Abstract
Introduction
The basaloid triple-negative breast cancer (B-TNBC) is one of the most aggressive, therapy-resistant, and metastatic tumors. Current models do not recapitulate the basaloid phenotype of TNBC, thus limiting the understanding of its biology and designing new treatments. We identified HCC1806 as a line expressing typical B-TNBC markers, engineered a subline with traceable reporters, and determined growth, drug sensitivity, recurrence, and vascular and metastatic patterns of orthotopic xenografts in immunodeficient mice.
Methods
mRNA and protein analyses showed that HCC1806 expresses basal but not luminal or mesenchymal markers. HCC1806-RR subline stably expressing red fluorescent protein and Renilla luciferase was generated and characterized for sensitivity to chemodrugs, orthotopic growth, vascular properties, recurrence, metastasis, and responsiveness in vivo.
Results
The HCC1806 cells were highly sensitive to paclitaxel, but cytotoxicity was accompanied by pro-survival vascular endothelial growth factor-A loop. In vivo, HCC1806-RR tumors display linear growth, induce peritumoral lymphatics, and spontaneously metastasize to lymph nodes (LNs) and lungs. Similarly to human B-TNBC, HCC1806-RR tumors were initially sensitive to taxane therapy but subsequently recur. Bevacizumab significantly suppressed recurrence by 50% and reduced the incidence of LN and pulmonary metastases by, respectively, 50% and 87%.
Conclusions
The HCC1806-RR is a new model that expresses bona fide markers of B-TNBC and traceable markers for quantifying metastases. Combination of bevacizumab with nab-paclitaxel significantly improved the outcome, suggesting that this approach can apply to human patients with B-TNBC. This model can be used for defining the metastatic mechanisms of B-TNBC and testing new therapies.
Introduction
Breast cancer is the second leading cause of cancer-related deaths in the United States with 40,000 deaths and 200,000 new cases diagnosed annually [1]. Approximately 15% to 19% of patients are diagnosed with triple-negative breast cancer (TNBC) [2–5], which are mammary tumors that lack receptors for estrogen (ER), progesterone (PR), and human epidermal growth factor receptor 2 (HER2) [4,6]. TNBC is most common in women who are obese, premenopausal, of low socioeconomic status, or of African American descent [7–9]. This is a particularly lethal subtype of breast cancer with a 5-year survival rate as low as 40% [10–12]. Patients with TNBC have a high frequency of lymphatic [6,12,13] and distant metastasis [11,14] and, consequently, a significantly greater risk for recurrence and shortened survival compared with patients with ER/PR-positive tumors [10,13]. The life expectancy after detection of visceral metastasis in TNBC patients is estimated as 3 to 22 months [12,15].
Although, biologically and genetically, TNBC is a heterogeneous group of tumors [16], the majority (∼80–90%) falls into the classification of basal-like subtype [5,17]. Basaloid TNBC (B-TNBC) is characterized by expression of cytokeratins 5, 6, 14, and 17 [3,6,18,19], epidermal growth factor receptor (EGFR) [3,20,21], c-Kit [3], mutated BRCA1 [3,14,21], and mutated or deleted p53 [22,23]. Patients with B-TNBC have higher tumor mitotic index [24] and a worse prognosis than patients with triple-negative tumors that do not express basal markers [3,17,25]. B-TNBC subgroup has a tendency to generate larger tumors [19] with frequent lymphovascular invasion [26,27] and metastasis to multiple sites, whereas nonbasaloid tumors typically metastasize only to one site [17]. Despite generally poor prognosis for B-TNBC patients, these tumors are sensitive to cytotoxic therapy [6,13,14,28] with one study demonstrating the highest response rate (85%) of all breast cancer subtypes [14]. However, despite the initial response, patients with TNBC tumors had the worst disease-free and overall survival of all subtypes [14,29], presumably because of the tendency of these tumors to recur at distant sites [14].
Despite the well-known challenges to successful treatment of B-TNBC, little is known about the unique properties of this cancer that predispose patients to metastasis and tumor recurrence. This is mainly because of paucity of reliable in vivo models that faithfully recapitulate major attributes of this disease, particularly those of the basaloid group. On the basis of hierarchical clustering analyses of microarray studies, several breast carcinoma cell lines are qualified to represent the ER/PR/HER2-negative TNBC group. The most frequently suggested lines in this list are HCC38 [30–32], HCC70 [30–32], HCC1937 [30–32], MDA-MB-468 [32], MDA-MB-231 [32–34], and HCC1806 [30–32,35]. However, the potential of these lines to serve as an animal TNBC model is still uncertain because, with the exception of MDA-MB-231 [36,37], most of these lines have not been tested for the ability to grow in vivo. Some lines (e.g., MDA-MB-468 and HCC1806) are known to generate tumors in mice; however, these tumor models have not been quantitatively assessed for recurrence and metastasis, the two main B-TNBC properties that cause patient death.
Of the aforementioned lines, MDA-MB-231 deserves special discussion because it is the most frequently used TNBC model and is considered to represent the basaloid subtype of TNBC [32–34,38–40]. In contrast to other lines, both unmodified and luciferase-tagged MDA-MB-231 sublines have been extensively characterized for growth patterns at the orthotopic site in vivo including quantitative assessment of kinetics, burden, and organ distribution of spontaneous metastasis to lymph node (LN) and lungs [36,37]. Although this metastatic behavior and lack of ER/PR/HER2 markers are both consistent with B-TNBC phenotype, neither the MDA-MB-231 cell line nor all other candidates for TNBC models have been previously tested for the expression of basal cytokeratins 5, 6, 14, and 17. Moreover, MDA-MB-231 cells express a broad variety of mesenchymal-specific proteins including vimentin [20], which places this line into the mesenchymal [20] or mesenchymal stem-like [31] TNBC category that has distinct molecular signature and drug sensitivity from those in the basaloid group [31]. The source of the MDA-MB-231 line is thought to be a rare type of breast cancer positive for myoepithelial markers [20,41] that is heterogeneously described as metaplastic [41], sarcomatoid, or spindle cell carcinomas [20,42]. The incidence of this tumor type is reportedly 0.02% [43] to 0.2% [44], which is a likely reason for the absence of mesenchymal samples among the typical collection of breast cancer specimens [20]. The rarity of this subtype among clinical samples and the distinct genetic profile of this line create a problem with classification of MDA-MB-231 as a B-TNBC prototype. Ultimately, information derived from studies using this model may not be applicable to clinical B-TNBC.
With these limitations in mind, we sought to establish a bona fide B-TNBC model that would comply with the following requirements: 1) lack of ER/PR/HER2 as a general marker of all TNBC subtypes; 2) lack of vimentin that is expressed in myoepithelial but not basal epithelial cells [45]; 3) expression of basal cytokeratins 5, 6, 14, and 17 as clear evidence for basal origin [46]; 4) ability to grow at the orthotopic site in mice [i.e., mammary fat pad (MFP)]; 5) ability to spontaneously metastasize from the orthotopic site to LNs and visceral organs mimicking the high metastatic potential of B-TNBC [6,11]; 6) and lastly, ability to initially respond to cytotoxic therapy followed by rapid recurrence, as a well-documented feature of clinical B-TNBC [47].
After extensive search and several pilot studies with different lines, we determined that the HCC1806 cell line complies with all of the above requirements. This line was established from a tumor of a patient with TNBC [30] and was subsequently confirmed to lack ER/PR/HER2 [30] as well as p53 [48], another marker of aggressive breast cancers. The consistency of the HCC1806 genetic profile with the basaloid subgroup of TNBC type has been recently confirmed by a microarray study classifying a heterogeneous TNBC group into seven subtypes with defined molecular signatures and sensitivity to drugs [31]. Here, after verifying the basaloid nature of HCC1806 cell line in our laboratory, we genetically modified this line for expression of bioluminescent and fluorescent reporter genes, validated in vitro and in vivo its growth patterns, and quantitatively determined the metastatic incidence, burden, and spread to normal organs in mice. We also characterized the sensitivity of HCC1806 cells to commonly used anticancer drugs and analyzed the unique properties of blood and lymphatic vessels in HCC1806 tumors in vivo that might contribute to biologic behavior of B-TNBC. Lastly, we assessed the responsiveness of this new B-TNBC model to a combination of nab-paclitaxel and bevacizumab, a therapy that was highly successful in other models of aggressive breast cancer [36,37]. Collectively, these studies provide a detailed in vitro and in vivo characterization of a novel B-TNBC model that should be useful for defining unique biologic mechanisms causing patient mortality and for assessing effective therapies to counteract them.
Materials and Methods
Materials
Dulbecco's modified Eagle's medium (DMEM) and standard additives were obtained from Lonza (Basel, Switzerland). Ketamine and xylazine were purchased from Phoenix Scientific (St Joseph, MO). Endotoxin-free sterile 0.9% NaCl solution (saline) and protease inhibitors were from Sigma-Aldrich (St Louis, MO). Matrigel was from BD Biosciences (Franklin Lakes, NJ).
Study Drugs
Paclitaxel formulated as albumin-bound particles for injectable suspension (nab-paclitaxel, Abraxane) was obtained from Celgene, formerly Abraxis BioScience (Los Angeles, CA). Paclitaxel (Taxol) was obtained from Bristol-Myers Squibb (New York, NY). Perifosine was obtained from Cayman Chemical Company (Ann Arbor, MI). Doxorubicin was obtained from Teva Parenteral Medicines, Inc. (Irvine, CA). Cis-diammineplatinum(II) dichloride (cisplatin) was purchased from Sigma. Erlotinib (Tarceva) was obtained from OSI Pharmaceuticals (Melville, NY). Bevacizumab (Avastin), humanized anti-vascular endothelial growth factor-A (VEGF-A) antibody, manufactured by Genentech (San Francisco, CA), was obtained from a local pharmacy. Drugs were reconstituted in saline, prepared fresh daily, and used within 1 hour of preparation.
Antibodies
We used the following primary antibodies: rabbit anti-cytokeratin 5, 6, and 14 (Abcam, Cambridge, MA); rat anti-mouse MECA-32 and mouse anti-β-actin (Developmental Studies Hybridoma Bank, Iowa City, IA); rat anti-mouse LYVE-1 (R&D Systems, Minneapolis, MN); and rabbit anti-Renilla luciferase (Rluc; MBL, Woburn, MA). Secondary IR680-conjugated donkey anti-mouse and anti-rabbit antibodies (LI-COR, Lincoln, NE) and secondary DyLight 488- and DyLight 549-conjugated donkey anti-rabbit and anti-rat antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA).
Culture of Human HCC1806 Breast Carcinoma Cell Line and Its Derivatives
HCC1806 cell line was purchased from ATCC (Rockville, MD) and cultured in DMEM containing 5% FBS, 2 mM glutamine, 1 mM sodium pyruvate, and 1 mM nonessential amino acids at 37°C in 10% CO2. Cells were passaged biweekly by incubating for 5 minutes at 37°C first in 0.5 mM EDTA dissolved in Dulbecco phosphate-buffered saline (PBS) followed by 0.5% of trypsin. Cells were routinely tested for mycoplasma using immunodetection kit from Roche Diagnostics GmbH (Penzberg, Germany).
Total RNA Extraction and Reverse Transcription-Polymerase Chain Reaction and Reverse Transcription-Quantitative Polymerase Chain Reaction Analyses
Total RNA extracted by using TRI-reagent (Sigma-Aldrich) was reverse transcribed with RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas, Glen Burnie, MD) according to the manufacturer's instructions. Endpoint reverse transcription-polymerase chain reaction (RT-PCR) analysis was performed as previously described [49] using Apex TaqDNA Polymerase (Genesee Scientific, San Diego, CA). Gels were visualized and analyzed using a FluroChem 5500 imager (AlphaInnotech, San Leandro, CA). RT-quantitative PCR (RT-qPCR) was performed using Go Taq Green Master Mix (Promega, Madison, WI). Products were analyzed using ABI 7500 Real-Time System (Applied BioSystems, Foster City, CA). Data were normalized to β-actin and relative mRNA expression was determined using the ΔΔCt method. All primers used in this study are listed in Table W1.
Western Blot Analysis
MCF-7, MDA-MB-231, and HCC1806 cell pellets were lysed in ice-cold buffer [50 mM tris(hydroxymethyl)aminomethane-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate, phenylmethylsulfonyl fluoride (1:100), and protease inhibitor cocktail (1:50)]. Proteins separated in 12% sodium dodecyl sulfate-polyacrylamide gel were transferred onto nitrocellulose membranes followed by overnight incubation with primary antibodies against cytokeratins 5, 6, and 14 and β-actin. After a 1-hour incubation with IR680-conjugated secondary antibodies, blots were washed and imaged using Odyssey Infrared Imager 2.0 (LI-COR).
