institutional storage facility, or c) using a commercial cloud storage solution. It is more convenient to store NGS data institutionally or commercially rather than locally. Cloud computing is an especially promising data storage solution, however, privacy and security must be ensured to meet rigid HIPAA requirements.
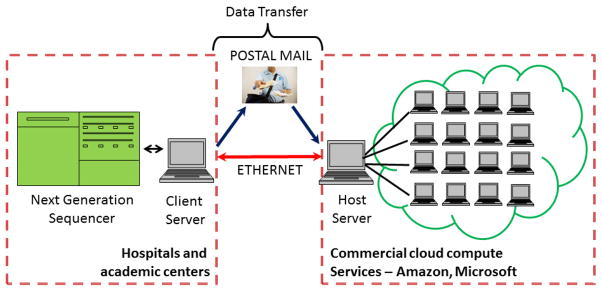
Online computer clusters have become commercially available for public “on demand” access in the form of “cloud computing” solutions. Amazon, Google and Microsoft have created centralized supercomputing facilities through virtualization of software, a process whereby a user can access an “image” of the operating system (Linux or Windows) residing on the server of the company hosting the cloud. This interface image is indistinguishable from an ordinary desktop interface. The difference is that the virtual operating system is hosted on a remote server (Fig. 2). The advantage of a cloud solution is access to supercomputing power without installation and maintenance of expensive hardware. Fees for cloud services are currently affordable for an average user with pay-as-you-go pricing. Private vendors such as Amazon S3 (Simple Storage Service) also provide long-term storage of datasets through networked storage facilities, a critically important issue given the scale of NGS datasets. Disadvantages with cloud services include satisfaction of HIPAA compliance and security of data transfer over a vulnerable internet network. Another critical variable is the insurance policy regarding long-term storage and protection of clinical data in a commercial environment where ownership is subject to change, merger or acquisition.
Figure 2. Prospective utilization of cloud computing in next generation sequencing.
The schematic illustrates the transfer of data from a sequencing instrument to a commercial cloud vendor service through the internet or using regular postal mail. Subsequent analysis may be performed remotely from the sequencing laboratory domain.
7. Training and Implementation of NGS in the Clinical Workplace
Analysis of NGS data requires multidisciplinary teams of clinical and biomedical/pathology informaticians, computational biologists, molecular pathologists, programmers, statisticians, biologists, as well as clinicians. Consequently, substantial institutional support for resources and personnel is needed for clinical implementation of NGS technology. Furthermore, physicians will have to be trained to interpret vast and comprehensive molecular datasets. At present, there are approximately 1,000 medical geneticists and 3,000 genetic counselors in the United States. These numbers are grossly inadequate to deal with the explosive growth of genomics testing. One solution is to form strategic collaborations between disciplines. For example, there are over 17,000 pathologists in the United States with broad education in anatomic pathology and laboratory medicine who could undergo further training to integrate large datasets with clinical findings (57). Specifically, there is need to create a subspecialty of “Computational Pathology” to train pathologists to manage and interpret high-throughput biological data including that derived from genomic, proteomic and metabolomics analysis.
8. Summary
The development of massively parallel sequencing methods has expanded genomic sequencing from delineating the prototypical reference human genome to characterization of the individual patient genome as the building block of personalized medicine. The accessibility of this technology has produced important results from the small research lab to enterprise level analysis regarding genomic changes associated with cancer. Benchtop systems incorporating NGS technology have been recently released and are being marketed to the clinical laboratory to meet the demand for personalized medicine applications. To achieve long term success in the clinical domain, critical requirements include the development of versatile, robust and affordable instrument platforms, the development of user friendly bioinformatics tools and support, and evolution of a workforce with pertinent knowledge of molecular biology and genomics. If successful, these systems will provide an incomparable level of diagnostic insight for patients in the future as novel genomic biomarkers and structural changes are identified for application to the clinical domain.
Key Points.
There are unique requirements and limitations critical to implementation of genomic sequencing instrumentation and analysis tools in a small research laboratory or clinical environment.
