SUMMARY
How cells coordinate size with growth and development is a major, unresolved question in cell biology. In previous work we identified the glucosyltransferase UgtP as a division inhibitor responsible for increasing the size of Bacillus subtilis cells under nutrient-rich conditions. In nutrient-rich medium, UgtP is distributed more or less uniformly throughout the cytoplasm and concentrated at the cell poles and/or the cytokinetic ring. Under these conditions, UgtP interacts directly with FtsZ to inhibit division and increase cell size. Conversely, under nutrient-poor conditions, UgtP is sequestered away from FtsZ in punctate foci, and division proceeds unimpeded resulting in a reduction in average cell size. Here we report that nutrient-dependent changes in UgtP's oligomerization potential serve as a molecular rheostat to precisely coordinate B. subtilis cell size with nutrient availability. Our data indicate UgtP interacts with itself and the essential cell division protein FtsZ in a high affinity manner influenced in part by UDP-glucose, an intracellular proxy for nutrient availability. These findings support a model in which UDP-glc dependent changes in UgtP's oligomerization potential shift the equilibrium between UgtP•UgtP and UgtP•FtsZ, fine tuning the amount of FtsZ available for assembly into the cytokinetic ring and with it cell size.
Keywords: nutrient-dependent cell size control, UDP-glucose, FtsZ assembly, UgtP oligomerization
INTRODUCTION
In all domains of life, growth and division must be coordinated to ensure that daughter cells are the correct size for a given condition or developmental fate. Despite the universal requirement for cell size control, specifically how cells are able to detect the achievement of a particular size is one of the major, unresolved questions in cell biology.
For single-celled organisms, nutrient availability is a primary determinant of cell size. Single-celled eukaryotes, including the classic cell cycle model system, Schizosaccharomyces pombe, have long been known to increase size in response to increases in nutrient availability (Cooper, 1969, Fantes & Nurse, 1977, Schaechter et al., 1958). Similarly, when cultured in a nutrient-rich medium such as Luria-Bertani (LB), both the Gram-negative bacteria Salmonella typhimurium and Escherichia coli as well as the Gram-positive bacterium Bacillus subtilis exhibit rapid mass doubling times and can be up to twice as large as their slower growing counterparts cultured in a nutrient-poor medium such as minimal salts supplemented with a poorly utilized sugar or amino acid (Donachie & Begg, 1989, Sargent, 1975, Schaechter et al., 1958).
Experiments in which growing cells are shifted between nutrient-poor and nutrient-rich medium suggest that nutrient-dependent cell size control is a rapid and cell intrinsic phenomenon. When moved from a poor nutrient source to a rich one, Escherichia coli cells immediately increase their growth rate, but delay division until they have achieved the appropriate size for the new growth condition (Cooper, 1969). Thus, bacteria must be able to sense changes in nutrient availability and growth rate, and transmit this information to the division apparatus to ensure that daughter cells are the appropriate size for a given condition.
In previous work we identified the glucosyltransferase UgtP as a metabolic sensor responsible for increasing B. subtilis cell size under nutrient-rich conditions (Weart et al., 2007). In nutrient-rich medium, UgtP is expressed at high levels and distributed relatively smoothly throughout the cytoplasm with some protein concentrated at cell poles and the nascent septum. Conversely, under nutrient-poor conditions UgtP levels are reduced ~six-fold and the remaining protein is localized away from the cytokinetic ring in randomly distributed punctate foci. UgtP localization and division inhibition is dependent on UDP-glucose (UDP-glc) biosynthesis, suggesting the nucleotide sugar might serve as an intracellular proxy for nutrient availability. UgtP accumulation, on the other hand, is controlled by a growth rate-dependent, but UDP-glc independent mechanism. UgtP interacts directly with the essential, tubulin-like cell division protein FtsZ to inhibit its assembly.
On the basis of these results, we proposed that during growth in nutrient-rich medium, high levels of UDP-glc stimulate UgtP interaction with FtsZ both at the cytokinetic ring and in the cytoplasm, delaying maturation of the cytokinetic ring and increasing cell size. Under nutrient-poor conditions, UgtP is prevented from interacting with FtsZ through at least two independent mechanisms: sequestration in punctate foci in response to low levels of UDP-glc, and UDP-glc independent reductions in the intracellular concentration of UgtP.
Here we dissect the mechanism by which changes in UDP-glc availability impact UgtP localization and activity, specifically clarifying the role of UDP-glc in interactions between UgtP and FtsZ. Our findings suggest that the overwhelming majority of UgtP is bound either to itself or to FtsZ regardless of growth rate. UDP-glc dependent changes in the equilibrium between UgtP•FtsZ and UgtP•UgtP serve as a molecular rheostat to help ensure that B. subtilis cell size is precisely coordinated with growth rate and nutrient availability.
RESULTS
UgtP forms higher order structures in vivo and in vitro
Electron micrographs of a purified Thioredoxin-UgtP (Thio-UgtP) fusion protein in low salt FtsZ assembly buffer suggest UgtP is able to form large, highly ordered structures of ~35 to 40 subunits arranged in closely stacked spirals of seven to eight Thio-UgtP monomers per turn (Weart et al., 2007). These structures are not visible in the presence of FtsZ, suggesting the cell division protein may interfere with UgtP's ability to oligomerize. UDP-glc did not have a significant impact on the ability of UgtP to form these higher order structures at a near physiological concentration of 1 μM. Based on these observations, we speculated that the UgtP foci observed in cells cultured in nutrient-poor medium were in fact UgtP oligomers.
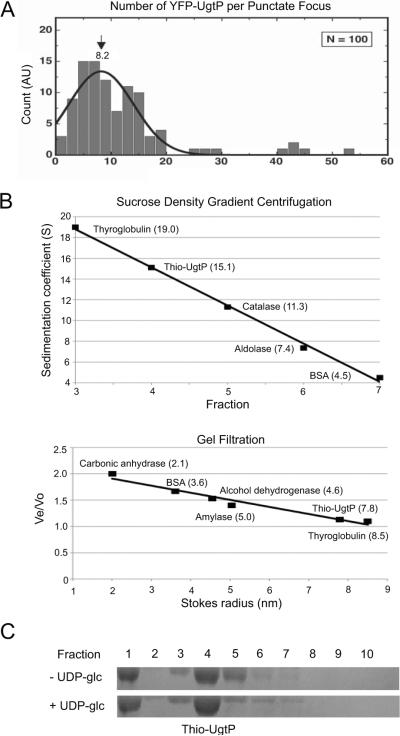
To determine if the UgtP foci we observed in vivo are themselves composed of UgtP oligomers, we employed a single molecule approach, integrated photon molecular counting (IPMC) (Wang et al., 2005). For these experiments, we imaged B. subtilis cells expressing a functional (Nishibori et al., 2005), xylose-inducible YFP-UgtP fusion, taking advantage of a strain defective in UDP-glc biosynthesis (PL2292 pgcA∷spc amyE∷Pxyl-yfp-ugtP). Although this strain is wild type for growth and viability, the absence of UDP-glc biosynthesis results in punctate UgtP localization pattern regardless of nutrient availability (Weart et al., 2007). This phenotype ensures a high frequency of UgtP foci and greatly simplifies analysis. IPMC indicated that UgtP forms oligomers in vivo that are almost identical in size to those formed in vitro. The lifetime photon count distribution of 100 YFP-UgtP punctate foci yielded an average of ~8.2 YFP-UgtP monomers per focus when fitted to a Gaussian distribution (Figure 1A), approximately equivalent to one turn of the spirals observed in our previous electron microscopy data. Intriguingly, the histogram of IPMC data also indicated the presence even larger UgtP structures consisting of more than 8 subunits, suggesting that some YFP-UgtP foci may contain even higher order UgtP oligomers.
