When plants are subjected to water stress such as drought and high salt, they develop more lateral roots through reducing miRNA167 levels and indirectly increasing IAR3 mRNA levels. The increased IAR3 enzyme converts inactive auxin to the active form, which promotes root branching. This stress adaptation miRNA-mRNA pair is evolutionarily conserved.
Abstract
The functions of microRNAs and their target mRNAs in Arabidopsis thaliana development have been widely documented; however, roles of stress-responsive microRNAs and their targets are not as well understood. Using small RNA deep sequencing and ATH1 microarrays to profile mRNAs, we identified IAA-Ala Resistant3 (IAR3) as a new target of miR167a. As expected, IAR3 mRNA was cleaved at the miR167a complementary site and under high osmotic stress miR167a levels decreased, whereas IAR3 mRNA levels increased. IAR3 hydrolyzes an inactive form of auxin (indole-3-acetic acid [IAA]-alanine) and releases bioactive auxin (IAA), a central phytohormone for root development. In contrast with the wild type, iar3 mutants accumulated reduced IAA levels and did not display high osmotic stress–induced root architecture changes. Transgenic plants expressing a cleavage-resistant form of IAR3 mRNA accumulated high levels of IAR3 mRNAs and showed increased lateral root development compared with transgenic plants expressing wild-type IAR3. Expression of an inducible noncoding RNA to sequester miR167a by target mimicry led to an increase in IAR3 mRNA levels, further confirming the inverse relationship between the two partners. Sequence comparison revealed the miR167 target site on IAR3 mRNA is conserved in evolutionarily distant plant species. Finally, we showed that IAR3 is required for drought tolerance.
INTRODUCTION
Arabidopsis thaliana plants accumulate a multitude of microRNAs (miRNAs), which are single-stranded RNA molecules 20 to 24 nucleotides in length (Reinhart et al., 2002; Carrington and Ambros, 2003). When bound to ARGONAUTE proteins, miRNAs guide RNA-induced silencing complexes to mRNAs that harbor complementary sequences (Chapman and Carrington, 2007; Montgomery and Carrington, 2008). The RNA-induced silencing complex then inhibits gene expression through translational repression or mRNA cleavage (Llave et al., 2002; Aukerman and Sakai, 2003; Bartel, 2004; Chen, 2004). One of the challenges in the field is to identify functional miRNA-mRNA pairs and elucidate their roles in the life of a plant.
Phenotypes of mutants defective in miRNA biogenesis components, such as dicer-like1, hua enhancer1, serrate, and argonaute1, provided the first indication that miRNAs are functional in developmental processes (Jacobsen et al., 1999; Lynn et al., 1999; Golden et al., 2002; Park et al., 2002; Schauer et al., 2002; Vaucheret et al., 2004; Lobbes et al., 2006; Yang et al., 2006; Sunkar et al., 2007). To date, functions of various miRNA-mRNA pairs have been clarified in the temporal and spatial development of flowers, leaves, and roots (Kutter et al., 2007; Chen, 2009; Poethig, 2009; Covarrubias and Reyes, 2010; Martin et al., 2010; Nonogaki, 2010; Nag and Jack, 2010; Khan et al., 2011).
Throughout their lifecycle, plants are challenged by highly variable environmental conditions, such as changes in nutrient levels and water availability. Plants exhibit adaptive adjustments in these conditions, allowing individual plants to maintain function and hence survival across a range of diverse environments (Sultan, 2000; Moldovan et al., 2010). The dynamic stress-responsive gene expressions, which underpin physiological adjustments, suggest that miRNAs are also likely to be involved in stress adaptations. Consistent with this notion, a mutation in HYPONASTIC LEAVES1 (HYL1), a component of the miRNA biogenesis machinery, caused altered perception of abscisic acid (ABA). Because ABA is involved in conditions of water limitation, such as salt and drought stress, this observation possibly implicates HYL1 in stress responses (Lu and Fedoroff, 2000; Han et al., 2004; Vazquez et al., 2004).
Potential involvement of a number of miRNAs has been suggested for responses to water-limiting conditions (e.g., salt, drought, and ABA responses). In particular, miR393, miR397b, and miR402, whose predicted targets are mRNAs encoding auxin receptor TRANSPORT INHIBITOR RESPONSE1, LACCASE, and DEMETER-LIKE PROTEIN3, respectively, were upregulated in response to cold, dehydration, NaCl, and ABA in Arabidopsis, rice (Oryza sativa), and Phaseolus vulgaris (Dharmasiri and Estelle, 2002; Sunkar and Zhu, 2004; Dharmasiri et al., 2005; Zhao et al., 2007; Liu et al., 2008; Arenas-Huertero et al., 2009). However, only a few miRNA-target mRNA pairs, such as miR169-NFYA5, have been functionally validated in cases of water stress (Li et al., 2008).
We hypothesized there may be additional new and functional miRNA-mRNA pairs to be identified if we analyze differential changes in miRNA and mRNA expression patterns between standard conditions and water-limiting conditions. To this end, we subjected hydroponically grown Arabidopsis plants to high osmotic stress and determined miRNA and mRNA expression patterns by parallel analyses via deep sequencing (for small RNAs) and microarray expression profiles (mRNA levels). We found the miR167-IAR3 (for IAA-Ala Resistant3) pair to be a modifying factor for root architecture under osmotic stress conditions. We showed that IAR3 is a regulator of root architecture changes in osmotic stress and drought tolerance. Consistent with its proposed role, the miR167-IAR3 relationship seems to be evolutionarily conserved in higher dicot and monocot species.
RESULTS
Reduced Accumulation of miR164a/b, miR167a/b, and miR172a/b under High Osmotic Stress
To identify new miRNA-mRNA pairs related to high osmotic conditions, we used a deep sequencing technique to profile small RNA populations in leaf and root tissues of plants under high osmotic stress and control conditions. We recovered around 20 million reads from each library, which corresponded to at least 18 million reads after adapter trimming, and more than 60% of these perfectly matched Arabidopsis genome sequences. This resulted in at least 11 million total reads for each library, which were used in subsequent analyses (see Supplemental Table 1 online). First, we searched for global changes in the population of small RNAs under high osmotic stress (see Supplemental Figure 1 online). Apart from minor changes observed among 21- and 24-nucleotide small RNAs, there were no significant differences in small RNA size distribution between the osmotic stress and control sample (see Supplemental Figures 1A and 1B online). To eliminate the noise generated by high-copy-number small RNA species, we asked whether the same pattern would be observed if small RNA species were only counted once, without any regard to copy number. Here, again, we did not detect any significant differences between the osmotic stress and control small RNA population (see Supplemental Figures 1C and 1D online). Since there were no major global changes in the small RNA population, we searched for specific small RNAs whose accumulation was affected by stress. All annotated miRNAs and trans-acting small interfering RNAs were queried in our sequencing data sets, and most of them did not change under stress. However, the accumulation of miR164a/b, miR167a/b, and miR172a/b was reduced under high osmotic stress (Table 1).
Table 1. Downregulated miRNAs with mRNA Targets in High Osmotic Stress.
| miRNA Annotation | Normalized Read Nos. |
|||||||
|---|---|---|---|---|---|---|---|---|
| Leaf |
Root |
Fold Change |
Target Genes |
|||||
| Control | Osmo | Control | Osmo | Leaf | Root | Previous Studies | This Study | |
| miR172a/b | 1,074 | 164 | 377 | 257 | 0.01 | 0.35 | AP2 | |
| miR164a/b | 1,547 | 811 | 2,404 | 1,771 | 0.27 | 0.38 | NAC1, CUC1, NAC2/ORE1 | |
| miR167a/b | 39,610 | 19,051 | 8,787 | 7,252 | 0.23 | 0.44 | ARF6/8 | IAR3 |
Normalized read numbers (reads per million) of miR164a/b, miR167a/b, and miR172a/b in leaf and root tissues of control and high osmotic stress (Osmo) plants, as well as fold changes. Thirty-day-old plants were subjected to high osmotic stress (300 mM mannitol, osmotic stress) or 0 mM mannitol (control) for 3 h and sampled. Previously identified and putative target mRNAs are shown. This study focused on IAR3.
