Summary
Expression of microRNAs (miRNAs) is under stringent regulation at both transcriptional and post-transcriptional levels. Disturbance at either level could cause dysregulation of miRNAs. Here we show that MLL fusion proteins negatively regulate production of miR-150, an miRNA widely repressed in acute leukemia, by blocking miR-150 precursors from being processed to mature miRNAs through MYC/LIN28 functional axis. Forced expression of miR-150 dramatically inhibited leukemic cell growth and delayed MLL-fusion-mediated leukemogenesis, likely through targeting FLT3 and MYB and thereby interfering with the HOXA9/MEIS1/FLT3/MYB signaling network, which in turn caused downregulation of MYC/LIN28. Collectively, we revealed a MLL-fusion/MYC/LIN28⊣miR-150⊣FLT3/MYB/HOXA9/MEIS1 signaling circuit underlying the pathogenesis of leukemia, where miR-150 functions as a pivotal gatekeeper and its repression is required for leukemogenesis.
Keywords: miR-150, MLL-associated leukemia, MYC, LIN28, FLT3, MYB, HOXA9, MEIS1, microRNA maturation, signaling axis, leukemogenesis
INTRODUCTION
MicroRNAs (miRNAs), a class of small, non-coding RNAs, are important for posttranscriptional gene regulation in both health and disease (He and Hannon, 2004; Xiao and Rajewsky, 2009). The stringent control of miRNAs, at both the transcriptional and posttranscriptional levels, is critical for maintaining a variety of important biological processes, including development, differentiation and hematopoiesis (Siomi and Siomi, 2010). After being transcribed by RNA polymerase II, the primary miRNA transcripts (pri-miRNAs) would be under a two-step bio-cleavage process. In the cell nucleus, the Microprocessor, containing an RNaseIII enzyme Drosha and its cofactor DGCR8, crops the pri-miRNA into a ~70 nucleotide (nt) hairpin structured precursor (pre-miRNA), which is then exported to the cytoplasm and cleaved by another RNaseIII enzyme, Dicer, to remove the “terminal loop region”, or pre-element (preE), and to yield the mature miRNA (Newman and Hammond, 2010). The transcriptional regulation of miRNAs has been extensively studied, but the current understanding of their post-transcriptional control in cancer is limited.
Lin28 is an RNA-binding protein known to block the maturation of let-7 and is an important modulator in the post-transcriptional regulation of miRNA maturation (Heo et al., 2008; Newman et al., 2008; Viswanathan et al., 2008). Lin28 itself is a direct downstream target of Myc (Chang et al., 2009). However, although MYC is aberrantly overexpressed in various types of cancer including lymphoma and leukemia (He et al., 2005; Hoffman et al., 2002; O’Donnell et al., 2005), the involvement of the MYC/LIN28 axis in the post-transcriptional regulation of miRNA maturation in hematopoietic malignancies, e.g., acute myeloid leukemia (AML), is poorly understood.
AML is a heterogeneous group of genetically diverse hematopoietic malignancies with variable response to treatment (Chen et al., 2010). Chromosome translocations are frequently observed in AML (Rowley, 2008). AML with chromosomal rearrangements involving the mixed lineage leukemia gene (MLL) is associated with a poor prognosis, especially in infants and individuals with leukemia that arises secondary to previous chemotherapy (Behm et al., 1996; Krivtsov and Armstrong, 2007; Rowley, 2008). More than 60 different loci that translocate to the MLL locus have been identified and cloned (Rowley, 2008). The critical feature of these chromosomal rearrangements is the generation of a chimeric transcript consisting of 5′ MLL and 3′ sequences of the partner gene, many of which are involved in transcriptional regulation. The human AF9 gene at 9p22 is one of the most common fusion partner genes with MLL (Krivtsov and Armstrong, 2007).
Several important oncogenes are known to be direct or indirect downstream targets of MLL-fusion proteins. Among those, homeobox A (HOXA) genes, MEIS1, FLT3, MYB and MYC are frequently up-regulated in MLL-associated leukemias (Armstrong et al., 2003; Armstrong et al., 2002; Schreiner et al., 2001; Zeisig et al., 2004). The HOXA cluster genes are direct targets of MLL (Milne et al., 2005a; Milne et al., 2005b; Yu et al., 1995) and MLL fusion proteins promote their expression by epigenetic mechanisms (e.g., H3K79 methylation) (Bernt et al., 2011; Faber et al., 2009; Krivtsov and Armstrong, 2007; Krivtsov et al., 2008). High expression of HOXA9 and its co-factor, MEIS1, was often found in MLL-associated leukemias (Armstrong et al., 2002; Bullinger et al., 2004; Faber et al., 2009). FLT3 and its downstream effectors (e.g., AKT, STAT5 and ERK) are broadly involved in multiple processes of hematopoiesis and leukemogenesis via regulating the expression of a group of targets, such as JUN and MYC (Takahashi, 2011). MYC is a transcription factor involved in cell proliferation and apoptosis and is upregulated in the FLT3-ITD-transduced CD34+ hematopoietic stem/progenitor cells (Li et al., 2007). MYB is an essential downstream target of HOXA9/MEIS1 signaling (Hess et al., 2006), and an autoregulatory feedback loop was reported recently in which MYB binds MLL through MENIN and regulates expression of HOXA9/MEIS1 directly (Jin et al., 2010).
As MYC is aberrantly overexpressed in various types of cancer including lymphoma and leukemia (He et al., 2005; Hoffman et al., 2002; O’Donnell et al., 2005), we tested the hypothesis that the MYC/LIN28 axis plays an essential role in AML in which MYC functions as an important downstream target of MLL fusions and FLT3 through post-transcriptional regulation of maturation of some critical tumor-suppressor miRNAs.
RESULTS
Expression of miR-150 is down-regulated in most AML
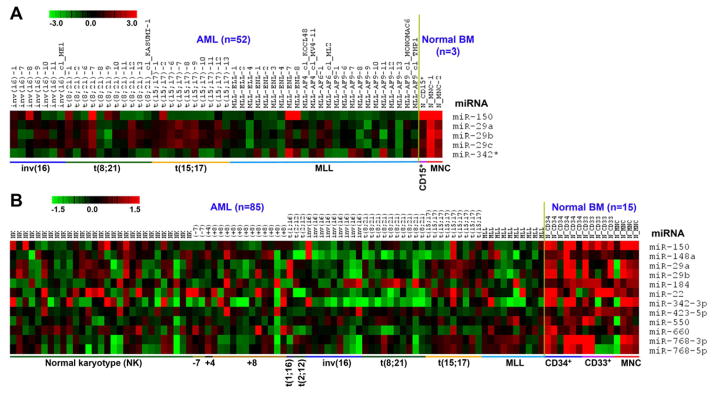
To identify potential tumor-suppressor miRNAs that are significantly down-regulated in AML, we performed a bead-based miRNA expression profiling assay of 52 AML samples (45 patient samples and 7 cell lines; all bearing chromosomal translocations) along with three normal control samples and an Exiqon miRNA array assay of 100 samples (including 85 AML and 15 normal control samples). In both profiling assays, we found that miR-150 was the most significantly and consistently down-regulated miRNA (q<0.01; significance analysis of microarrays (SAM) (Tusher et al., 2001)) in most of the AML samples, including those bearing t(8;21), inv(16), t(15;17) and MLL rearrangements, compared to normal controls (Figure 1).
Figure 1. Expression of miRNAs that are downregulated in most AML relative to normal controls.
(A) miRNAs that are significantly down-regulated (q<0.001; FDR<0.0001; SAM (Tusher et al., 2001)) in AML samples (n=52) compared to normal bone marrow (BM) controls (n=3; one CD15+ myeloid progenitor and two mononuclear (MNC) cell samples) as detected by bead-based method. (B) miRNAs that are significantly down-regulated (q<0.0001; FDR<0.0001; SAM) in AML (n=85) samples than in normal BM controls (n=15; 6 CD34+ hematopoietic stem/progenitor, 5 CD33+ myeloid progenitor, and 4 MNC cell samples) as detected by Exiqon arrays. Expression data was mean-centered and the relative value for each sample is represented by a color, with red and green representing a high and low expression, respectively (scale shown in the upper left). MNC, mononuclear cell; MLL, MLL-associated leukemia.