Generation and Characterization of HCC1806 Cells Tagged with Red Fluorescent Protein and Rluc
HCC1806 cells were seeded in a six-well plate at the density of 100,000 cells per well. Cells were allowed to attach overnight, washed, and fed with fresh medium before transfection. Cells were transfected with a mixture of plasmids in which pKT2/red fluorescent protein (RFP)-Renilla-neomycin, pKT2/CAAGS-ires-puro, and Sleeping Beauty (SB) transposase-encoding vectors were present at the ratio of 1:1:4, respectively. Plasmids were mixed with lipofectamine and the transfection mixture was incubated with cells overnight. The following day, cells were washed with Dulbecco PBS, fed with fresh growth medium, and allowed to recover for 48 hours before selection with puromycin (1 µg/ml) for 7 to 10 days. Puromycin-resistant cells were assessed for RFP expression by direct fluorescent microscopy and subcloned to obtain monoclonal isolates. HCC1806 clones demonstrating identical morphology and growth potential (calculated by doubling time) to the parental line were selected and designated as HCC1806-RR to indicate the expression with both RFP and Rluc. The selected clones were expanded and extensively tested for growth and metastatic distribution in vivo. Only those clones that had identical growth and metastatic patterns to the parental line were selected for further studies.
Determination of Rluc Activity in HCC1806-RR Subclones
Viable cells identified by trypan blue exclusion were serially diluted in 1.5-ml tubes to generate a standard curve ranging from 780 to 100,000 cells per tube. Cells were pelleted by centrifugation at 13,000 rpm for 10 minutes, resuspended in 150 µl of the passive lysis buffer (Promega), and placed on ice for 10 minutes before being cleared of insoluble material by additional centrifugation at 13,000 rpm for 10 minutes. Ten microliters of whole-cell lysate was added to 50 µl of Rluc substrate (Biosynth, Rietlistr, Switzerland), and luciferase activity was measured for 10 seconds using a single-tube luminometer (Berthold Technologies, Oakridge, TN). All measurements were done in triplicate and the results are presented as mean of relative light units per second (RLU/s) ± SD. Luciferase expression per cell was calculated by dividing the mean RLU/s by cell number in each tube.
Proliferation Assay of Parental and HCC1806-RR Lines In Vitro
HCC1806 parental or HCC1806-RR cells (15,000/well) were seeded in triplicate into 24-well plates supplemented with 500 µl of growth medium in triplicate wells for each time point. Cells were trypsinized and counted every 24 hours for 96 hours. The results are presented as the mean values ± SD of the two independent experiments performed. The difference in proliferation between the parental and HCC1806-RR cell lines was assessed by a Student's t test considering P values less than .05 as significant.
Determination of In Vitro Sensitivity of MDA-MB-231 and HCC1806 Cell Lines to Commonly Used Anticancer Drugs
MDA-MB-231 and HCC1806-RR cells were seeded into a 24-well plate at a density of 50,000 cells per well in a volume of 500 µl of growth medium. After overnight incubation to allow for attachment without drugs, cells were treated with a range of doses of nab-paclitaxel, paclitaxel, perifosine, doxorubicin, cisplatin, erlotinib, or bevacizumab. Viable cell counts were determined after 48 hours of exposure to the drugs using trypan blue exclusion method. Each point was performed in duplicate and all experiments were repeated twice. The dose-response curve was constructed on the basis of the number of cells that survived after a 48-hour exposure to each drug concentration. The half-maximal inhibitory concentration (IC50) of each drug that was calculated on the basis of this curve represents a drug concentration that kills 50% of the cells as compared with untreated or vehicle-treated control cells.
Characterization of Tumor Growth of HCC1806 and HCC1806-RR Lines In Vivo
Four million HCC1806 parental or HCC1806-RR cells suspended in a solution of 50% Matrigel in DMEM (100 µl) were orthotopically implanted in the MFP of 4- to 6-week-old female nu/nu mice (Harlan, Indianapolis, IN), as previously described [36,37,49]. Every 2 to 3 days, perpendicular tumor diameters were measured by digital caliper and used to calculate tumor volume according to the formula: volume = Dd2π/6, where D equals the larger diameter and d equals the smaller diameter. The care, drug treatment, and all procedures including sacrifice of the animals were performed in accordance with the protocols approved by the Laboratory Animal Care and Use Committee of Southern Illinois University School of Medicine.
Assessment of Bioluminescence of Primary Tumors and Metastatic Lesions
Mice were injected intravenously (i.v.) with 2 mg/kg Rluc substrate, colenterazine (Biotium, Hayward, CA). The substrate was allowed to circulate for approximately 5 minutes before anesthetizing mice with a ketamine/xylazine cocktail. Bioluminescence was detected using Xenogen Ivis Lumina (Caliper Life Sciences, Hopkinton, MA) for 3 to 10 minutes. After imaging, mice were killed, and all major organs were collected for quantitative assessment of metastatic burden.
Determination of Metastatic Burden
Tissues were homogenized in ice-cold passive lysis buffer (Promega) containing protease inhibitors. Rluc substrate (50 µl) was mixed with lysates (10 µl) followed by luminescence detection using a luminometer (Berthold Technologies). Luminescence detected for the buffer alone was subtracted from the results. Data are expressed as the mean RLU/s ± SD from duplicate readings normalized per milligram of total protein determined by Bradford assay. Extracts with luciferase activity of 1500 RLU/s above background were considered positive for metastasis. This value was based on established correlation between the luciferase activity of the lysates and the ability to identify metastatic lesions by tissue staining with tumor-specific markers.
Immunohistochemical Analysis of Blood Vessels, Lymphatic Vessels, and Rluc-tagged Tumor Cells
Staining was performed on 8-µm-thick sections that were produced from snap-frozen HCC1806-RR tumors. Slides were equilibrated to room temperature for 20 minutes, fixed in acetone for 10 minutes, and rehydrated in PBS with 0.1% Tween 20. Antibodies against Rluc and mouse LYVE-1 were diluted 1:100, and MECA-32 hybridoma conditioned medium was used undiluted. All primary and secondary antibodies were incubated on the slides for 1 hour at 37°C and washed for 10 minutes in PBS with 0.1% Tween 20 after each incubation. After the last wash, slides were mounted in VectaShield medium containing 4,6′-diamidino-2-phenylindole nuclear stain (Vector Labs, Orton, Southgate, United Kingdom). Images were acquired on an Olympus upright BX41 microscope equipped with a DP70 digital camera and DP Controller software (Olympus, Center Valley, PA).
Measurement of Mean Vascular Area
The mean vascular area of MECA-32-positive staining per field was calculated as described [50], with slight modifications. Briefly, 200x magnified fluorescent images were acquired from MECA-32-stained tumor and normal MFP sections at a constant exposure using an Olympus BX41 microscope, as described above. Each experimental group was represented by four tumors or MFPs derived from individual mice (n = 4) from which four randomly selected images were acquired. The area of five to eight blood vessels in each image were measured using Image J software (http://rsbweb.nih.gov/ij/), and the values for each group were averaged. Twenty to thirty-two blood vessels were analyzed per group. The results are presented as the mean vascular area ± SEM.
Kinetics of VEGF-A Protein Secretion In Vitro Determined by ELISA
HCC1806-RR cells were seeded in a six-well plate at a density of 500,000 cells in 2 ml of growth medium. The cells were allowed to attach overnight before treatment with 10 nM nab-paclitaxel for 0, 2, 6, 24, 48, and 72 hours. Collected conditioned medium was concentrated 2.7-fold by low-speed centrifugation through Amicon concentrators with 10-kDa cutoff membrane (Millipore, Bellerica, MA). VEGF-A ELISA Kit was purchased from Peprotech (Rocky Hill, NJ) and performed as directed except that visualization of VEGF-A binding to antibody was detected by using the TMB development solution (Scytek, Logan, UT). All measurements were reproduced twice. Results are presented as the mean pg ± SD of VEGF-A normalized per 106 cells of four independently obtained values derived from two experiments performed in duplicate.
Assessment of Pro-survival Effect of VEGF-A on Paclitaxel-treated HCC1806-RR Cells
HCC1806-RR cells were seed in a 24-well plate at a density of 50,000 cells in 0.5 ml of growth medium. The cells were allowed to attach for 2 hours and then treated with VEGF-A isoform 121 (VEGF-A121 or V121; 10–200 ng/ml), VEGF-A isoform 165 (VEGF-A165 or V165; 10–200 ng/ml), or bevacizumab (5 µg/ml) overnight. Cells were then treated with nab-paclitaxel (0.19–25 nM) for 72 hours. Cells were then trypsinized and enumerated. Each point was performed in duplicate and all experiments were repeated twice. Data were presented as the mean percent of untreated control ± SD.
Tumor Treatment with Nab-Paclitaxel and Bevacizumab In Vivo
Mice bearing established HCC1806-RR tumors with a mean volume of 150 mm3 were randomized into four groups (six to eight mice per group) and treated with saline (control), nab-paclitaxel alone (10 mg/kg, i.v., daily for five consecutive days), bevacizumab alone [4 mg/kg, intraperitoneally (i.p.), biweekly], or nab-paclitaxel combined with bevacizumab. Nab-paclitaxel course was given for one cycle. Bevacizumab treatment began 24 hours after the first nabpaclitaxel injection and continued for the study's duration. The control group received saline (0.1 ml), injected i.v. or i.p. on the same days as nab-paclitaxel and bevacizumab treatments, respectively. Mice were sacrificed when tumor volumes in the control group reached 1800 mm3. Metastasis was quantified by determining protein-normalized luciferase activity in tissue extracts of ipsilateral LNs (ILNs), contralateral LN (CLN), and lungs.
Statistical Analysis
Statistical analysis was performed using SPSS 14.0 (SPSS, Chicago, IL). Results are expressed as mean ± SE. Statistically significant differences in tumor volume among experimental groups were determined by Student's t test. Statistically significant differences in incidence and burden of metastases were determined by Fisher exact and Wilcoxin ranked sums tests, respectively. The P values less than .05 were considered as significant for each test.
Results
HCC1806 Cell Line Expresses Typical Markers of B-TNBC
Few cell models that effectively recapitulate clinical B-TNBC are available, limiting our understanding of TNBC biology and ability to assess potential therapies. Despite findings that several cell lines display the clinical hallmarks of TNBC [31] (i.e., the absence of ER, PR, and HER2), none of the available lines have been characterized in details for consistency with other markers of B-TNBC as well as the ability to mimic typical clinical manifestations of this subgroup. We sought to establish a new model that both faithfully represented the unique molecular features of B-TNBC and mimics its biologic behavior in vivo.
During the search for this model, we identified HCC1806 as a cell line that had been established from a patient with TNBC and subsequently confirmed to lack hormonal receptors and HER2 [30]. To determine whether the HCC1806 line displayed distinct markers of the B-TNBC subgroup, we analyzed HCC1806 cells by RT-PCR and RT-qPCR for the expression of hormonal receptors, HER2, and luminal, basal, and mesenchymal markers (Figure 1, A–E). Additionally, protein expression of basal cytokeratins 5, 6, and 14 was assessed by Western blot (Figure 1F). The profile of HCC1806 was compared with profiles of MCF-7 line representing luminal A subtype [2] and MDA-MB-231 line currently considered as a representative of human B-TNBC [32,33,38].
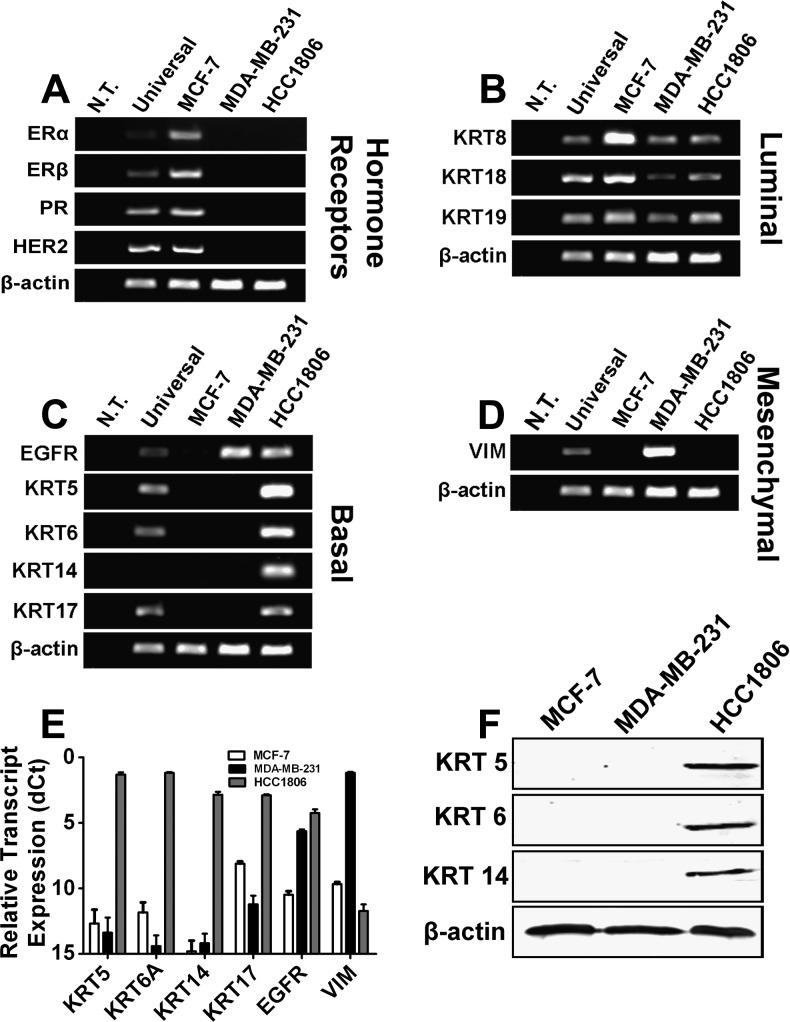
Figure 1.
