Abstract
Lipoic acid is a coenzyme that is essential for the activity of enzyme complexes such as those of pyruvate dehydrogenase and glycine decarboxylase. We report here the isolation and characterization of LIP1 cDNA for lipoic acid synthase of Arabidopsis. The Arabidopsis LIP1 cDNA was isolated using an expressed sequence tag homologous to the lipoic acid synthase of Escherichia coli. This cDNA was shown to code for Arabidopsis lipoic acid synthase by its ability to complement a lipA mutant of E. coli defective in lipoic acid synthase. DNA-sequence analysis of the LIP1 cDNA revealed an open reading frame predicting a protein of 374 amino acids. Comparisons of the deduced amino acid sequence with those of E. coli and yeast lipoic acid synthase homologs showed a high degree of sequence similarity and the presence of a leader sequence presumably required for import into the mitochondria. Southern-hybridization analysis suggested that LIP1 is a single-copy gene in Arabidopsis. Western analysis with an antibody against lipoic acid synthase demonstrated that this enzyme is located in the mitochondrial compartment in Arabidopsis cells as a 43-kD polypeptide.
Lipoic acid (6,8-thioctic acid) is a sulfur-containing coenzyme that is required for the activity of enzyme complexes involved in the oxidative decarboxylation of α-ketoacids (Reed and Hackert, 1990; Perham, 1991; Mattevi et al., 1992) and in the Gly-cleavage system (Fujiwara et al., 1990; Kim and Oliver, 1990; Macherel et al., 1990). There are five lipoyl proteins: the dihydrolipoamide acyltransferase subunits of pyruvate, α-ketoglutarate, branched-chain α-ketoacid dehydrogenase complexes, protein X of the pyruvate dehydrogenase complex, and the H-protein of the Gly-cleavage system. Lipoic acid is covalently bound to these proteins via an amide linkage to the ε-amino group of specific Lys residues (Reed and Hackert, 1966). The lipoyl-Lys arm functions as a carrier of reaction intermediates, and reducing equivalents interact with the active sites of the components of the complexes (Yeaman, 1989; Douce et al., 1994).
Despite the importance of the lipoyl-prosthetic group in the functioning of several enzyme complexes, the biosynthesis of lipoic acid is not well understood in any organism. Molecular genetic studies (Vanden Boom et al., 1991; Reed and Cronan, 1993; Morris et al., 1994, 1995) of Escherichia coli have identified three genes, lipA, lipB, and lplA that are involved in the biosynthesis and transfer of lipoic acid. lipA encodes a lipoic acid synthase that is required for an insertion of the first sulfur atom into the octanoate backbone (Reed and Cronan, 1993). lplA and lipB encode lipoate ligases that are involved in the transfer of lipoic acid to cognate proteins (Morris et al., 1994, 1995). LplA and LipB proteins primarily function in the utilization of lipoic acid exogenously added to the growth medium and endogenously synthesized lipoic acid in E. coli cells, respectively (Morris et al., 1994, 1995). LplA protein is involved not only in biosynthesis of lipoyl-AMP with lipoic acid and ATP but also in the transfer of the lipoyl group from lipoyl-AMP to cognate proteins (Morris et al., 1995). Although LipB protein also transfers the lipoyl group to cognate protein, this transfer involves the lipoyl group in lipoyl-ACP (Jordan and Cronan, 1997).
Biosynthesis of lipoic acid in E. coli has also been studied using labeling experiments. Parry (1983) demonstrated that octanoic acid is a direct precursor of lipoic acid and 8-thiooctanoic and 6-thiooctanoic acids are possible intermediates in lipoic acid biosynthesis. This finding is consistent with the fact that the E. coli lipA mutant is complemented by the addition of both thiooctanoic acids (Reed and Cronan, 1993) and suggests the presence of a second enzyme involved in insertion of a second sulfur atom into thiooctanoic acids.
Although the biosynthesis of lipoic acid in E. coli has been studied, much less is known about biosynthesis of lipoic acid in eukaryotes. Sulo and Martin (1993) isolated the LIP5 gene from the yeast Saccharomyces cerevisiae by functional complementation of a mutant defective in biosynthesis of lipoic acid. DNA-sequence analysis of LIP5 revealed that it encodes a protein homologous to lipoic acid synthase of E. coli. A gene for lipoyltransferase, which catalyzes the transfer of the lipoic acid to cognate proteins, has recently been cloned from another yeast, Kluyveromyces lactis (Chen, 1997).
Fujiwara et al. (1994, 1996) purified two isoforms of lipoyltransferase from bovine liver mitochondria using the apoH protein of the Gly-decarboxylase complex as an acceptor of lipoate. Both isoforms of lipoyltransferase catalyze the transfer of the lipoyl group from lipoyl-AMP to apoH protein but have no activity to activate lipoate to lipoyl-AMP, in contrast to the E. coli lipoyltransferase encoded by the lplA gene (Morris et al., 1995). Furthermore, these researchers cloned a cDNA for the bovine lipoyltransferase and found that both isoforms are derived from the same translated product but are processed differently (Fujiwara et al., 1997). We recently studied fatty acid synthesis in mitochondria isolated from pea and found that a major part of the de novo-synthesized fatty acids may be used for the biosynthesis of lipoic acid (Wada et al., 1997). Together these results have begun to define the biosynthesis of lipoic acid and its intracellular organization in eukaryotes.
We describe the isolation of an Arabidopsis cDNA, LIP1, that encodes a lipoic acid synthase and show that this cDNA complements the E. coli lipA mutant defective in lipoic acid synthase. To our knowledge, this is the first cDNA for lipoic acid synthase that has been cloned and characterized in a higher organism. We also report the intracellular localization of the enzyme in Arabidopsis cells and demonstrate that lipoic acid synthase is located in the mitochondrial compartment.
MATERIALS AND METHODS
Plant Material
Arabidopsis ecotype Columbia was grown under continuous light (40 μmol photons m−2 s−1) at 25°C in trays containing vermiculite. Leaves, roots, and flowers were harvested from 4-week-old plants and used for RNA, DNA, protein, and lipoic acid extractions.
Cell Fractionation of Arabidopsis
Twenty grams of leaves from 4-week-old Arabidopsis plants was used for cell fractionation. Plants were placed in the dark for 12 h prior to harvesting the leaves. All of the procedures described below were carried out at 4°C. The leaves were homogenized with a grinder in 40 mL of medium containing 50 mm Tes-NaOH, pH 7.2, 0.3 m mannitol, 1 mm EDTA, 1 mm MgCl2, 0.2% (w/v) BSA, 0.5% (w/v) PVP-40, 4 mm Cys, and 10 mm 2-mercaptoethanol. The brei was filtered through eight layers of gauze and centrifuged at 3,300g for 5 min, and then the supernatant was centrifuged at 10,500g for 20 min. The supernatant obtained after the 10,500g centrifugation was centrifuged at 100,000g for 2 h, and the supernatant and pellet obtained were used for western analysis as the cytosol and microsome fractions, respectively.