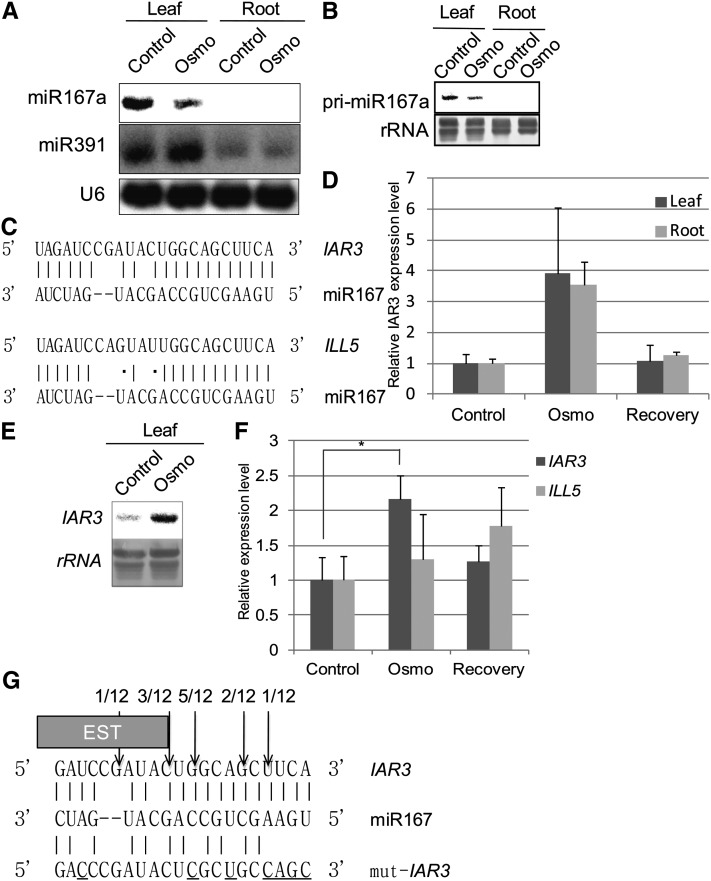
RNA gel blot analyses confirmed miR167a/b accumulation was reduced under high osmotic stress compared with control in leaf samples, whereas miR391 remained at similar levels in both tissues (Figure 1A). Using a specific probe, we found that the accumulation of pri-miR167a, which is the most highly expressed miR167 precursor, was also reduced under high osmotic stress (Figure 1B). Therefore, the reduced accumulation of miR167a/b under stress was mainly due to changes at the pre-miR167 level and not due to regulation of precursor processing.
Figure 1.
The IAR3 mRNA Accumulation Pattern Is Inversely Correlated with That of miR167a/b.
(A) RNA gel blot analysis of miR167a/b and miR391 expressions in high osmotic stress (Osmo) and control leaf and root tissues. Conditions were identical to those in Table 1. U6 accumulation is shown as a loading control and each lane contained 20 μg total RNA.
(B) RNA gel blot analysis of pre-miR167a expression in control and high osmotic stress (Osmo) leaf and root tissues. 25S rRNA was used as a loading control and each lane contained 10 μg total RNA.
(C) Sequence complementarity of miR167a/b with IAR3 and ILL5 mRNAs. Hydrogen bond is shown by a vertical line, G⋅U wobble is shown by a dot, and gap is shown by a dash.
(D) Relative expression levels of IAR3 mRNA in high osmotic stress samples (Osmo) compared with control from ATH1 microarray analyses. Microarray analyses were performed using identical samples as deep sequence analyses. In addition, materials that were sampled 24 h after stress conditions were analyzed to observe transient changes. These samples were designated “Recovery.” Bars show se (n = 3).
(E) RNA gel blot analysis of the IAR3 mRNA expression pattern in leaf tissue. 25S rRNA was used as a loading control and each lane contained 10ug RNA.
(F) Relative expression patterns of IAR3 and ILL5 mRNAs using quantitative RT-PCR with gene-specific primers. Bars show se (n = 3). Asterisks indicate a significant difference between Osmo and mock treatment, based on t test (P < 0.05).
(G) The 5′ end of the cleaved product determined by sequencing is indicated by an arrow in the miRNA:mRNA base-pairing diagram, along with the number of clones analyzed. The box shows the end of a truncated IAR3 EST clone (accession number EBENXNS01CL7RN). The bottom sequence shows a mutation strategy to generate the miR167-resistant form of IAR3 without changing its amino acid sequence. Substituted nucleotides are underlined.
A miR167a/b mRNA Target, IAR3, Is Upregulated in High Osmotic Stress
Like animal small RNAs, the inhibitory effects of plant miRNAs on target gene expression may involve translation inhibition, but plant miRNAs also mediate mRNA cleavage at the complementary site (Brodersen et al., 2008; Lin et al., 2008; Pant et al., 2008). We focused on the mRNA cleavage function, which could be monitored by changes in mRNA levels.
To find new target mRNAs, we investigated expression levels of putative target mRNAs in microarrays using samples identical to those used in small RNA deep sequencing experiments. Our computer analysis predicted nine genes that were potential miR167 targets, and seven of them were represented in the ATH1 array (see Supplemental Table 2 online). Only IAR3 showed increased mRNA levels of at least twofold under water stress (Figures 1C and 1D; see Supplemental Data Set 1 online). IAR3 encodes an indole-3-acetic acid (IAA)-Ala hydrolase, which releases bioactive auxin (IAA) from inactive auxin storage (IAA-Ala; Davies et al., 1999). To evaluate a possible transient mode of mRNA accumulation, we harvested recovery tissues from plants that were returned to the normal medium for 24 h after stress treatment. In this sample, IAR3 expression returned to control levels, strengthening our hypothesis that IAR3 mRNA was transiently upregulated in high osmotic stress. Therefore, our subsequent work focused on this putative miR167a/b-IAR3 pair. To validate this experiment, we verified that genes mediating ABA signaling, such as ABA INSENSITIVE1/2, and downstream marker genes, such as KIN1, RESPONSIVE TO DESSICATION29A/B, and COLD-REGULATED15A/B, were upregulated (see Supplemental Figure 2 online).
RNA gel blot analyses confirmed increased accumulation of the putative target IAR3 mRNA (Figure 1E). The Arabidopsis genome, however, also encodes IAA-LEUCINE RESISTANT-LIKE GENE5 (ILL5), which is located next to IAR3 on chromosome 1 and is 85% identical to IAR3 at the nucleotide sequence level (Davies et al., 1999; LeClere et al., 2002). Sequence analysis of the miR167 complementary site uncovered a three-nucleotide difference between IAR3 and ILL5, one occurring in the mismatched nucleotide between miR167 and IAR3 and the other two resulting in the hydrogen bond to G⋅U wobble changes (Figure 1C). Our initial computer prediction did not identify ILL5 as a potential target of miR167a/b because the total mismatch number exceeded the specified threshold.
To distinguish between IAR3 and ILL5 mRNAs, we designed discriminating gene primers and analyzed mRNA levels using quantitative RT-PCR. We reproduced microarray and RNA gel blot results for IAR3 but did not observe meaningful changes in ILL5 in response to water stress (Figure 1F). Although we cannot completely rule out the possibility that ILL5 is also a target, our results indicated that IAR3 was the preferred target of miR167a/b.
We noted that annealing between miR167 and IAR3 at the target site would generate an exceptional two-nucleotide asymmetric bulge. To determine if there were alternative forms of miR167, or miR167-like small RNAs that could better align with IAR3 mRNAs, we aligned all small RNAs in our osmotic stress and control data set to IAR3. We found eight miR167a-like small RNAs, which were one to three nucleotides longer than miR167a (see Supplemental Table 3 online). These small RNAs had a similar alignment to IAR3 as miR167a. However, their read numbers were too low to be functionally significant (see Supplemental Table 4 online). Thus, miR167a seemed to be the main mediator of the IAR3 cleavage.