Selection of a cell line that represents B-TNBC. MCF-7, MDA-MB-231, and HCC1806 were screened by endpoint RT-PCR for expression of (A) hormone receptors and HER2, (B) luminal cytokeratins, (C) basal cytokeratins, and (D) vimentin, a mesenchymal marker. (E) Basal markers and vimentin were confirmed by RT-qPCR and normalized to β-actin presented as dCt ± SE. (F) Protein expression of basal cytokeratins 5, 6, and 14 was also confirmed by Western blot analysis in which β-actin demonstrated equal protein loading.
Consistent with prior report [31], MCF-7 expressed hormonal receptors and HER2, whereas both MDA-MB-231 and HCC1806 lines lacked these receptors (Figure 1A). Luminal cytokeratins 8, 18, and 19 were expressed in all three lines with MCF-7 demonstrating the highest expression compared with MDA-MB-231 and HCC1806 (Figure 1B). EGFR, a signaling receptor with propensity to be overexpressed in B-TNBC [3], was detected in both MDA-MB-231 and HCC1806 lines (Figure 1C), indicating some commonality in gene expression between two lines. In sharp contrast, well-documented markers of B-TNBC such as basal cytokeratins 5, 6, 14, and 17 [17–19,51] were present exclusively in HCC1806 cells but not in MCF-7 or MDA-MB-231 cells (Figure 1, C and E). This has been confirmed in both using RT-PCR and RT-qPCR assays and independent pairs of primers (Table W1), suggesting that HCC1806 (but not MDA-MB-231) cell line is derived from the basaloid origin. This was further confirmed by analysis of the expression of a bona fide mesenchymal marker vimentin that was present at high levels in MDAMB- 231 cells but not in HCC1806 or MCF-7 (Figure 1D). This was further confirmed by Western blot analysis demonstrating protein expression of basal cytokeratins 5, 6, and 14 in HCC1806 but not in MDA-MB-231 cells (Figure 1F). These data show that although MDA-MB-231 and HCC1806 lines share some features of TNBC, only the latter expresses all typical markers of the B-TNBC. Conversely, MDA-MB-231 is more accurately classified as a mesenchymal or mesenchymal stem-like TNBC line [31] rather than basal-like, as suggested by some previous studies [33,38].
Generation of HCC1806 Sublines Co-expressing RFP and Rluc
Because we confirmed that HCC1806 cell line displays all molecular features of B-TNBC, we selected this line for establishing an in vivo model. The main goal of this part of the study was to assess the potential of HCC1806 cells to linearly expand at the orthotopic site (i.e., MFP in mouse), spontaneously metastasize from the MFP, respond initially to cytotoxic therapy [29,47], and recur at high rate [52].
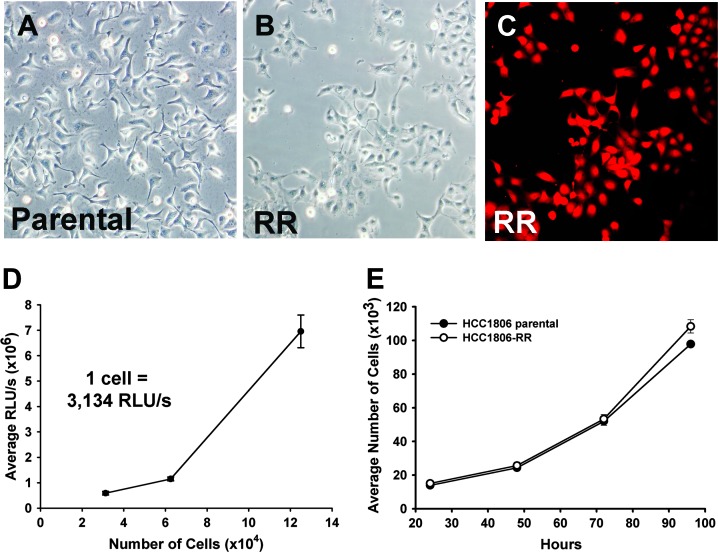
Because the sensitivity of tumor cell detection is the key parameter for establishment of a metastatic model, we aimed to employ stable expression of two sensitive fluorescent and bioluminescent reporters achieved using nonviral, DNA integrating vector system, SB transposon [53]. To this end, a transgenic cell line was engineered to co-express RFP and Rluc and referred to as HCC1806-RR. We preferred to engineer the subline with Rluc as alternative to the frequently used Firefly luciferase because the light production by Rluc is ATP-independent [54], which eliminates a potential problem with inaccurate reflection of tumor and metastatic burden because of necrosis and reduced tumor metabolism [55]. HCC1806 cells (1 x 105 per well) were co-transfected with the following plasmids: a SB transposon encoding for RFP and Rluc both transcriptionally regulated by a synthetic bidirectional promoter (pKT2/RBiL, 500 ng); SB transposase (500 ng), and a limited amount of a second transposon that conferred resistance to puromycin (pKT2/Puro, 10 ng) to permit clonal selection. Two days after transfection, cells were plated into puromycin supplemented medium, and selection continued for 2 weeks with medium changes every 3 to 4 days. At this point, RFP-expressing and puromycin-resistant clones were isolated and replated at limiting dilution to derive 10 to 12 subclones that were determined to have typical HCC1806 cellular morphology (Figure 2, A and B), uniform expression of RFP (Figure 2C), and very high expression of Rluc activity as reflected by >3000 RLU/s per cell (Figure 2D). Several monoclonal isolates with these characteristics were then tested for cell proliferation in vitro. Only those sublines that showed an identical pattern and doubling time to the parental line (Figure 2E) were selected for further studies.
Figure 2.
Generation and characterization of HCC1806-RR in vitro. HCC1806-RR line was generated using the SB transposon system by a co-transfection of transposon plasmids encoding for dual expression of RFP and Rluc or a puromycin resistance gene in combination with SB transposase. (A, B) Morphology of expanded monoclonal cell populations was compared to the parental cell line by bright-field microscopy. (C) Monoclonal isolates with identical morphology were then assessed for homogenous RFP expression using fluorescent microscopy. (D) Average RLU/s per cell was calculated to ensure a linear correlation with luciferase activity measured in triplicate and expressed as the mean RLU/s per cell ± SD. (E) All selected clones were assessed for unaltered growth pattern as compared with the parental cell line. Each time point was performed in triplicate, repeated twice, and presented as the mean cell number ± SD. Student's t test analysis showed no statistical difference between the proliferation rates of the HCC1806-RR clone and the parental line.
HCC1806-RR Cells Exhibit Varying Levels of Sensitivity to Commonly Used Chemotherapeutic Drugs
After verifying the basal nature of HCC1806 and characterizing its tagged cell derivatives, we sought to establish the profile of drug sensitivity of this model in vitro. Clinical studies indicate that different TNBC subgroups are initially sensitive to cytotoxic drugs; however, the recurrence rate of B-TNBC is typically higher than that of other groups [14,52]. Hence, it is of interest to compare the drug sensitivity of basaloid with other TNBC subtypes because this information could be useful for tailoring therapeutic approaches specifically for this cancer type.
To initiate these studies, we compared the drug sensitivity of a mesenchymal and a basaloid subtype of TNBC represented by MDA-MB-231 and HCC1806 (and -RR derivatives) cell lines, respectively. To determine responsiveness to commonly used anticancer drugs, 5 x 104 MDA-MB-231 or HCC1806-RR cells were seeded into wells of 24-well plates and allowed to adhere overnight before supplementing the medium with escalating doses of (A) nanoparticle-bound paclitaxel (nab-paclitaxel), (B) paclitaxel, (C) perifosine, (D) doxorubicin, (E) cisplatin, or (F) erlotinib (Figure 3). In addition, we also tested the sensitivity to anti-VEGF-A antibody, bevacizumab, as it is used in conjunction with some cytotoxic drugs. Viable cells remaining after 48 hours were enumerated to construct dose-response curves (Figure 3, A–F) that were used to determine the IC50 for each agent (Table 1).
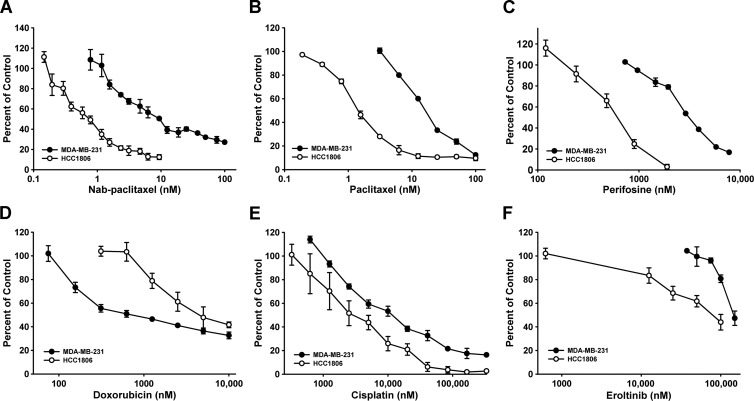
Figure 3.
Sensitivity of HCC1806-RR to various chemotherapeutics. MDA-MB-231 and HCC1806-RR lines were assessed for sensitivity to the following chemotherapeutic drugs: (A) nab-paclitaxel, (B) paclitaxel, (C) perifosine, (D) doxorubicin, (E) cisplatin, and (F) erlotinib. Each point represents the average percent of control ± SD derived from two independent experiments performed in duplicates.
Table 1.
In Vitro Sensitivity of TNBC Lines to Chemotherapeutic Drugs.
| Drug | Mode of Action | IC50 (nM) | Fold Change in IC50 | |
| MDA-MB-231 | HCC1806* | MDA-MB-231 vs HCC1806 | ||
| Nab-paclitaxel | Microtubule stabilizer | 10 | 0.78 | 12.82 |
| Paclitaxel | Microtubule stabilizer | 12 | 0.78 | 15.38 |
| Perifosine | MAPK inhibitor | 3,500 | 600 | 5.83 |
| Doxorubicin | DNA cross linker | 625 | 5,000 | 0.12 |
| Cisplatin | DNA intercalator | 10,000 | 2,500 | 4.00 |
| Erlotinib | EGFR inhibitor | 150,000 | 75,000 | 2.00 |
| Bevacizumab | Anti-VEGF-A antibody | No effect | No effect | N/A† |
MAPK indicates mitogen-activated protein kinase.
HCC1806 and the line derivatives HCC1806-RR isolates showed identical sensitivity to all tested drugs.
N/A denotes not applicable.
The parental HCC1806 and -RR lines showed no differences in drug sensitivity, indicating that tagging this line with RFP and Rluc did not alter natural responsiveness to chemotherapeutic agents. In contrast, comparison of MDA-MB-231 and HCC1806 lines showed drastic differences in this regard. Although both lines were sensitive to paclitaxel and nab-paclitaxel, the IC50 for taxanes of MDA-MB-231 cells exceeded that of HCC1806 by 12.8-fold (Table 1), suggesting that B-TNBC might be more sensitive to taxanes than mesenchymal breast cancer subtype. The trend for increased sensitivity to cytotoxic drugs of B-TNBC line was evident for all tested drugs with the exception of doxorubicin (Figure 3D and Table 1). HCC1806 cells were also fairly sensitive to perifosine (the mitogen-activated protein kinase inhibitor) with IC50 of 600 nM (Figure 3C and Table 1). In contrast, two other cytotoxic drugs, anthracycline doxorubicin and the alkylating agent cisplatin, were significantly less potent for both mesenchymal and B-TNBC lines with IC50 ranging between 2500 and 5000 nM (Figure 3, D and F, and Table 1). These IC50 were 1000- to 3000-fold higher than IC50 for taxane-based drugs. Finally, erlotinib and bevacizumab, the inhibitors of EGFR and VEGF-A, respectively, were largely ineffective in all tested concentrations at inhibiting growth of either TNBC lines in culture (Figure 3F and Table 1).