Down-regulation of miR-150 is not related to DNA copy number changes, methylation, or mutations
To understand how miR-150 is down-regulated in AML, we first examined the DNA copy number of the miR-150 locus at 19q13.33 in 33 samples, including 29 AML samples and 4 normal controls. As shown in Figure S1A, there was no significant amplification or deletion of the genomic locus of miR-150 in MLL-associated or other subtypes of leukemia samples relative to normal controls. We also assessed the DNA methylation status of a 475-bp CpG-enriched, immediate upstream region of the miR-150 gene locus in 19 samples, including 17 AML samples and 2 controls, using bisulfate genomic DNA sequencing methods. No significant changes in the DNA methylation level of the miR-150 CpG-enriched region were observed in leukemia samples relative to normal controls (Figure S1B). Moreover, no mutation was found in either the precursor miRNA sequence or the CpG island of miR-150. Together, our data suggest that other mechanism(s) might be underlying the down-regulation of miR-150 in AML.
Expression of miR-150 is regulated by MLL fusion proteins at both the transcriptional and posttranscriptional levels
In contrast to fusion proteins with transcriptional repressing functions (e.g. t(8;21), inv(16), t(15;17), etc.) (Chen et al., 2010), MLL fusion proteins have been shown to act as transcriptional activators that directly up-regulate expression of a group of oncogenes (e.g., HOXA genes and MEIS1) and miRNAs (e.g., miR-17-92 and miR-196b) (Faber et al., 2009; Li et al., 2012b; Mi et al., 2010; Popovic et al., 2009; Zeisig et al., 2004). Thus, it is of great interest to understand how miR-150 expression is down-regulated in MLL-associated leukemia.
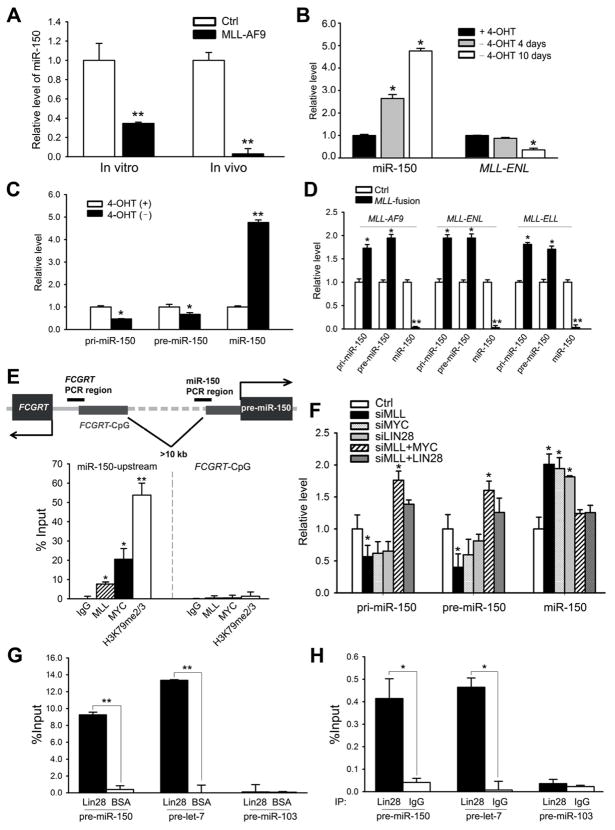
We first investigated whether forced expression of MLL fusion genes can cause down-regulation of miR-150. Indeed, we found that the level of miR-150 was dramatically down-regulated by ectopic expression of MLL-AF9 in normal mouse bone marrow (BM) progenitor cells (i.e., lineage negative; Lin−) both in vitro and in vivo (Figure 2A). In HEK293T cells, the level of miR-150 correlated with the overexpression level of MLL-AF9 in a dose-dependent manner (Figure S1C). Further studies were done using the MLL-ENL-ERtm cell line, a cell line stably expressing a conditional MLL-ENL derivative (Zeisig et al., 2004). As shown in Figure 2B, the level of miR-150 continued to increase after withdrawal of 4-Hydroxytamoxifen (4-OHT) for the indicated number of days, along with the decreasing level of MLL-ENL.
Figure 2. MLL-fusion regulates the level of miR-150 via the MYC/LIN28 axis.
(A) Relative expression levels of mature miR-150 in MLL-AF9-transduced colony-forming cells (in vitro) and BM cells of MLL-AF9 leukemic mice (in vivo; n=3). (B) Expression changes of miR-150 and MLL-ENL during withdrawal of 4-Hydroxy-tamoxifen (4-OHT) (days 0, 4, and 10 are shown) in MLL-ENL-ERtm cells. (C) Expression changes of pri-, pre-, and mature miR-150 transcripts 10 days post 4-OHT withdrawal in MLL-ENL-ERtm cells. (D) Expression changes of pri-, pre-, and mature miR-150 in BM cells of mice with MLL-fusion-mediated leukemia. (E) ChIP assays of potential binding of MLL fusion proteins and MYC to the miR-150 gene locus and the control locus (i.e., CpG island of FCGRT, a neighboring gene) in MONOMAC-6 cells. (F) Expression changes of pri-, pre-, and mature miR-150 in MONOMAC-6 cells when the expression of MLL, MYC, and LIN28 was altered. (G, H) Assessing potential enrichments of pre-miR-150 and pre-let-7 in the LIN28-protein-RNA complex in THP-1 and MONOMAC-6 cells by RNA pull-down assay (G) and RNA-binding protein immunoprecipitation (RIP) assay (H), coupled with RT-qPCR. Pre-miR-103 is used as a negative control. Mean±SD values are shown. *, p<0.05; **, p<0.01. See also Figure S1.
In order to determine whether the inhibitory effect of MLL fusions on miR-150 occurs at the transcriptional level, miR-150 primary (i.e., pri-miR-150) and precursor (i.e., pre-miR-150) transcripts were measured in the MLL-ENL-ERtm cell line. Strikingly, the miRNA primary and precursor transcripts decreased to 47%~67% after 4-OHT withdrawal, while the level of mature miR-150 increased 4.8 fold, compared with cells with 4-OHT treatment (Figure 2C), suggesting that the MLL fusions might regulate the expression of miR-150 at both the transcriptional and posttranscriptional levels. Consistently, in the BM cells of MLL-associated leukemic mice, the levels of both pri-miR-150 and pre-miR-150 were up-regulated, while the level of mature miR-150 was decreased (Figure 2D). Similarly, analysis of leukemic patient samples showed that mature miR-150 was consistently down-regulated while the levels of pri-miR-150 and pre-miR-150 were up-regulated in samples with MLL fusions (Figure S1D).
It was reported that MLL wild-type or MLL fusion proteins regulate the transcription of a variety of target genes, such as HOX genes, by directly binding to their promoter region (Milne et al., 2005a; Milne et al., 2005b; Zeisig et al., 2004; Zeisig et al., 2003). As shown in Figure 2E, we determined through chromatin immunoprecipitation (ChIP) assay that MLL fusion proteins bind directly to an immediate upstream region (<1kb) of the miR-150 locus, and this binding is associated with increased level of histone H3 lysine 79 di- and tri-methylation (i.e., H3K79me2/3), a marker for active transcription in MLL-associated leukemic cells (Bernt et al., 2011; Krivtsov et al., 2008). There is no enrichment of MLL-fusion binding nor H3K79me2/3 at the control locus that is >10 kb upstream of the miR-150 locus (Figure 2E). Therefore miR-150 is likely a direct downstream target of MLL fusions.
MLL fusion proteins inhibit miR-150 maturation through the MYC/LIN28 functional axis
Recently, miR-150 was identified as one of the 3 miRNAs other than let-7 that could be repressed by both Lin28 and Myc (Chang et al., 2009). Interestingly, Myc has been implicated as a direct target gene of MLL and is frequently up-regulated in MLL-associated leukemia (Schreiner et al., 2001; Zeisig et al., 2003), while Lin28 is a direct target gene of Myc (Chang et al., 2009). We then sought to investigate whether MYC and LIN28 participate in MLL-fusion-protein-mediated post-transcriptional regulation of miR-150. As shown in Figure 2F, knockdown of MLL in MONOMAC-6 cells by siRNA reduced the levels of both pri-miR-150 and pre-miR-150 to 57% and 40%, respectively, while mature miR-150 was raised 2 fold, compared to scrambled controls. Knockdown of MYC or LIN28 resulted in effects similar to knockdown of MLL. Overexpression of either MYC or LIN28 efficiently reversed the effects of MLL knockdown on these cells (Figure 2F), indicating that MYC and LIN28 might function as downstream effectors of MLL in regulating the process of miR-150 maturation. Notably, forced expression of MYC in cells with MLL knock-down also significantly up-regulated both pri- and pre-miR-150 transcripts (Figure 2F), implying that MYC itself may regulate miR-150 primary transcription directly as well as serving as a downstream effector of MLL fusions. Indeed, our ChIP assay revealed that MYC also binds directly to the immediate upstream region of miR-150 (Figure 2E).