Figure 1. UgtP forms higher order structures in vivo and in vitro.
(A) The lifetime photon count distribution of 100 YFP-UgtP punctate foci in amyE∷Pxyl-yfp-ugtP pgcA∷spc cells yielded an average of ~8.2 YFP-UgtP monomers per focus when fitted to a Gaussian distribution. The experimental setup was described previously (DeSantis et al., 2010). The single molecule experiments were performed using prism-type total internal reflection fluorescence (TIRF) microscopy and the number of YFPUgtP molecules per punctate focus was determined using the IPMC method (Wang et al., 2005), in which the ratio between the mean life time photon count of YFP-UgtP foci and the mean lifetime photon count of individual YFP molecules were obtained by in vitro imaging of YFP adsorbed on fused-silica surfaces and counting their total emission photons before bleaching.
(B) Top: Standard curve of sedimentation coefficients of Thio-UgtP and markers generated by sucrose density gradient centrifugation. Purified Thio-UgtP and markers were loaded onto 4 ml 20%–50% sucrose gradients in UgtP buffer (100 mM NaCl, 10% glycerol, 20 mM Tris pH 8.0) for centrifugation. Ten fractions were collected from the top to the bottom of the gradients and examined by SDS-PAGE. The greater the fraction number, the lower the sucrose concentration of the fraction (ie. fraction 1 contained 50% sucrose whereas fraction 10 contained 20% sucrose). Bottom: Standard curve of Stokes radii of Thio-UgtP and markers generated by gel filtration analysis. Purified Thio-UgtP and markers in UgtP buffer were applied to an S300 gel filtration column and 2 ml fractions were collected and examined by SDS-PAGE. Combining these two approaches, Thio-UgtP has an apparent molecular weight of ~495 kDa in vitro, equivalent to ~8.8 monomers, consistent with the oligomeric state of UgtP in vivo.
(C) Sucrose density gradient centrifugation of Thio-UgtP in UgtP buffer in the absence and presence of 2 mM UDP-glc indicated that the addition of UDP-glc does not change the oligomeric state of Thio-UgtP in vitro. Thio-UgtP still peaked at the same fraction (fraction 4) regardless of the presence of UDP-glc. Thio-UgtP peaked at the fraction containing the highest sucrose concentration likely represents even higher order UgtP oligomers (fraction 1).
We next examined the ability of Thio-UgtP to oligomerize in vitro by sucrose density gradient centrifugation and gel filtration analysis, as a means of validating our previous electron microscopy data (Weart et al., 2007). To reduce non-specific interactions, we employed a buffer that was higher in pH and salt (see Experimental Procedures for all buffer compositions). In support of our previous work, these experiments indicated that UgtP has a strong propensity to oligomerize. In vitro Thio-UgtP has a sedimentation coefficient of ~15.1 and a Stokes radius of ~7.8 (Figure 1B). Combining these data, the apparent molecular weight of Thio-UgtP oligomer is ~495 kDa, equivalent to ~8.8 Thio-UgtP monomers, a number on par with our IPMC data, and importantly a size approximately equivalent to a single turn of the large, helical oligomers we observed in our previous study. The difference in oligomer size between our previous work and this one likely reflects a reduction in the ability of UgtP oligomers to stack in the more stringent buffer conditions. We did observe a secondary concentration of UgtP in the highest density fraction (Lane 1 in Figure 1C). Although not the predominant species, it is likely that UgtP in this fraction corresponds to the stacked oligomers we observed by electron microscopy and the brighter foci IPMC identified as containing greater than 8 subunits of UgtP (Weart et al., 2007) (Figure 1A).
Despite the impact of UDP-glc biosynthesis on UgtP puncta formation in vivo (Weart et al., 2007), in vitro the addition of the nucleotide sugar did not have a measurable impact on UgtP oligomerization under the conditions we employed for sucrose density gradient centrifugation (Figure 1C). The relative concentration of UgtP in the various fractions was more or less identical in experiments conducted in the presence and absence of UDP-glc. This finding is consistent with our previous electron microscopy data, suggesting that in vitro UDP-glc does not inhibit UgtP oligomerization in the absence of FtsZ (Weart et al., 2007).
UDP-glc reduces UgtP's affinity for itself ~four-fold
The lack of impact of UDP-glc on UgtP oligomerization in vitro contrasts starkly with our finding that UDP-glc biosynthesis prevents the formation of UgtP foci in vivo (Weart et al., 2007). One explanation for this discrepancy is that UgtP's affinity for itself is too high for us to detect gross changes in UgtP oligomerization under our experimental conditions, even in the presence of UDP-glc. To clarify this issue we took advantage of an Octet Red96 instrument (FortéBio) to measure the affinity of UgtP for itself in the presence and absence of UDP-glc. The Octet Red96 instrument employs a label-free, optical analytical technology that measures protein interactions based on Bio-Layer Interferometry (BLI). Any interaction between UgtP immobilized on the biosensor tip surface and UgtP in the solution produces an increase in optical thickness, resulting in a wavelength shift indicative of the change in the biological layer thickness.
Affinity data indicate that UDP-glc has a modest but significant effect on UgtP·UgtP interaction. At the same time, UgtP's affinity for itself is in the nanomolar range even in the presence of UDP-glc, suggesting its default state is oligomeric during growth in nutrient rich medium where its physiologically concentration is ~1.45 μM. {UgtP concentration was calculated using calculations of UgtP per cell and cell diameter and width data from our previous work (Weart et al., 2007). Cell volume was calculated using the methods described here (Heldal et al.) and here (Loferer-Krossbacher et al.).} UgtP's affinity for itself is ~four-fold higher in the absence of UDP-glc (KD = 17 nM in the absence of UDP-glc versus 69 nM in the presence of UDP-glc), supporting the idea that interaction with UDP-glc reduces UgtP's oligomerization potential (Table 1). UgtP's high affinity for itself, means that UDP-glc dependent changes in UgtP's oligomerization potential would only be apparent when UgtP concentration is lower than ~7 nM, well below our limit of detection. This finding explains our inability to observe differences in UgtP oligomerization in the presence and absence of UDP-glc in vitro (Weart et al., 2007) (Figure 1C).