The pellet obtained after the 10,500g centrifugation was used for fractionation of chloroplasts and mitochondria as described by Day et al. (1985) with the following modifications. The pellet was suspended in 2 mL of medium A containing 20 mm Tes-NaOH, pH 7.2, 2 mm KH2PO4, 1 mm EDTA, 2 mm MgCl2, 0.1% BSA, and 14 mm 2-mercaptoethanol, applied onto a stepwise gradient composed of 2 mL of 21%, 4 mL of 26%, and 2 mL of 47% (w/v) Percoll solutions, and centrifuged at 65,000g for 45 min in a swinging-bucket rotor (model RPS-40T, Hitachi, Tokyo, Japan). All of the Percoll solutions used in the gradient centrifugation were made up in 10 mm Tes-NaOH, pH 7.2, 0.25 m Suc, and 0.1% BSA.
After the centrifugation chloroplasts and mitochondria were separated as a green band at the interface between the 21% and 26% Percoll layers and as a white band at the interface between the 26% and 47% Percoll layers, respectively. Both chloroplast and mitochondria fractions were separately recovered from the gradient and diluted six times with medium B, which contained the same components as medium A except that it had 2 mm DTT instead of 2-mercaptoethanol and 2 mm KH2PO4, and then centrifuged at 12,500g for 20 min. The obtained pellets were suspended in medium B and used in western analysis as the chloroplast and mitochondria fractions.
cDNA Cloning and Analysis
An Arabidopsis EST clone, 193K14, which contains an open reading frame encoding a polypeptide homologous to lipoic acid synthase of Escherichia coli, was obtained from the Arabidopsis Biological Resource Center (The Ohio State University, Columbus). The 5′-terminal region of Arabidopsis LIP1 cDNA was amplified by the 5′-RACE method (5′-Full RACE Core Set, Takara, Shiga, Japan). A phosphorylated primer, 5′-GTGTGTAATCTACGG-3′, was used for the synthesis of cDNA using total RNAs isolated from Arabidopsis leaves. The following two primer sets were also used for first and second PCR in the 5′ RACE, respectively: a set of 5′-CAGCCGCGTGTACATGTATCCCCA-3′ and 5′-CGGAATTCGAAATGGGATTCAGAT-3′ and another set of 5′-GACATTTCGCTTCCTCGCAGACGG-3′ and 5′-CGAAGCTTGATAGAAGCCGATCGA-3′.
The amplified DNA was subcloned into a pCRII vector (Original TA Cloning Kit, Invitrogen, San Diego, CA). The obtained plasmid was designated pRACE-5′ and its nucleotide sequence was determined. The 3′-terminal region of LIP1 was also amplified by PCR with 193K14 as the template and subcloned into pBluescript II KS(+) (Stratagene). This clone, designated pBlue-3′, was used to determine the sequence of both strands in the 3′-terminal region of LIP1 cDNA.
DNA-sequencing reactions were performed (BcaBest dideoxy sequencing kit, Takara, Shiga, Japan; Thermo sequenase sequencing kit, Amersham) and sequences obtained (ALF Red DNA sequencer, Pharmacia Biotech, Tokyo, Japan; DNA sequencer DSQ-1000, Shimadzu, Kyoto, Japan). Double-stranded DNAs were used as templates, and the sequence of each strand was determined. Nucleotide and deduced amino acid sequences were analyzed by suitable software (GENETYX-MAC, Software Development Co., Tokyo, Japan).
Nucleic Acid Extraction and Analysis
Genomic DNA used for Southern analysis was extracted from Arabidopsis leaves using a DNA-extraction kit (ISOPLANT, Nippon Gene, Tokyo, Japan). The genomic DNA was digested with the appropriate restriction enzymes, separated by electrophoresis on a 0.8% (w/v) agarose gel, and transferred to a nylon membrane (Hybond-N+, Amersham). The membrane was hybridized using a DNA-labeling and -detection system (ECL kit, Amersham).
Total RNAs used for cDNA synthesis and RT-PCR were extracted from leaves, roots, and flowers of Arabidopsis using an RNA-extraction kit (RNeasy plant kit, Qiagen, Chatsworth, CA). RT-PCR analysis of the LIP1 gene was performed with an RT-PCR kit (SuperScript OneStep System, Life Technologies) using the following two primers: 5′-TTGGGGATACATGTACACGC-3′ and 5′-CTGAATCCCATTTCCATGCC-3′. Two micrograms of total RNAs from each organ was used for RT-PCR analysis. As a control, the expression of the RCO1 gene for the cytosolic form of cyclophilin was also checked by RT-PCR with the same RNA preparations using the following two primers: 5′-ACTTCGACATGACCATCGAC-3′ and 5′-TTCCCATGAGAACACACACC-3′.
Functional Complementation of the E. coli lipA Mutant
Deletions of the LIP1 cDNA were created using PCR with specific primers designed to remove varying numbers of residues from the N terminus of the LIP1 protein. The eight-nucleotide sequence 5′-CGCCATGG-3′ including an NcoI site was added to the 5′ end of each primer. PCR products were digested with NcoI and ligated into the NcoI site of an expression vector, pKK233-2 (Clontech, Palo Alto, CA), to give an in-frame desired product. The obtained plasmids were designated pLIP1-Δ0, pLIP1-Δ18, and pLIP1-Δ26, where the number after the Δ represents the number of amino acid residues deleted from the N terminus of LIP1 protein. These plasmids were used for transformation of the E. coli lipA mutant KER176 (Reed and Cronan, 1993). The transformants were plated on rich-broth medium (Davis et al., 1992) supplemented with 50 μg mL−1 ampicillin, 10 μg mL−1 kanamycin, and 50 ng mL−1 lipoic acid.
To determine the effect of the deletions on the function of the resulting LIP1 proteins, colonies of the transformants on the rich-broth plate were streaked onto other plates, which contained M9 medium supplemented with 50 μg mL−1 ampicillin, 10 μg mL−1 kanamycin, 1 mm MgSO4, 1% (w/v) succinate, 5 mm acetate, 5 × 10−5 % (w/v) vitamin B1, 0.4 mm IPTG, and either 0 or 50 ng mL−1 lipoic acid, and then the growth of the transformants on the plates was checked. The transformant of the E. coli lipA mutant with pKK233-2 was used as a negative control in these experiments.