Furthermore, Lin28 is likely to target the maturation processing of precursor miR-150 directly. In MLL-ENL-inducible cell lines, knockdown of Lin28 resulted in down-regualtion of pri- and pre-miR-150, and up-regulation of mature miR-150 (Figure S1E), while overexpression of Lin28 in MONOMAC-6 cells prevented the maturation of miR-150 (Figure S1F). To investigate whether the let-7 pathway is implicated in Lin28-mediated blockade of miR-150 maturation, we transfected let-7 alone or together with Lin28 into MONOMAC6 cells and found that forced expression of let-7 (40–70 fold increased expression) did not result in a significantly increased miR-150 maturation (Figure S1F), suggesting that let-7 is not an essential mediator of this blockade. To determine whether LIN28 proteins can bind to miR-150 precursor RNA, we performed both RNA pull-down assay and RNA-binding protein immunoprecipitation (RIP) assay, coupled with qPCR. We first used His-tagged recombinant LIN28 protein to pull down target RNAs from total RNA isolated from THP-1 cells and the LIN28-RNA complex was purified with Ni-NTA beads. We found that pre-miR-150 transcripts, along with let-7 precursor (as a positive control), are significantly enriched in the LIN28-RNA complex (Figure 2G), suggesting a potential direct binding between LIN28 and pre-miR-150. Furthermore, we used an anti-LIN28 antibody to precipitate endogenous LIN28-RNA complex from MONOMAC-6 cell lyses via RIP assay, and found that both miR-150 and let-7 precursors are highly enriched in LIN28 immunoprecipitation complex (Figure 2H), indicating an association between the miRNA precursors and endogenous LIN28 protein. In contrast, as a negative control, precursor of miR-103, a miRNA whose maturation was not influenced by Lin28 (Viswanathan et al., 2008), was not enriched in either assay (Figures 2G and H).
Taken together, our data suggest that both MLL fusions and MYC can promote primary transcription of miR-150, whereas the MYC/LIN28 axis inhibits its posttranscriptional maturation.
miR-150 inhibits cell proliferation/transformation in vitro, and leukemogenesis in vivo
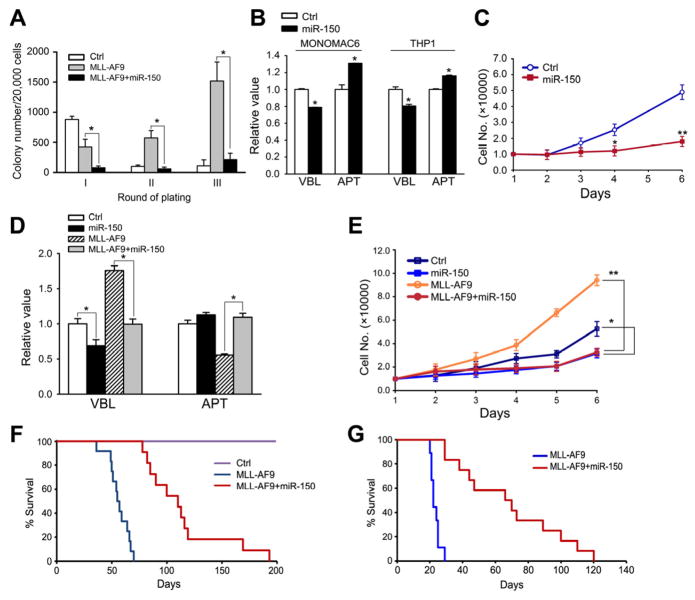
In order to investigate whether miR-150 has an antagonistic action on the oncogenicity of MLL fusions, we performed colony-forming/replating assays. Mouse BM progenitor cells transduced with MSCV-PIG (bearing a PGK-puromycin-IRES-GFP cassette (He et al., 2005)) or MSCV-PIG-miR-150, together with MSCVneo or MSCVneo-MLL-AF9, were plated on methylcellulose medium. The colonies were replated every 7 days under the same conditions. As shown in Figure 3A, co-transduction of miR-150 and MLL-AF9 caused a significant reduction in colonies compared to transduction of MLL-AF9 alone, indicating that forced expression of miR-150 (Figure S2A) strongly inhibits the colony-forming capacity induced by MLL-AF9.
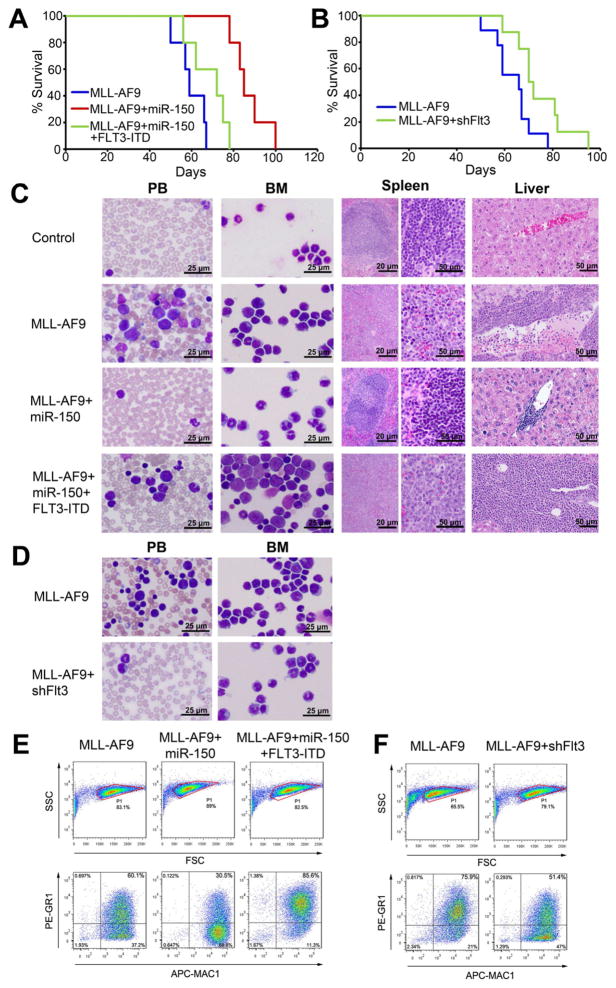
Figure 3. miR-150 inhibits cell growth in vitro and leukemogenesis in vivo.
(A) Colony-forming/replating assays of mouse BM progenitor cells co-transduced with MSCV-PIG+MSCVneo (Ctrl), MSCV-PIG+MSCVneo-MLL-AF9 (MLL-AF9), or MSCV-PIG-miR-150+MSCVneo-MLL-AF9 (MLL-AF9+miR-150). (B) Analyses of viability (VBL) and apoptosis (APT) of MONOMAC-6 or THP-1 cells 48 hours post-transfection with MSCV-PIG (Ctrl) or MSCV-PIG-miR-150 (miR-150). (C) Effects of miR-150 on the proliferation of MONOMAC-6 cells. (D, E) Effects of miR-150 on viability/apoptosis (D) and proliferation (E) of mouse BM progenitor cells in the presence or absence of MLL-AF9. Mean±SD values are shown. *, p<0.05; **, p<0.01. (F, G) Effects of miR-150 in MLL-AF9-mediated leukemogenesis in vivo. Kaplan-Meier curves are shown for three groups of transplanted mice including control (n=10), MLL-AF9 (n=12), and MLL-AF9+miR-150 (n=11) in a primary BMT assay (F), and for two groups of transplanted mice including MLL-AF9+miR-150 (n=12) and MLL-AF9 (n=9) in secondary BMT assay (G). See also Figure S2.