Table 1.
Affinities of wild type and mutant UgtPs for themselves
| Protein on Biosensor | Protein in Solution | Dissociation Constant (KD) |
|---|---|---|
| UgtP WT | UgtP WT − UDP-glc | 17 nM |
| UgtP WT | UgtP WT + UDP-glc | 69 nM |
| UgtP (F112A V117A E306A N309A) | UgtP (F112A V117A E306A N309A) − UDP-glc | 16 nM |
| UgtP (F112 A V117A E306A N309A) | UgtP (F112A V117A E306A N309A) + UDP-glc | 21 nM |
FtsZ inhibits UgtP oligomerization
Our discovery that UgtP retained a nanomolar affinity for itself even in the presence of UDP-glc, is at odds with our previous work indicating that biosynthesis of the nucleotide sugar has a relatively dramatic impact on UgtP localization and activity in vivo (Weart et al., 2007). Based on the observation that higher order UgtP structures are not detectable by electron microscopy in the presence of FtsZ (Weart et al., 2007), we speculated that the cell division protein might serve as a modulator of UgtP oligomerization in vivo. To test this possibility, we repeated the sucrose gradient density centrifugation experiments in the presence of a 10:1 molar ratio of FtsZ to UgtP, and also examined the impact of changes in the intracellular concentration of FtsZ on UgtP localization in vivo.
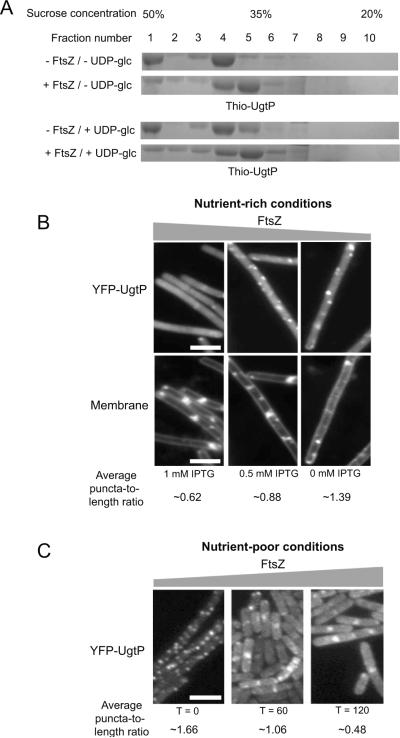
Sucrose gradient density centrifugation data (Figure 2A) indicate that FtsZ inhibits UgtP oligomerization, in agreement with our previous findings (Weart et al., 2007). The addition of FtsZ shifted the Thio-UgtP peak towards lower sucrose concentrations, indicative of a reduction in UgtP oligomerization. Significantly, the presence of FtsZ significantly reduced the amount of UgtP in the fraction containing highest sucrose concentration (Fraction 1, Figure 2A), suggesting FtsZ strongly interferes with the formation of the extremely large UgtP oligomers.
Figure 2. FtsZ influences UgtP oligomerization in vitro and in vivo.
(A) Sucrose density gradient centrifugation indicates that the presence of FtsZ lowers the oligomeric state of Thio-UgtP. In the presence of FtsZ, Thio-UgtP peaked at a fraction with lower sucrose concentration (fraction 5) compared to in the absence of FtsZ (fraction 4), suggesting that FtsZ prevents Thio-UgtP oligomerization in vitro. Moreover, the presence of FtsZ prevented Thio-UgtP from concentrating in the fraction containing the highest sucrose concentration (fraction 1), indicating that FtsZ prevented higher order UgtP oligomerization. FtsZ influenced UgtP oligomerization in vitro regardless of the presence of 2 mM UDP-glc. Thio-UgtP was at 3 μM and the molar ratio of FtsZ to UgtP was 10:1. UgtP buffer (100 mM NaCl, 10% glycerol, 20 mM Tris pH 8.0) was used.
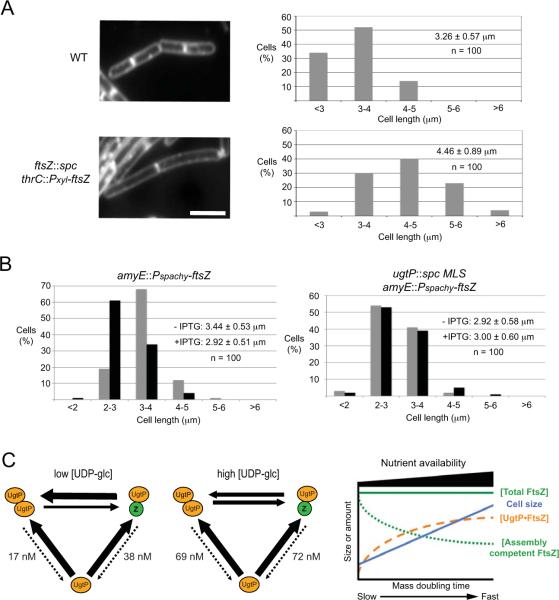
(B) Depletion of intracellular FtsZ in PL2430 ftsZ∷Pspachy-ftsZ amyE∷Pxyl-yfp-ugtP cells cultured under nutrient-rich conditions forced YFP-UgtP to re-localize from cytoplasm to punctate foci. The greater the reductions in the intracellular FtsZ levels (the lower the concentration of the inducer IPTG), the higher the average YFP-UgtP puncta-to-length ratio. Different amounts of IPTG were added to the culture to generate different levels of FtsZ in the cells. In the bottom panel, cells were stained with membrane probe TMA-DPH. Bar = 3 μm.
(C) Approximately three-fold over-expression of ftsZ inhibited YFP-UgtP oligomerization and puncta formation in JC115 thrC∷Pxyl-ftsZ amyE∷Pspachy-yfp-ugtP cells during growth in nutrient-poor S750 minimal sorbitol, suggesting that FtsZ inhibits UgtP oligomerization in vivo. Cells were cultured in minimal sorbitol at 30°C and imaged 60 minutes and 120 minutes after inducing the over-expression of ftsZ. Bar = 3 μm.
We next determined if UgtP oligomerization in vivo was influenced by changes in the intracellular concentration of FtsZ. Specifically, we wondered if depleting FtsZ would be sufficient to promote the formation of UgtP puncta in cells cultured under nutrient-rich conditions, and if increasing the intracellular concentration of FtsZ would inhibit the formation of UgtP puncta in cells cultured under nutrient-poor conditions. For these experiments we localized YFP-UgtP in rapidly growing cells extensively depleted for FtsZ, as well as in slow growing cells where ftsZ was over-expressed ~three-fold. For all experiments FtsZ levels were monitored by quantitative immunoblot (Figure S1).