This analysis demonstrated that both subtypes of TNBC are most sensitive to taxanes, relatively resistant to doxorubicin and cisplatin, and completely insensitive to erlotinib and bevacizumab. This study also showed that mesenchymal and basaloid prototypes of TNBC significantly differ in sensitivity to chemodrugs as evidenced by 2- to 12-fold differences in IC50 to individual agents (Table 1). This finding further underscores the importance of characterizing TNBC subgroups to identify drug sensitivity and chemoresistance mechanisms that could be specific for each group.
Orthotopic HCC1806-RR Tumors Show Linear Kinetics and Spontaneous Metastasis to LN and Lungs
After establishing the basic parameters of HCC1806-RR line in vitro, we next determined the growth pattern and metastatic potential of these cells in vivo. To this end, we first screened several monoclonal HCC1806-RR isolates for the ability to grow at the MFP at a similar rate to the parental line. This was done to ensure that high expression levels of RFP and Rluc did not alter in vivo growth properties. Only those clones that showed an identical growth pattern to unmodified HCC1806 line were selected for the further studies. One example of such clone is shown in Figure 4A.
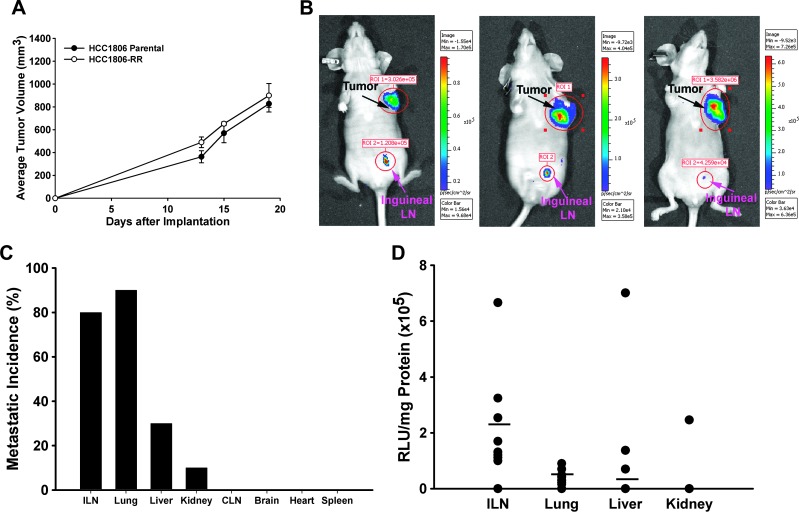
Figure 4.
Characterization of HCC1806-RR tumor growth and metastasis in vivo. (A) Parental HCC1806 and HCC1806-RR tumors were implanted into the MFP of female nu/nu mice. Tumors were measured two to three times per week, and each point represents the average tumor volume ± SEM. Analysis of tumor growth of parental and modified lines by Student's t test showed no statistically significant differences. (B) Tumors bearing orthotopic HCC1806-RR were injected i.v. with 2 mg/kg Rluc substrate (coelenterazine) that was allowed to circulate for 5 minutes. Then, mice were immobilized and imaged for 3 to 5 minutes. The black and pink arrows indicate the primary tumor and the positive inguinal LN, respectively. (C) All major organs were tested for the presence of metastasis by measuring Rluc activity. Each bar represents the percentage of metastatic incidence to respective organ in the group of 10 mice. (D) The metastatic burden was measured by Rluc activity normalized per mg of total protein, as described under Materials and Methods. Each black dot represents an animal testing positive for tumor burden from the indicated organs. The black bar indicates the mean burden in each organ for the entire group (n = 10).
Modification of HCC1806-RR cells with Rluc provided an opportunity to use whole-body imaging to monitor tumor burden and metastasis in live animals or a luminometer to quantify metastasis in organ homogenates after sacrifice. The in vivo growth potential of HCC1806-RR was monitored by injecting animals with coelenterazine (2 mg/kg) i.v. and exposing them to a charged-coupled device (CCD) camera (Xenogen-100) 5 minutes later. Rluc activity was detected as light emitted from the tumor cells and acquired as a pseudo-color image superimposed over a black and white photograph of the animal. All mice (n = 5) demonstrated very high Rluc activity at the primary site with the majority of mice simultaneously showing metastases to inguinal ILNs. Representative images of mice bearing HCC1806-RR tumors are shown in Figure 4B.
We next determined metastatic distribution by measuring protein-normalized Rluc activity in all major organs including LN, heart, lung, liver, kidney, spleen, and brain. Consistent with clinical observations, the highest metastatic incidence was in ILNs and lungs that were positive in 80% and 90% of the mice (n = 10), respectively (Figure 4C). Liver and kidney were minor metastatic sites detected in 20% and 10% of the mice. Other organs were negative at the time of analysis when the average tumor volume was approximately 1100 mm3. Analysis of metastatic burden per organ showed the highest burden in ILN (2.34 x 105 ± 0.60 x 105 RLU/mg), followed by pulmonary metastases with the mean value of 0.43 x 105 ± 0.08 x 105 RLU/mg of total lung tissue protein (Figure 4D).
These studies demonstrate that the HCC1806-RR model mimics the major clinical manifestations of metastatic breast cancer in human patients including the ability to linearly expand at the mammary site and spontaneously spread to ipsilateral regional LNs and to lungs from the orthotopic site.
HCC1806 Tumors and Their Derivatives Induce Highly Abnormal Vasculature
As recapitulated by the HCC1806-RR model (Figure 4), breast cancers have the propensity to metastasize to the proximate LNs followed by metastasis to distant organs. The lymphatic metastasis is influenced by tumor-induced lymphangiogenesis [56], whereas systemic metastasis is determined, in part, by specific properties of tumor blood vessels [57]. To the best of our knowledge, no previous study has examined the structural properties of B-TNBC although such analysis can provide insights into mechanisms of the high metastatic potential and frequent recurrence associated with this cancer group [29,47]. The novel HCC1806-RR B-TNBC model established here provided us with an opportunity to characterize the type, location, and structural properties of blood and lymphatic vessels induced by this tumor in vivo.
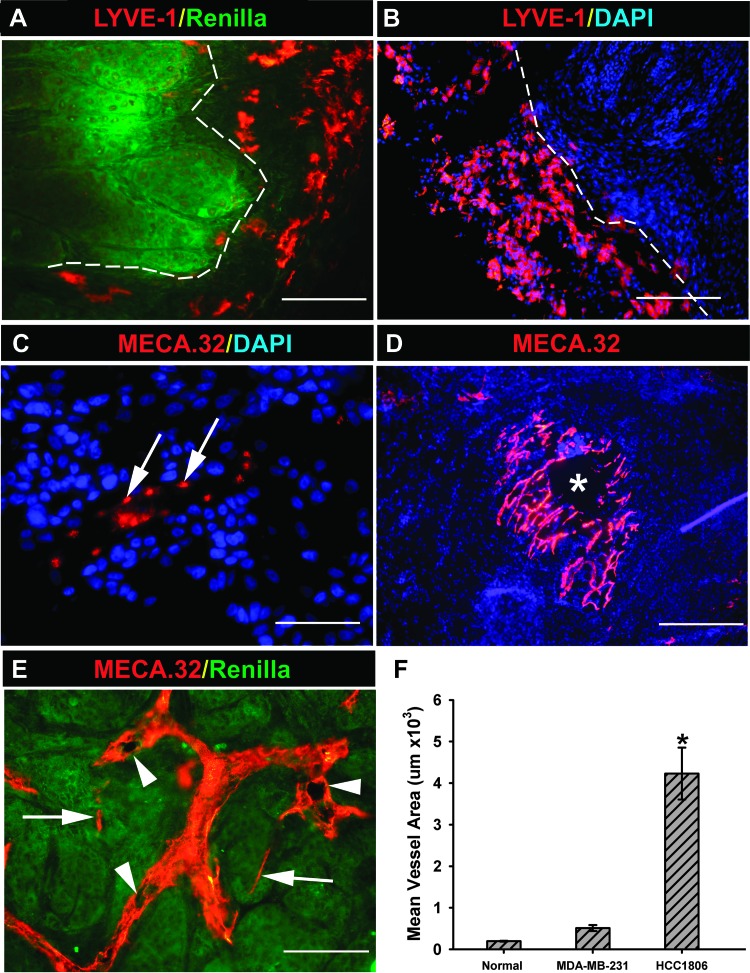
Because the lymphatic metastasis is dominant in this model, we first characterize lymphatic vessels using an antibody against a lymphatic marker, LYVE-1. Some tumor sections were co-stained with antibody against Rluc to determine the sustainability of this marker expression in vivo and for demarcation of tumor-host borders. Figure 5A shows that Rluc was homogeneously expressed in all tumor cells (green), which permitted clear identification of tumor margins. Co-staining with anti-LYVE-1 antibody revealed the absence of intratumoral lymphatic vessels in all HCC1806 or HCC1806-RR tumors examined (n = 10), although high density of LYVE-1+ structures was found in the host tissue adjacent to the tumor border (Figure 5A). Additionally, the peritumoral space was populated by numerous single cells and clusters positive for LYVE-1 (Figure 5B). These observations suggest that breast tumors modeled by HCC1806 metastasize to LNs exclusively through peritumoral lymphatic vessels whose generation might be aided by LYVE-1-positive progenitors mobilized to the tumor-host interface.
Figure 5.
Characterization of lymphatic and blood vessels in HCC1806-RR model. (A) The staining for Rluc expression (green) homogeneously identified all HCC1806-RR tumor cells and clearly demarcated a tumor-host interface border denoted by the white dotted line. Co-staining for LYVE-1 showed that all lymphatic vessels (red) are located in the peritumoral space and do not penetrate the tumor mass. This image was acquired at 200x. (B) In addition to lymphatic vascular structures, the peritumoral space was heavily populated by LYVE-1+ single cells (red) that were mobilized to tumor margins but did not penetrate the tumor. This image was acquired at 100x. (C) The center of the tumor was devoid of blood vascular structures, but individual cells stained positively for MECA-32 (red and denoted by white arrows). This image was acquired at 400x. (D) MECA-32 vascular structures were mainly located at the periphery of the tumor and were composed of large disorganized plexuses. These vessels may have reduced functionality, as suggested by necrosis frequently observed at the center of most plexuses (indicated by lack of blue 4,6′-diamidino-2-phenylindole staining and denoted by a white asterisk). This low-magnification image was acquired at 40x. (E) Additionally, MECA-32.positive blood vessels (red) were hyperdilated and exhibited various structural malformations including multiple lumens (white arrowheads). Note the substantial differences in size and shape between the hyperdilated and malformed vessel (white arrowheads) and few “normal type” blood capillaries (white arrows) that co-exist in the same tumor region. This image was taken at magnification of 200x. (F) The total area of blood vessels was measured using Image J software based on four images acquired from four different tissue samples of normal MFP, MDA-MB-231 tumors, and HCC1806-RR tumors. The asterisk indicates a statistically significant difference measured by Student's t test (P < .01) between the mean total vascular area in HCC1806-RR tumors and that in MDA-MB-231 tumors or normal MFP.
We next examined the blood vessels induced by HCC1806 tumors using a pan-endothelial marker of blood vascular endothelium, MECA-32. This staining revealed three striking abnormalities of the HCC1806-induced blood vasculature. First, in all examined tumors (n = 10), the tumor center was nearly devoid of MECA-32+ vascular structures being replaced by clusters of MECA-32+ isolated cells sporadically appearing throughout the central region (Figure 5C, white arrows). Second, the extensive blood vascular network present at the tumor periphery was chaotically arranged in the loosely connected plexuses (Figure 5D) that neither resembled the organized network in the normal MFP nor vascular pattern in other tumors (e.g., MDA-MB-231). These plexuses were surrounded by avascular space and might only be partly functional, as indicated by a necrotic spot appearing at its center (Figure 5D, asterisk). Third, many blood vessels were extremely dilated and have various morphologic deformities including multiple lumens (Figure 5E, white arrowheads). The severe structural disturbances in the HCC1806 tumor-induced blood vasculature were particularly conspicuous when compared side by side with normally sized and shaped capillaries that occasionally were present in the same field (Figure 5E, compare structures indicated by arrows and arrowheads). Although dilation of blood vessels is a known feature of tumor vasculature [58], HCC1806 tumors induce extraordinarily dilated vessels. This is demonstrated by the fact that the total area of vascular network of HCC1806 tumors was 8-fold larger than that of MDA-MB-231 tumors (4.16 ± 0.62 µm2 vs 0.51 ± 0.07 µm2) and 21-fold larger than of the normal MFP (Figure 5F). Collectively, these observations point out to clear pathophysiological differences between HCC1806 and MDA-MB-231 tumor vasculature and underscore the unique vascular properties of B-TNBC that might be critical for promoting metastatic spread, impeding drug delivery, and supporting tumor recurrence after initial responsiveness to the drugs.