Overexpression of LIP1 in E. coli and Antibody Production
The region of LIP1 cDNA encoding a putative mature protein was amplified by PCR using the following two primers: 5′-CGCCATGGGCTTCTCCTCTTCCTC-3′ and 5′-CGCCATGGAGTGTGTAATCTACGG-3′. The eight-nucleotide sequence 5′-CGCCATGG-3′ including an NcoI site was added to the 5′ end of both primers. The amplified DNA fragment was digested with NcoI and ligated into the NcoI site of an expression vector, pET-30a(+) (Novagen, Madison, WI). The resultant plasmid, designated pET-LIP1, was used for transformation of E. coli BL21(DE3) (Studier et al., 1990). The transformant of BL21(DE3) with pET-LIP1 was grown at 37°C in Luria-Bertani medium supplemented with 200 μg mL−1 ampicillin. When A600 of the E. coli culture reached 0.5, IPTG was added to the medium at a final concentration of 0.4 mm to induce expression of LIP1, and the culture was incubated for a further 26 h at 25°C.
Overexpressed LIP1 fusion protein, which contains six His residues at the N terminus as a tag, was purified by nickel-affinity chromatography (His-Bind Resin and Buffer Kit, Novagen). Polyclonal antibody was generated by injecting the purified LIP1 fusion protein into a rabbit. The antiserum was prepared from the rabbit and the anti-Arabidopsis LIP1 fusion protein IgG was purified using a kit (ImmunoPure, Pierce).
SDS-PAGE and Western Analysis
SDS-PAGE was performed according to the method of Laemmli (1970) using a 12% (w/v) polyacrylamide gel. Protein concentration was determined by the method of Bradford (1976) using BSA as the standard. Western analysis was carried out according to the method of Post-Beittenmiller et al. (1991).
Biological Assay of Lipoic Acid
For extraction of lipoic acid, leaves, roots, and flowers of Arabidopsis were collected from 4-week-old plants, frozen in liquid nitrogen, and ground with a mortar and pestle. The obtained powders were suspended in 6 n H2SO4 and autoclaved for 2 h to release lipoic acids from proteins. After autoclaving, the suspension was adjusted to pH 7.0 with 4 n NaOH, made up to a known volume, and filtered to remove any insoluble materials. The obtained solution was used for biological assay by the turbidimetric method (Herbert and Guest, 1970) using the E. coli lipA mutant (Reed and Cronan, 1993) as the test organism.
RESULTS
Identification of the cDNA Coding for Arabidopsis Lipoic Acid Synthase
A BLAST search (Pearson and Lipman, 1988) of the database of the National Center for Biotechnology Information using the amino acid sequence of E. coli lipoic acid synthase (LipA) identified an Arabidopsis EST clone containing an open reading frame encoding a polypeptide homologous to lipoic acid synthase of E. coli. This clone, 193K14, was obtained from the Arabidopsis Biological Resource Center, and the complete sequence of the 1224-bp cDNA in the EST clone was determined. The cDNA contains an open reading frame that codes for a protein with an amino acid sequence similar to those of lipoic acid synthases of E. coli (Reed and Cronan, 1993) and yeast (Sulo and Martin, 1993).
Since this open reading frame began at the 5′ terminus of the cDNA and did not begin with a Met residue, it appeared that the cDNA was not a full-length copy of the corresponding mRNA. Therefore, the 5′-terminal region of the corresponding mRNA was amplified by the 5′-RACE method using cDNAs prepared with total RNAs from leaves. The PCR products obtained by the 5′-RACE method were subcloned into the vector pCRII. Five cloned PCR products were isolated and sequenced. The sequences of all the clones overlapped with each other and with the 5′-terminal region of 193K14. The sequence of the longest clone, pRACE-5′, extended the sequence of 193K14 by 105 nucleotides to a total length of 1330 nucleotides.
The 3′ region of LIP1 cDNA was also amplified by PCR and subcloned into pBluescript II KS(+) to determine the nucleotide sequence of both strands of this region. The obtained clone was designated pBlue-3′. The sequence of pBlue-3′ perfectly overlapped with the 3′-terminus region of LIP1 cDNA in 193K14. The nucleotide sequence of full-length LIP1 cDNA was obtained by a combination of sequences of 193K14, pRACE-5′, and pBlue-3′. This entire nucleotide sequence of the LIP1 cDNA was deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases with the accession no. AB007987. This LIP1 cDNA encodes a polypeptide of 374 amino acids, which corresponds to a molecular mass of 41,344 D.
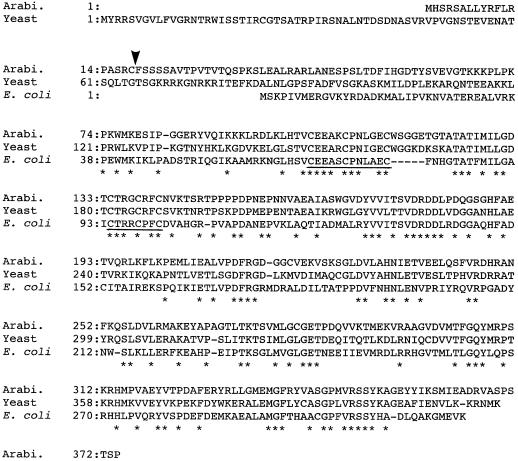
The amino acid sequence of LIP1 protein is compared in Figure 1 with those of lipoic acid synthases of E. coli and yeast. The amino acid sequence identity between the Arabidopsis LIP1 protein and the lipoic acid synthases of E. coli and Saccharomyces cerevisiae are 44% and 67%, respectively. Several stretches of conserved residues that may be important for the function of this enzyme were found. Two conserved C motifs, C-E-E-A-X-C-P-N-X-X-E-C and C-T-R-X-C-X-F-C, were found at positions from 103 to 114 and 135 to 142 in the Arabidopsis LIP1 protein, respectively. The second C motif is also conserved in biotin synthases of Arabidopsis (Patton et al., 1996; Weaver et al., 1996) and microorganisms (Otsuka et al., 1988; Zhang et al., 1994; Bower et al., 1996). Since lipoic acid synthase and biotin synthase catalyze a similar reaction that inserts a sulfur atom into a hydrocarbon chain, the conserved C residues may play an important role in this process. Comparison of the N-terminal region of the sequences clearly indicates that Arabidopsis LIP1 protein contains a 36-residue extension relative to the lipoic acid synthase of E. coli. The amino acid sequence of this region has some characteristics in common with transit peptides for targeting to mitochondria (von Heijne et al., 1989; von Heijne, 1992): an overall positive charge due to the relatively high proportion of R residues, the lack of negatively charged residues, and the relatively high proportion of hydroxylated residues. In addition, the sequence R17-C-F-S is similar to a motif (R-X-↓-X-S), which may represent the potential cleavage site for a mitochondrial transit peptide (von Heijne, 1992). If this sequence motif is the actual cleavage site, the mature form of LIP1 protein is a polypeptide of 356 amino acid residues with a molecular mass of 39,197 D.