IAR3 mRNA Is Cleaved at the miR167a/b Complementary Site
We used 5′ rapid amplification of cDNA ends (RACE) to determine the cleavage site of IAR3 mRNA. Sequence analysis of 12 independent clones placed the 5′ end of the cleaved fragment in the middle of the miR167/IAR3 mRNA complementary site (Figure 1G). We also found an EST clone (accession number EBENXNS01CL7RN), whose 3′ end is located at the second highest cleavage position (Figure 1G). Together, these results demonstrate cleavage of IAR3 mRNA at the center of the miR167a/b complementary site.
iar3 Mutants Are Insensitive to High Osmotic Stress
The upregulation of IAR3 mRNA in osmotic stress indicated that IAR3 was a potential new positive regulator in the high osmotic stress pathway. The iar3 mutant was originally isolated by Davies et al. (1999) in their effort to identify a gene product that can convert IAA-Ala to IAA using the inhibitory effect of IAA-Ala on Arabidopsis root growth. Whereas primary root growth was inhibited in the wild type, iar3 mutant primary roots continued to grow on IAA-Ala–supplemented media; hence, the name IAA-Ala Resistant3 (IAR3). The root growth inhibition in both IAA-Ala–supplemented media and high osmotic conditions led us to hypothesize that, under high osmotic stress, the upregulated IAR3 expression might contribute to primary root growth inhibition by releasing biologically active IAA. To test this hypothesis, we obtained two knockout lines harboring independent T-DNA insertions in the IAR3 locus (Alonso et al., 2003). These lines were designated as iar3-5 and iar3-6 in sequence after iar3-1 to iar3-4 mutants that were previously characterized by Davies et al. (1999). The two mutants, iar3-5 and iar3-6, contain T-DNA insertions in the 5′ untranslated region and the promoter of IAR3, respectively, and IAR3 mRNA expression levels were significantly reduced in both mutants compared with the wild type (see Supplemental Figure 3 online).
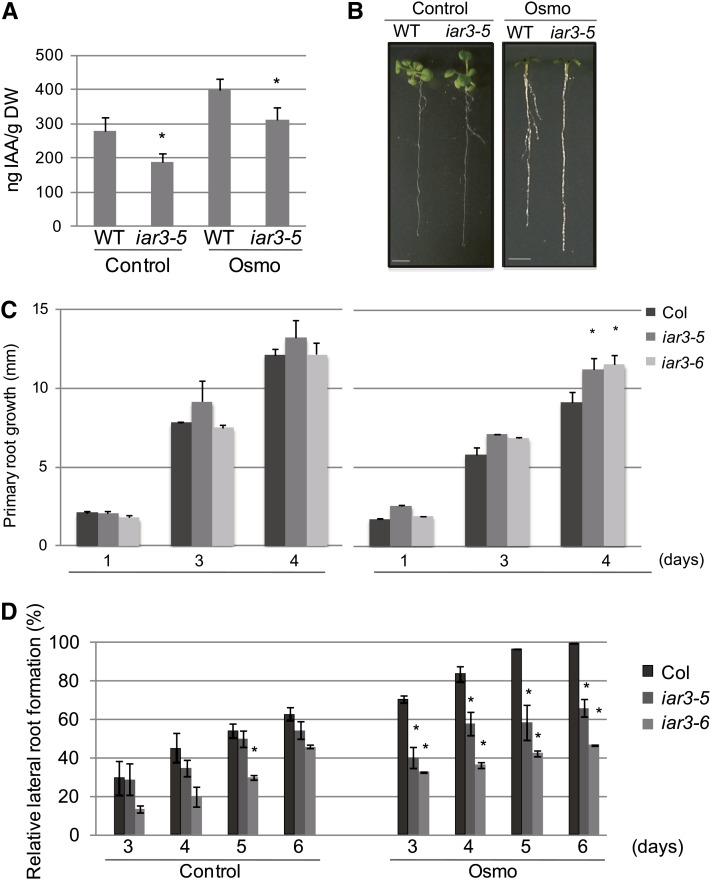
Because IAR3 is a hydrolase that releases free IAA from an inactive form (IAA-Ala), we analyzed free IAA accumulation levels in the wild type and iar3-5 mutants. Figure 2A shows that under control conditions, iar3-5 accumulated less IAA than the wild type, as previously reported (Rampey et al., 2004). Under high osmotic conditions, the wild type accumulated increased amounts of IAA compared with control conditions, whereas iar3-5 accumulated reduced amounts compared with the wild type in both control and high osmotic conditions. These results suggest that upregulated IAR3 expression levels under high osmotic conditions may contribute to increased amounts of free IAA in Arabidopsis.
Figure 2.
iar3 Mutants Are Insensitive to Osmotic Stress.
(A) Measurement of endogenous free IAA levels by liquid chromatography–electrospray ionization–tandem mass spectrometry. Increase in dry weight (DW) due to mannitol uptake was adjusted by dividing the dry weight by the average increase in dry weight under mannitol treatment. Bars show se (n = 23). Asterisks indicate a significant difference between the wild type (WT) and the mutant, based on t test (P < 0.05).
(B) Morphology of the wild type (Col) and iar3-5 under high osmotic stress (right) and control (left). Wild-type and iar3-5 plants were grown in control media for 8 d and transferred to the Osmo plate (MS containing 0.25 M mannitol) or control plate (MS alone). Pictures were taken after 1 week (Osmo) or 10 days (Control). Bar = 5 mm.
(C) Time-course experiments of the wild type, iar3-5, and iar3-6, quantifying primary root growth (mm) after seedlings were transferred to Osmo or control. Root lengths were analyzed using ImageJ (National Institutes of Health). Bars show se (n ≥ 12). Asterisks indicate a significant difference between the wild type and the mutant, based on t test (P < 0.05).
(D) Time-course experiment quantifying lateral root numbers in the wild type, iar3-5, and iar3-6. Experiments were done as in (B). Lateral roots were counted under a stereomicroscope (Leica). Lateral root numbers are represented as percentage of wild-type lateral root numbers in Osmo media at day 6. Bars show se (n ≥ 20). Asterisks indicate a significant difference between the wild type and mutant, based on t test (P < 0.01).
Since IAA is a major factor regulating primary root growth and lateral root development, we compared root morphologies of iar3 mutant and wild-type plants. Under high osmotic conditions, wild-type plants showed reduced primary root growth but increased lateral root development. By contrast, iar3 mutants were insensitive to the effect of high osmotic stress on root development (Figure 2B). We observed reduced miR167 expression levels under stress conditions primarily in leaves, but not as clearly in roots (Figures 1A and 1B), whereas IAR3 expression was enhanced in both leaf and root samples (Figures 1D to 1F). It could be, in osmotic stressed root samples, the reduction of miR167 levels was tissue specific and, therefore, not detectable by RNA gel blot analyses. Another possibility is that as the leaf miR167 level decreased under stress conditions, the resulting increase in IAR3 mRNA and protein levels led to an increased amount of IAA, which was transported to the root system. Quantitative analyses showed no obvious differences in primary root growth between the wild type and iar3 mutants under control conditions. By contrast, under high osmotic stress, both iar3 mutant alleles displayed marked insensitivity, whereas wild-type primary root growth was severely inhibited (Figure 2C).
With respect to lateral root formation, iar3-6 produced slightly fewer lateral roots compared with the wild type only on day 5 of control conditions; however, we did not observe any consistent and significant difference between iar3 mutant alleles and the wild type at other time points under the same conditions (Figure 2D). Upon transfer to high osmotic conditions, the wild type exhibited robust lateral root formation, whereas iar3 mutants were insensitive to the stress and formed fewer lateral roots (Figure 2D). In addition, iar3-6 mutants produced fewer lateral roots than iar3-5 mutants under stress conditions. This was probably due to the tendency of iar3-6 to form fewer lateral roots compared with iar3-5 under control conditions. Therefore, although the two alleles seemed to produce slightly different numbers of lateral roots, their responses to stress were very similar. These results suggest that IAR3 is a new positive regulator of high osmotic stress signaling and iar3 mutants are compromised in their response to this stress. The simplest explanation for the underlying mechanism is that bioactive IAA released by IAR3 contributes to the developmental changes in roots. Only one of the two alleles tested here showed a slight decrease in lateral root formation under control condition. This is consistent with the notion that auxin is also involved in the normal course of lateral root development. Under this condition, IAR3 is expressed at low levels but presumably plays a functional role. On the other hand, there is a greater demand for auxin under high osmotic stress conditions when plants need to develop more lateral roots. The contribution of IAR3 to plant development becomes much greater under stress, which explains its stress-induced expression (Figures 1D to 1F).
Direct Involvement of miR167 and IAR3
To obtain evidence that miR167 directly mediates IAR3 mRNA cleavage, we introduced seven nucleotide mismatches into the miR167 complementary site in the IAR3 mRNA sequence without changing the encoded amino acids, and the mutant gene version was designated mut-IAR3 (Figure 1G).