HCC1806-RR Cells Express Low Levels of VEGF-A and VEGF-A Receptors that Are Upregulated by Nab-Paclitaxel
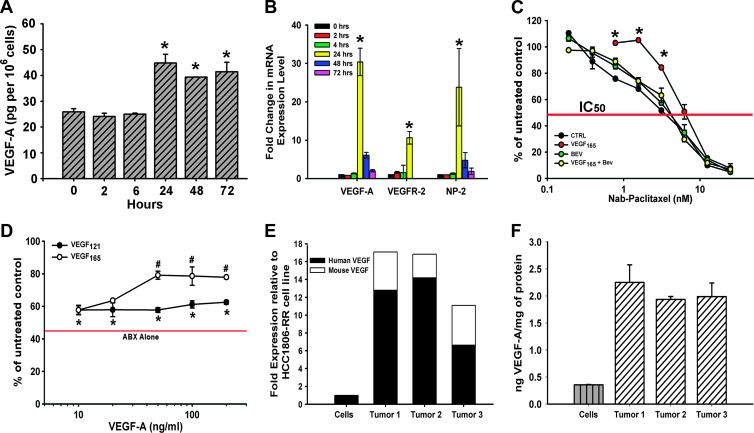
The structural abnormalities of HCC1806-induced blood vessels suggested that this tumor line might be deficient in the production of essential angiogenic factors such as VEGF-A [59]. To test this hypothesis, we determined secretion of VEGF-A protein in HCC1806 cells cultured for 24 hours in growth medium. We found that both HCC1806 and HCC1806-RR produce an average of 26 ± 3 pg of VEGF-A normalized per 106 cells (Figure 6A, time 0). This value is 12-fold lower than that determined for MDA-MB-231 cells under identical conditions [36], suggesting that a relatively low level of VEGF-A might be partly responsible for vascular deficiencies observed in HCC1806 tumors in vivo (Figure 5, C–E).
Figure 6.
The effect of nab-paclitaxel on VEGF-A expression in HCC1806-RR cells in vitro. (A) ELISA was used to measure VEGF-A protein in conditioned medium of untreated HCC1806-RR cells (0 hours) and those treated for 2, 6, 24, 48, and 72 hours with nab-paclitaxel (10 nM). Statistically significant increases in secreted VEGF-A protein were detected at 24, 48, and 72 hours after initiation of cytotoxic treatment. VEGF-A secretion was measured in pg and normalized per 1 million cells. Each ELISA was performed in duplicate, and the experiment was reproduced twice yielding the mean presented values ± SD. (B) Untreated and nab-paclitaxel.treated HCC1806-RR cells were also quantitatively assessed for VEGF-A, VEGFR-2, and NP-2 transcript levels by RT-qPCR. Asterisks at 24 hours indicate a significant up-regulation of mRNA for both VEGF-A and its receptors, VEGFR-2, and NP-2 at a 24-hour time point. RT-qPCR assays were performed in duplicate wells and repeated three times. Results are presented as the mean values from three experiments ± SD. (C) HCC1806-RR cells were treated with nab-paclitaxel (0.19.25 nM) alone or in combination with 10 ng/ml VEGF-A165, 5 µg/ml bevacizumab, or a combination of all three. Statistically significant pro-survival shift by exogenous VEGF-A165 is indicated by asterisk (P < .001). Each point was performed in duplicate wells in two independent experiments and is represented as the mean value ± SD. (D) The pro-survival benefit of VEGF-A121 and VEGF-A165 was compared under constant nab-paclitaxel treatment (5 nM). The red bar indicates the IC50 of nab-paclitaxel alone. Asterisk indicates a statistically significant increase in survival from nab-paclitaxel alone to nab-paclitaxel treated with either VEGF-A121 or VEGF-A165. The symbol “#” indicates a statistically significant increase in enhanced survival between VEGF-A121 and VEGF-A165. Each assay was performed in duplicate wells and in two independent experiments, and the results are represented as the mean value ± SD. (E) Human and mouse VEGF-A levels were measured by RT-qPCR from HCC1806-RR cells and from three HCC1806-RR tumors. The black portion of the bar indicates the fold increase in human VEGF-A expression compared to the cells. The white portion indicates the increase in mouse transcript levels compared to the human VEGF-A expression from cultured cells. Each assay was performed in duplicate wells and repeated three times. (F) Protein expression for VEGF-A was compared from HCC1806-RR cells and from three HCC1806-RR tumors. Each bar represents the mean ng/mg protein of VEGF-A. Each ELISA was performed in duplicate and was repeated twice. Each bar is the mean VEGF-A expression ± SD.
We previously showed that some breast carcinoma lines not only express VEGF-A receptors but also upregulate both VEGF-A and VEGFR-2 or neuropilins in response to paclitaxel therapy [36,37]. We, therefore, determined whether HCC1806 followed the similar response to cytotoxic drugs. To this end, we treated cells with 10 nM nab-paclitaxel and collected medium and mRNA at 2, 6, 24, 48, and 72 hours after drug addition, for VEGF-A ELISA and RT-qPCR assays, respectively. Both assays showed that VEGF-A and VEGFR-2 as well as neuropilin-2 (NP-2) were drastically increased after addition of the drug, in particular, at the 24-hour time point (Figure 6). At this time, we observed 10- to 30-fold increase in mRNA for VEGF-A and both receptors and nearly a two-fold increase in secreted VEGF-A protein (Figure 6B). This finding suggests that, analogous to our findings in other breast carcinoma lines [36,37], taxane therapy, while effective in killing the tumor cells, also induces coincident expression of VEGF-A and its receptors, thus potentially creating an autocrine prosurvival loop. This observation suggests that eliminating a blunting effect of VEGF-A with a neutralizing antibody may significantly improve the response of B-TNBC tumors to taxanes in vivo, as reported in other breast cancer models [37]. To further explore this hypothesis, we tested whether the effect of nab-paclitaxel is modulated by the addition of VEGF-A and negated by the addition of bevacizumab. We found that, indeed, addition of 10 ng/ml VEGF-A165 shifts the IC50 by approximately 300%, whereas bevacizumab (2 µg/ml) shifts the curve back to control demonstrating its capability to neutralize exogenous VEGF-A (Figure 6C).
We then asked the question whether this pro-survival effect is dose dependent and is equally endowed by the two major isoforms of VEGF-A, V165 and V121. The results showed that the pro-survival effect is dose dependent with maximal effect achieved at concentrations above 1 ng/ml and that V165 is significantly more efficacious than V121. At concentrations above 25 ng/ml, exogenous V165 afforded survival of 2.5-fold more cells as compared with nab-paclitaxel alone (Figure 6D). Cultured HCC1806 cells, however, produce significantly less VEGF-A (Figure 6, A and B), raising the question whether the pro-survival effect promoted by 10 to 100 ng/ml VEGF-A is relevant to in vivo conditions. To answer this question, we compared the amounts of mRNA and protein produced by HCC1806 cells in culture and by HCC1806 tumors in vivo. The results showed a significant difference in production of VEGF-A at both mRNA and protein levels in culture as opposed to in situ settings (Figure 6, E and F). When normalized per expression of mRNA of β-actin (Figure 6E) or mg of total protein (Figure 6F), in vivo grown tumors transcribed 11.2-fold more VEGF-A transcripts (the average of three tumor samples) and increased correspondingly VEGF-A protein production by 16.8-fold (n = 3). Given the nab-paclitaxel capacity to increase VEGF-A expression nearly 30-fold (Figure 6B), these results suggest that the local concentration of VEGF-A in the local environment of HCC1806-treated tumors can reach 20 to 50 ng/mg total protein. Therefore, it is plausible that VEGF-A induced by paclitaxel can play a deciding role in negating drug-mediated death leading to the hypothesis that anti-VEGF-A antibody can increase the efficacy of cytotoxic therapy.
Combination Nab-Paclitaxel/Bevacizumab Therapy Is Significantly More Efficacious in Suppression of HCC1806 Tumor Growth and Metastases Compared to Either Agent Alone
Because we observed paclitaxel-mediated up-regulation of VEGF-A in HCC1806 cells in vitro (Figure 6), we hypothesized that bevacizumab, an anti-VEGF-A antibody, should improve suppression of HCC1806 tumors when combined with cytotoxic therapy in vivo. It should be noted that expression of VEGF-A in cultured cells is limited by the absence of host cells and other elements of the tumor environment that increase VEGF-A production nearly 20-fold compared to cells cultured in vitro (Figure 6). This observation strengthens our hypothesis that is further supported by previously reported success using this combined treatment in both MDA-MB-231 and MDA-MB-435 models [36,37]. Moreover, synergistic effect of bevacizumab and paclitaxel therapy has been widely reported in a variety of other cancer models [37,60,61] and clinical trials [62,63]. However, combination of nab-paclitaxel and bevacizumab has not been previously assessed in a B-TNBC experimental model, and currently, it is not a routine therapeutic approach to patients with TNBC in clinics.
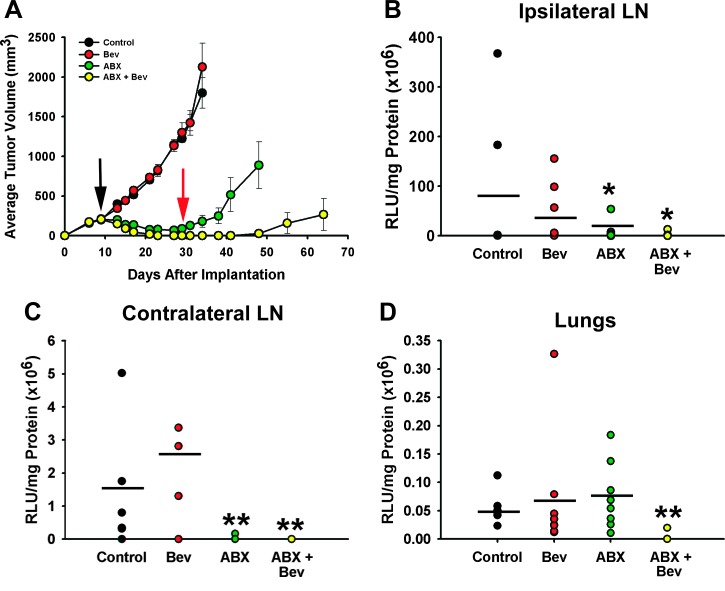
To determine the efficacy of the nab-paclitaxel/bevacizumab treatment in the HCC1806 model, we allowed HCC1806-RR tumors to reach ∼150 mm3 in volume before randomizing mice to four groups (seven to eight mice each) and treating them with saline (control), nab-paclitaxel alone (10 mg/kg, i.v., daily for five consecutive days), bevacizumab alone (4 mg/kg, i.p., biweekly), or nab-paclitaxel in combination with bevacizumab. Drug response was monitored by monitoring tumor volume measured every 3 days (Figure 7A and Table 2). Tumors formed in saline-treated mice reached maximal volume in 38 days with a growth rate of 56.2 ± 4.4 mm3 per day. Mimicking the IC50 study in vitro, bevacizumab alone had no effect on tumor growth, whereas nab-paclitaxel alone reduced the growth rate by nearly five-fold (Figure 7A and Table 2). Nab-paclitaxel reduced tumor volume up to 90% as compared with saline-treated controls (P < .001, Table 2). However, 100% of tumors (n = 8) recurred upon cessation of therapy (Figure 7A, second red arrow), a pattern reminiscent of clinical behavior of TNBC [14,29,52]. Significantly, nab-paclitaxel combined with bevacizumab resulted in a substantially better outcome as evidenced by 50% of complete responses (CRs) in a combination group (Table 2) as compared with 0% CR (i.e., 100% recurrence) in all other groups.
Figure 7.
The effect of nab-paclitaxel and bevacizumab combination therapy on tumor growth and metastasis in HCC1806-RR model in vivo. (A) HCC1806-RR tumors were implanted into the MFP and monitored two to three times per week. Each line represents the mean daily tumor volume ± SE. The black and red arrows indicate the beginning of treatment and recurrence after nab-paclitaxel treatment, respectively. (B) ILN, (C) CLN, and (D) lungs were assessed for metastatic burden. Each dot represents the metastatic burden for individual mice in each group as measured by luciferase activity normalized per mg of total protein. The results are expressed as mean RLU/mg protein. The black bar indicates the mean metastatic burden for each group (n = 10). Statistically significant differences in metastatic burden were identified by a Wilcoxon ranked sums test (*P < .05, **P < 0).