To deplete FtsZ, we took advantage of a strain in which ftsZ expression is dependent on an IPTG-inducible promoter (PL2430 ftsZ∷Pspachy-ftsZ amyE∷Pxyl-yfp-ugtP). Cells were subcultured in nutrient-rich LB in the presence of varying amounts of IPTG to generate a range of intracellular concentrations of FtsZ. We previously employed this approach to establish that UgtP localization to the cytokinetic ring is FtsZ-dependent (Weart et al., 2007). However, beyond confirming the loss of UgtP localization to the cytokinetic ring in the absence of FtsZ, we did not monitor the changes in UgtP localization closely. For the complementary experiment, we monitored YFP-UgtP localization during growth in nutrient-poor medium (S750 minimal medium supplemented with 1% sorbitol) in a strain engineered to over-produce FtsZ ~three-fold in the presence of 0.5% xylose (JC115 thrC∷Pxyl-ftsZ amyE∷Pspachy-ayfp-ugtP). Mass doubling times at 30°C were ~40 min in LB and ~120 min in S750 minimal medium supplemented with 1% sorbitol.
Our findings support the idea that FtsZ is a direct inhibitor of UgtP oligomerization in vivo. As shown in Figure 2B, the average UgtP puncta-to-length ratio in PL2430 (ftsZ∷Pspachy-ftsZ amyE∷Pxyl-yfp-ugtP) cells cultured in the presence of saturating levels of IPTG (1 mM) was ~0.62 μm−1. In contrast the average puncta-to-length ratio in PL2430 cells cultured in the absence of IPTG for two hours was ~1.39 μm−1. Partial depletion of FtsZ, achieved by adding sub-saturating amounts of IPTG (0.5 mM) resulted in an intermediate puncta-to-length ratio of ~0.88 μm−1. For comparison, cells defective in UDP-glc biosynthesis have an average puncta-to-length ratio of ~1.36 μm−1, and wild type cells have an average puncta-to-length ratio of ~0.11 μm−1.
Depleting FtsZ leads to a reduction in cytoplasmic YFP-UgtP fluorescence, a finding that supports the idea that UgtP interacts with FtsZ in the cytoplasm as well as at the cytokinetic ring. Measurements of fluorescence intensity using ImageJ (Schneider et al., 2012) indicated that YFP-UgtP intensity in the cytoplasm decreased by ~60% in extensively FtsZ-depleted cells, concurrent with the increase in the number of punctate foci.
In the complementary experiment, we determined that increasing intracellular FtsZ levels ~three-fold was sufficient to force UgtP from punctate foci to the cytokinetic ring in cells cultured under nutrient-poor conditions (Figure 2C). Within two hours of ftsZ induction, the majority of YFP-UgtP was either cytoplasmic and/or concentrated at mid-cell or poles, a pattern normally observed only under nutrient-rich conditions. The average puncta-to-length ratio was ~1.66 μm−1 prior to ftsZ induction, ~1.06 μm−1 one hour after ftsZ induction, and ~0.48 μm−1 two hours after ftsZ induction. Consistent with the idea that UgtP interacts with FtsZ in the cytoplasm as well as at the cytokinetic ring, measurements of fluorescence intensity using ImageJ indicate that YFP-UgtP intensity in the cytoplasm increased by ~four-fold after two hours of FtsZ overproduction, concurrent with the decrease in the number of punctate foci.
UgtP foci are dynamic structures influenced by changes in intracellular UDP-glc availability
Our finding that UgtP re-localized in response to the changes in intracellular FtsZ levels, raised the possibility that UgtP foci are dynamic structures, capable of dissociating in response to the changes in nutrient availability and intracellular UDP-glc levels. To explore this possibility, we monitored UgtP localization in cells engineered to increase or decrease the availability of UDP-glc in response to an inducer. To increase intracellular UDP-glc levels, we employed a strain producing the B. subtilis pyrophosphorylase GtaB only in the presence of IPTG (JC313 gtaB∷spc thrC∷Pspachy-gtaB-cmyc amyE∷Pxyl-yfp-ugtP). GtaB is required for the intracellular conversion of glucose-1-phosphate (glc-1-P) to UDP-glc. Induction of gtaB expression in cells cultured in nutrient-rich medium, which presumably have a sizeable pool of glc-1-P, should thus result in a rapid increase in intracellular UDP-glc concentration. Conversely, reducing intracellular UDP-glc levels was achieved by over-expressing B. subtilis ywqF from an IPTG-inducible promoter (JC330 thrC∷Pspachy-ywqF amyE∷Pxyl-yfp-ugtP). YwqF is a UDP-glc dehydrogenase responsible for UDP-glucuronate synthesis that has not been implicated in either cell division or cell cycle control in B. subtilis (Mijakovic et al., 2003). By virtue of its ability to bind UDP-glc, ywqF induction should lead to the rapid depletion of available UDP-glc without directly impacting cell division.
Taking advantage of these strain constructs, we determined that UgtP can re-localize from punctate foci to the cytokinetic ring and vice versa relatively rapidly in response to changes in intracellular UDP-glc levels. (Figures 3A and 3B). The average puncta-to-length ratio of JC313 cells (gtaB∷spc thrC∷Pspachy-gtaB-cmyc amyE∷Pxyl-yfp-ugtP) was ~0.65 μm−1 prior to gtaB induction, ~0.24 μm-1 10 minutes, and ~0.18 μm−1 20 minutes after gtaB induction. Conversely, the average puncta-to-length ratio of JC330 cells (thrC∷Pspachy-ywqF amyE∷Pxyl-yfp-ugtP) was ~0.18 μm−1 prior to ywqF induction, ~0.36 μm−1 10 minutes, and ~0.60 μm−1 20 minutes after ywqF induction. Together these data support the idea that UgtP oligomerization is a dynamic, UDP-glc sensitive process and that intermediate levels of UDP-glc lead to intermediate UgtP localization, and almost certainly intermediate UgtP activity.
Figure 3. UgtP oligomerization is sensitive to the changes in UDP-glc availability.