Figure 1.
Comparison of the amino acid sequences of lipoic acid synthases. The deduced amino acid sequence of lipoic acid synthase encoded by Arabidopsis (Arabi.) LIP1 cDNA is compared with that of lipoic acid synthases of S. cerevisiae (Yeast) (Sulo and Martin, 1993) and E. coli (Reed and Cronan, 1993). The amino acid residues conserved in all sequences are indicated by asterisks. Hyphens represent gaps to maximize the alignment of the sequences. The conserved Cys motifs are underlined. The putative cleavage site for the mitochondrial transit peptide is indicated by an arrowhead.
Complementation of the E. coli lipA Mutant
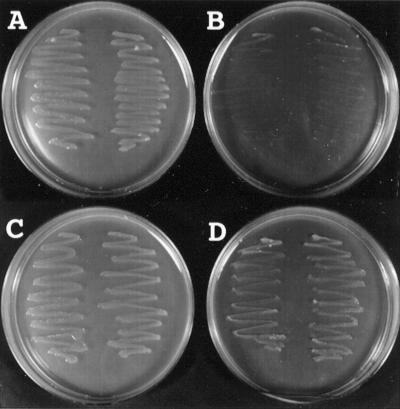
To confirm that LIP1 protein is a lipoic acid synthase of Arabidopsis, the LIP1 cDNA was expressed in the E. coli lipA mutant, which is defective in lipoic acid synthase (Reed and Cronan, 1993). As shown in Figure 2, when the E. coli lipA mutant was transformed with pKK233-2 (control vector) and plated on lipoic-acid-free medium, no growth of the transformant was observed, although the transformant could grow on medium containing lipoic acid. By contrast, the transformant with the plasmid (pLIP1-Δ0) carrying the full-length LIP1 cDNA could grow on lipoic-acid-free medium. These findings clearly demonstrate that Arabidopsis LIP1 cDNA functionally complements the E. coli lipA mutant and that the LIP1 cDNA encodes a lipoic acid synthase.
Figure 2.
Complementation of the lipA mutant of E. coli by expression of Arabidopsis LIP1 cDNA. The E. coli lipA mutant (KER176), which is defective in lipoic acid synthase (Reed and Cronan, 1993), was transformed with the plasmids pKK233-2 (control, plates A and B) and pLIP1-Δ0 (plates C and D). The colonies of the each transformant were streaked onto plates containing 50 μg mL−1 ampicillin, 50 μg mL−1 kanamycin, and either 50 ng mL−1 lipoic acid (plates A and C) or no lipoic acid (plates B and D), and the plates were incubated at 37°C for 3 d.
Deletions of N-terminal residues in the LIP1 protein also resulted in functional complementation of the E. coli lipA mutant (data not shown). However, the deletion of 18 residues, which gives a putative, mature LIP1 protein, enhanced the growth of the mutant, and the deletion of 26 residues slightly inhibited the growth of the transformant compared with the full-length clone. These results suggest that the deleted amino acid residues are not required for catalytic activity of LIP1 protein.
Expression and Organization of the LIP1 Gene for Arabidopsis Lipoic Acid Synthase
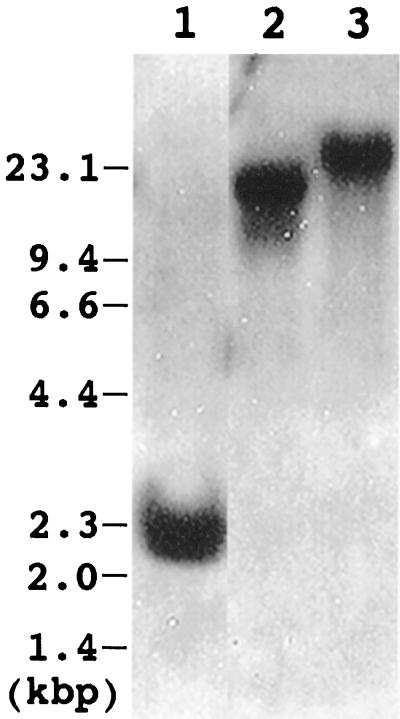
Figure 3 shows genomic Southern analysis of the LIP1 gene of Arabidopsis. The 1.1-kb EcoRI fragment containing Arabidopsis LIP1 cDNA was obtained by digestion of 193K14 with EcoRI and was used as a probe. When the genomic DNA was digested with EcoRI, BamHI, or SalI, a single hybridizing band was detected in all cases, indicating that in Arabidopsis lipoic acid synthase is encoded by a single gene.
Figure 3.
Genomic Southern analysis of the Arabidopsis LIP1 gene. Genomic DNA was extracted from Arabidopsis leaves and digested with EcoRI (lane 1), BamHI (lane 2), and SalI (lane 3). Three micrograms of DNA was applied to each lane. The 1.1-kb EcoRI fragment containing LIP1 cDNA was prepared from the 193K14 clone and used as a probe. The positions of the DNA size markers (in kb) are indicated on the left.
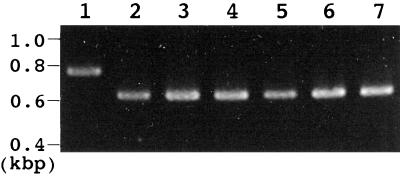
To investigate the organ-specific expression of the LIP1 gene, northern analysis was carried out using total RNAs prepared from leaves, roots, and flowers. However, no clear signal was detected in the analysis (data not shown), suggesting that the level of expression of LIP1 gene was low. Therefore, RT-PCR analysis was used to estimate the level of expression of LIP1 gene in each organ. Figure 4 shows the results of RT-PCR. In all tested organs a 628-bp DNA fragment corresponding to LIP1 cDNA was detected at the same level (lanes 2–4). When the genomic DNA was used as the template instead of RNA (lane 1), a 700-bp DNA fragment was amplified. The difference between the 628- and 700-bp DNA fragments was most likely caused by the presence of an intron in this region of the LIP1 gene. The 700-bp DNA fragment detected with genomic DNA was not detected with total RNAs prepared from any of the tested organs, suggesting that the total RNAs used for RT-PCR were not contaminated with genomic DNA. As a control the level of expression of the RCO1 gene for the cytosolic form of cyclophilin, which is expressed at the same level in leaves, roots, and flowers (Lippuner et al., 1994), was also checked by RT-PCR of the same RNA samples (lanes 5–7). In all tested organs, 628 bp, which corresponds to RCO1 cDNA, was detected at same level. These results suggest that the LIP1 gene is expressed at the same level in all organs of Arabidopsis.
Figure 4.