Table 2.
Effect of Combination Therapy on Growth of HCC1806-RR Tumors In Vivo.
| Treatment | N | Drug Dose (mg/kg) | Mean Tumor Volume (mm3) | % Inhibition of Control | TGD* vs Control | % CR† | P value vs | |
| Control | ABX | |||||||
| Control | 7 | - | 1799 ± 169 | - | - | 0 | - | - |
| Bev‡ | 8 | 4 | 2126 ± 297 | 0 | 0 | 0 | NS‡ | - |
| ABX‡ | 8 | 10 | 180 ± 55 | 90 | >28 | 0 | <.001 | - |
| ABX + Bev | 8 | 10/4 | 0 | 100 | >28 | 50 | <.001 | .024 |
Tumor growth delay (TGD) is defined as number of days that delayed the mean tumor volume per group reaching 1000 mm3 as compared with the saline-treated control groups.
CR was defined as the absence of palpable tumor at the original tumor injection site at the end of the experiment.
Bev, ABX, and ABX + Bev denote treatment with bevacizumab, nab-paclitaxel, and combination of the two drugs, respectively. NS stands for not significant.
We next compared the efficacy of combined drugs and monotherapies on metastatic incidence and burden at two major metastatic sites identified in the earlier studies (Figure 4). ILN and pulmonary metastases were detected in 100% of saline-treated mice, whereas CLN metastases were detected in 71% of the control group. ILN in the control mice had the highest metastatic burden (79 x 106 ± 143 x 106 RLU/mg protein), followed by burden in CLN and lungs with 1.6 x 106 ± 1.9 x 106 and 0.05 x 106 ± 0.02 x 106 RLU/mg protein, respectively. This burden was largely unaffected by bevacizumab alone, whereas nab-paclitaxel alone significantly reduced the amount of metastases in LNs but not in lungs (Figure 7). Once again, combination therapy was more efficacious than a cytotoxic or an antiangiogenic drug alone as demonstrated by a 20.7-fold and a 2.7-fold reduction in burden in the ILN and lungs, respectively. In comparison, nab-paclitaxel alone reduced burden in ILN by 7.1-fold but had no effect on the burden on pulmonary site. Bevacizumab alone reduced ILN burden by less than 2-fold and had no impact on metastatic growth in other sites.
Importantly, combination therapy was efficacious not only in reducing the burden of metastases but also their incidence. The incidence in ILN, CLN, and lungs was reduced to 50%, 0%, and 13% with P values ranging from .007 to .001 (Table 3), indicating a statistically significant benefit of nab-paclitaxel/bevacizumab therapy in eradicating both regional and distant metastatic lesions. In comparison, bevacizumab alone had no effect on incidence in any of the three metastatic sites, and nab-paclitaxel alone was only effective in suppressing metastasis to CLNs but not to other sites (Table 3).
Table 3.
The Effect of Combination Therapy on Metastatic Incidence of HCC1806-RR Tumors.
| Treatment | Drug Dose (mg/kg) | Incidence N/Total (%) | P Value vs | |
| Control | ABX | |||
| ILNs | ||||
| Control | - | 7/7 (100) | - | - |
| Bev* | 4 | 8/8 (100) | NS* | - |
| ABX* | 10 | 6/8 (75) | NS | - |
| ABX + Bev | 10/4 | 4/8 (50) | .077 | NS |
| CLN | ||||
| Control | - | 5/7 (71) | - | - |
| Bev | 4 | 3/8 (38) | NS | - |
| ABX | 10 | 1/7 (13) | .041 | - |
| ABX + Bev | 10/4 | 0/8 (0) | .007 | NS |
| Lung | ||||
| Control | - | 7/7 (100) | - | - |
| Bev | 4 | 8/8 (100) | NS | - |
| ABX | 10 | 8/8 (100) | NS | - |
| ABX + Bev | 10/4 | 1/8 (13) | .001 | .001 |
Bev, ABX, and ABX + Bev denote treatment with bevacizumab, nab-paclitaxel, and combination of the two drugs, respectively. NS stands for not significant.
In summary, we showed here for the first time that a moderate dose of nab-paclitaxel (10 mg/kg) in combination with bevacizumab (4 mg/kg) was efficacious against B-TNBC as demonstrated by complete sustainable regressions of primary tumors in half of the treated animals and 50% to 100% reduction in incidence of mice with metastatic organs. These data suggest that a similar strategy can be beneficial for treating human patients with B-TNBC.
Discussion
B-TNBC is one of the most aggressive subtypes of breast tumors associated with high incidence of recurrence and metastasis and, consequently, with reduced rates of patient survival [10,11,13,14]. The main challenges to successful B-TNBC treatment are the lack of surface targets (i.e., ER and HER2) for available molecular therapies and subtype-specific genetic alterations that promote rapid recurrence after a seemingly successful therapy [6,13,14,28]. Despite the undisputable need for better understanding of the unique B-TNBC biology and identification of new targets for treatment, most currently available animal models do not adequately mimic this phenotype. We, therefore, sought to establish a new B-TNBC model that faithfully recapitulates both the genetic signature of B-TNBC and its main clinical manifestations.
To this end, we first identified a bona fide human TNBC cell line that expresses basal markers and exhibits biologic behavior in vivo consistent with the phenotype of clinical B-TNBC. Next, we engineered a subline with stable expression of RFP and Rluc (i.e., HCC1806-RR) to enable sensitive in vivo detection and quantification of tumor growth and metastases. We validated that the HCC1806-RR line had identical morphology, growth, and metastasis patterns to those of the parental line. Next, we compared the drug sensitivity of this line with MDA-MB-231, a line that is currently thought to represent TNBC [33,34,38,39]. This was followed by a detailed characterization of the growth, metastasis, and vascular patterns in orthotopic HCC1806-RR tumors in vivo. Lastly, using the validated B-TNBC model, we discovered that this type of tumor may recover from cytotoxic therapy through induction of autocrine pro-survival VEGF-A·VEGFR loop, a finding that provides evidence-based rationale for combining cytotoxic and anti-VEGF-A therapies. Collectively, this study presents a new metastasis-quantifiable B-TNBC model that displays essential features of the corresponding human disease and is suitable for in-depth molecular analyses as well as for testing new treatments.
Currently, the MDA-MB-231 cell line is the most frequently described TNBC model in more than 50 publications (e.g., as presented in [39,40,64,65]). This is based on evidence showing that this line lacks ER, PR, and HER2 [32,66] while being positive for a TNBC marker, EGFR [20,66]. Both of these properties have been confirmed in the present study (Figure 1). However, on the basis of independent hierarchical clustering analyses [20], MDA-MB-231 cells express vimentin and a variety of other mesenchymal-specific markers [20], classifying this line as mesenchymal [20] or mesenchymal stem-like [31] subgroup. This classification is supported by our findings demonstrating lack of expression of bona fide basal markers such as cytokeratins 5, 6, 14, and 17 (Figure 1), a cluster of basal epithelial proteins typically detected in clinical specimens of B-TNBC [3,6,18,19]. Moreover, several studies suggested that the genetic profile of MDA-MB-231 line is consistent with a rare group of breast tumors occurring in less than 0.2% of patients [44] that is classified as metaplastic, sarcomatoid, or spindle carcinoma of the breast [20,41,42]. On the basis of this information and independent marker analysis shown here (Figure 1), we concluded that MDA-MB-231, albeit being a convenient line for studies of tumor lymphangiogenesis and lymphatic metastasis [36,37], may not be an ideal model for analyzing biologic behavior of clinical B-TNBC.
Hence, the primary goal of this work was to establish a new model that reproduces both the molecular signature and the principal clinical manifestations of B-TNBC. HCC1806 was identified in our preliminary studies as a line that complied with the criteria for establishing such a model (Table 4). Not only does the HCC1806 line lack ER, PR, and HER2, an essential, but insufficient requirement for modeling B-TNBC, it also does not express mesenchymal markers (e.g., vimentin) while being positive for all four basal cytokeratins 5, 6, 14, and 17 (Figure 1). In contrast, MDA-MB-231 line is positive for vimentin and lacks all basal cytokeratins (Figure 1 and Table 4). Moreover, biologically, these two lines display drastic differences in sensitivity to chemotherapeutic drugs (Figure 3 and Table 1) with HCC1806 cells being 2- to 12-fold more sensitive compared with MDA-MB-231. This is consistent with paradoxical behavior of clinical B-TNBC that is reported to be fairly sensitive to cytotoxic treatments despite the subsequent heightened recurrence rate [6,13,14,28]. Both of these parameters were reproduced in the in vivo HCC1806 model that, albeit being more sensitive to taxanes than MDA-MB-231 by three to four-fold [36,37], quickly recurs after chemotherapy within 4 to 5 days (Figure 7A). The fact that these biologic characteristics are consistent with the behavior of clinical B-TNBC [29,47] supports the validity of the selected line and indicates the potential to use this model for dissecting mechanisms of recurrence.
Table 4.
Representation of Salient Features of Clinical B-TNBC by Cell Culture Models.
| Hallmarks of Clinical B-TNBC | MDA-MB-231 | HCC1806-RR |
| Marker Expression | ||
| Negative for ER, PR, HER2/Neu |  |
 |
| Positive for EGFR |  |
 |
| Negative for vimentin |  |
 |
| Positive for basal cytokeratins 5, 6, 14, and 17 |  |
 |
| Clinical Parameters | ||
| Lymphatic metastasis |  |
 |
| Hematogenous metastasis |  |
 |
| Initial sensitivity to chemotherapy |  |
 |
| Recurrence after treatment |  |
 |
Another nonclinical advantage of the HCC1806-RR model is its dual labeling with Rluc and RFP (Figures 2, 4, and 5), which provides a unique tool set for assessing growth, metastasis, identification, and quantification of human tumor cells in vivo by immunohistochemistry, fluorescence, and bioluminescence methodologies. We validated in both culture and in vivo settings that HCC1806-RR had unaltered morphology and growth identical to the parental cells (Figures 2E and 4A). We regard this as an important, and often overlooked, requirement for a reliable experimental cancer model because, in our hands, cell line modifications with stable expression of nonmammalian proteins such as GFP, RFP, or luciferases may interfere with basic properties of these lines. We, therefore, took precautions to ensure that the selected derivatives do not deviate in any observable properties from the parental cells. Some advantages of HCC1806-RR model are illustrated in Figures 2, 4, and 5: Double labeling provides two independent measures for monitoring tumor growth, Rlucmediated ATP-independent [54] noninvasive monitoring of metastases (Figure 4B), their quantifiable burden, incidence and organ distribution, and a clear demarcation of tumor border juxtaposed to peritumoral tissue of the host.
We took an advantage of this model to initiate studies on HCC1806 tumor vasculature whose unique features may contribute to chemoresistance, recurrence, and metastasis associated with B-TNBC. To our knowledge, no other study had examined the specific properties of TNBC-induced vessels and compared them with those in other tumors or normal tissues. We discovered that although the HCC1806 model features prominent lymphatic metastasis (Figure 4 and Table 3), none of the examined tumors had intratumoral LYVE-1+ lymphatic vessels (Figure 5, A and B). This is consistent with growing evidence from preclinical [67,68] and clinical studies [69,70], indicating the sufficiency of peritumoral lymphatic vessels to deliver tumor cells to regional LNs [71]. Most tumors displayed not only LYVE-1+ vascular structures but also accumulation of LYVE-1+ single cells at the tumor-host interface (Figure 5B). Although the identity of these cells would require an additional study, their peritumoral location and interaction with lymphatic vessels suggest tumor-induced recruitment of lymphatic endothelial cell progenitors reminiscent of macrophage-derived progenitors mobilized to sites of cancer [72] and inflammation [73]. If this observation is generalized to clinical B-TNBC, it might provide an opportunity to repress lymphatic metastasis by interfering with generation of peritumoral lymphatics.
We also examined the blood vasculature in HCC1806 and HCC1806-RR tumors. Both types of tumors induce an identical and highly abnormal blood vascular network characterized by dissociated MECA-32+ endothelial cells (Figure 5C), highly heterogeneous distribution of blood vessels within the tumor mass, highly irregular vascular plexuses located intermittently with long stretches of avascular areas (Figure 5D), structural deformities illustrated by multilumen channels (Figure 5E, arrowheads), and extraordinarily dilated vessels with area exceeding that of the normal MFP vasculature by 21-fold (Figure 5F). These abnormalities supersede even those induced by MDA-MB-231 tumor vessels that, in turn, considerably differ from vessels in normal mammary tissue. Because this is the first time that B-TNBC vasculature has been analyzed in either clinical specimens or an experimental model, we do not know whether HCC1806 vascular abnormalities are typical for clinical B-TNBC or an idiosyncrasy of HCC1806 tumor model. However, if vascular dilation and other structural vascular irregularities are confirmed in clinical B-TNBC, this might explain the propensity to metastasize [14], which can be enhanced by disruption of the endothelial barrier. Irregular vessel distribution and vascular abnormalities can also explain, in part, the initial sensitivity to chemodrugs because of vulnerability of poorly perfused tumor tissue that develops necrosis even without treatment with cytotoxic drugs (Figure 5D, asterisk). Clearly, the comparison of the described vascular features to those in clinical B-TNBC might open a new platform for understanding unique patterns of chemoresistance, recurrence, and metastasis in this type of tumors.