We then explored the effects of ectopic expression of miR-150 on human MLL-associated leukemic cells. As shown in Figure 3B, forced expression of miR-150 (Figure S2B) significantly decreased cell viability and increased apoptosis in MONOMAC-6/t(9;11) and THP-1/t(9;11) cells. Moreover, overexpression of miR-150 consistently inhibited the proliferation of MONOMAC-6 cells (Figure 3C). Similarly, in retrovirally transduced mouse BM progenitor cells, we found that in the presence of MLL-AF9, forced expression of miR-150 significantly decreased cell viability and increased apoptosis (Figure 3D), as well as inhibited cell growth (Figure 3E).
In order to elucidate the in vivo role of miR-150 in leukemogenesis, we performed primary BM transplantation (BMT) assay and found that forced expression of miR-150 (Figure S2C) significantly delayed leukemogenesis mediated by MLL-AF9 (median overall survival, 110 days versus 56 days; p<0.001, log-rank test) (see Figure 3F). We then performed secondary BMT and showed that miR-150+MLL-AF9 leukemic cells developed AML in secondary recipient mice remarkably slower than MLL-AF9 leukemic cells (median overall survival, 70 days vs. 42 days; p<0.001; Figure 3G). The miR-150-alone group did not show visible differences compared to the control group, either in survival or in the morphology of cells and tissues (data not shown). These findings suggest that miR-150 does play a critical tumor suppressor role in preventing MLL-associated leukemogenesis.
miR-150 targets both MYB and FLT3
To identify potential target genes of miR-150 in MLL-associated leukemia, we performed Agilent custom-design arrays of the 8 human de novo MLL-associated AML and 9 normal control (including 3 CD34+, 2 CD33+, and 4 MNC) samples that were used in Exiqon miRNA profiling assay (see Figure 1B). By correlating expression of predicted target genes and miR-150 across the above 17 samples, we identified 158 putative target genes that exhibited a significant inverse correlation of expression with miR-150 (p<0.05; Pearson Correlation). Analysis of Affymetrix Exon arrays of 15 additional human MLL-associated leukemia samples and 9 normal controls (including 3 each of CD34+, CD33+, and MNC) (Li et al., 2012b) revealed that 22 of the above 158 candidate target genes were significantly overexpressed (q<0.05; False Discovery Rate, FDR<0.01; SAM (Tusher et al., 2001)) in MLL-associated leukemia samples relative to normal controls. Furthermore, in the analysis of Affymetrix gene arrays of 9 MLL-AF9-mediated mouse leukemia samples and 6 control samples (Li et al., 2012b), we observed that 4 (i.e., Flt3, Myb, Hdhd2, and Letmd1) of the above 22 candidate target genes were significantly overexpressed (q<0.05; FDR<0.01; SAM) in MLL-AF9 mouse leukemia samples relative to normal controls. The expression patterns of these four genes in the above three sample sets are shown in Figure S3.
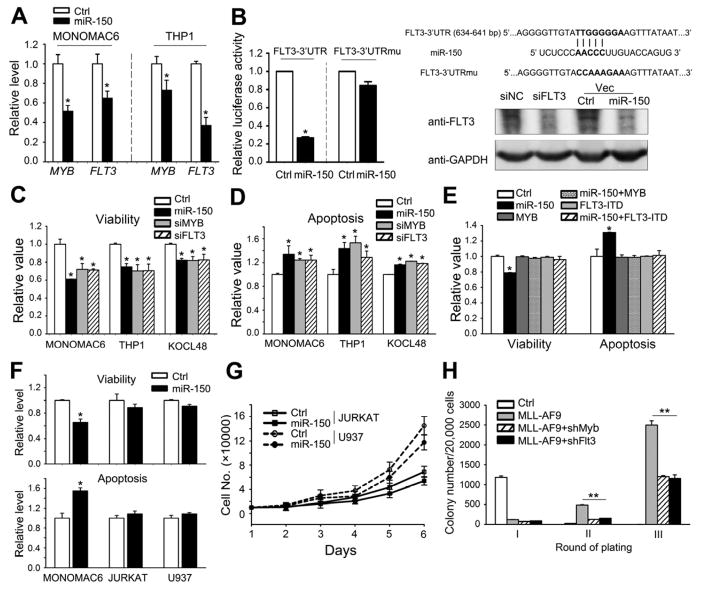
As FLT3 and MYB have been shown to be broadly involved in multiple processes of hematopoiesis and leukemogenesis (Ramsay et al., 2003; Takahashi, 2011) and be critical oncogenes in MLL-associated leukemia (Armstrong et al., 2003; Armstrong et al., 2002; Hess et al., 2006; Zuber et al., 2011), we focused on these two genes for further studies. Forced expression of miR-150 reduced the levels of both MYB and FLT3 to 53~74% and 40~65%, respectively, in MONOMAC-6 and THP-1 cells (Figure 4A). MYB has been reported to be a direct target gene of miR-150 (Lu et al., 2008; Xiao et al., 2007). Luciferase reporter and mutagenesis assays showed that miR-150 also targeted the 3′UTR of FLT3 directly (Figure 4B).
Figure 4. miR-150 targets both MYB and FLT3.
(A) Effects of overexpression of miR-150 on endogenous expression of MYB and FLT3. (B) Luciferase reporter and mutagenesis assays. Left panel: effects of miR-150 on luciferase activity of the reporter gene bearing wild-type or mutant 3′UTR of FLT3 in HEK293T cells. Right upper panel: Putative miR-150 target site and mutant in the 3′UTR of FLT3. Right lower panel Western blot analysis of FLT3 in MONOMAC-6 cells that were transfected with MSCV-PIG (Ctrl), MSCV-PIG-miR-150 (miR-150), scrambled siRNA control oligos (siNC), or FLT3 siRNAs (siFLT3). (C, D) Effects of depletion of MYB or FLT3 by siRNAs on viability (C) and apoptosis (D) of MONOMAC-6, THP-1, or KOCL-48 cells. (E) Effects of forced expression of MYB-CDS or FLT3-ITD-CDS alone, or together with miR-150, on viability and apoptosis of MONOMAC-6 cells. (F) Effect of miR-150 on cell viability and apoptosis of Jurkat, U937 and MONOMAC-6 cells. (G) Effect of miR-150 on the proliferation of Jurkat and U937 cells. (H) Effects of knockdown of Myb or Flt3 on MLL-AF9-induced colony-formation of mouse BM progenitor cells. Mean±SD values are shown. *, p<0.05; **, p<0.01. See also Figure S3.
Knockdown of the expression of MYB or FLT3 by siRNAs can mimic the effects of miR-150 overexpression such as decreasing cell viability (Figure 4C) and increasing apoptosis (Figure 4D) in MONOMAC-6, THP-1, and KOCL-48 cells. Co-transfection of MYB or FLT3-ITD, a constitutively active mutant of FLT3, with miR-150 completely reversed the effects of miR-150 on cell viability and apoptosis (Figure 4E). Overexpression of MYB or FLT3-ITD alone did not show noticeable effects on cell viability or apoptosis, likely due to the high expression level of endogenous MYB and FLT3 in MLL-associated leukemic cells (Somervaille et al., 2009) (Figure 4E). In FLT3-expression-negative cell lines (e.g. Jurkat and U937) (Shankar et al., 2007; Yao et al., 2003), overexpression of miR-150 (Figure S2B) showed slight, if any, inhibitory effects on cell viability, apoptosis, or proliferation (see Figures 4F and G). Results of colony-forming/replating assays showed that co-transduction of Myb or Flt3 shRNA and MLL-AF9 caused a significant reduction in colonies compared to transduction of MLL-AF9 alone (Figure 4H), mimicking the inhibitory effect of miR-150 on colony formation (see Figure 3A). These results indicate that both Myb and Flt3 function as direct targets of miR-150 in regulating leukemic cell self-renewal.
Repression of Flt3 activity is required for the inhibitory effect of miR-150 in MLL-associated leukemogenesis
The requirement of Myb function in MLL-associated cell transformation and leukemogenesis has been well documented (Hess et al., 2006; Zuber et al., 2011), suggesting that the inhibitory effects of forced expression of miR-150 on leukemogenesis would be, at least in part, related to its repression of Myb expression.