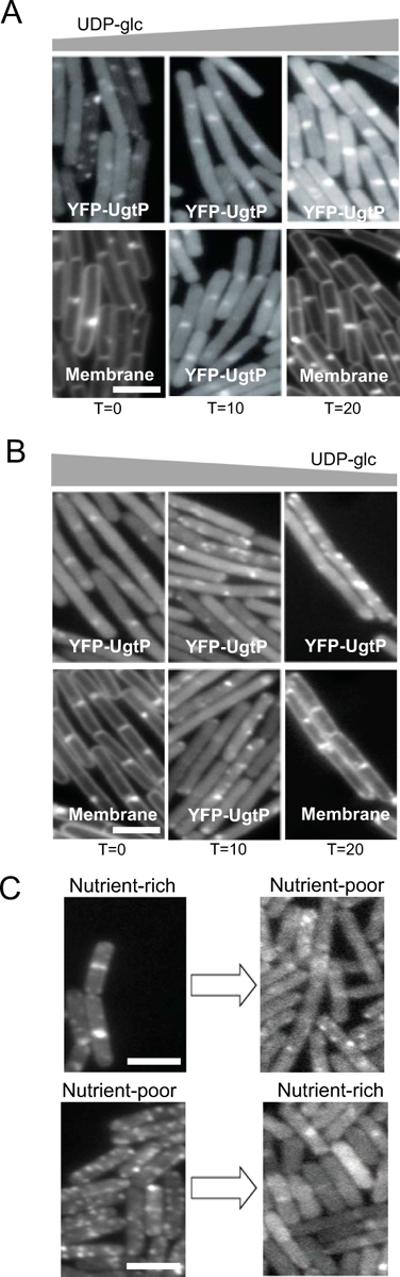
(A) YFP-UgtP re-localized from punctate foci to the cytoplasm and medial/polar rings in response to increases in intracellular UDP-glc levels within 20 minutes of expression of the UDP-glc synthesizing protein GtaB. JC313 gtaB∷spc thrC∷Pspachy-gtaB-cmyc amyE∷Pxyl-yfp-ugtP cells were cultured in nutrient-rich LB at 30°C. For T = 0 and T = 20 time points, membrane probe TMA-DPH was used.
(B) YFP-UgtP re-localized from the cytoplasm and medial/polar rings to punctate foci in response to decreases in intracellular UDP-glc levels within 20 minutes of expression of the UDP-glc binding protein YwqF. JC330 thrC∷Pspachy-ywqF amyE∷Pxyl-yfp-ugtP cells were cultured in nutrient-rich LB at 30°C. For T = 0 and T = 20 time points, membrane probe TMA-DPH was used.
(C) Shifting PL2423 amyE∷Pxyl-yfp-ugtP cells between nutrient-rich and nutrient-poor medium led to the protein synthesis independent re-localization of YFP-UgtP. Top: YFP-UgtP re-localized from the cytokinetic ring to punctate foci within 30 minutes of a shift from nutrient-rich LB to nutrient-poor minimal sorbitol. Bottom: YFP-UgtP re-localized from punctate foci to the cytokinetic ring within 30 minutes of a shift from nutrient-poor minimal sorbitol to nutrient-rich LB. Both experiments were conducted in the presence of 200 μg ml−1 chloramphenicol to inhibit new protein synthesis and cells were cultured at 30°C with a mass doubling time ~40 minutes.
Note that exposure times are identical in A–B. In C, exposure times were increased to compensate for the lack of new protein synthesis in the latter image. Bar = 3 μm.
Shifts between nutrient-rich and nutrient-poor medium stimulate re-localization of UgtP independent of protein synthesis
We next determined if shifts in nutrient availability are accompanied by changes in UgtP localization on a time scale similar to that observed with changes in intracellular UDP-glc levels. We reasoned that if UgtP foci are in fact dynamic structures capable of dissociating rapidly in response to changes in nutrient and UDP-glc availability, changes in UgtP localization should be independent of new protein synthesis. To test this possibility, we examined YFP-UgtP localization following abrupt shifts in nutrient availability in the absence of new protein synthesis.
In support of a model in which the nutrient-dependent changes in UgtP localization and oligomerization are dynamic and protein synthesis independent, UgtP re-localized to punctate foci within 30 minutes of a shift from nutrient-rich to nutrient-poor medium in the presence of 200 μg/ml protein synthesis inhibitor chloramphenicol (Figure 3C). In the complementary experiment, UgtP re-localized from punctate foci to the cytoplasm, cell poles and cytokinetic ring within 30 minutes of a shift from nutrient-poor to nutrient-rich medium in the presence of chloramphenicol. Shifting cells between the same nutrient conditions in the presence of chloramphenicol had no effects on UgtP localization (Figure S2).
Defects in UgtP's putative UDP-glc binding sites render it nutrient-blind with regard to localization and division inhibition in vivo but do not abolish its interaction with FtsZ
Data presented here (Figure 3 and Table 1) and in our previous work (Weart et al., 2007) support a model in which interactions with UDP-glc alter UgtP′s ability to oligomerize and to interact with FtsZ. As a genetic test of this model, we generated UgtP mutants defective in residues implicated in interactions with UDP-glc. Mutations were made based on an alignment between UgtP and a related glycosyltransferase, MGDG synthase from Spinacia oleracea, for which there is significant structural data (Botte et al., 2005). The two proteins show 25% identity and 60% similarity across their entire length, and 50% identity and 75% similarity in their nucleotide sugar binding sites. Like MGDG synthase, UgtP is predicted to bind its donor substrate UDP-glc at the interface of its N- and C-terminal domains indicated by 3DLigandSite (Wass et al., 2010). Based on alignments between UgtP and MGDG synthase (Figure S3A), we generated two sets of putative UDP-glc binding mutants of UgtP: F112A V117A and E306A N309A. The first two residues are located in UgtP′s putative N-terminal nucleotide-binding site whereas the latter are located in the putative C-terminal hexose-binding site.
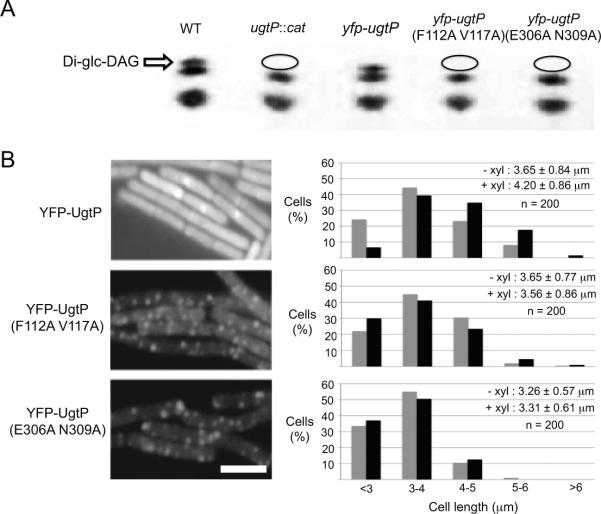
Thin layer chromatography (TLC) indicated that mutations in either UgtP′s predicted nucleotide- or hexose-binding site disrupt its glucosyltransferase activity (Figure 4A). Lipid extracts from ugtP null cells expressing a wild type yfp-ugtP fusion from an ectopic locus (PL2295 ugtP∷spc MLS amyE∷Pxyl-yfp-ugtP) contained diglucosyl-diacylglycerol (Di-glc-DAG), confirming it is a functional fusion with regard to its glucosyltransferase activity. In contrast, lipid extracts from ugtP null cells encoding YFP-fusions to either the putative nucleotide- (JC47 ugtP∷spc MLS amyE∷Pxyl-yfp-ugtP F112A V117A) or hexose-binding mutant (JC183 ugtP∷spc MLS amyE∷Pxyl-yfp-ugtP E306A N309A) did not contain any Di-glc-DAG,suggesting that these mutations disrupt UgtP′s glucosyltransferase activity, most likely by preventing interactions with UDP-glc.