Organ-specific expression of the LIP1 gene. The level of LIP1 mRNA for lipoic acid synthase was analyzed by RT-PCR with total RNAs extracted from leaves (lane 2), roots (lane 3), and flowers (lane 4). In lane 1, genomic DNA was used as the template for RT-PCR instead of RNA. The level of mRNA for the cytosolic form of cyclophilin was also analyzed as a control with the same total RNAs extracted from leaves (lane 5), roots (lane 6), and flowers (lane 7). The positions of the DNA size markers (in kb) are indicated on the left.
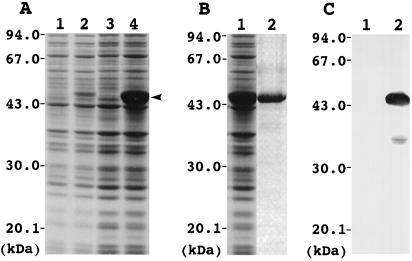
To prepare polyclonal antibody against Arabidopsis LIP1 protein, LIP1 was overexpressed in E. coli BL21(DE3) as a fusion protein containing a His tag at the N terminus. Figure 5A shows SDS-PAGE analysis of proteins from BL21(DE3) cells that were transformed with pET-30a(+) and pET-LIP1. In the transformant with pET-LIP1, a 46-kD protein was detected, and the level of the protein was dramatically increased upon addition of IPTG. The molecular mass of the protein was close to that of the Arabidopsis LIP1 fusion protein encoded in pET-LIP1. By contrast, in the transformant with pET-30a(+), the 46-kD protein was not detected and the protein composition did not significantly change upon addition of IPTG. These findings suggest that the 46-kD protein is an Arabidopsis LIP1 fusion protein.
Figure 5.
Overexpression of Arabidopsis LIP1 cDNA in E. coli. A, Changes in protein composition in E. coli BL21(DE3) cells transformed with pET-30a(+) and pET-LIP1. Total proteins from BL21(DE3)/pET-30a(+) (lanes 1 and 3) and BL21(DE3)/pET-LIP1 (lanes 2 and 4) were analyzed by SDS-PAGE on a 12% (w/v) polyacrylamide gel. IPTG (0.4 mm) was added to the growth medium to induce expression of LIP1, and then the culture was incubated for 0 h (lanes 1 and 2) and 26 h (lanes 3 and 4) at 25°C. An arrowhead indicates the position of the LIP1 protein. The positions of protein molecular mass markers are indicated on the left (in kD). B, Purification of LIP1 fusion protein from the homogenates of BL21(DE3)/pET-LIP1. Total proteins before (lane 1) and after (lane 2) purification with nickel-affinity chromatography were analyzed by SDS-PAGE. C, Western analysis of total proteins from BL21(DE3)/pET-30a(+) (lane 1) and BL21(DE3)/pET-LIP1 (lane 2). The samples corresponding to lanes 3 and 4 in A were applied to SDS-PAGE, blotted to the nitrocellulose membrane, and used for western analysis. The blot was probed with anti-Arabidopsis LIP1 IgG.
The overexpressed LIP1 fusion protein containing a His tag was purified to near homogeneity by nickel-affinity chromatography from the E. coli homogenate (Fig. 5B) and the purified protein was used for preparation of polyclonal antibody against Arabidopsis LIP1 protein. Figure 5C shows the results of western analysis of total proteins from BL21(DE3)/pET-30a(+) and BL21(DE3)/pET-LIP1 with the anti-Arabidopsis LIP1 IgG. In BL21(DE3)/pET-LIP1, signals were detected at the position of 46 and 37 kD, whereas no signal was detected in BL21(DE3)/pET30a(+). The strong signal at 46 kD corresponds to the position of LIP1 fusion protein. The weak signal at 37 kD may have been caused by the degradation of the LIP1 fusion protein in E. coli cells, because the size of the signal was lower than that of LIP1 fusion protein and no signal was detected in the control BL21(DE3)/pET30a(+). These findings demonstrate that the reactivity of the prepared IgG is specific to the Arabidopsis LIP1 protein.
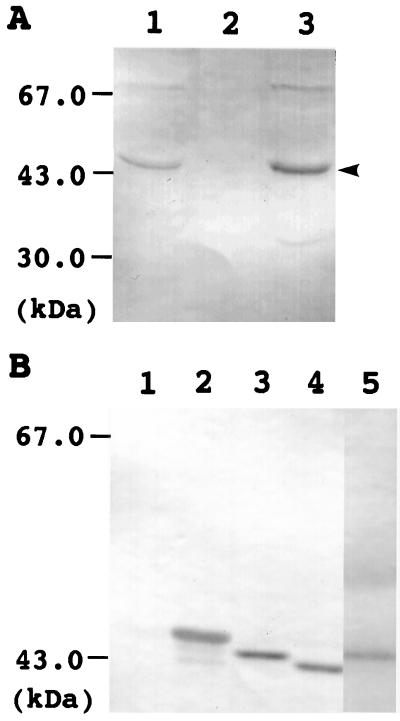
To investigate the organ-specific localization of Arabidopsis lipoic acid synthase, total proteins were prepared from leaves, roots, and flowers and used for western analysis. Figure 6A shows the results of western analysis with the anti-Arabidopsis LIP1 IgG. Two major signals were detected at positions 43 and 67 kD in leaves and flowers, but not in roots. The size of the signal at 43 kD is close to the molecular mass of mature lipoic acid synthase calculated from the deduced amino acid sequence, and the signal was not detected when the same sample was analyzed using a rabbit preimmune serum instead of anti-LIP1 IgG (data not shown). The signal at 67 kD was not always detected, and the size of the signal was much higher than the molecular mass of mature lipoic acid synthase calculated from the deduced amino acid sequence. These results suggest that the signal at 43 kD but not the one at 67 kD corresponds to lipoic acid synthase and the content of lipoic acid synthase is higher in leaves and flowers than in roots.
Figure 6.
Western analysis of lipoic acid synthase in Arabidopsis. A, Organ-specific localization of lipoic acid synthase. Total proteins prepared from leaves (lane 1), roots (lane 2), and flowers (lane 3) were applied to SDS-PAGE, blotted to a nitrocellulose membrane, and used for western analysis with anti-Arabidopsis LIP1 IgG. The arrowhead indicates the position of lipoic acid synthase. The positions of protein molecular mass markers (in kD) are indicated on the left. Fifty-five micrograms of protein was applied to each lane. B, Processing of lipoic acid synthase. Total proteins prepared from the E. coli lipA mutant cells transformed with plasmids pKK233-2 (lane 1), pLIP1-Δ0 (lane 2), pLIP1-Δ18 (lane 3), and pLIP1-Δ26 (lane 4) and Arabidopsis leaves (lane 5) were used for western analysis with anti-Arabidopsis LIP1 IgG.