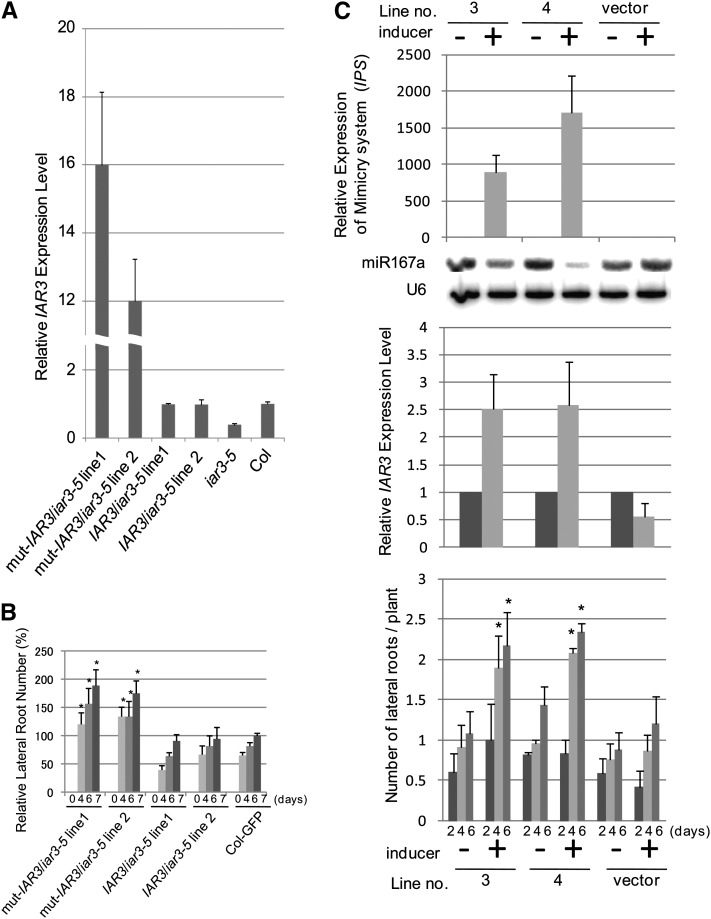
We transformed iar3-5 mutants with either IAR3:mut-IAR3 or IAR3:WT-IAR3. mRNA analyses of two independent T1 lines for each construct showed that mut-IAR3/iar3-5 lines expressed much higher IAR3 levels than WT-IAR3/iar3-5 lines (Figure 3A). For this experiment, we used control conditions to allow higher miR167 accumulation levels, thus providing a more sensitive condition than that of high osmotic stress. To determine the physiological consequence of the accumulated IAR3 mRNA levels, we examined lateral root formation in these lines under the same conditions as in Figure 3A. Plants expressing mut-IAR3 developed more lateral roots than wild-type IAR3 transgenic plants (Figure 3B). Under this condition, we did not observe consistent and significant difference between the wild type and iar3-5 (Figure 2D). Similar results were obtained when the native IAR3 promoter was replaced by a 35S promoter (see Supplemental Figure 4 online). Together, these results indicate that the higher IAR3 mRNA level in mut-IAR3 transgenic plants compared with wild-type IAR3 transgenic lines was due to increased stabilization through mutations at the miR167a complementary site.
Figure 3.
Relationships between miR167, IAR3, and Lateral Root Numbers.
(A) Relative expression levels of IAR3 mRNAs using quantitative RT-PCR in IAR3:IAR3/iar3-5, IAR3:mut-IAR3/iar3-5, the wild type, and iar3-5. Bars show se (n = 3). Asterisks indicate a significant difference between the wild type and iar3-5 or transgenic lines based on t test (P < 0.05).
(B) Lateral root numbers were analyzed in IAR3:IAR3/iar3-5, IAR3:mut-IAR3/iar3-5, and 35S:GFP (for green fluorescent protein) control plants. Average lateral root numbers of T2 population derived from two independent T1 lines are shown. Antibiotic-resistant plants were transferred to a vertical plate after 1 week and lateral root numbers scored. At day 0 (d0) there were no lateral roots. Bars show se (n ≥ 12). Asterisks indicate a significant difference between IAR3 promoter:mut-IAR3 and 35S:GFP, based on t test (P < 0.05).
(C) Transcript levels of target mimicry against miR167 (MIM167, top), miR167a (RNA gel blot), IAR3 (middle panel), and lateral root numbers (bottom) were assessed in two independent MIM167 transgenic lines. Vector control or 2-week-old seedlings were grown in control media for 2 weeks and transferred to media containing 0 or 50 μM β-estradiol. Seedlings were harvested after 5 d for RNA gel blot and quantitative PCR analyses. Bars indicate se (n = 3). Lateral root numbers were counted under a stereomicroscope at 2, 4, and 6 d after seedlings were transferred to media containing 0 or 50 μM β-estradiol. Bars indicate se (n ≥ 12). Asterisks indicate a significant difference between MIM167 and vector control transgenic lines based on t test (P < 0.05).
To confirm further that IAR3 mRNA accumulation is directly regulated by miR167, we introduced an inducible target mimicry construct against miR167a (MIM167a), so that we could sequester and downregulate miR167a activity in an inducer-dependent manner (Zuo et al., 2000; Franco-Zorrilla et al., 2007). Consistent with our expectations, two independent lines with high levels of MIM167a expression downregulated miR167 levels in an inducer-dependent manner. In the presence of inducer, these lines showed significantly increased accumulation of endogenous IAR3 mRNA compared with untreated controls (Figure 3C, top and middle panels). We also observed that these lines exhibited increased numbers of lateral roots in an inducer-dependent manner (Figure 3C, bottom panel). Thus MIM167a transgenic plants phenocopied IAR3:mut-IAR3 transgenic lines, which is in good agreement with our hypothesis. As a control, we monitored ILL5 mRNA levels and found no clear mRNA upregulation in the presence of inducer (see Supplemental Figure 5A online, left panel). This is consistent with our data that ILL5 mRNA levels did not change under stress (Figure 1F).
Using these lines, we also examined AUXIN RESPONSE FACTOR6 (ARF6) and ARF8 mRNA levels as these mRNAs harbor sequence complementarity to miR167 and have been shown to be targets of miR167 (Wu et al., 2006). We found that ARF6 and ARF8 mRNA levels were also elevated in an inducer-dependent manner (see Supplemental Figure 5A online, middle and right panels). This result prompted us to examine ARF6/8 involvement in osmotic stress. First, we confirmed by an independent analysis that ARF6/8 mRNA expression patterns did not change under osmotic stress (see Supplemental Figure 5B online). Second, we examined single and double mutant phenotypes under stress. Single mutants arf6-2 and arf8-3 showed no clear difference compared with the wild type in control and stress conditions with respect to lateral root development and primary root growth (see Supplemental Figures 5C and 5D online). However, to rule out functional redundancies, we also examined phenotypes of the arf6-2arf8-3 double mutant. The double mutant showed only a marginal phenotype under osmotic stress conditions in terms of primary root growth and lateral root development compared with the arf6-2 single mutant, which segregated from the same population as the arf6-2 arf8-3 double mutant is infertile (see Supplemental Figures 5E and 5F online). Mutant plants of arf8-3 developed more lateral roots than wild-type and arf6-2 plants under stress at day 6. This was probably due to a faster growth rate of arf8-3, as this mutant also produced more lateral roots at day 3 under control conditions. These results suggest that although ARF6/8 mRNA levels responded to reduction of miR167 levels in artificial systems like the MIM167 lines, under osmotic stress, miR167 was preferentially targeted to IAR3 over ARF6/8 by an unknown mechanism.
The miR167-IAR3 Relationship Is Evolutionarily Conserved in Vascular Plants
Since IAR3 is a positive regulator of high osmotic stress responses, the miR167- IAR3 interaction might have an important role in this process. Thus, we hypothesized that this interaction had evolved under selective pressure to conserve a responsive mechanism in high osmotic conditions, such as long drought.
We verified that the sequence of miR167a has been almost completely conserved among vascular plant species (see Supplemental Table 5 online). In all nine analyzed species, the only difference in miR167a was either an additional 22nd nucleotide or a substitution at the last nucleotide (miRBase).