The lessons learned in the clinical integration of massively parallel sequencing technologies in genomics may provide useful information for the establishment of similar emerging technologies for proteomics and metabolomics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Campbell PJ, Stephens PJ, Pleasance ED, O’Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, Teague JW, Menzies A, Goodhead I, Turner DJ, Clee CM, Quail MA, Cox A, Brown C, Durbin R, Hurles ME, Edwards PA, Bignell GR, Stratton MR, Futreal PA. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008 Jun;40(6):722–9. doi: 10.1038/ng.128. Epub 2008 Apr 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008 Oct;26(10):1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 3.Harismendy O, Ng PC, Strausberg RL, Wang X, Stockwell TB, Beeson KY, Schork NJ, Murray SS, Topol EJ, Levy S, Frazer KA. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10(3):R32. doi: 10.1186/gb-2009-10-3-r32. Epub 2009 Mar 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010 Jan;11(1):31–46. doi: 10.1038/nrg2626. Epub 2009 Dec 8. Review. [DOI] [PubMed] [Google Scholar]
- 5.Ross JS, Cronin M. Whole cancer genome sequencing by next-generation methods. Am J Clin Pathol. 2011 Oct;136(4):527–39. doi: 10.1309/AJCPR1SVT1VHUGXW. Review. [DOI] [PubMed] [Google Scholar]
- 6.West M, Ginsburg GS, Huang AT, Nevins JR. Embracing the complexity of genomic data for personalized medicine. Genome Res. 2006 May;16(5):559–66. doi: 10.1101/gr.3851306. Review. [DOI] [PubMed] [Google Scholar]
- 7.Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, Steidl C, Holt RA, Jones S, Sun M, Leung G, Moore R, Severson T, Taylor GA, Teschendorff AE, Tse K, Turashvili G, Varhol R, Warren RL, Watson P, Zhao Y, Caldas C, Huntsman D, Hirst M, Marra MA, Aparicio S. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009 Oct 8;461(7265):809–13. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 8.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. Performance comparison of bench top high-throughput sequencing platforms. Nat Biotechnol. 2012 Apr 22; doi: 10.1038/nbt.2198. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 9.Horn S. Target enrichment via DNA hybridization capture. Methods Mol Biol. 2012;840:177–88. doi: 10.1007/978-1-61779-516-9_21. [DOI] [PubMed] [Google Scholar]
- 10.Feero WG, Guttmacher AE, Collins FS. Genomic medicine--an updated primer. N Engl J Med. 2010 May 27;362(21):2001–11. doi: 10.1056/NEJMra0907175. Review. [DOI] [PubMed] [Google Scholar]
- 11.Lee JT, Li L, Brafford PA, van den Eijnden M, Halloran MB, Sproesser K, Haass NK, Smalley KS, Tsai J, Bollag G, Herlyn M. PLX4032, a potent inhibitor of the B-Raf V600E oncogene, selectively inhibits V600E-positive melanomas. Pigment Cell Melanoma Res. 2010 Dec;23(6):820–7. doi: 10.1111/j.1755-148X.2010.00763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly CM, Bernard PS, Krishnamurthy S, Wang B, Ebbert MT, Bastien RR, Boucher KM, Young E, Iwamoto T, Pusztai L. Agreement in Risk Prediction Between the 21-Gene Recurrence Score Assay (Oncotype DX(R)) and the PAM50 Breast Cancer Intrinsic Classifier™ in Early-Stage Estrogen Receptor-Positive Breast Cancer. Oncologist. 2012;17(4):492–8. doi: 10.1634/theoncologist.2012-0007. Epub 2012 Mar 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002 Dec 19;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 14.Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci Am. 2007 Mar;296(3):50–7. [PubMed] [Google Scholar]
- 15.International Cancer Genome Consortium. International network of cancer genome projects. Nature. 2010 Apr 15;464(7291):993–8. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008 Nov;18(11):1851–8. doi: 10.1101/gr.078212.108. Epub 2008 Aug 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lippert RA, Mobarry CM, Walenz BP. A space-efficient construction of the Burrows-Wheeler transform for genomic data. J Comput Biol. 2005 Sep;12(7):943–51. doi: 10.1089/cmb.2005.12.943. Review. [DOI] [PubMed] [Google Scholar]
- 18.Pop M, Salzberg SL. Bioinformatics challenges of new sequencing technology. Trends Genet. 2008 Mar;24(3):142–9. doi: 10.1016/j.tig.2007.12.007. Epub 2008 Feb 11. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008 Mar 1;24(5):713–4. doi: 10.1093/bioinformatics/btn025. Epub 2008 Jan 28. [DOI] [PubMed] [Google Scholar]