Lastly, we used this new model for assessing potential benefits of nab-paclitaxel/bevacizumab combination therapy. We have previously shown in two aggressive models of breast cancer (MDA-MB-231 and MDA-MB-435) that addition of anti-VEGF-A antibody to nabpaclitaxel significantly increases the curative potential of cytotoxic therapy [37]. This combined therapy (but not each component alone) eradicated large MDA-MB-231 tumors (>500 mm3) along with preexisting LN and lung metastases in more than 70% of the mice [37]. We showed that the likely mechanism for synergistic action may involve taxane-induced stress response that hyperactivates the NF-κB pathway leading to upregulated VEGF-A and its receptors [37]. We hypothesized that a similar mechanism may exist in HCC1806 tumor cells. Examination of VEGF-A and its receptor transcripts showed that while nab-paclitaxel kills some tumor cells (Figure 3) it coincidently upregulates VEGF-A, VEGFR-2, and NP-2 in surviving cells by 10- to 30-fold (Figure 6B). Accordingly, VEGF-A protein in the conditioned medium of nab-paclitaxel-treated tumor cells was nearly doubled as compared with controls (Figure 6A). These data are in line with our previous findings in MDA-MB-231 and MDA-MB-435 models [37], suggesting a common mechanism for DNA damage stress response responsible for generating a VEGF-A-mediated autocrine loop that blunts response to cytotoxic therapy. The autocrine mechanism is, however, difficult to analyze in culture because in the absence of host cells and three-dimensional environment, HCC1806 cells manipulated in vitro secrete 15- to 20-fold less VEGF-A than corresponding tumors grown in vivo (Figure 6, D–F). In addition, the host cells add roughly a third to the half of VEGF-A measured in tumors (Figure 6E), further complicating the distinction between the autocrine and paracrine mechanisms of VEGF-A-dependent promotion of tumor growth. This is because VEGF-A receptors expressed on human malignant cells equally bind both human and mouse-produced protein. Given these experimental challenges, we can only suggest that both autocrine and paracrine modes of VEGF-A action are likely to contribute to resistance to paclitaxel therapy although the exact contribution of each of these mechanisms cannot be deduced from these studies.
Nevertheless, we demonstrate here in the new HCC1806-RR model that the combination therapy reduced the recurrence rate by half as compared with all other groups (Figure 7 and Table 2). Importantly, the incidence of positive LNs and lungs was also reduced in the combined therapy group, respectively, by 50% and 87% (Table 3). These findings obtained in the validated B-TNBC model strongly advocate for including bevacizumab, or similar VEGF-A neutralizing agent, as a routine adjuvant for taxane-based therapy.
Conclusions
In summary, we present evidence for generation of a novel model for human B-TNBC that grows at the MFP and spontaneously metastasizes from the orthotopic site in mice. We provide a detailed characterization of this model in both in vitro and in vivo settings. Our data show many advantages of this model including close representation of molecular signature of clinical B-TNBC, its propensity to recur, and high metastatic potential to LNs and lungs. These properties that create significant challenges in clinics might be addressed in the future by analyzing the biologic mechanisms underlying the aggressive phenotype of TNBC. Additionally, a double-labeled HCC1806 system is fully validated for quantitative analysis of tumor growth and metastasis in vivo, which should facilitate assessment of new therapies. We, therefore, believe that the HCC1806-RR model will be broadly used for unraveling unique B-TNBC biology and designing effective strategies for eradication of this disease.
Supplementary Material
Acknowledgments
We thank Dr Vuong Trieu for supplying nab-paclitaxel and Shelly Reeter for tissue sectioning and immunostaining.
Abbreviations
- ABX
abraxane, nanoparticle albumin-embedded paclitaxel
- BEV
bevacizumab, anti-human VEGF-A antibody
- B-TNBC
basaloid triple-negative breast cancer
- CLN
contralateral lymph node
- CR
complete response
- DPBS
Dulbecco phosphate-buffered saline
- ER
estrogen receptor
- EGFR
epidermal growth factor receptor
- HCC1806-RR
HCC1806 breast carcinoma line tagged with red fluorescent protein and Renilla luciferase
- IC50
half-maximal inhibitory concentration
- ILN
ipsilateral lymph nodes
- LN
lymph node
- MFP
mammary fat pad
- NT
no template control
- PBS
phosphate-buffered saline
- PBST
PBS with 0.1% Tween 20
- RFP
red fluorescent protein
- Rluc
Renilla luciferase
- RLU/s
relative light units per second
- SD
standard deviation
- SEM
standard error of the mean
- V121 (VEGF-A121)
vascular endothelial growth factor-A isoform 121
- V165 (VEGF-A165)
vascular endothelial growth factor-A isoform 165
Footnotes
This study was supported in part by Celgene (former Abraxis BioScience, Los Angeles, CA; generation of tagged HCC1806 cell line) and by NIH/NCI grant 2R01CA140732 awarded to Sophia Ran. The authors declare no competing interests.
This article refers to supplementary material, which is designated by Table W1 and is available online at www.neoplasia.com.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, vandeRijn RM, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 4.Dawson SJ, Provenzano E, Caldas C. Triple negative breast cancers: clinical and prognostic implications. Eur J Cancer. 2009;45(suppl 1):27–40. doi: 10.1016/S0959-8049(09)70013-9. [DOI] [PubMed] [Google Scholar]
- 5.Dolle JM, Daling JR, White E, Brinton LA, Doody DR, Porter PL, Malone KE. Risk factors for triple-negative breast cancer in women under the age of 45 years. Cancer Epidemiol Biomarkers Prev. 2009;18:1157–1166. doi: 10.1158/1055-9965.EPI-08-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nofech-Mozes S, Trudeau M, Kahn HK, Dent R, Rawlinson E, Sun P, Narod SA, Hanna WM. Patterns of recurrence in the basal and non-basal subtypes of triple-negative breast cancers. Breast Cancer Res Treat. 2009;118:131–137. doi: 10.1007/s10549-008-0295-8. [DOI] [PubMed] [Google Scholar]
- 7.Trivers KF, Lund MJ, Porter PL, Liff JM, Flagg EW, Coates RJ, Eley JW. The epidemiology of triple-negative breast cancer, including race. Cancer Causes Control. 2009;20:1071–1082. doi: 10.1007/s10552-009-9331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawood S, Broglio K, Kau SW, Green MC, Giordano SH, Meric-Bernstam F, Buchholz TA, Albarracin C, Yang WT, Hennessy BT, et al. Triple receptor-negative breast cancer: the effect of race on response to primary systemic treatment and survival outcomes. J Clin Oncol. 2009;27:220–226. doi: 10.1200/JCO.2008.17.9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stead LA, Lash TL, Sobieraj JE, Chi DD, Westrup JL, Charlot M, Blanchard RA, Lee JC, King TC, Rosenberg CL. Triple-negative breast cancers are increased in black women regardless of age or body mass index. Breast Cancer Res. 2009;11:R18. doi: 10.1186/bcr2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mersin H, Yildirim E, Berberoglu U, Gulben K. Triple negative phenotype and N-ratio are important for prognosis in patients with stage IIIB non-inflammatory breast carcinoma. J Surg Oncol. 2009;100:681–687. doi: 10.1002/jso.21411. [DOI] [PubMed] [Google Scholar]
- 11.Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res. 2009;7:4–13. doi: 10.3121/cmr.2009.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heitz F, Harter P, Lueck HJ, Fissler-Eckhoff A, Lorenz-Salehi F, Scheil-Bertram S, Traut A, du Bois BA. Triple-negative and HER2-overexpressing breast cancers exhibit an elevated risk and an earlier occurrence of cerebral metastases. Eur J Cancer. 2009;45:2792–2798. doi: 10.1016/j.ejca.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 13.Dent R, Hanna WM, Trudeau M, Rawlinson E, Sun P, Narod SA. Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res Treat. 2009;115:423–428. doi: 10.1007/s10549-008-0086-2. [DOI] [PubMed] [Google Scholar]
- 14.Anders C, Carey LA. Understanding and treating triple-negative breast cancer. Oncology (Williston Park) 2008;22:1233–1239. [PMC free article] [PubMed] [Google Scholar]
- 15.Dawood S, Broglio K, Esteva FJ, Yang W, Kau SW, Islam R, Albarracin C, Yu TK, Green M, Hortobagyi GN, et al. Survival among women with triple receptor-negative breast cancer and brain metastases. Ann Oncol. 2009;20:621–627. doi: 10.1093/annonc/mdn682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rakha EA, Ellis IO. Triple-negative/basal-like breast cancer: review. Pathology. 2009;41:40–47. doi: 10.1080/00313020802563510. [DOI] [PubMed] [Google Scholar]
- 17.Rakha EA, Elsheikh SE, Aleskandarany MA, Habashi HO, Green AR, Powe DG, El-Sayed ME, Benhasouna A, Brunet JS, Akslen LA, et al. Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res. 2009;15:2302–2310. doi: 10.1158/1078-0432.CCR-08-2132. [DOI] [PubMed] [Google Scholar]
- 18.van de Rijn RM, Perou CM, Tibshirani R, Haas P, Kallioniemi O, Kononen J, Torhorst J, Sauter G, Zuber M, Kochli OR, et al. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol. 2002;161:1991–1996. doi: 10.1016/S0002-9440(10)64476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasa M, Bando Y, Takahashi M, Hirose T, Nagao T. Screening for basal marker expression is necessary for decision of therapeutic strategy for triple-negative breast cancer. J Surg Oncol. 2008;97:30–34. doi: 10.1002/jso.20906. [DOI] [PubMed] [Google Scholar]
- 20.Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adelaide J, Cervera N, Fekairi S, Xerri L, Jacquemier J, Birnbaum D, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–2284. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 21.Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, Perou CM. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 22.Grushko TA, Blackwood MA, Schumm PL, Hagos FG, Adeyanju MO, Feldman MD, Sanders MO, Weber BL, Olopade OI. Molecular-cytogenetic analysis of HER-2/neu gene in BRCA1-associated breast cancers. Cancer Res. 2002;62:1481–1488. [PubMed] [Google Scholar]
- 23.Armes JE, Trute L, White D, Southey MC, Hammet F, Tesoriero A, Hutchins AM, Dite GS, McCredie MR, Giles GG, et al. Distinct molecular pathogeneses of early-onset breast cancers in BRCA1 and BRCA2 mutation carriers: a population-based study. Cancer Res. 1999;59:2011–2017. [PubMed] [Google Scholar]
- 24.Umemura S, Takekoshi S, Suzuki Y, Saitoh Y, Tokuda Y, Osamura RY. Estrogen receptor-negative and human epidermal growth factor receptor 2-negative breast cancer tissue have the highest Ki-67 labeling index and EGFR expression: gene amplification does not contribute to EGFR expression. Oncol Rep. 2005;14:337–343. [PubMed] [Google Scholar]
- 25.Jacquemier J, Ginestier C, Rougemont J, Bardou VJ, Charafe-Jauffret E, Geneix J, Adelaide J, Koki A, Houvenaeghel G, Hassoun J, et al. Protein expression profiling identifies subclasses of breast cancer and predicts prognosis. Cancer Res. 2005;65:767–779. [PubMed] [Google Scholar]
- 26.Sabatier R, Jacquemier J, Bertucci F, Esterni B, Finetti P, Azario F, Birnbaum D, Viens P, Goncalves A, Extra JM. Peritumoural vascular invasion: amajor determinant of triple-negative breast cancer outcome. Eur J Cancer. 2011;47:1537–1545. doi: 10.1016/j.ejca.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Liu HT, Ma R, Yang QF, Du G, Zhang CJ. Lymphangiogenic characteristics of triple negativity in node-negative breast cancer. Int J Surg Pathol. 2009;17:426–431. doi: 10.1177/1066896909337505. [DOI] [PubMed] [Google Scholar]