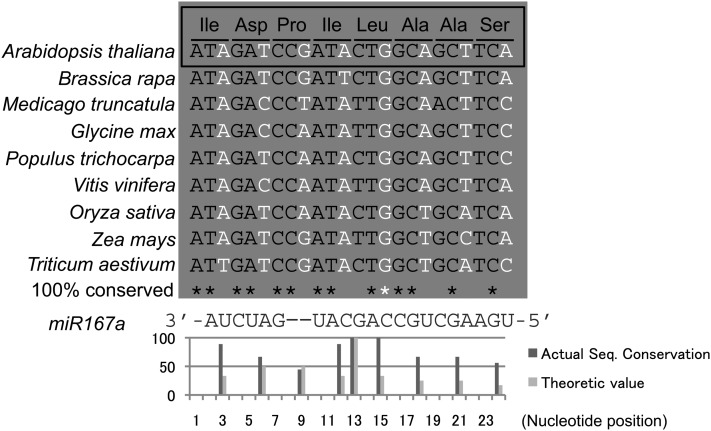
The miR167 target site on IAR3 mRNA is also conserved at both the amino acid and nucleotide levels (Campanella et al., 2003). Since the miR167 complementary site is located in an exon, this conservation is most likely due to the selective pressure on the amino acid sequence and not its relationship with miR167a/b. To observe sequence conservation at the miR167 complementary site in a manner that is affected by miR167 targeting, we took advantage of synonymous substitutions, which often occurred in the 3rd nucleotide of triplet codons. Figure 4 shows IAR3 sequences from different vascular plant species. Although there is no clear conservation of the 3rd nucleotide of amino acids located in 3′ and 5′ ends of the target site, those at the center of the complementary site, which is thought to be crucial for the cleavage, are almost completely conserved. We quantified the conservation rate by calculating actual and theoretical nucleotide conservation ratios, which confirmed that the actual conservation ratios were consistently higher than the theoretical values at the center (Figure 4, bottom). In fact, the 3rd nucleotide of Leu was most conserved and this position was exactly where the most frequent cleavage occurred in our 5′ RACE analyses (Figure 1G). The first nucleotide of Leu did not seem to be conserved since there were synonymous substitutions. However, the corresponding nucleotide of the miR167a/b was G, which meant that both C and T were complementary nucleotides. Thus, this nucleotide was almost completely conserved as well.
Figure 4.
IAR3 Sequence of the miR167 Target Site Is Evolutionarily Conserved.
IAR3 sequences of the miR167 target site from Arabidopsis, Brassica rapa, M. truncatula, soybean, Populus trichocarpa, grape (Vitis vinifera), rice, maize, and wheat (Triticum aestivum) are shown. The amino acid sequence is shown on top. Third nucleotide of triplet codon is indicated in white; 100% conserved nucleotide in first or second nucleotide of triplet codon is indicated by a black asterisk; that of a 3rd nucleotide is indicated by a white asterisk. miR167a/b sequence is shown at the bottom. The graph shows the percentage of nucleotide sequence conservation for the third nucleotide of each amino acid, calculated from the species shown in this figure.
Given that IAR3 but not ILL5 responded to miR167, we predicted that there was no similar sequence conservation of the miR167 target site between IAR3 and other members of the IAR3 family. Indeed, there was no significant sequence conservation at the synonymous substitution sites (see Supplemental Table 6 online). Taken together, the miR167a-IAR3 relationship appears to be evolutionarily conserved.
IAR3 Is a Positive Regulator of Drought Stress Tolerance
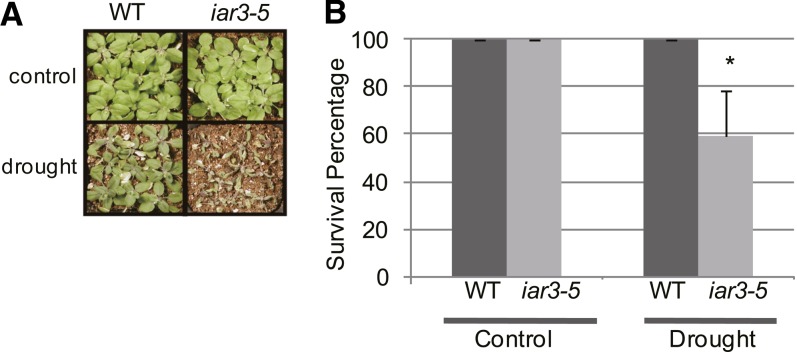
That IAR3-dependent increase in lateral root development occurred during high osmotic stress and that IAR3 was an evolutionarily conserved target of miR167a suggested a possible role of IAR3 in drought stress tolerance. To explore this possibility, we analyzed the drought stress tolerance of iar3 mutants and the wild type by measuring their capacity to survive for 2 weeks after withholding water. We found that the iar3 mutants were significantly more sensitive to drought stress than the wild type (Figure 5). Thus, the function of IAR3 was indispensable in drought tolerance.
Figure 5.
IAR3 Is a Positive Regulator of Drought Tolerance.
(A) iar3-5 and wild-type (WT; Col) plants were grown for 2 weeks after soil transfer, with half of the population subjected to drought stress by withholding the water supply and the other half watered normally. Pictures were taken 2.5 weeks after withholding water.
(B) Col and iar3-5 plants that survived the drought stress were scored after resuming the water supply. Bars show se (n = 27). Asterisks indicate a significant difference between the wild type and iar3-5, based on t test (P < 0.05).
DISCUSSION
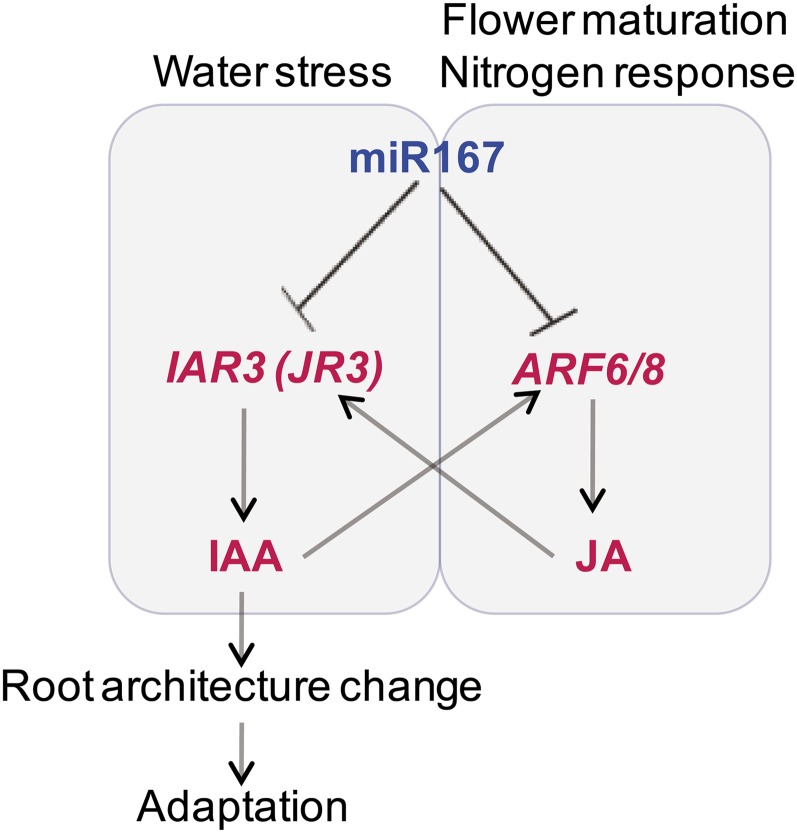
By a combination of deep sequencing of small RNAs and microarray profiling of mRNAs, we showed that plants responded to high osmotic stress by downregulating miR167 expression, which in turn, upregulated IAR3 mRNA abundance (Figure 6). We identified a new and functional miRNA-mRNA pair by showing (1) an inverse correlation in expression pattern between the two partners; (2) IAR3 mRNA cleavage at the miR167 complementary site; (3) transgenic plants expressing a miR167-resistant form of IAR3 without changing its encoded amino acid sequence exhibited a constitutive stress response, and these plants showed high levels of IAR3 mRNA accumulation and increased lateral root development compared with plants expressing the wild type form of IAR3; and (4) miR167a downregulation by target mimicry technique resulted in the upregulation of IAR3 transcripts as well as lateral root development.
Figure 6.
A Working Model of an Integrated View of the First and Secondary Circuits of miR167 Pathways.
Blue letters represent lower expression levels, and red letters represent increased mRNA levels. Dotted arrows show possible secondary effects, in which jasmonic acid produced from the ARF6/8 pathway stimulates accumulation of IAR3 mRNA, which is also called JR3. Likewise, auxin released from IAR3 stimulates ARF6/8.
We also showed that IAR3, which releases bioactive IAA from an inactive precursor, is a previously unknown positive factor in root architecture changes in this process (Figure 6). This is consistent with the rich genetic and experimental evidence that auxin is the morphogenic trigger for lateral root formation (Benková et al., 2009) and that ABA and auxin interact in seedling stage (Belin et al., 2009). Consistent with its function in root architecture changes under high osmotic stress, IAR3 is also important for drought tolerance. Our observation is also consistent with the result of Liu et al. (1992) using excised segments of Vigna hypocotyl that showed that IAA applied simultaneously with osmotic stress enhanced the adaptive recovery of elongation growth by enhancing wall extensibility.