To determine whether repression of Flt3 expression is also required for the inhibitory effects of miR-150 in MLL-associated leukemogenesis, we performed a series of in vivo functional studies. As shown in Figure 5A, the delay in MLL-AF9-induced leukemogenesis mediated by forced expression of miR-150 (Figure S2C) could be largely reversed by co-expression of FLT3-ITD-CDS. Our loss-of-function study showed that knockdown of endogenous Flt3 by shRNA significantly delayed primary leukemogenesis mediated by MLL-AF9 (median overall survival, 76 days versus 65 days; p<0.05) (Figure 5B), though not as significantly as did forced expression of miR-150 (see Figure 3F). Leukemia in MLL-AF9+miR-150 mice is much less aggressive than that of the MLL-AF9 group, while ectopic expression of FLT3-ITD remarkably reversed the phenotype caused by miR-150 overexpression (Figure 5C). Knockdown of endogenous Flt3 by shRNA resulted in reduction of leukemic blast cells in both BM and peripheral blood (PB), similar to that found in miR-150 overexpressing mice (Figure 5D). There was a decrease in the proportion of Mac-1/Gr-1 double-positive cells in MLL–AF9+miR-150 mice, compared to the MLL-AF9 group, while co-expression with FLT3-ITD significantly increased the Mac-1/Gr-1 double-positive cell ratio (Figure 5E). Knockdown of Flt3 by Flt3-shRNA (i.e., shFlt3) showed an effect similar to forced expression of miR-150 on reducing the percentage of Mac-1/Gr-1 double-positive cells (Figure 5F). These results demonstrate that the repression of Flt3 activity by miR-150 is, at least in part, responsible for the inhibitory effects of forced expression of miR-150 on leukemogenesis.
Figure 5. miR-150 inhibits MLL-fusion-mediated leukemogenesis via regulating Flt3.
(A) Kaplan-Meier curves are shown for three groups of transplanted mice (n=5 for each group) in a primary BMT assay. (B) Kaplan-Meier curves are shown for two groups of transplanted mice including MLL-AF9 (n=9) and MLL-AF9+shFlt3 (n=8) in an additional primary BMT assay. (C, D) Wright-Giemsa staining of mouse peripheral blood (PB) and BM, or hematoxylin and eosin (H&E) staining of mouse spleen and liver. (E, F) Flow cytometry analyses of mouse BM cells. The “blast” population was gated by FSC/SSC (in a red frame; upper panel) and then the proportions of Gr1+ and/or Mac1+ cells were analyzed (lower panel).
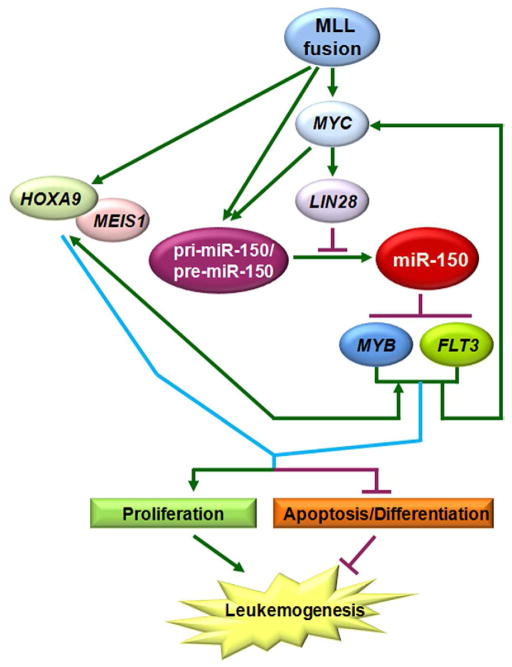
The MLL-fusion/MYC/LIN28⊣miR-150⊣FLT3/MYB/HOXA9/MEIS1 regulatory circuit
HOXA9 and its co-factor MEIS1 are two of the best studied critical downstream target genes of MLL fusion proteins (Krivtsov et al., 2008; Milne et al., 2005a; Zeisig et al., 2004). MYB has been reported to be a direct target of HOXA9/MEIS1 signaling (Hess et al., 2006), while a recent study showed that MYB can also regulate expression of HOXA9/MEIS1 at the transcription level (Jin et al., 2010), suggesting there is an autoregulatory feedback loop between them. Interestingly, FLT3 has been identified as a direct target gene of MEIS1 (Wang et al., 2005) as well as an upstream regulator of MYC (Li et al., 2007; Takahashi, 2011). MYC is also a downstream target of MLL fusion proteins (Dawson et al., 2011; Schreiner et al., 2001) and an upstream regulator of Lin28 (Chang et al., 2009). These findings together with the data we showed above suggest that there is a critical MLL-fusion/MYC/LIN28⊣miR-150⊣FLT3/MYB/HOXA9/MEIS1 regulatory circuit in MLL-associated leukemia (see Figure 6).
Figure 6. A schematic model of the MLL-fusion/MYC/LIN28⊣miR-150⊣FLT3/MYB/HOXA9/MEIS1 regulatory circuit in MLL-associated leukemia.
MLL fusion proteins inhibit the maturation of miR-150 through MYC/LIN28, release miR-150 inhibition on its target genes (FLT3 and MYB), then subsequently upregulate expression of MYC, LIN28, HOXA9, and MEIS1. The upregulation of MYC/LIN28 further represses the level of mature miR-150 and enhances/maintains the up-regulation of the six genes in this regulatory circuit. These changes, in turn, result in increased cell proliferation and decreased apoptosis/ differentiation, and eventually lead to leukemogenesis.
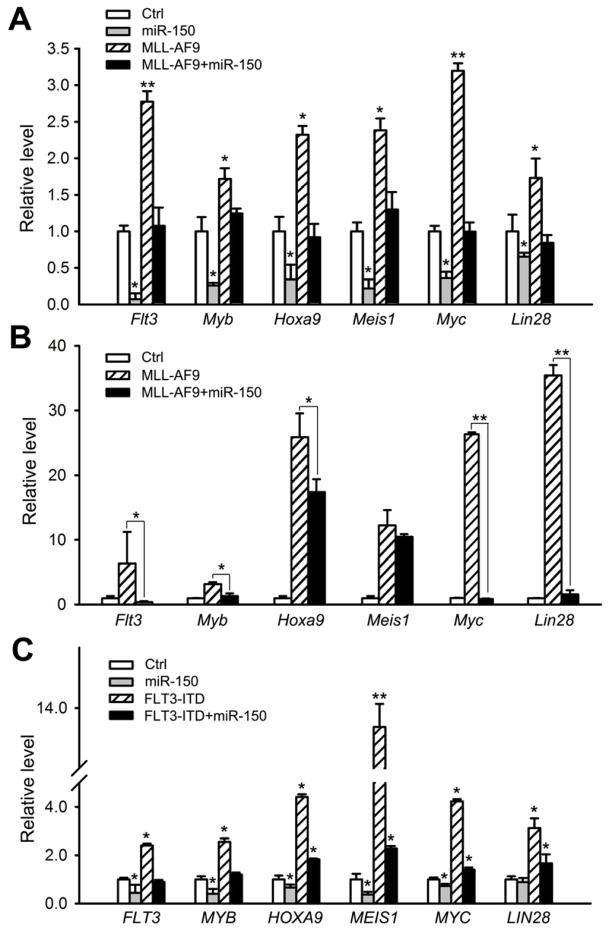
In this regulatory circuit, MLL fusion proteins function as the driver. Indeed, in human leukemia samples bearing MLL rearrangements, levels of expression of all downstream genes including MYC, LIN28, HOXA9, MEIS1, MYB, and FLT3 are significantly increased (p<0.05, t-test) relative to normal control samples (see Figure S4A). MLL-AF9 retrovirally transduced into mouse normal BM progenitor cells significantly induced expression of all six genes both in vitro (Figure 7A) and in vivo (Figure 7B), confirming that they are downstream target genes of MLL fusion proteins. In the MLL-ENL-ERtm cell line, their expression levels were significantly reduced (p<0.05, t-test) in cells 10 days after withdrawal of 4-OHT (Figure S4B), indicating their dependence on the presence of MLL fusion proteins.
Figure 7. The effects of MLL fusion proteins, miR-150, and FLT3 on the expression of the six MLL-fusion downstream genes.