Figure 4. Defects in UgtP's putative UDP-glc binding sites render it nutrient-blind with regard to localization and division inhibition in vivo.
(A) Thin layer chromatography of wild type cells and cells expressing variants of yfp-ugtP in ugtP∷cat background. While wild type yfp-ugtP fully complemented the ugtP null strain with regard to Di-glc-DAG synthesis (uppermost band), lipid extracts of cells expressing either putative UDP-glc binding mutant of yfp-ugtP indicated that both mutants are defective in their ability to synthesize Di-glc-DAG. Ovals indicate the absence of Di-glc-DAG.
(B) Left: Defects in UgtP's putative UDP-glc binding sites results in YFP-UgtP punctate localization during growth in nutrient-rich medium. YFP-fusions to the putative UDP-glc binding mutants, UgtP (F112A V117A) and UgtP (E306A N309A), exhibit a punctate localization pattern during growth in LB. In contrast, wild type YFP-UgtP displays both a smooth cytoplasmic staining and a concentration at the cytokinetic ring. Bar = 3 μm. Right: Putative UDP-glc binding mutants of YFP-UgtP are functionally null with regard to division inhibition. Size distribution of 200 ugtP∷cat cells (gray bars) expressing either wild type yfp-ugtP, yfp-ugtP (F112A V117A), or yfp-ugtP (E306A E309A) (black bars) from an ectopic locus. In contrast to wild type yfp-ugtP whose expression increases the average length of ugtP∷cat cells by ~10% (p < 0.001), putative UDP-glc binding mutants of yfp-ugtP do not rescue the short cell phenotype of ugtP∷cat cells. Bar = 3 μm.
Analysis of the putative UDP-glc binding mutants of UgtP in vivo supports the idea that UDP-glc dependent changes in UgtP oligomerization play a critical role in the regulatory circuit governing nutrient-dependent cell size control. YFP-fusions to either the putative nucleotide- or hexose-binding mutant of UgtP localized in punctate foci regardless of nutrient availability, a pattern indistinguishable from wild type YFP-UgtP in cells defective in UDP-glc biosynthesis (Figure 4B). Under nutrient-rich conditions, the average puncta-to-length ratio was ~0.11 μm−1 for cells expressing wild type YFP-UgtP, while the average puncta-to-length ratios were ~1.20 μm−1 and μ1.42 μm−1 for cells expressing the putative nucleotide- and hexose-binding mutants respectively. Sub-cellular localization of all three fusions was independent of the presence of the wild type ugtP allele (Figure S3B). Consistent with UDP-glc dependent changes in UgtP oligomerization serving as an essential element of nutrient-dependent cell size control, the putative nucleotide- and hexose-binding mutants were unable to complement the cell size defect of ugtP null cells during growth in nutrient-rich medium (Figure 4B). Moreover, using the Octet system described before, we determined the affinity of a UgtP mutant defective in both the putative nucleotide- and hexose-binding sites for itself was essentially unchanged regardless of the presence of UDP-glc (KD = 16 nM in the absence of UDP-glc versus KD = 21 nM in the presence of UDP-glc), consistent with our proposed model where interactions with UDP-glc alter UgtP′s ability to oligomerize (Table 1).
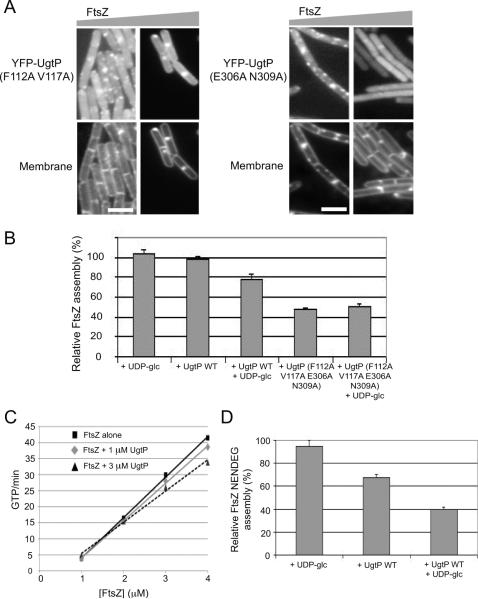
Although both mutants, UgtP (F112A V117A) and UgtP (E306A N309A), were insensitive to changes in nutrient availability with regard to localization and oligomerization, both re-localized in response to a ~three-fold increase in intracellular FtsZ levels, suggesting they retain a high affinity for FtsZ (Figure 5A). As before we took advantage of the IPTG-inducible construct of ftsZ described above (ftsZ∷Pspachy-ftsZ) and FtsZ levels were measured by quantitative immunoblot (Figure S4). Under nutrient-rich conditions, within 30 minutes of ftsZ induction, YFP-fusions to both the putative UDP-glc binding mutants of UgtP exhibited smooth cytoplasmic staining with some of the cells displaying a medial and/or polar YFP-UgtP localization pattern. The average puncta-to-length ratio declined ~four-fold from ~1.20 μm−1 to~0.31 μm−1 for cells expressing the putative nucleotide-binding mutant of UgtP, and ~seven-fold from ~1.42 μm−1 to ~0.20 μm−1 for cells expressing the putative hexose-binding mutant of UgtP. This finding is consistent with our observation that increasing FtsZ levels inhibits UgtP oligomerization in cells cultured under nutrient-poor conditions (Figure 2C).
Figure 5. UDP-glc stimulates UgtP mediated inhibition of FtsZ assembly in vitro.
(A)Approximately three-fold over-expression of ftsZ is sufficient to stimulate the relocalization of YFP-fusions to the putative UDP-glc binding mutants of UgtP from punctate foci to the cytokinetic ring, indicating both mutants still interact with FtsZ. Cells were cultured in nutrient-rich LB at 30°C. In the bottom panel, cells were stained with membrane probe TMA-DPH. Bar = 3 μm.
(B) Pooled results of 90° angle light-scattering assays conducted in a higher salt buffer indicate that UDP-glc stimulates UgtP mediated inhibition of FtsZ assembly in vitro. Using this buffer system, UgtP only inhibits FtsZ assembly in the presence of UDP-glc. UgtP mutant defective in both the putative UDP-glc binding sites inhibits FtsZ assembly better than wild type UgtP but is insensitive to the presence of UDP-glc. FtsZ is at 5 μM, UgtP is at 1.67 μM and UDP-glc is at 2 mM. Assembly buffer contains 50 mM MES pH 6.5, 5 mM MgCl2, 1 mM EGTA and 75 mM NaCl. Percent inhibition was determined by peak assembly relative to that of 5 μM FtsZ alone. Error bars = standard deviations. N = 3.