Taken together, our results suggest that the miR167–IAR3 interaction plays a role in root architecture changes under high osmotic stress as well as in drought stress tolerance. In agreement with this notion, the miR167-IAR3 interaction appears to be evolutionarily conserved.
Localization of the miR167-IAR3 Circuit
In addition to IAR3 mRNA, ARF6/8 mRNAs are also targets of miR167 (Wu et al., 2006). Gifford et al. (2008) showed that the miR167-ARF6/8 circuit regulates nitrogen-responsive root development by fine-tuning the ratio of initiating/emerging lateral roots. In support of this role, both pre-miR167a and ARF8 were expressed in the pericycle, where lateral roots emerge (Dubrovsky et al., 2000; Hardtke, 2006; De Smet et al., 2006; Parizot et al., 2008; Petricka and Benfey, 2008). Since these authors performed global cellular expression profiling (Gifford et al., 2008), we searched their database to see if IAR3 was also expressed in the pericycle. Indeed, we found that among different root tissues, the highest IAR3 expression level was detected in the pericycle (see Supplemental Figure 6 online). This is consistent with our finding that miR167 and IAR3 interact directly and that IAR3 enhances lateral root development (Figure 6).
However, miR167 does not seem to target ARF6/8 under high osmotic stress (see Supplemental Figures 5B to 5E online). In the future, it would be interesting to investigate how plants use a common regulator (miR167) to perceive different inputs (osmotic stress or nitrogen application) but have distinct outputs as a result.
Although miR167-ARF6/8 and miR167-IAR3 circuits do not interact directly, we note that ARF6/8 promote jasmonic acid production (Nagpal et al., 2005), and jasmonic acid is a strong inducer of IAR3, which is identical to JASMONATE RESPONSIVE3 (JR3). On the other hand, bioactive auxin that is released by IAR3 could easily stimulate the expression of ARF6/8, which are auxin-responsive factors. Thus, although these two pathways are not linked in the initial response, they may interact in secondary responses (Figure 6).
Evolutionary Conservation of miR167-IAR3
The miR167-IAR3 relationship appears to be evolutionarily conserved because sequences of synonymous substitution sites are conserved in evolutionarily distant plant species. This is consistent with the recent findings of Wang et al. (2011) that the functions of miR156 and SPL mRNAs encoding transcription factors in juvenile-to-adult transition are conserved between Arabidopsis, maize (Zea mays), and trees (Wu et al., 2009; Wang et al., 2009). The observed versus actual conservation rate revealed that the 5′ end of miR167 is more conserved than the 3′ end, in agreement with the result of Lin et al. (2009), who reported that the 5′ half of artificial miRNA is crucial for its recognition. Furthermore, the complete conservation of the guanine residue at the cleavage site is also consistent with the result of artificial miRNA-targeted virus sequence evolution studies (Lafforgue et al., 2011). Analyses of virus sequence mutations in the lineages that broke artificial miRNA suppression showed that the nucleotide sequence at the cleavage site was most frequently mutated, suggesting also that this cleavage site is crucial in mRNA cleavage by miRNA.
What makes the miR167-IAR3 relationship advantageous during the course of evolution is not known for certain. Perhaps, developmental flexibility—short and highly branched root architecture—in water-limiting conditions contributes to plant survival in long droughts.
While most of the angiosperm species evolved one to four orthologs of IAR3, legume species such as Medicago truncatula and soybean (Glycine max) evolved seven orthologs (www.phytozome.net). Perhaps these IAR3 orthologs contribute to better fitness through optimizing root architecture for maximum nitrogen assimilation. Another point is that M. truncatula is much more drought tolerant than other leguminous species (Gonzalez et al., 1995; Gálvez et al., 2005). It would be interesting to explore the functional divergence of M. truncatula IAR3s in terms of legume-rhizobium symbiosis and drought tolerance.
PIN3 Possibly Functions Downstream of IAR3
Our microarray analyses showed that auxin-related genes, such as PIN-FORMED3 (PIN3), INDOLE-3-ACETIC ACID INDUCIBLE30 (IAA30), MYB DOMAIN PROTEIN14, and HOMEOBOX PROTEIN40, were upregulated in high osmotic conditions (see Supplemental Figure 7 online). These genes could be responding to auxin released by IAR3. PIN3, which controls the direction and rate of cellular auxin efflux, is of particular interest as it is a key factor in the establishment of auxin gradient in adaptation responses such as gravitropism, phototropism, and shade avoidance (Friml et al., 2002; Ding and Friml, 2010; Keuskamp et al., 2010).
In roots, PIN3 has been reported to mediate the early phase of root gravity response and this is inhibited by cold stress (Friml et al., 2002; Shibasaki et al., 2009). This auxin transporter is localized in pericycle and columella cells (Friml et al., 2002; Blilou et al., 2005), and the former cell type is where miR167 and IAR3 are expressed (see above). PIN3 proteins in columella cells contribute to the reestablishment of the auxin gradient in gravitropism (Friml et al., 2002), and in pericycle cells, PIN3 proteins are involved in the lateral root development (Benková et al., 2003). Under normal growth conditions, pin3 mutants produce fewer lateral root initials, and the double mutant pin1 pin3 shows defects in lateral root primordium development (Benková et al., 2003). Expressed in the inner cells of lateral root primordia, PIN1 subcellular localization changes polarity from transverse to lateral membranes toward the primordium tip. This correlates with the establishment of an auxin gradient with its maxima at primordium tip (Benková et al., 2003). Interestingly, PIN3 is expressed at the base of lateral root primordia (Benková et al., 2003).
It is possible that, under high osmotic stress, free auxin released by IAR3 stimulates lateral root development through PIN3 where the latter contributes to auxin gradient establishment in the lateral root primordial by an unknown mechanism. Future experiments should test if the PIN3 subcellular localization changes under high osmotic conditions, particularly in relation to lateral root primordia.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used as the wild type. iar3-5 (SALK_042101) and iar3-6 (SALK_090805) were obtained from the ABRC (Alonso et al., 2003). For hydroponic culture, wild-type seeds after a 3-d cold treatment at 4°C were sowed on rock wool (Grodan) presoaked in media containing 1.75 mM sodium phosphate buffer (pH5.8), 1.5 mM MgSO4, 2.0 mM Ca(NO3)2, 3.0 mM KNO3, 67 μM Na2EDTA, 8.6 μM FeSO4, 10.3 μM MnSO4, 30μM H3BO3, 1.0 μM ZnSO4, 24 nM (NH4)6Mo7O24, 130 nM CoCl2, and 1 μM CuSO4 (Fujiwara et al., 1992; Taniguchi et al., 1998). In short, plants were grown at 22°C with a 150 μmol m−2 s−1 fluorescence rate and under 12-h/12-h light/dark cycle in MGRL hydroponic medium. At this time, three separate populations of seedlings were defined. Two populations of 30-d-old plants were then subjected to high osmotic stress by adding 0.3 M mannitol to the basal MGRL medium for 3 h in the light (designated: Osmo treatment). The remaining population was treated without 0.3 M mannitol (designated: Mock). Recovery plants were returned to the MGRL medium for 1 d after the stress treatment and sampled (designated: Recovery). This procedure was used to treat three independent biological replicates. Leaf and root samples were collected separately from the same set of plants. There were a total of 18 samples (two tissues × three samples × three replicas).
RNA Isolation and sRNA Library Construction
For small RNA library construction, three biological replicates were pooled and total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. Small RNAs (18 to 28 nucleotides) from 40 μg total RNA were separated and then fractionated using 15% polyacrylamide urea gel. Subsequent steps were performed by the protocol of Digital Gene Expression for Small RNA Sample Preparation (Illumina). Qualities and quantities of individual sRNA libraries were confirmed using a bioanalyzer. Solexa sequencing was performed at the Genomics Resource Center in the Rockefeller University following the manufacturer’s instructions (Illumina).