Arabidopsis lipoic acid synthase contains a leader sequence, presumably required for import into mitochondria. To determine whether this leader sequence is processed in Arabidopsis cells, we analyzed total proteins prepared from Arabidopsis leaves by western analysis with anti-Arabidopsis LIP1 IgG and compared the size of mature lipoic acid synthase in leaves with those of LIP1 proteins expressed in the E. coli lipA mutant cells transformed with pLIP1-Δ0, pLIP1-Δ18, and pLIP1-Δ26. As shown in Figure 6B, the size of mature lipoic acid synthase in leaves was close to that of the LIP1 protein expressed in the E. coli cells transformed with pLIP1-Δ18, which encodes a LIP1 protein in which 18 amino acid residues were deleted at the N terminus. This result suggests that lipoic acid synthase is N-terminally processed in Arabidopsis cells.
Intracellular Localization of Lipoic Acid Synthase in Arabidopsis
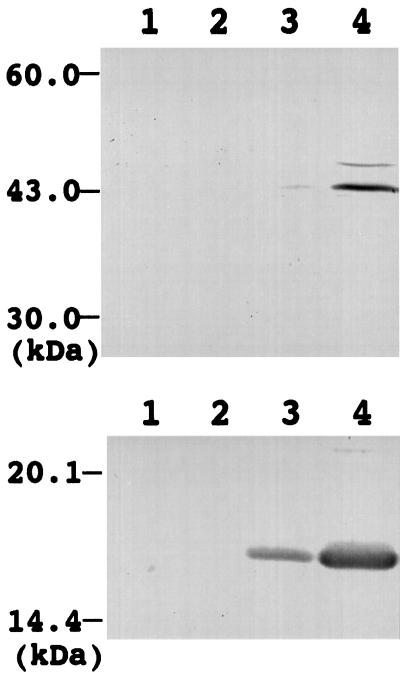
As described above, Arabidopsis lipoic acid synthase contains a putative mitochondria-targeting transit peptide and is N-terminally processed in Arabidopsis cells. These findings raise the possibility that lipoic acid synthase is located in mitochondria. To check this possibility we prepared cytosol, microsome, chloroplast, and mitochondria fractions from Arabidopsis leaves and investigated the intracellular localization of lipoic acid synthase by western analysis with anti-Arabidopsis LIP1 IgG. Figure 7, top, shows the results of the western analysis. A strong band at 43 kD, corresponding to lipoic acid synthase, was detected in the mitochondrial fraction (lane 4), whereas a faint band at the same position was detected in the chloroplast fraction (lane 3), which suggests that lipoic acid synthase is mainly located in the mitochondria.
Figure 7.
Intracellular localization of lipoic acid synthase in Arabidopsis. Proteins prepared from cytosol (lanes 1), microsome (lanes 2), chloroplast (lanes 3), and mitochondria (lanes 4) fractions were applied to SDS-PAGE, blotted to a nitrocellulose membrane, and used for western analysis with anti-Arabidopsis LIP1 IgG (top) and with an antibody against pea H-protein (bottom). The positions of protein molecular mass markers (in kD) are indicated on the left. One-hundred micrograms of protein was applied to each lane.
To confirm the localization of lipoic acid synthase in mitochondria, the same fractionated samples were analyzed by western analysis with an antibody against pea H-protein, which is a subunit of the Gly-decarboxylase complex and is present in the matrix of mitochondria (Oliver, 1994). As shown in Figure 7, bottom, the H-protein was detected in the mitochondrial and chloroplast fractions, although the intensity of the H-protein band in the mitochondrial fraction was much stronger than that of the band in the chloroplast fraction. This suggests that the mitochondrial fraction is abundant in mitochondria, that the chloroplast fraction is slightly contaminated with mitochondria, and that the faint band of lipoic acid synthase detected in the chloroplast fraction is caused by the contamination of mitochondria into the chloroplast fraction. Together, these results demonstrate that lipoic acid synthase is located in mitochondria in Arabidopsis cells.
The content of lipoic acids in leaves, roots, and flowers was determined to be 170, 90, and 290 ng g−1 fresh weight, respectively. These results indicate that the content of lipoic acid is correlated with the content of lipoic acid synthase in each organ but not with the level of the expression of the LIP1 gene.
DISCUSSION
In this study we cloned the LIP1 cDNA from Arabidopsis and demonstrated that it encodes a lipoic acid synthase based on the sequence similarity to lipoic acid synthases of E. coli and yeast and on its functional complementation of an E. coli lipA mutant defective in lipoic acid synthase. In all eukaryotes except plants, all of the known lipoic-acid-containing proteins and lipoyltransferases are located in the mitochondria (Fujiwara et al., 1990; Kim and Oliver, 1990; Macherel et al., 1990; Reed and Hackert, 1990; Perham, 1991; Mattevi et al., 1992). Therefore, it is reasonable to assume that the biosynthesis of lipoic acid takes place in the mitochondria. In fact, lipoic acid synthases of yeast (Sulo and Martin, 1993) and Arabidopsis (this study) contain leader sequences at the N terminus that are similar to transit peptides for targeting to mitochondria. However, it is worthwhile to mention that in plants the pyruvate-dehydrogenase complex, which contains lipoic acid as an enzyme cofactor, is present both in mitochondria and in plastids (Lernmark and Gardeström, 1994), raising the possibility that lipoic acid is also synthesized in plastids, or it is synthesized in mitochondria and then transported to plastids. In this study we prepared a polyclonal antibody specific to lipoic acid synthase of Arabidopsis and investigated its intracellular localization in Arabidopsis cells. The obtained results demonstrated that lipoic acid synthase is present in mitochondria, suggesting that the biosynthesis of lipoic acid takes place in mitochondria, which is consistent with the previous observation that a major part of de novo synthesized fatty acids in mitochondria may be used for biosynthesis of lipoic acid (Wada et al., 1997).
Although lipoic acid is an essential cofactor for several enzymes involved in central metabolism and is synthesized in most organisms, the mechanism of lipoic acid synthesis is not well understood. Octanoic acid has been shown to be the precursor of the carbon chain (White, 1980; Parry, 1983), but neither the origin nor the mechanism whereby the sulfur atoms are inserted into the carbon chain is known. We recently investigated the function of mitochondrial ACP, which was discovered in the mitochondria of several organisms (Shintani and Ohlrogge, 1994; Schneider et al., 1997), and found that it is involved in the biosynthesis of fatty acids in mitochondria of pea and Neurospora (Wada et al., 1997). We also found that octanoyl-ACP is synthesized in mitochondria as an intermediate of fatty acid synthesis and may be used for the biosynthesis of lipoic acid (Wada et al., 1997).