(A) Effects of miR-150 on expression of endogenous Flt3, Myb, Hoxa9, Meis1, Myc, and Lin28 in mouse BM progenitor cells in the presence or absence of MLL-AF9. RNAs were collected from the first passage of colony-forming cells. (B) Effects of miR-150 on the expression of above six genes in mouse MLL-AF9 leukemic BM cells (n=5 for each group). (C) Effects of miR-150 and/or FLT3-ITD on expression of the six genes in MONOMAC-6 cells. Mean±SD values are shown. *, p<0.05, t-test. See also Figure S4.
On the other hand, this circuit also highlights the importance of the post-transcriptional regulation of the miR-150 maturation process in MLL-associated leukemogenesis. Co-transduction of miR-150 with MLL-AF9 reduced the MLL-AF9-mediated induction of expression of the six genes both in vitro (Figure 7A) and in vivo (Figure 7B), leading to a significant reduction (p<0.05) in colony-formation (Figure 3A) and a delay in leukemogenesis (Figures 3F and G). Similarly, ectopic expression of miR-150 in MONOMAC-6 cells also down-regulated the expression of all six genes (see Figure 7C), leading to a significant decrease in cell viability/proliferation and an increase in apoptosis (Figures 3B and 3C). These results indicate that mature miR-150 has an influence on the expression of all six genes in this regulatory circuit through directly targeting FLT3 and MYB, suggesting that post-transcriptional repression of miR-150 is an essential and required event in the pathogenesis of MLL-associated leukemia.
In addition, co-transfected FLT3-ITD could completely reverse the effects of ectopic expression of miR-150 in MONOMAC-6 cells on the expression of the six genes (Figure 7C) and on cell viability and apoptosis (see Figure 4E). Notably, forced expression of FLT3-ITD alone could significantly increase expression (p<0.05) of all six genes (Figure 7C). These data indicate that FLT3 is an essential target gene of miR-150 and a critical component of this regulatory circuit.
The strong and widely existing inhibitory effect of miR-150 on cell transformation
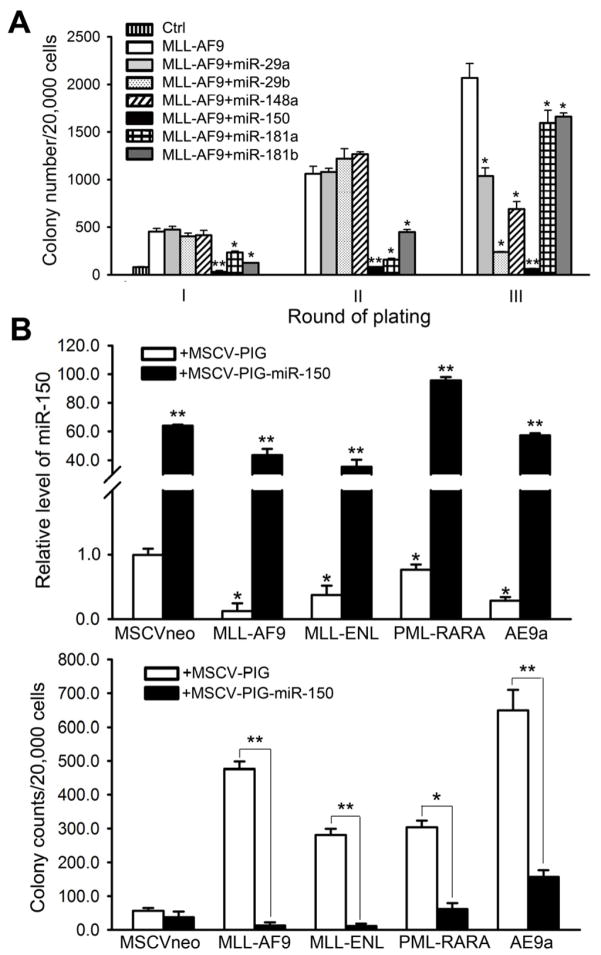
We have also conducted in vitro colony-forming/replating assays to investigate the functions of several other miRNAs (e.g., miR-29a, miR-29b, miR-148a, miR-181a, and miR-181b) in MLL-fusion-mediated cell transformation since these miRNAs are also significantly down-regulated in human MLL-associated AML compared to normal controls (Figure 1) or other subtypes of AML (Li et al., 2008). However, none of these miRNAs exhibited an inhibitory effect as significant and as consistent as miR-150 in inhibiting the cell transformation (Figure 8A).
Figure 8. The strong and widely existing inhibitory effect of miR-150 on cell transformation.
(A) Comparison of inhibitory effect of miR-150 with that of miR-29a, miR-29b, miR-148a, miR-181a, or miR-181b on MLL-AF9-induced colony forming capacity of mouse BM progenitor cells. (B) Relative expression level (upper panel) of ectopically expressed miR-150 in mouse BM progenitor cells transduced with common leukemic fusion genes such as MLL-AF9, MLL-ENL, PML-RARA, and AML1-ETO9a/AE9a, and its effects (lower panel) on the colony-forming/replating capacity of the transduced cells. Mean±SD values are shown. *, p<0.05; **, p<0.01.
In addition, as miR-150 was significantly downregulated in almost all subtypes of AML (Figure 1), in order to test the role of miR-150 in other subtypes of AML, we performed colony-forming/replating assays and found that forced expression of miR-150 could significantly inhibit both MLL fusion (e.g., MLL-AF9 and MLL-ENL) and non-MLL fusion (e.g., PML-RARA resulting from t(15;17) and AML1-ETO9a/AE9a resulting from t(8;21))-induced cell transformation (Figure 8B). Thus, miR-150 likely also plays a critical tumor suppressor role in other subtypes of AML.
DISCUSSION
Expression of miRNAs is under stringent regulation at both the transcriptional and posttranscriptional levels. In the present study we showed that mature miR-150 level was significantly down-regulated in most AML samples, including MLL-associated AML. Strikingly, we found that despite the decreased abundance of mature transcripts of miR-150, its primary and precursor transcript abundance is increased in human MLL-associated AML, relative to normal controls. We further showed that MLL fusion proteins could bind to the genomic locus of miR-150 and significantly promote its primary transcription, leading to increased abundance of its primary and precursor transcripts. In contrast, the mature miR-150 abundance was significantly decreased when MLL fusion genes were expressed, and the opposite is true when MLL fusion gene expression was silenced or knocked down. Thus, an important question arises: What caused such a discrepancy between primary/precursor and mature miR-150 transcript levels in cells bearing MLL fusion genes?
A recent study showed that Myc binds to the promoter region of Lin28 and activates its transcription, which is essential for Myc-mediated let-7 repression (Chang et al., 2009). Interestingly, besides let-7, three additional miRNAs (miR-150, miR-146a, and miR-210) can also be repressed by both Myc and Lin28. MYC, a well-known oncogene, is a direct downstream target gene of MLL fusion proteins and thereby is frequently up-regulated in MLL-associated leukemia (Schreiner et al., 2001; Zeisig et al., 2003), while Lin28 is a direct target gene of Myc. We therefore investigated whether MYC and LIN28 participate in MLL-fusion-protein-mediated post-transcriptional repression of miR-150. Indeed, here we showed that the maturation of miR-150 is also regulated by MYC/LIN28, and revealed an interesting function of MLL fusions in the regulation of miRNA maturation through activation of the MYC/LIN28 axis.
MiR-150 has been implicated as either an oncogene or a tumor suppressor in various types of solid tumors (Li et al., 2012a; Srivastava et al., 2011; Wu et al., 2010; Zhang et al., 2011). However, its function in the pathogenesis of AML is unknown. The abundance of miR-150 in different populations of normal hematopoietic cells, including CD34+ hematopoietic stem/progenitor cells, CD33+ or CD15+ myeloid cells, and mononuclear (MNC) cells, is rather consistent, but such abundance is significantly down-regulated in almost all subtypes of AML samples, including MLL-associated leukemia, suggesting that miR-150 might be important in maintaining normal myelopoiesis and in inhibiting leukemogenesis. Indeed, we showed that miR-150 plays a significant inhibitory role on MLL-fusion-mediated cell transformation and leukemogenesis, and the inhibitory effect of miR-150 on cell transformation is much stronger than other miRNAs.