Small RNA Gel Blot Hybridization and 5′ RACE
RNA gel blot and 5′ RACE were performed essentially according to Reyes and Chua (2007). Briefly, total RNA was separated on 15% polyacrylamide urea gels and then transferred onto Amersham Hybond-N+ (GE Healthcare). The membrane was prehybridized with Ultrahyb-oligo buffer (Ambion) at 42°C for at least 1 h after UV cross-linking. For probe labeling, 15 pmol oligo DNA complementary to a mature miRNA sequence was radioactively labeled by T7 polynucleotide kinase reaction (NEB). [32γ]ATP (40 μCi) was used for the phosphorylation reaction at 37°C for 1 h. Radioactive ATP unincorporated was removed by a mini quick spin oligo columns (Roche), and the purified probe was used for hybridization at 42°C overnight. The membrane was washed with 2× SSC and 0.1% SDS. The First Choice 5′ RLM-RACE Kit (Ambion) was used for 5′ RACE according to the manufacturer’s instructions.
Phenotypic Assays
For root growth assays, plants were grown for 1 week (Murashige and Skoog [MS], 0 or 0.5% Suc, and 0.8% agar) and transferred to vertical plates containing MS medium (half-strength MS, 0.5% Suc, and 0.6% agar) supplemented with 0.25 M mannitol for high osmotic stress treatments. Growth rates of primary roots were measured using ImageJ software (National Institutes of Health). Drought assays were done according to Catala et al. (2007). All experiments were performed with at least three biological replicates.
Small RNA Sequencing Data
Adapter sequence was clipped by the Perl program. All trimmed reads were mapped to the Arabidopsis genome (TAIR9) using local C program. Only reads with a perfect hit(s) were selected for further analysis.
Prediction and Identification of Arabidopsis miRNAs and Their mRNA Targets
The method of Wang et al. (2004) was used.
GeneChip Arrays
In short, targets for Arabidopsis GeneChip ATH1 were synthesized from 1 μg total RNA extracted by RNA easy extraction kit (Qiagen). In total, there were 18 individual GeneChip targets comprising three biological replicates for CL (Col leaf), CR (Col root), OL (osmotic-stressed leaf), OR (osmotic-stressed root), RL (recovery leaf), and RR (recovery root). Target synthesis, hybridization, and scanning of all arrays were performed at the Genomics Resource Center, Rockefeller University, following the manufacturer’s instructions (Affymetrix). Background correction was performed as described by Irizarry et al. (2003). Quantile normalization was performed as described by Bolstad et al. (2003). Finally, targets were summarized into single-gene expression (probe set) values on a per chip basis using the median polish algorithm (Tukey, 1977). Differential gene expression was determined for each of the following comparisons: OR/CR, OR/RR, OL/CL, and OL/RL. Significance was determined at P values below 0.05, adjusted for multiplicity using the Bonferroni correction method (22,811 probe sets). All comparisons were made using the ebayes function implemented under the limma package in the R statistical environment (Clayton and Kaldor, 1987; Smyth, 2005). Adjusted P values for all the genes represented in the ATH1 array can be found in Supplemental Data Set 1 online.
Plasmid Construction, Plant Transformation, and RNA Isolation
Plant transformation, plasmid construction, and RNA gel blot analyses were done as described by Kinoshita et al. (2010). Promoter fusions were constructed by QuikChange Lightning Site-Directed Mutagenesis (Stratagene) and In-Fusion Cloning Kit (Clontech) according to the manufacturers’ instructions using oligonucleotides in Supplemental Table 7 online. pBA002a was used as a vector without promoter with EcoRV and MluI as restriction enzymes (Møller et al., 2001). Gateway (Invitrogen)–compatible inducible vector and pBA002 vector were used for inducible MIM167 cloning and 35S:IAR3 cloning, respectively (Zuo et al., 2000).
Endogenous Auxin Measurement
Free IAA was measured in three biological replicates according to Sugawara et al. (2009).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: IAR3 (AT1G51760), ILL5 (AT1g51780), ARF6 (AT1G30330), ARF8 (AT5G37020), miR167a (AT3G22886) and PIN3 (AT1G70940). Raw data are available from the Gene Expression Omnibus under accession numbers GSE36560 (small RNA) and GSE36789 (microarray).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Size Distribution of Small RNAs from Leaves and Roots in Control and High Osmotic Stress.
Supplemental Figure 2. Relative Expression Levels of High Osmotic Stress-Responsive Genes.
Supplemental Figure 3. IAR3 mRNA Levels in iar3-5 and iar3-6.
Supplemental Figure 4. Relationships between miR167, IAR3, and Lateral Root Numbers in Constitutive Promoter System.
Supplemental Figure 5. Involvement of ILL5, ARF6, and ARF8 in the MIM167 System.
Supplemental Figure 6. Tissue-Specific Expression of IAR3 mRNA in Root Tissue.
Supplemental Figure 7. Relative Expression Patterns of Auxin-Responsive Genes.
Supplemental Table 1. Summary of Deep Sequencing Results.
Supplemental Table 2. List of miR167 Putative Targets.
Supplemental Table 3. miR167-Like Small RNAs That Might Target IAR3 mRNA.
Supplemental Table 4. Normalized Expression Levels of miR167 and miR167-Like RNAs in Stress.
Supplemental Table 5. Sequence Conservation of miR167 among Vascular Plants.
Supplemental Table 6. miR167a Target Site Conservation among IAR3 Family Members.
Supplemental Table 7. List of Oligonucleotide Sequences.
Supplemental Data Set 1. Significance Tests for All Comparisons.
Acknowledgments
We thank Jason Reed for the generous gifts of arf6-2 and arf8-3 single mutant and arf6-2 arf8-3 double mutant seeds, our laboratory members for discussions, Nagarajan Chandramouli of Rockefeller Proteomics Resource Center for sample preparation relating to free IAA measurement, and Scott Dewell of the Genomics Resource Center for deep sequencing. N.K. was supported by postdoctoral fellowships from the Uehara Memorial Foundation, Swiss National Science Foundation (for Prospective Researchers), and the Japan Society for the Promotion of Science. This work was supported in part by Bayer Crop Science and in part by the Cooperative Research Program for Agricultural Science and Technology Development (PJ906910), Rural Development Administration, Republic of Korea.
AUTHOR CONTRIBUTIONS
N.K., H.W., and N.-H.C. designed research. N.K. performed most of the experiments. H.K. and Y.K. measured endogenous auxin. H.W., C.M., and J.L. carried out bioinformatics analyses. Results were discussed with all authors. The article was written by N.K. and N.-H.C. and read by all the authors.