(C) The addition of UgtP has no significant impact on FtsZ mediated GTP hydrolysis. FtsZ alone (black rectangles) or in the presence of 1 μM UgtP (gray diamonds) and 3 μM UgtP (dark gray triangles). FtsZ is either at 1 μM, 2 μM, 3μM or 4 μM. UDP-glc is at 2 mM.
(D)UgtP inhibits assembly of an FtsZ mutant that is unable to form stabilizing lateral interactions between protofilaments (FtsZ NENDEG) (Buske and Levin, 2012). FtsZ NENDEG is at 5 μM, UgtP is at 2.5 μM and UDP-glc is at 2 mM. N = 3. Error bars = standard deviations. Reaction buffer contains 50 mM MES pH 6.5, 5 mM MgCl2, 1 mM EGTA.
In support of the idea that UDP-glc modulates but is not required for the interaction between UgtP and FtsZ, the affinity of wild type and mutant UgtPs for FtsZ was high in both the presence and absence of UDP-glc. Using the Octet system, we determined the affinity of wild type UgtP for FtsZ to be 38 nM in the absence of UDP-glc and 72 nM in the presence of UDP-glc (Table 2). A version of UgtP encoding mutations in both the putative nucleotide- and hexose-binding sites exhibited high affinities for FtsZ regardless of the presence of UDP-glc (16 nM in the absence of UDP-glc versus 20 nM in the presence of UDP-glc.).
Table 2.
Affinities of wild type and mutant UgtPs for FtsZ
| Protein on Biosensor | Protein in Solution | Dissociation Constant (KD) |
|---|---|---|
| UgtP WT | FtsZ − UDP-glc | 38 nM |
| UgtP WT | FtsZ + UDP-glc | 72 nM |
| UgtP (F112A V117A E306A N309A) | FtsZ − UDP-glc | 16 nM |
| UgtP (F112 A V117A E306A N309A) | FtsZ + UDP-glc | 20 nM |
Interactions with UDP-glc stimulate UgtP mediated inhibition of FtsZ assembly
The nanomolar affinity of UgtP for itself and FtsZ suggests that UgtP is essentially always in complex with either itself or FtsZ at its physiological concentration of ~1.45 μM. Instead of increasing the pool of free UgtP, increases in intracellular UDP-glc levels shift the equilibrium from UgtP•UgtP towards UgtP•FtsZ, reducing the amount of FtsZ available for assembly into the cytokinetic ring and thereby increasing cell size. Wild type UgtP is thus expected to inhibit FtsZ assembly better in the presence of UDP-glc when UgtP′s affinity for itself is more or less equivalent to its affinity for FtsZ (69 nM and 72 nM respectively) than in the absence of the nucleotide sugar when UgtP′s affinity for FtsZ is ~two-fold lower than its affinity for itself (17 nM versus 38 nM) (Tables 1 and 2). In contrast, given its greater than wild type, UDP-glc independent affinity for FtsZ, the putative UDP-glc binding mutant of UgtP should inhibit FtsZ assembly as well or better than wild type UgtP.
To clarify the role of UDP-glc in mediating the interaction between UgtP and FtsZ, we determined the ability of wild type and mutant UgtPs to inhibit FtsZ assembly in the presence of UDP-glc using an in vitro, 90° angle light-scattering assay. Although we had seen a slight enhancement of UgtP mediated inhibition of FtsZ assembly in the presence of UDP-glc, the difference was near the margin of error (Weart et al., 2007). These reactions were conducted in the low salt buffer we typically used in our FtsZ assembly assays, the same conditions that stimulate the formation of extremely large UgtP oligomers observed in our previous electron microscopy data. We thus repeated these experiments, increasing NaCl concentration from 25 mM to 75 mM to ensure that UgtP oligomers were of physiologically relevant size. This small increase in NaCl concentration lowers total FtsZ assembly ~two-fold, due to a reduction in stabilizing lateral interactions between protofilaments (Buske & Levin, 2012).
90° angle light-scattering assay indicates that UDP-glc had a strong impact on the ability of UgtP to inhibit FtsZ assembly, suggesting that relatively small changes in UgtP′s affinity for itself and FtsZ are sufficient for UgtP mediated division inhibition in vivo. Consistent with UDP-glc dependent changes in UgtP′s oligomerization potential being a primary determinant of UgtP mediated inhibition of FtsZ assembly, the addition of UDP-glc enhanced UgtP's ability to inhibit FtsZ assembly by ~20 % in vitro (Figure 5B). Specifically, while UgtP alone had a negligible effect on FtsZ assembly under these buffer conditions, the addition of UDP-glc stimulated UgtP mediated inhibition of FtsZ assembly, reducing the total light-scattering signal by ~20% at a 1:3 ratio of UgtP to FtsZ. This ratio is physiologically relevant as it is close to the experimentally determined 1:2 ratio of UgtP to FtsZ in vivo (Weart et al., 2007). Thus, although UDP-glc does not have a significant impact on UgtP oligomerization as measured by sucrose density gradient centrifugation (Figure 1C), its impact is more than sufficient to significantly enhance UgtP mediated inhibition of FtsZ assembly as indicated by 90° angle light-scattering assay.
A mutant version of UgtP defective in both the putative nucleotide- and hexose-binding sites, UgtP (F112A V117A E306A N309A), was a better inhibitor of FtsZ assembly than wild type UgtP, independent of UDP-glc. This finding is consistent with affinity data indicating that the mutant has a ~four-fold higher affinity for FtsZ than wild type UgtP (KD = 20 nM in the absence of UDP-glc versus KD = 72 nM in the presence of UDP-glc) as well as data indicating its affinity for FtsZ is essentially equivalent to its affinity for itself regardless of the presence of UDP-glc (KD = 20 nM versus KD = 21 nM in the presence of UDP-glc and KD = 16 nM for both interactions in the absence of UDP-glc).
UgtP inhibits FtsZ single filament formation
Although biochemical data presented in this work (Figure 5B) and in our previous study (Weart et al., 2007) indicate that UgtP interacts directly with FtsZ, it remains unclear how UgtP modulates FtsZ assembly at the molecular level. To clarify the molecular mechanism by which UgtP inhibits FtsZ assembly, we first tested the effects of UgtP on the GTPase activity of FtsZ. Like its homolog tubulin, GTP binding stimulates FtsZ assembly, which in turn leads to the formation of a GTPase active site between subunits, and ultimately hydrolysis and disassembly depending on the conditions (Graumann, 2007). Under our buffer conditions, FtsZ alone hydrolyzed GTP at an average rate of 9.5 ± 1.2 GTP μM−1 min−1. Although the addition of 3 μM UgtP slightly reduced FtsZ's GTP hydrolysis rate (8.3 ± 0.5 GTP μM−1 min−1), the reduction was within the margin of error. Moreover, in contrast to E. coli SulA which binds to and sequesters FtsZ monomers (Erickson et al., 2010, Dajkovic et al, 2008), the presence of UgtP did not increase the critical concentration of FtsZ mediated GTPase hydrolysis (Figure 5C).