This involvement of mitochondrial ACP in lipoic acid production was recently demonstrated in yeast by Brody et al. (1997). Jordan and Cronan (1997) reported that fatty acid synthesis in E. coli is directly linked to the biosynthesis of lipoic acid and that lipoate is donated from lipoyl-ACP to the pyruvate-dehydrogenase complex by lipoyl-ACP-protein N-lipoyltransferase. This enzyme activity was also discovered in mitochondria of both plants and fungi. These findings suggest that lipoic acid synthesis may proceed through ACP-bound intermediates and that lipoic acid synthase may insert a sulfur atom into octanoic acid that has been bound to ACP.
Biotin synthase also catalyzes a reaction similar to that catalyzed by lipoic acid synthase, the insertion of a sulfur atom into dethiobiotin (Eisenberg, 1987; Baldet et al., 1993). Birch et al. (1995) studied the in vitro assay system for biotin synthase and found that this enzyme requires several low-molecular-mass cofactors and protein components. Although it can be expected that lipoic acid synthase requires similar enzyme cofactors as those required for biotin synthase, an in vitro assay system for lipoic acid synthase has not yet been established; therefore, it has not been possible to measure the activity of lipoic acid synthase in vitro. Biotin synthase is a [2Fe-2S] cluster protein (Sanyal et al., 1994) and the Cys residues may coordinate to the Fe atom of the Fe-S cluster. In this study we found that two conserved Cys motifs are present in lipoic acid synthases of E. coli, yeast, and Arabidopsis, and one of the Cys motifs is also conserved in the biotin synthases of Arabidopsis and microorganisms. Although it is not known whether lipoic acid synthase is an [Fe-S] cluster protein, it can be assumed that the conserved Cys residues may play a role in the enzyme activity, possibly in the coordination of the Fe atom. In this study we overexpressed Arabidopsis lipoic acid synthase in E. coli and purified the overexpressed protein from the E. coli homogenate. This system may help to establish an assay system for lipoic acid synthase to aid in the study of the function of the conserved Cys residues and how lipoic acid synthase inserts a sulfur atom into octanoic acid.
ACKNOWLEDGMENTS
This work would not have been possible without the support and helpful suggestions of John Ohlrogge and the technical assistance of Linda Savage (Michigan State University). We acknowledge Sean Jordan and John E. Cronan, Jr. (University of Illinois at Urbana-Champaign), for providing the E. coli lipA mutant (KER176) and the Arabidopsis Biological Resource Center (The Ohio State University) for providing the Arabidopsis EST clone used in this study. We also thank Dr. David J. Oliver (Iowa State University) for providing the antibody against pea H-protein.
Abbreviations:
- ACP
acyl carrier protein
- EST
expressed sequence tag
- IPTG
isopropyl-1-thio-β-d-galactoside
- RACE
rapid amplification of cDNA ends
- RT-PCR
reverse transcription-PCR
Footnotes
This work was supported by a Grant-in-Aid for Scientific Research (no. 10640636) from the Ministry of Education, Science, Sports, and Culture (Japan) and by a grant from the Asahi Glass Foundation to H.W.
LITERATURE CITED
- Baldet P, Gerbling H, Axiotis S, Douce R. Biotin biosynthesis in higher plant cells. Identification of intermediates. Eur J Biochem. 1993;217:479–485. doi: 10.1111/j.1432-1033.1993.tb18267.x. [DOI] [PubMed] [Google Scholar]
- Birch OM, Fuhrmann M, Shaw NM. Biotin synthase from Escherichia coli, an investigation of the low molecular weight and protein components required for activity in vitro. J Biol Chem. 1995;270:19158–19165. doi: 10.1074/jbc.270.32.19158. [DOI] [PubMed] [Google Scholar]
- Bower S, Perkins JB, Yocum RR, Howitt CL, Rahaim P, Pero J. Cloning, sequencing, and characterization of the Bacillus subtilis biotin biosynthetic operon. J Bacteriol. 1996;178:4122–4130. doi: 10.1128/jb.178.14.4122-4130.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of dye-binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brody S, Oh C, Hoja U, Schweizer E. Mitochondrial acyl carrier protein is involved in lipoic acid synthesis in Saccharomyces cerevisiae. FEBS Lett. 1997;408:217–220. doi: 10.1016/s0014-5793(97)00428-6. [DOI] [PubMed] [Google Scholar]
- Chen XJ. Cloning and characterization of the lipoyl-protein ligase gene LIPB from the yeast Kluyveromyces lactis: synergetic respiratory deficiency due to mutations in LIPB and mitochondrial F1-ATPase subunits. Mol Gen Genet. 1997;255:341–349. doi: 10.1007/s004380050505. [DOI] [PubMed] [Google Scholar]
- Davis TN, Muller ED, Cronan JE., Jr The virion of the lipid-containing bacteriophage PR4. Virology. 1992;120:287–306. doi: 10.1016/0042-6822(82)90031-9. [DOI] [PubMed] [Google Scholar]
- Day DA, Neuburger M, Douce R. Biochemical characterization of chlorophyll-free mitochondria from pea leaves. Aust J Plant Physiol. 1985;12:219–228. [Google Scholar]
- Douce R, Bourguignon J, Macherel D, Neuburger M. The glycine decarboxylase system in higher plant mitochondria: structure, function and biogenesis. Biochem Soc Trans. 1994;22:184–188. doi: 10.1042/bst0220184. [DOI] [PubMed] [Google Scholar]
- Eisenberg M (1987) Biosynthesis of biotin and lipoic acid. In FC Neidhardt, JL Ingraham, KB Low, B Magasanik, M Schaechter, HE Umbarger, eds, Escherichia coli and Salmonella typhimurium, Vol 1. American Society for Microbiology, Washington, DC, pp 544–550
- Fujiwara K, Okamura-Ikeda K, Motokawa Y. cDNA sequence, in vitro synthesis, and intramitochondrial lipoylation of H-protein of the glycine cleavage system. J Biol Chem. 1990;265:17463–17467. [PubMed] [Google Scholar]
- Fujiwara K, Okamura-Ikeda K, Motokawa Y. Purification and characterization of lipoyl-AMP: Nε-lysine lipoyltransferase from bovine liver mitochondria. J Biol Chem. 1994;269:16605–16609. [PubMed] [Google Scholar]
- Fujiwara K, Okamura-Ikeda K, Motokawa Y. Lipoylation of acyltransferase components of α-ketoacid dehydrogenase complexes. J Biol Chem. 1996;271:12932–12936. doi: 10.1074/jbc.271.22.12932. [DOI] [PubMed] [Google Scholar]
- Fujiwara K, Okamura-Ikeda K, Motokawa Y. Cloning and expression of a cDNA encoding bovine lipoyltransferase. J Biol Chem. 1997;272:31974–31978. doi: 10.1074/jbc.272.51.31974. [DOI] [PubMed] [Google Scholar]
- Herbert AA, Guest JR. Turbidimetric and polarographic assays for lipoic acid using mutants of Escherichia coli. Methods Enzymol. 1970;12:269–272. [Google Scholar]