More importantly, we identified the MLL-fusion/MYC/LIN28⊣miR-150⊣FLT3/MYB/ HOXA9/MEIS1 regulatory circuit in MLL-associated leukemia. In this circuit, MLL fusion proteins function as the driver, and their presence leads to the significant up-regulation of all six downstream genes, MYC, LIN28, FLT3, MYB, HOXA9, and MEIS1, as well as the primary transcription of miR-150. The up-regulation of MYC/LIN28 results in the blockade of the miR-150 maturation process, which in turn leads to the release of miR-150 inhibition on FLT3 and MYB expression. The release of FLT3 and MYB would enhance the expression of HOXA9, MEIS1, MYC, and LIN28, and further enhance/maintain the blockade of miR-150 maturation. As a result, the cells reach and maintain high levels of MYC/LIN28/FLT3/MYB/HOXA9/MEIS1, and thereby transform the cells and lead to leukemogenesis.
Our finding that FLT3 is a critical target gene of miR-150 and an essential component of the regulatory circuit provides further mechanistic evidence to support the notion that FLT3 is a promising therapeutic target in the treatment of MLL-associated leukemia (Armstrong et al., 2003; Stubbs and Armstrong, 2007). On the other hand, miR-150 functions as a pivotal gatekeeper in inhibiting cell transformation and leukemogenesis by directly targeting FLT3/MYB and thereby inactivating the positive feedback loops within this circuit. The addition of MLL fusion proteins disrupts the balance between miR-150 and the other six genes. As the repression of miR-150 maturation is an essential event in MLL-fusion-mediated cell transformation and leukemogenesis, restoration of mature miR-150 would be an attractive strategy to treat MLL-associated leukemia alone, or in combination with other strategies, in the future.
Finally, as miR-150 was significantly downregulated in almost all subtypes of AML and exhibited a significant inhibitory effect on cell transformation mediated by various types of leukemic fusion genes, it would be important in the future to systematically investigate miR-150s pathologenic role and critical target genes/pathways in other subtypes of AML, and to reveal the relevant molecular mechanisms underlying its down-regulation. It would also be important to determine whether the MYC/LIN28⊣miR-150⊣FLT3/MYB/HOXA9/MEIS1 regulatory circuit also exists in other subtypes of AML as a whole or at least in part. Indeed, given the pivotal oncogenic functions of the MYC/LIN28 axis in various types of cancer including AML (Chang et al., 2009; He et al., 2005; Hoffman et al., 2002; O’Donnell et al., 2005; Viswanathan et al., 2008), it is highly likely that miR-150, as an essential downstream target and antagonist of MYC/LIN28, plays a critical tumor-suppressor role in not only MLL-rearranged AML but also other subtypes of AML and even in other types of cancer.
Taken together, we revealed a regulatory circuit, namely MLL-fusion/MYC/LIN28⊣miR-150⊣FLT3/MYB/HOXA9/MEIS1 in MLL-associated leukemia (Figure 6). Our findings may advance our understanding of the complex molecular mechanisms underlying the development and maintenance of MLL-associated leukemia, and may also provide effective strategies to treat MLL-associated leukemia, a diesase that is presently treatment resistant, and probably also other subtypes of AML, or even other types of cancer that also utilize at least part of the signaling circuit we have described herein.
EXPERIMENTAL PROCEDURES
Leukemic samples and treatment protocols
All of the AML patient samples were obtained at the time of diagnosis or relapse and with informed consent at the University of Chicago Hospital (UCH), and were approved by the institutional review board of the hospital. All patients were treated according to the protocols of the hospital.
miRNA and mRNA expression profiling assays
The miRNA expression profiling assays of the 55 human (52 AML and 3 normal control) sample set and the 100 human (85 AML and 15 normal control) samples were conducted by use of a bead-based method (Li et al., 2008) and Exiqon miRCURY LNA™ arrays (v10.0; covering 757 human miRNAs; Exiqon, Woburn, MA), respectively. The mRNA microarrays of the 8 MLL-associated AML and 9 normal control sample set, the 15 MLL-rearranged AML and 9 normal control sample set, and the 15 mouse sample set were conducted by use of Agilent’s custom-design microarrays (Agilent Technologies Inc., Santa Clara, CA), Affymetrix GeneChip Human Exon 1.0 ST arrays, and Affymetrix GeneChip Mouse Gene 1.0 ST arrays, respectively.
Cell culture and transfection
THP-1, KOCL-48, U937 and MONOMAC-6 cells were grown in RPMI medium 1640 and transfected using the Amaxa® Nucleofector® Technology (Amaxa Biosystems, Berlin, Germany). See Supplementary Materials and Methods for more details.
Cell apoptosis and viability assay
Cell apoptosis and viability were assessed 48 hours post-transfection using ApoLive-Glo Multiplex Assay Kit (Promega, Madison, WI) following the manufacturer’s manuals.
Luciferase reporter and mutagenesis assays
Luciferase reporter and mutagenesis assays were conducted as described previously (Li et al., 2012b), with some modifications (see Supplementary Methods).
Chromatin Immunoprecipitation (ChIP)
ChIP assay was performed with SABiosciences Corporation’s ChampionChiP™ One-Day kit (Qiagen, Frederick, MD) following the manufacturer’s protocol with some modifications (see Supplementary Methods).
Colony-forming/replating assays
These experiments were conducted as described previously (Li et al., 2008) with some modifications (see Supplementary Methods).
Primary and secondary bone marrow transplantation (BMT)
All experiments on mice were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Chicago. For primary BMT assays, normal BM cells of B6.SJL (CD45.1) mice were retrovirally transduced with corresponding constructs, through two rounds of spinoculation (Li et al., 2012b), and then injected by tail vein into lethally irradiated (960 rads) 8- to 10-week-old C57BL/6 (CD45.2) recipient mice with 3×105 donor cells per mouse plus a radioprotective dose of 1×106 whole BM cells. For secondary BMT assays, leukemic BM cells isolated from primary recipient mice were further transplanted into lethally irradiated C57BL/6 secondary recipient mice with the same dosage as primary BMT.
Flow Cytometry
Cells from PB, BM, spleen or liver were harvested for analysis of immunophenotypes. After washing with phosphate-buffered saline (PBS) and blocking unspecific binding with Affinity Purified anti-mouse CD16/32 (eBioscience, San Diego, CA), cells were stained at 4°C with various antibodies diluted in Flow Cytometry Staining Buffer (eBioscience) for 30 min. Subsequently, cells were washed with PBS and resuspended in IC Fixation Buffer for flow cytometric analysis. APC-conjugated anti-mouse CD11b (Mac-1) and PE-conjugated anti-mouse Ly-6G (Gr-1) antibodies (eBioscience) were used for the analyses.
Histopathology and Immunohistochemistry
Tissue samples were fixed in formalin, embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E). Cytospins of PB and BM were stained with Wright-Giemsa.
In vitro RNA pull-down and RNA-binding protein immunoprecipitation (RIP) assays
THP-1 and MONOMAC-6 cells were used for the RNA pull-down and RIP assays, respectively, to investigate whether Lin28 protein binds directly to precursor miR-150 (see Supplementary Methods).
Supplementary Material
Significance.
Although altered expression of many miRNAs has been reported in various cancers, including leukemia, their dysregulational mechanisms and pathologic functions remain less understood. Here we show that miR-150 is significantly down-regulated in most cases of acute myeloid leukemia (AML). In AML with MLL rearrangements, the maturation of miR-150 is inhibited by the MLL-fusion/MYC/LIN28 functional axis. Furthermore, miR-150 itself functions as a pivotal tumor suppressor in MLL-associated leukemogenesis through repressing the expression of FLT3 and MYB, two essential oncogenes, and subsequently interfering the HOXA9/MEIS1/FLT3/MYB/MYC/LIN28 signaling network. Taken together, we report here a signaling circuit in leukemogenesis, in which the post-transcriptional repression of miR-150 maturation is a critical and necessary event. Thus, our results may provide optional strategies for anti-leukemia therapy.
Highlights.
miR-150 is down-regulated in most cases of AML.
Maturation of miR-150 is inhibited by the MLL-fusion/MYC/LIN28 functional axis.
miR-150 inhibits MLL-associated leukemogenesis by down-regulating FLT3 and MYB.
Discovery of a MLL-fusion/MYC/LIN28 ⊣miR-150⊣FLT3/MYB/HOXA9/MEIS1 signaling circuit.