- 28.Kaplan HG, Malmgren JA, Atwood M. T1N0 triple negative breast cancer: risk of recurrence and adjuvant chemotherapy. Breast J. 2009;15:454–460. doi: 10.1111/j.1524-4741.2009.00789.x. [DOI] [PubMed] [Google Scholar]
- 29.Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 30.Gazdar AF, Kurvari V, Virmani A, Gollahon L, Sakaguchi M, Westerfield M, Kodagoda D, Stasny V, Cunningham HT, Wistuba II, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer. 1998;78:766–774. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 31.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu IM, Michalowski AM, Hoenerhoff M, Szauter KM, Luger D, Sato M, Flanders K, Oshima A, Csiszar K, Green JE. GATA3 inhibits lysyl oxidase-mediated metastases of human basal triple-negative breast cancer cells. Oncogene. 2011:2017–2027. doi: 10.1038/onc.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murrow LM, Garimella SV, Jones TL, Caplen NJ, Lipkowitz S. Identification of WEE1 as a potential molecular target in cancer cells by RNAi screening of the human tyrosine kinome. Breast Cancer Res Treat. 2010;122:347–357. doi: 10.1007/s10549-009-0571-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buchholz S, Seitz S, Schally AV, Engel JB, Rick FG, Szalontay L, Hohla F, Krishan A, Papadia A, Gaiser T, et al. Triple-negative breast cancers express receptors for luteinizing hormone-releasing hormone (LHRH) and respond to LHRH antagonist cetrorelix with growth inhibition. Int J Oncol. 2009;35:789–796. doi: 10.3892/ijo_00000391. [DOI] [PubMed] [Google Scholar]
- 36.Volk LD, Flister MJ, Bivens CM, Stutzman A, Desai N, Trieu V, Ran S. Nab-paclitaxel efficacy in the orthotopic model of human breast cancer is significantly enhanced by concurrent anti-vascular endothelial growth factor A therapy. Neoplasia. 2008;10:613–623. doi: 10.1593/neo.08302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volk LD, Flister MJ, Chihade D, Desai N, Trieu V, Ran S. Synergy of nab-paclitaxel and bevacizumab in eradicating large orthotopic breast tumors and preexisting metastases. Neoplasia. 2011;13:327–338. doi: 10.1593/neo.101490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Wang H, Oosterwijk E, Tu C, Shiverick KT, Silverman DN, Frost SC. Expression and activity of carbonic anhydrase IX is associated with metabolic dysfunction in MDA-MB-231 breast cancer cells. Cancer Invest. 2009;27:613–623. doi: 10.1080/07357900802653464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dolai S, Xu Q, Liu F, Molloy MP. Quantitative chemical proteomics in small-scale culture of phorbol ester stimulated basal breast cancer cells. Proteomics. 2011;11:2683–2692. doi: 10.1002/pmic.201000801. [DOI] [PubMed] [Google Scholar]
- 40.Li Q, Chen C, Kapadia A, Zhou Q, Harper MK, Schaack J, LaBarbera DV. 3D models of epithelial-mesenchymal transition in breast cancer metastasis: high-throughput screening assay development, validation, and pilot screen. J Biomol Screen. 2011;16:141–154. doi: 10.1177/1087057110392995. [DOI] [PubMed] [Google Scholar]
- 41.Leibl S, Moinfar F. Metaplastic breast carcinomas are negative for Her-2 but frequently express EGFR (Her-1): potential relevance to adjuvant treatment with EGFR tyrosine kinase inhibitors? J Clin Pathol. 2005;58:700–704. doi: 10.1136/jcp.2004.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaguchi R, Horii R, Maeda I, Suga S, Makita M, Iwase T, Oguchi M, Ito Y, Akiyama F. Clinicopathologic study of 53 metaplastic breast carcinomas: their elements and prognostic implications. Hum Pathol. 2010;41:679–685. doi: 10.1016/j.humpath.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Singal R, Singh P, Sahu P, Mittal A, Naredi B, Gupta S. Metaplastic carcinoma of the breast with high grade spindle cell component with osteoid formation—a rare case report. Acta Chir Belg. 2011;111:243–245. [PubMed] [Google Scholar]
- 44.Honda M, Saji S, Horiguchi S, Suzuki E, Aruga T, Horiguchi K, Kitagawa D, Sekine S, Funata N, Toi M, et al. Clinicopathological analysis of ten patients with metaplastic squamous cell carcinoma of the breast. Surg Today. 2011;41:328–332. doi: 10.1007/s00595-009-4276-2. [DOI] [PubMed] [Google Scholar]
- 45.Guelstein VI, Tchypysheva TA, Ermilova VD, Litvinova LV, Troyanovsky SM, Bannikov GA. Monoclonal antibody mapping of keratins 8 and 17 and of vimentin in normal human mammary gland, benign tumors, dysplasias and breast cancer. Int J Cancer. 1988;42:147–153. doi: 10.1002/ijc.2910420202. [DOI] [PubMed] [Google Scholar]
- 46.Gusterson BA, Ross DT, Heath VJ, Stein T. Basal cytokeratins and their relationship to the cellular origin and functional classification of breast cancer. Breast Cancer Res. 2005;7:143–148. doi: 10.1186/bcr1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carey LA. Directed therapy of subtypes of triple-negative breast cancer. Oncologist. 2011;16(suppl 1):71–78. doi: 10.1634/theoncologist.2011-S1-71. [DOI] [PubMed] [Google Scholar]
- 48.Fan LZ, Cherian MG. Potential role of p53 on metallothionein induction in human epithelial breast cancer cells. Br J Cancer. 2002;87:1019–1026. doi: 10.1038/sj.bjc.6600549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitehurst B, Flister MJ, Bagaitkar J, Volk L, Bivens CM, Pickett B, Castro-Rivera E, Brekken RA, Gerard RD, Ran S. Anti-VEGF-A therapy reduces lymphatic vessel density and expression of VEGFR-3 in an orthotopic breast tumor model. Int J Cancer. 2007;121:2181–2191. doi: 10.1002/ijc.22937. [DOI] [PubMed] [Google Scholar]
- 50.Flister MJ, Volk LD, Ran S. Characterization of Prox1 and VEGFR-3 expression and lymphatic phenotype in normal organs of mice lacking p50 subunit of NF-κB. Microcirculation. 2011;18:85–101. doi: 10.1111/j.1549-8719.2010.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korsching E, Packeisen J, Agelopoulos K, Eisenacher M, Voss R, Isola J, van Diest PJ, Brandt B, Boecker W, Buerger H. Cytogenetic alterations and cytokeratin expression patterns in breast cancer: integrating a new model of breast differentiation into cytogenetic pathways of breast carcinogenesis. Lab Invest. 2002;82:1525–1533. doi: 10.1097/01.lab.0000038508.86221.b3. [DOI] [PubMed] [Google Scholar]
- 52.Linderholm BK, Hellborg H, Johansson U, Elmberger G, Skoog L, Lehtio J, Lewensohn R. Significantly higher levels of vascular endothelial growth factor (VEGF) and shorter survival times for patients with primary operable triple-negative breast cancer. Ann Oncol. 2009;20:1639–1646. doi: 10.1093/annonc/mdp062. [DOI] [PubMed] [Google Scholar]
- 53.Izsvak Z, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J Mol Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 54.Luker GD, Luker KE. Optical imaging: current applications and future directions. J Nucl Med. 2008;49:1–4. doi: 10.2967/jnumed.107.045799. [DOI] [PubMed] [Google Scholar]
- 55.Hockel M, Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst. 2001;93:266–276. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 56.Ran S, Volk L, Hall K, Flister MJ. Lymphangiogenesis and lymphatic metastasis in breast cancer. Pathophysiology. 2009;17:229–251. doi: 10.1016/j.pathophys.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hall K, Ran S. Regulation of tumor angiogenesis by the local environment. Front Biosci. 2010;15:195–212. doi: 10.2741/3615. [DOI] [PubMed] [Google Scholar]
- 58.Goel S, Duda DG, Xu L, Munn LL, Boucher Y, Fukumura D, Jain RK. Normalization of the vasculature for treatment of cancer and other diseases. Physiol Rev. 2011;91:1071–1121. doi: 10.1152/physrev.00038.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carmeliet P. VEGF as a key mediator of angiogenesis in cancer. Oncology. 2005;69(suppl 3):4–10. doi: 10.1159/000088478. [DOI] [PubMed] [Google Scholar]
- 60.Hu L, Hofmann J, Zaloudek C, Ferrara N, Hamilton T, Jaffe RB. Vascular endothelial growth factor immunoneutralization plus paclitaxel markedly reduces tumor burden and ascites in athymic mouse model of ovarian cancer. Am J Pathol. 2002;161:1917–1924. doi: 10.1016/S0002-9440(10)64467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fox WD, Higgins B, Maiese KM, Drobnjak M, Cordon-Cardo C, Scher HI, Agus DB. Antibody to vascular endothelial growth factor slows growth of an androgen-independent xenograft model of prostate cancer. Clin Cancer Res. 2002;8:3226–3231. [PubMed] [Google Scholar]
- 62.Sirohi B, Smith K. Bevacizumab in the treatment of breast cancer. Expert Rev Anticancer Ther. 2008;8:1559–1568. doi: 10.1586/14737140.8.10.1559. [DOI] [PubMed] [Google Scholar]
- 63.Lobo C, Lopes G, Silva O, Gluck S. Paclitaxel albumin-bound particles (abraxane) in combination with bevacizumab with or without gemcitabine: early experience at the University of Miami/Braman Family Breast Cancer Institute. Biomed Pharmacother. 2007;61:531–533. doi: 10.1016/j.biopha.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 64.Wong SW, Tiong KH, Kong WY, Yue YC, Chua CH, Lim JY, Lee CY, Quah SI, Fow C, Chung C, et al. Rapamycin synergizes cisplatin sensitivity in basal-like breast cancer cells through up-regulation of p73. Breast Cancer Res Treat. 2011;128:301–313. doi: 10.1007/s10549-010-1055-0. [DOI] [PubMed] [Google Scholar]
- 65.Morgan K, Meyer C, Miller N, Sims AH, Cagnan I, Faratian D, Harrison DJ, Millar RP, Langdon SP. GnRH receptor activation competes at a low level with growth signaling in stably transfected human breast cell lines. BMC Cancer. 2011;11:476. doi: 10.1186/1471-2407-11-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jonsson G, Staaf J, Olsson E, Heidenblad M, Vallon-Christersson J, Osoegawa K, de Jong JP, Oredsson S, Ringner M, Hoglund M, et al. High-resolution genomic profiles of breast cancer cell lines assessed by tiling BAC array comparative genomic hybridization. Genes Chromosomes Cancer. 2007;46:543–558. doi: 10.1002/gcc.20438. [DOI] [PubMed] [Google Scholar]
- 67.Bjorndahl MA, Cao R, Burton JB, Brakenhielm E, Religa P, Galter D, Wu L, Cao Y. Vascular endothelial growth factor-a promotes peritumoral lymphangiogenesis and lymphatic metastasis. Cancer Res. 2005;65:9261–9268. doi: 10.1158/0008-5472.CAN-04-2345. [DOI] [PubMed] [Google Scholar]
- 68.Dadiani M, Kalchenko V, Yosepovich A, Margalit R, Hassid Y, Degani H, Seger D. Real-time imaging of lymphogenic metastasis in orthotopic human breast cancer. Cancer Res. 2006;66:8037–8041. doi: 10.1158/0008-5472.CAN-06-0728. [DOI] [PubMed] [Google Scholar]
- 69.Ejlertsen B, Jensen MB, Rank F, Rasmussen BB, Christiansen P, Kroman N, Kvistgaard ME, Overgaard M, Toftdahl DB, Mouridsen HT. Population-based study of peritumoral lymphovascular invasion and outcome among patients with operable breast cancer. J Natl Cancer Inst. 2009;101:729–735. doi: 10.1093/jnci/djp090. [DOI] [PubMed] [Google Scholar]
- 70.Kwon Y, Ro J, Kang HS, Kim SK, Hong EK, Khang SK, Gong G, Ro JY. Clinicopathological parameters and biological markers predicting nonsentinel node metastasis in sentinel node-positive breast cancer patients. Oncol Rep. 2011;25:1063–1071. doi: 10.3892/or.2011.1157. [DOI] [PubMed] [Google Scholar]
- 71.Williams CS, Leek RD, Robson AM, Banerji S, Prevo R, Harris AL, Jackson DG. Absence of lymphangiogenesis and intratumoural lymph vessels in human metastatic breast cancer. J Pathol. 2003;200:195–206. doi: 10.1002/path.1343. [DOI] [PubMed] [Google Scholar]
- 72.Zumsteg A, Baeriswyl V, Imaizumi N, Schwendener R, Ruegg C, Christofori G. Myeloid cells contribute to tumor lymphangiogenesis. PLoS One. 2009;4:e7067. doi: 10.1371/journal.pone.0007067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hall KL, Volk-Draper LD, Flister MJ, Ran S. New model of macrophage acquisition of the lymphatic endothelial phenotype. PLoS One. 2012;7:e31794. doi: 10.1371/journal.pone.0031794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.