- Jordan SW, Cronan JE., Jr A new metabolic link. The acyl carrier protein of lipid synthesis donates lipoic acid to the pyruvate dehydrogenase complex in Escherichia coli and mitochondria. J Biol Chem. 1997;272:17903–17906. doi: 10.1074/jbc.272.29.17903. [DOI] [PubMed] [Google Scholar]
- Kim Y, Oliver DJ. Molecular cloning, transcriptional characterization, and sequencing of cDNA encoding the H-protein of the mitochondrial glycine decarboxylase complex in peas. J Biol Chem. 1990;265:848–853. [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lernmark U, Gardeström P. Distribution of pyruvate dehydrogenase complex activities between chloroplasts and mitochondria from leaves of different species. Plant Physiol. 1994;106:1633–1638. doi: 10.1104/pp.106.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippuner V, Chou IT, Scott SV, Ettinger WF, Theg SM, Gasser CS. Cloning and characterization of chloroplast and cytosolic forms of cyclophilin from Arabidopsis thaliana. J Biol Chem. 1994;269:7863–7868. [PubMed] [Google Scholar]
- Macherel D, Lebrun M, Gagnon J, Neuburger M, Douce R. cDNA cloning, primary structure and gene expression for H-protein, a component of the glycine-cleavage system (glycine decarboxylase) of pea (Pisum sativum) leaf mitochondria. Biochem J. 1990;268:783–789. doi: 10.1042/bj2680783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattevi A, de Kok A, Perham RN. The pyruvate dehydrogenase multienzyme complex. Curr Opin Struct Biol. 1992;2:877–887. [Google Scholar]
- Morris TW, Reed KE, Cronan JE., Jr Identification of the gene encoding lipoate-protein ligase A of Escherichia coli. J Biol Chem. 1994;269:16091–16100. [PubMed] [Google Scholar]
- Morris TW, Reed KE, Cronan JE., Jr Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. J Bacteriol. 1995;177:1–10. doi: 10.1128/jb.177.1.1-10.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver DJ. The glycine decarboxylase complex from plant mitochondria. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:323–337. [Google Scholar]
- Otsuka AJ, Buoncristiani MR, Howard PK, Flamm J, Johnson C, Yamamoto R, Uchida K, Cook C, Ruppert J, Matsuzaki J. The Escherichia coli biotin biosynthetic enzyme sequences predicted from the nucleotide sequence of the bio operon. J Biol Chem. 1988;263:19577–19585. [PubMed] [Google Scholar]
- Parry RJ. Biosynthesis of some sulfur-containing natural products. Investigations of the mechanism of carbon-sulfur bond formation. Tetrahedron. 1983;39:1215–1238. [Google Scholar]
- Patton DA, Johnson M, Ward ER. Biotin synthase from Arabidopsis thaliana. cDNA isolation and characterization of gene expression. Plant Physiol. 1996;112:371–378. doi: 10.1104/pp.112.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perham RN. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry. 1991;30:8501–8512. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- Post-Beittenmiller D, Jaworski JG, Ohlrogge JB. In vivo pools of free and acylated acyl carrier proteins in spinach: evidence for sites of regulation of fatty acid biosynthesis. J Biol Chem. 1991;266:1858–1865. [PubMed] [Google Scholar]
- Reed KE, Cronan JE., Jr Lipoic acid metabolism in Escherichia coli: sequencing and functional characterization of the lipA and lipB genes. J Bacteriol. 1993;175:1325–1336. doi: 10.1128/jb.175.5.1325-1336.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Hackert ML. Structure-function relationship in dihydrolipoamide acyltransferase. J Biol Chem. 1990;265:8971–8974. [PubMed] [Google Scholar]
- Reed LJ, Hackert ML (1966) Chemistry and function of lipoic Acid. In M Florkin, EM Stotz, eds, Comprehensive Biochemistry, Vol 14. Elsevier Publishing, New York, pp 99–126
- Sanyal I, Cohen G, Flint DH. Biotin synthase: purification, characterization as a [2Fe-2S] cluster protein, and in vitro activity of the Escherichia coli bioB gene product. Biochemistry. 1994;33:3625–3631. doi: 10.1021/bi00178a020. [DOI] [PubMed] [Google Scholar]
- Schneider R, Brors B, Massow M, Weiss H. Mitochondrial fatty acid synthesis: a relic of endosymbiontic origin and a specialized means for respiration. FEBS Lett. 1997;407:249–252. doi: 10.1016/s0014-5793(97)00360-8. [DOI] [PubMed] [Google Scholar]
- Shintani DK, Ohlrogge JB. The characterization of a mitochondrial acyl carrier protein isoform isolated from Arabidopsis thaliana. Plant Physiol. 1994;104:1221. doi: 10.1104/pp.104.4.1221. 1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Sulo PS, Martin NC. Isolation and characterization of LIP5. A lipoate biosynthetic locus of Saccharomyces cerevisiae. J Biol Chem. 1993;268:17634–17639. [PubMed] [Google Scholar]
- Vanden Boom TJ, Reed KE, Cronan JE., Jr Lipoic acid metabolism in Escherichia coli: isolation of null mutants defective in lipoic acid biosynthesis, molecular cloning and characterization of the E. coli lip locus, and identification of the lipoylated protein of the glycine cleavage system. J Bacteriol. 1991;173:6411–6420. doi: 10.1128/jb.173.20.6411-6420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Cleavage-site motifs in protein targeting sequences. Genetic Engin. 1992;14:1–11. doi: 10.1007/978-1-4615-3424-2_1. [DOI] [PubMed] [Google Scholar]
- von Heijne G, Steppuhn J, Hermann RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Wada H, Shintani D, Ohlrogge J. Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proc Natl Acad Sci USA. 1997;94:1591–1596. doi: 10.1073/pnas.94.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Yu F, Wurtele ES, Nokolau BJ. Characterization of the cDNA and gene coding for the biotin synthase of Arabidopsis thaliana. Plant Physiol. 1996;110:1021–1028. doi: 10.1104/pp.110.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RH. Stable isotope studies on the biosynthesis of lipoic acid in Escherichia coli. Biochemistry. 1980;19:15–19. doi: 10.1021/bi00542a003. [DOI] [PubMed] [Google Scholar]
- Yeaman SJ. The 2-oxo acid dehydrogenase complexes: recent advances. Biochem J. 1989;257:625–632. doi: 10.1042/bj2570625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Sanyal I, Bulboaca GH, Rich A, Flint D. The gene for biotin synthase from Saccharomyces cerevisiae: cloning, sequencing, and complementation of Escherichia coli strains lacking biotin synthase. Arch Biochem Biophys. 1994;309:29–35. doi: 10.1006/abbi.1994.1079. [DOI] [PubMed] [Google Scholar]