- 20.Lin H, Zhang Z, Zhang MQ, Ma B, Li M. ZOOM! Zillions of oligos mapped. Bioinformatics. 2008 Nov 1;24(21):2431–7. doi: 10.1093/bioinformatics/btn416. Epub 2008 Aug 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M. SHRiMP: accurate mapping of short color-space reads. PLoS Comput Biol. 2009 May;5(5):e1000386. doi: 10.1371/journal.pcbi.1000386. Epub 2009 May 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao S, Jiang R, Kwan W, Wang B, Ma X, Song YQ. Evaluation of next-generation sequencing software in mapping and assembly. J Hum Genet. 2011 Jun;56(6):406–14. doi: 10.1038/jhg.2011.43. Epub 2011 Apr 28. Review. [DOI] [PubMed] [Google Scholar]
- 23.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008 May;18(5):821–9. doi: 10.1101/gr.074492.107. Epub 2008 Mar 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zerbino DR. Using the Velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinformatics. 2010 Sep;Chapter 11(Unit 11.5) doi: 10.1002/0471250953.bi1105s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaisson MJ, Pevzner PA. Short read fragment assembly of bacterial genomes. Genome Res. 2008 Feb;18(2):324–30. doi: 10.1101/gr.7088808. Epub 2007 Dec 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez D, François P, Farinelli L, Osterås M, Schrenzel J. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 2008 May;18(5):802–9. doi: 10.1101/gr.072033.107. Epub 2008 Mar 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bryant DW, Jr, Wong WK, Mockler TC. QSRA: a quality-value guided de novo short read assembler. BMC Bioinformatics. 2009 Feb 24;10:69. doi: 10.1186/1471-2105-10-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009 Jun;19(6):1117–23. doi: 10.1101/gr.089532.108. Epub 2009 Feb 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004 Jun 4;304(5676):1497–500. doi: 10.1126/science.1099314. Epub 2004 Apr 29. [DOI] [PubMed] [Google Scholar]
- 30.Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, Shlien A, Cooke SL, Hinton J, Menzies A, Stebbings LA, Leroy C, Jia M, Rance R, Mudie LJ, Gamble SJ, Stephens PJ, McLaren S, Tarpey PS, Papaemmanuil E, Davies HR, Varela I, McBride DJ, Bignell GR, Leung K, Butler AP, Teague JW, Martin S, Jönsson G, Mariani O, Boyault S, Miron P, Fatima A, Langerød A, Aparicio SA, Tutt A, Sieuwerts AM, Borg A, Thomas G, Salomon AV, Richardson AL, Børresen-Dale AL, Futreal PA, Stratton MR, Campbell PJ Breast Cancer Working Group of the International Cancer Genome Consortium. The life history of 21 breast cancers. Cell. 2012 May 25;149(5):994–1007. doi: 10.1016/j.cell.2012.04.023. Epub 2012 May 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goya R, Sun MG, Morin RD, Leung G, Ha G, Wiegand KC, Senz J, Crisan A, Marra MA, Hirst M, Huntsman D, Murphy KP, Aparicio S, Shah SP. SNVMix: predicting single nucleotide variants from next-generation sequencing of tumors. Bioinformatics. 2010 Mar 15;26(6):730–6. doi: 10.1093/bioinformatics/btq040. Epub 2010 Feb 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012 Mar;22(3):568–76. doi: 10.1101/gr.129684.111. Epub 2012 Feb 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, Ley TJ, Mardis ER, Wilson RK, Ding L. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012 Feb 1;28(3):311–7. doi: 10.1093/bioinformatics/btr665. Epub 2011 Dec 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009 Aug 15;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. Epub 2009 Jun 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009 Nov 1;25(21):2865–71. doi: 10.1093/bioinformatics/btp394. Epub 2009 Jun 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011 May;43(5):491–8. doi: 10.1038/ng.806. Epub 2011 Apr 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal SA, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Griffin CA, Burton J, Swerdlow H, Quail MA, Stratton MR, Iacobuzio-Donahue C, Futreal PA. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010 Oct 28;467(7319):1109–13. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiang DY, Getz G, Jaffe DB, O’Kelly MJ, Zhao X, Carter SL, Russ C, Nusbaum C, Meyerson M, Lander ES. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009 Jan;6(1):99–103. doi: 10.1038/nmeth.1276. Epub 2008 Nov 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, Shi X, Fulton RS, Ley TJ, Wilson RK, Ding L, Mardis ER. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009 Sep;6(9):677–81. doi: 10.1038/nmeth.1363. Epub 2009 Aug 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortés ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareño C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011 Aug 26;333(6046):1157–60. doi: 10.1126/science.1208130. Epub 2011 Jul 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010 Jan 19;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berger MF, Levin JZ, Vijayendran K, Sivachenko A, Adiconis X, Maguire J, Johnson LA, Robinson J, Verhaak RG, Sougnez C, Onofrio RC, Ziaugra L, Cibulskis K, Laine E, Barretina J, Winckler W, Fisher DE, Getz G, Meyerson M, Jaffe DB, Gabriel SB, Lander ES, Dummer R, Gnirke A, Nusbaum C, Garraway LA. Integrative analysis of the melanoma transcriptome. Genome Res. 2010 Apr;20(4):413–27. doi: 10.1101/gr.103697.109. Epub 2010 Feb 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, Shah K, Sato M, Thomas RK, Barletta JA, Borecki IB, Broderick S, Chang AC, Chiang DY, Chirieac LR, Cho J, Fujii Y, Gazdar AF, Giordano T, Greulich H, Hanna M, Johnson BE, Kris MG, Lash A, Lin L, Lindeman N, Mardis ER, McPherson JD, Minna JD, Morgan MB, Nadel M, Orringer MB, Osborne JR, Ozenberger B, Ramos AH, Robinson J, Roth JA, Rusch V, Sasaki H, Shepherd F, Sougnez C, Spitz MR, Tsao MS, Twomey D, Verhaak RG, Weinstock GM, Wheeler DA, Winckler W, Yoshizawa A, Yu S, Zakowski MF, Zhang Q, Beer DG, Wistuba II, Watson MA, Garraway LA, Ladanyi M, Travis WD, Pao W, Rubin MA, Gabriel SB, Gibbs RA, Varmus HE, Wilson RK, Lander ES, Meyerson M. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007 Dec 6;450(7171):893–8. doi: 10.1038/nature06358. Epub 2007 Nov 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009 May 1;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. Epub 2009 Mar 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moxon S, Schwach F, Dalmay T, Maclean D, Studholme DJ, Moulton V. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics. 2008 Oct 1;24(19):2252–3. doi: 10.1093/bioinformatics/btn428. Epub 2008 Aug 19. [DOI] [PubMed] [Google Scholar]
- 46.Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008 Apr;26(4):407–15. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 47.Yang X, Li L. miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics. 2011 Sep 15;27(18):2614–5. doi: 10.1093/bioinformatics/btr430. Epub 2011 Jul 19. [DOI] [PubMed] [Google Scholar]
- 48.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010 Oct 28;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004 Oct 21;431(7011):931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 50. [Accessed May 21, 2012];Personal Genome Project. http://www.personalgenomes.org.
- 51.Venter JC. Multiple personal genomes await. Nature. 2010 Apr 1;464(7289):676–7. doi: 10.1038/464676a. [DOI] [PubMed] [Google Scholar]
- 52.International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010 Sep 2;467(7311):52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gulcher J, Stefansson K. Population genomics: laying the groundwork for genetic disease modeling and targeting. Clin Chem Lab Med. 1998 Aug;36(8):523–7. doi: 10.1515/CCLM.1998.089. [DOI] [PubMed] [Google Scholar]
- 54.Perkel JM. Sequence Analysis 101: A newbie’s guide to crunching next-generation sequencing data. The Scientist. 2011;25:60. [Google Scholar]
- 55.Maxmen A. Harnessing the cloud. The Scientist. 2010;24:71. [Google Scholar]
- 56.Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, Bao W, Fang H, Kawasaki ES, Hager J, Tikhonova IR, Walker SJ, Zhang L, Hurban P, de Longueville F, Fuscoe JC, Tong W, Shi L, Wolfinger RD. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat Biotechnol. 2006 Sep;24(9):1140–50. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- 57.Haspel RL, Arnaout R, Briere L, Kantarci S, Marchand K, Tonellato P, Connolly J, Boguski MS, Saffitz JE. A call to action: training pathology residents in genomics and personalized medicine. Am J Clin Pathol. 2010 Jun;133(6):832–4. doi: 10.1309/AJCPN6Q1QKCLYKXM. [DOI] [PubMed] [Google Scholar]