Glossary
- miRNA
microRNA
- ABA
abscisic acid
- IAA
indole-3-acetic acid
- RACE
rapid amplification of cDNA ends
- MGRL
to be defined
- MS
Murashige and Skoog
- Col
Columbia
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero C., Pérez B., Rabanal F., Blanco-Melo D., De la Rosa C., Estrada-Navarrete G., Sanchez F., Covarrubias A.A., Reyes J.L. (2009). Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol. Biol. 70: 385–401 [DOI] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Belin C., Megies C., Hauserová E., Lopez-Molina L. (2009). Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell 21: 2253–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Ivanchenko M.G., Friml J., Shishkova S., Dubrovsky J.G. (2009). A morphogenetic trigger: Is there an emerging concept in plant developmental biology? Trends Plant Sci. 14: 189–193 [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Campanella J.J., Larko D., Smalley J. (2003). A molecular phylogenomic analysis of the ILR1-like family of IAA amidohydrolase genes. Comp. Funct. Genomics 4: 584–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington J.C., Ambros V. (2003). Role of microRNAs in plant and animal development. Science 301: 336–338 [DOI] [PubMed] [Google Scholar]
- Catala R., Ouyang J., Abreu I.A., Hu Y., Seo H., Zhang X., Chua N.H. (2007). The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19: 2952–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.J., Carrington J.C. (2007). Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 8: 884–896 [DOI] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D., Kaldor J. (1987). Empirical Bayes estimates of age-standardized relative risks for use in disease mapping. Biometrics 43: 671–681 [PubMed] [Google Scholar]
- Covarrubias A.A., Reyes J.L. (2010). Post-transcriptional gene regulation of salinity and drought responses by plant microRNAs. Plant Cell Environ. 33: 481–489 [DOI] [PubMed] [Google Scholar]
- Davies R.T., Goetz D.H., Lasswell J., Anderson M.N., Bartel B. (1999). IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I., Vanneste S., Inzé D., Beeckman T. (2006). Lateral root initiation or the birth of a new meristem. Plant Mol. Biol. 60: 871–887 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri S., Estelle M. (2002). The role of regulated protein degradation in auxin response. Plant Mol. Biol. 49: 401–409 [PubMed] [Google Scholar]
- Ding Z., Friml J. (2010). Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 107: 12046–12051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky J.G., Doerner P.W., Colón-Carmona A., Rost T.L. (2000). Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol. 124: 1648–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Hirai M.Y., Chino M., Komeda Y., Naito S. (1992). Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 99: 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford M.L., Dean A., Gutierrez R.A., Coruzzi G.M., Birnbaum K.D. (2008). Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden T.A., Schauer S.E., Lang J.D., Pien S., Mushegian A.R., Grossniklaus U., Meinke D.W., Ray A. (2002). SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 130: 808–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E., Gordon A., James C., Arreseigor C. (1995). The role of sucrose synthase in the response of soybean nodules to drought. J. Exp. Bot. 46: 1515–1523 [Google Scholar]
- Gálvez L., González E.M., Arrese-Igor C. (2005). Evidence for carbon flux shortage and strong carbon/nitrogen interactions in pea nodules at early stages of water stress. J. Exp. Bot. 56: 2551–2561 [DOI] [PubMed] [Google Scholar]
- Han M.H., Goud S., Song L., Fedoroff N. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 101: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S. (2006). Root development—Branching into novel spheres. Curr. Opin. Plant Biol. 9: 66–71 [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jacobsen S.E., Running M.P., Meyerowitz E.M. (1999). Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126: 5231–5243 [DOI] [PubMed] [Google Scholar]
- Keuskamp D.H., Pollmann S., Voesenek L.A., Peeters A.J., Pierik R. (2010). Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc. Natl. Acad. Sci. USA 107: 22740–22744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan G.A., Declerck M., Sorin C., Hartmann C., Crespi M., Lelandais-Brière C. (2011). MicroRNAs as regulators of root development and architecture. Plant Mol. Biol. 77: 47–58 [DOI] [PubMed] [Google Scholar]
- Kinoshita N., Berr A., Belin C., Chappuis R., Nishizawa N.K., Lopez-Molina L. (2010). Identification of growth insensitive to ABA3 (gia3), a recessive mutation affecting ABA signaling for the control of early post-germination growth in Arabidopsis thaliana. Plant Cell Physiol. 51: 239–251 [DOI] [PubMed] [Google Scholar]
- Kutter C., Schöb H., Stadler M., Meins F., Jr, Si-Ammour A. (2007). MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 19: 2417–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafforgue G., Martínez F., Sardanyés J., de la Iglesia F., Niu Q.W., Lin S.S., Solé R.V., Chua N.H., Daròs J.A., Elena S.F. (2011). Tempo and mode of plant RNA virus escape from RNA interference-mediated resistance. J. Virol. 85: 9686–9695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere S., Tellez R., Rampey R.A., Matsuda S.P., Bartel B. (2002). Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277: 20446–20452 [DOI] [PubMed] [Google Scholar]
- Li W.X., Oono Y., Zhu J., He X.J., Wu J.M., Iida K., Lu X.Y., Cui X., Jin H., Zhu J.K. (2008). The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20: 2238–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.I., Chiang S.F., Lin W.Y., Chen J.W., Tseng C.Y., Wu P.C., Chiou T.J. (2008). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 147: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.S., Wu H.W., Elena S.F., Chen K.C., Niu Q.W., Yeh S.D., Chen C.C., Chua N.H. (2009). Molecular evolution of a viral non-coding sequence under the selective pressure of amiRNA-mediated silencing. PLoS Pathog. 5: e1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.H., Tian X., Li Y.J., Wu C.A., Zheng C.C. (2008). Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14: 836–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Katou K., Okamoto H. (1992). Effects of exogenous auxin on the regulation of elongation growth of excised segments of vigna hypocotyls under osmotic-stress. Plant Cell Physiol. 33: 915–919 [Google Scholar]
- Llave C., Xie Z., Kasschau K.D., Carrington J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056 [DOI] [PubMed] [Google Scholar]
- Lobbes D., Rallapalli G., Schmidt D.D., Martin C., Clarke J. (2006). SERRATE: A new player on the plant microRNA scene. EMBO Rep. 7: 1052–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Fedoroff N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn K., Fernandez A., Aida M., Sedbrook J., Tasaka M., Masson P., Barton M.K. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126: 469–481 [DOI] [PubMed] [Google Scholar]
- Martin R.C., Liu P.P., Goloviznina N.A., Nonogaki H. (2010). microRNA, seeds, and Darwin?: Diverse function of miRNA in seed biology and plant responses to stress. J. Exp. Bot. 61: 2229–2234 [DOI] [PubMed] [Google Scholar]
- Moldovan D., Spriggs A., Yang J., Pogson B.J., Dennis E.S., Wilson I.W. (2010). Hypoxia-responsive microRNAs and trans-acting small interfering RNAs in Arabidopsis. J. Exp. Bot. 61: 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller S.G., Kunkel T., Chua N.H. (2001). A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev. 15: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery T.A., Carrington J.C. (2008). Splicing and dicing with a SERRATEd edge. Proc. Natl. Acad. Sci. USA 105: 8489–8490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., Jack T. (2010). Sculpting the flower; the role of microRNAs in flower development. Curr. Top. Dev. Biol. 91: 349–378 [DOI] [PubMed] [Google Scholar]
- Nagpal P., Ellis C.M., Weber H., Ploense S.E., Barkawi L.S., Guilfoyle T.J., Hagen G., Alonso J.M., Cohen J.D., Farmer E.E., Ecker J.R., Reed J.W. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118 [DOI] [PubMed] [Google Scholar]
- Nonogaki H. (2010). MicroRNA gene regulation cascades during early stages of plant development. Plant Cell Physiol. 51: 1840–1846 [DOI] [PubMed] [Google Scholar]
- Pant B.D., Buhtz A., Kehr J., Scheible W.R. (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parizot B., et al. (2008). Diarch symmetry of the vascular bundle in Arabidopsis root encompasses the pericycle and is reflected in distich lateral root initiation. Plant Physiol. 146: 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka J.J., Benfey P.N. (2008). Root layers: Complex regulation of developmental patterning. Curr. Opin. Genet. Dev. 18: 354–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S. (2009). Small RNAs and developmental timing in plants. Curr. Opin. Genet. Dev. 19: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey R.A., LeClere S., Kowalczyk M., Ljung K., Sandberg G., Bartel B. (2004). A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 135: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. (2002). MicroRNAs in plants. Genes Dev. 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J.L., Chua N.H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49: 592–606 [DOI] [PubMed] [Google Scholar]
- Schauer S.E., Jacobsen S.E., Meinke D.W., Ray A. (2002). DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 7: 487–491 [DOI] [PubMed] [Google Scholar]
- Shibasaki K., Uemura M., Tsurumi S., Rahman A. (2009). Auxin response in Arabidopsis under cold stress: Underlying molecular mechanisms. Plant Cell 21: 3823–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. (2005). Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor, C.V.R. Gentleman, S. Dudoit, R. Irizarry, and W. Huber, eds (New York: Springer), pp. 397–420 [Google Scholar]
- Sugawara S., Hishiyama S., Jikumaru Y., Hanada A., Nishimura T., Koshiba T., Zhao Y., Kamiya Y., Kasahara H. (2009). Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 5430–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan S.E. (2000). Phenotypic plasticity for plant development, function and life history. Trends Plant Sci. 5: 537–542 [DOI] [PubMed] [Google Scholar]
- Sunkar R., Chinnusamy V., Zhu J., Zhu J.K. (2007). Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12: 301–309 [DOI] [PubMed] [Google Scholar]
- Sunkar R., Zhu J.K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M., Kiba T., Sakakibara H., Ueguchi C., Mizuno T., Sugiyama T. (1998). Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett. 429: 259–262 [DOI] [PubMed] [Google Scholar]
- Tukey J.W. (1977). Some thoughts on clinical trials, especially problems of multiplicity. Science 198: 679–684 [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez F., Crété P., Bartel D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Gasciolli V., Crété P., Vaucheret H. (2004). The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14: 346–351 [DOI] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wang J.W., Park M.Y., Wang L.J., Koo Y., Chen X.Y., Weigel D., Poethig R.S. (2011). miRNA control of vegetative phase change in trees. PLoS Genet. 7: e1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.J., Reyes J.L., Chua N.H., Gaasterland T. (2004). Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 5: R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.F., Tian Q., Reed J.W. (2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133: 4211–4218 [DOI] [PubMed] [Google Scholar]
- Yang L., Liu Z., Lu F., Dong A., Huang H. (2006). SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47: 841–850 [DOI] [PubMed] [Google Scholar]
- Zhao B., Liang R., Ge L., Li W., Xiao H., Lin H., Ruan K., Jin Y. (2007). Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 354: 585–590 [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]