Acknowledgments
The authors thank Dr. Janet D. Rowley for her support and constructive suggestions/comments, and thank Dr. Peter Breslin for critical reading and constructive comments. We are also grateful to Drs. Gregory Hannon, Scott Hammond, Lin He, Scott Armstrong, and Michael Thirman for providing retroviral constructs. This work was supported in part by the National Institutes of Health (NIH) R01 grant CA127277 (J.C.), American Cancer Society (ACS) Research Scholar grant (J.C.), The University of Chicago Committee on Cancer Biology (CCB) Fellowship Program (X.J.), LLS Special Fellowship (Z.L.), Gabrielle’s Angel Foundation for Cancer Research (J.C.; Z.L.; H.H.), Leukemia & Lymphoma Society (LLS) Translational Research Grant (J.D.R. & J.C.), the Fidelity Foundation (J.D.R. & J.C.), R01 HL95896 (J.Z.), and the Intramural Research Program of the National Human Genome Research Institute, NIH (A.E. & P.P.L.), NIH P01 CA40046 (M.M.L.B), and P30 CA014599 (CCSG) (M.M.L.B).
Abbreviatives
- BM
bone marrow
- BMT
bone marrow transplantation
- PB
peripheral blood
- MLL
mixed lineage leukemia
- AML
acute myeloid leukemia
- CDS
coding region
- Lin−
lineage-negative
- FLT3-ITD
internal tandem duplications of FLT3
- FDR
False Discovery Rate
- SAM
Significance Analysis of Microarrays
- ChIP
chromatin immunoprecipitation
- RIP
RNA-binding protein immunoprecipitation
Footnotes
ACCESSION NUMBERS
The microarray data have been deposited in the Gene expression Ominibus (GEO) repository with the accession numbers GSE30258, GSE34184, and GSE34185.
Supplemental Information includes Supplemental Experimental Procedures and four figures can be found with this article at doi: xxx..
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong SA, Kung AL, Mabon ME, Silverman LB, Stam RW, Den Boer ML, Pieters R, Kersey JH, Sallan SE, Fletcher JA, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3:173–183. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- Behm FG, Raimondi SC, Frestedt JL, Liu Q, Crist WM, Downing JR, Rivera GK, Kersey JH, Pui CH. Rearrangement of the MLL gene confers a poor prognosis in childhood acute lymphoblastic leukemia, regardless of presenting age. Blood. 1996;87:2870–2877. [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullinger L, Dohner K, Bair E, Frohling S, Schlenk RF, Tibshirani R, Dohner H, Pollack JR. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350:1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106:3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nat Rev Cancer. 2010;10:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M, Zwaan CM, Kung AL, Armstrong SA. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Hess JL, Bittner CB, Zeisig DT, Bach C, Fuchs U, Borkhardt A, Frampton J, Slany RK. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108:297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B, Amanullah A, Shafarenko M, Liebermann DA. The proto-oncogene c-myc in hematopoietic development and leukemogenesis. Oncogene. 2002;21:3414–3421. doi: 10.1038/sj.onc.1205400. [DOI] [PubMed] [Google Scholar]
- Jin S, Zhao H, Yi Y, Nakata Y, Kalota A, Gewirtz AM. c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. The Journal of clinical investigation. 2010;120:593–606. doi: 10.1172/JCI38030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Piloto O, Kim KT, Ye Z, Nguyen HB, Yu X, Levis M, Cheng L, Small D. FLT3/ITD expression increases expansion, survival and entry into cell cycle of human haematopoietic stem/progenitor cells. Br J Haematol. 2007;137:64–75. doi: 10.1111/j.1365-2141.2007.06525.x. [DOI] [PubMed] [Google Scholar]
- Li YJ, Zhang YX, Wang PY, Chi YL, Zhang C, Ma Y, Lv CJ, Xie SY. Regression of A549 lung cancer tumors by anti-miR-150 vector. Oncol Rep. 2012a;27:129–134. doi: 10.3892/or.2011.1466. [DOI] [PubMed] [Google Scholar]
- Li Z, Huang H, Chen P, He M, Li Y, Arnovitz S, Jiang X, He C, Hyjek E, Zhang J, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun. 2012b;2:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Guo S, Ebert BL, Zhang H, Peng X, Bosco J, Pretz J, Schlanger R, Wang JY, Mak RH, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Developmental cell. 2008;14:843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Li Z, Chen P, He C, Cao D, Elkahloun A, Lu J, Pelloso LA, Wunderlich M, Huang H, et al. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc Natl Acad Sci U S A. 2010;107:3710–3715. doi: 10.1073/pnas.0914900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci U S A. 2005a;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005b;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010;24:1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Popovic R, Riesbeck LE, Velu CS, Chaubey A, Zhang J, Achille NJ, Erfurth FE, Eaton K, Lu J, Grimes HL, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113:3314–3322. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay RG, Barton AL, Gonda TJ. Targeting c-Myb expression in human disease. Expert Opin Ther Targets. 2003;7:235–248. doi: 10.1517/14728222.7.2.235. [DOI] [PubMed] [Google Scholar]
- Rowley JD. Chromosomal translocations: revisited yet again. Blood. 2008;112:2183–2189. doi: 10.1182/blood-2008-04-097931. [DOI] [PubMed] [Google Scholar]
- Schreiner S, Birke M, Garcia-Cuellar MP, Zilles O, Greil J, Slany RK. MLL-ENL causes a reversible and myc-dependent block of myelomonocytic cell differentiation. Cancer research. 2001;61:6480–6486. [PubMed] [Google Scholar]
- Shankar DB, Li J, Tapang P, Owen McCall J, Pease LJ, Dai Y, Wei RQ, Albert DH, Bouska JJ, Osterling DJ, et al. ABT-869, a multitargeted receptor tyrosine kinase inhibitor: inhibition of FLT3 phosphorylation and signaling in acute myeloid leukemia. Blood. 2007;109:3400–3408. doi: 10.1182/blood-2006-06-029579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, Chang HY, Shurtleff SA, Downing JR, Cleary ML. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava SK, Bhardwaj A, Singh S, Arora S, Wang B, Grizzle WE, Singh AP. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis. 2011 doi: 10.1093/carcin/bgr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs MC, Armstrong SA. FLT3 as a therapeutic target in childhood acute leukemia. Current drug targets. 2007;8:703–714. doi: 10.2174/138945007780830782. [DOI] [PubMed] [Google Scholar]
- Takahashi S. Downstream molecular pathways of FLT3 in the pathogenesis of acute myeloid leukemia: biology and therapeutic implications. J Hematol Oncol. 2011;4:13. doi: 10.1186/1756-8722-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Pasillas MP, Kamps MP. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005;106:254–264. doi: 10.1182/blood-2004-12-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Jin H, Yang Z, Luo G, Lu Y, Li K, Ren G, Su T, Pan Y, Feng B, et al. MiR-150 promotes gastric cancer proliferation by negatively regulating the pro-apoptotic gene EGR2. Biochemical and biophysical research communications. 2010;392:340–345. doi: 10.1016/j.bbrc.2009.12.182. [DOI] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Yao Q, Nishiuchi R, Li Q, Kumar AR, Hudson WA, Kersey JH. FLT3 expressing leukemias are selectively sensitive to inhibitors of the molecular chaperone heat shock protein 90 through destabilization of signal transduction-associated kinases. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9:4483–4493. [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- Zeisig BB, Milne T, Garcia-Cuellar MP, Schreiner S, Martin ME, Fuchs U, Borkhardt A, Chanda SK, Walker J, Soden R, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisig BB, Schreiner S, Garcia-Cuellar MP, Slany RK. Transcriptional activation is a key function encoded by MLL fusion partners. Leukemia. 2003;17:359–365. doi: 10.1038/sj.leu.2402804. [DOI] [PubMed] [Google Scholar]
- Zhang J, Luo N, Luo Y, Peng Z, Zhang T, Li S. microRNA-150 inhibits human CD133-positive liver cancer stem cells through negative regulation of the transcription factor c-Myb. Int J Oncol. 2011;40:747–756. doi: 10.3892/ijo.2011.1242. [DOI] [PubMed] [Google Scholar]
- Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, Shi J, Weissmueller S, Fellmann C, Taylor MJ, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25:1628–1640. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.