In light of the negligible impact of UgtP on FtsZ mediated GTP hydrolysis, we next investigated the ability of UgtP to inhibit the formation of single stranded FtsZ polymers. Analysis of in vitro assembly reactions indicates that B. subtilis FtsZ has a high propensity to form stabilizing lateral interactions between single stranded polymers. Our GTPase data are consistent with a mechanism in which UgtP disrupts lateral interactions or one in which it inhibits FtsZ assembly by capping or severing single stranded polymers. To differentiate between these mechanisms, we took advantage of an FtsZ mutant (FtsZ NENDEG) that is unable to form significant numbers of lateral interactions in vitro (Buske & Levin, 2012).
In support of a mechanism in which UgtP inhibits FtsZ assembly by capping or severing single stranded polymers, UgtP reduced FtsZ NENDEG assembly by ~60% at a UgtP to FtsZ ratio of 1:2 in the presence of UDP-glc (Figure 5D). This finding is consistent with the short multimers of FtsZ we observed in the presence of UgtP in electron microscopy (Weart et al., 2007). This result is also in agreement with UgtP localization data that suggest UgtP can interact with FtsZ both at the cytokinetic ring and in the cytoplasm where FtsZ is believed to exist in small multimers and short polymers.
Small changes in the pool of FtsZ have a significant impact on B. subtilis cell size
Together our results support a model in which increases in intracellular UDP-glc levels shift the equilibrium between UgtP•UgtP and UgtP•FtsZ towards the latter complex, reducing the amount of FtsZ available for assembly into the cytokinetic ring, and ultimately increasing cell size. This model makes two predictions. First, because UDP-glc has only a modest impact on the interaction between UgtP and FtsZ in vitro (Table 2 and Figure 5B), we would also expect it to have a similarly modest impact on FtsZ assembly dynamics in vivo. In other words, given the relatively small impact of UDP-glc on the ability of UgtP to inhibit FtsZ assembly in vitro (~20%), small reductions in the intracellular concentration of FtsZ should have a significant impact on cell size under steady state conditions, to account for the relatively large impact of UDP-glc on B. subtilis cell size. [B. subtilis cells defective in UDP-glc biosynthesis are ~35% smaller than wild type during growth in nutrient-rich medium (Weart et al., 2007)]. While small reductions in intracellular FtsZ concentration have been shown to have significant impacts on E. coli cell size (Palacios et al., 1996), this has not been tested directly in B. subtilis. Second, because the model proposes that UgtP increases cell size by reducing the amount of FtsZ available for assembly into the cytokinetic ring, increasing the intracellular concentration of FtsZ should reduce the size of wild type cells but not ugtP null mutants during growth in nutrient-rich medium.
Consistent with the first prediction, small changes in intracellular FtsZ levels have a significant impact on B. subtilis cell size under steady state conditions. Taking advantage of a strain that expresses ftsZ under the control of a xylose-inducible promoter as the only copy of ftsZ (PL2084 ftsZ∷spc thrC∷Pxyl-ftsZ) (Weart & Levin, 2003), we identified conditions (0.1% xylose) at which FtsZ concentration was reduced by ~15% but growth remained normal (Figure S5). Under these conditions, the partially FtsZ-depleted cells were on average ~40% longer than wild type B. subtilis cells (Figure 6A). This finding is similar to that observed by Palacios et al., who determined that a similarly small reduction in total FtsZ levels, increases E. coli cell size by ~50% under steady state conditions (Palacios et al., 1996).
Figure 6. The pool of FtsZ available for assembly dictates cell size at division.
(A) ~15% reduction in intracellular FtsZ levels leads to a ~40% increase in average cell length (p < 0.001). Left: Wild type and PL2084 ftsZ∷spc thrC∷Pxyl-ftsZ cells plus 0.1% xylose to induce ftsZ expression to ~85% of wild type levels stained with membrane probe FM4-64. Bar = 3 μm. Right: Size distribution of 100 wild type cells and 100 PL2084 ftsZ∷spc thrC∷Pxyl-ftsZ cells plus 0.1% xylose.
(B) ~13% over-expression of ftsZ reduces the average length of wild type cells by ~15% but has no impact on ugtP null mutants (p < 0.001). Left: Size distribution of 100 PL950 amyE∷Pspachy-ftsZ cells cultured in the presence 20 μM IPTG (black bars) and no inducer control (gray bars). Right: Size distribution of 100 JC438 ugtP∷spc MLS amyE∷Pspachy-ftsZ cells cultured in the presence of 20 μM IPTG (black bars) and no inducer control (gray bars).
(C) Left: The equilibrium between UgtP•UgtP and UgtP•FtsZ influenced by UDP-glc functions as a molecular rheostat to coordinate cell size with nutrient availability. Under nutrient-poor conditions, low intracellular UDP-glc levels favor UgtP•UgtP interaction, increasing the amount of FtsZ available for assembly into the cytokinetic ring, allowing division to proceed unimpeded and reducing cell size. In contrast, under nutrient-rich conditions, high intracellular UDP-glc levels shift the balance from UgtP•UgtP towards UgtP•FtsZ, reducing the amount of FtsZ available for assembly into the cytokinetic ring, delaying division and increasing cell size. UgtP's nanomolar affinity for itself and for FtsZ means that free UgtP is a minor species at its physiological concentration of 1.45 μM, indicated by the dotted lines. Numbers indicate dissociation constants. Note that we have drawn UgtP binding to FtsZ as a monomer for simplicity. However, biochemical data presented in Figure 5 suggest UgtP most likely interacts with single stranded FtsZ polymers. Right: Graphic model for the growth rate and nutrient-dependent control of B. subtilis cell size. Cytoplasmic FtsZ concentration is constant regardless of growth rate and cell size (green line). Increases in intracellular UDP-glc levels coupled with growth-rate-dependent increases in UgtP levels, increase the amount of UgtP available to interact with FtsZ (orange line). Reductions in levels of assembly competent FtsZ (green dotted line), lead to transient delays in division and increases in average cell size (blue line).
Consistent with the second prediction, a modest increase in the intracellular concentration of FtsZ reduced the size of wild type cells but not ugtP null cells (Figure 6B). For this experiment, FtsZ levels were increased by ~13% through induction of an ectopic allele of ftsZ (amyE∷Pspachy-ftsZ) (Figure S6). In wild type cells a ~13% increase in total FtsZ levels led to a ~15% decrease in average cell length during growth in nutrient-rich medium. In contrast, the same increase in FtsZ concentration had no significant impact on the size of ugtP null cells, indicating the reduction in wild type cell size is dependent on the presence of ugtP.
DISCUSSION
Our findings support a model in which intracellular UgtP is predominantly oligomeric: bound either to itself or to FtsZ. Increases in the intracellular concentration of UDP-glc during growth in nutrient-rich medium shift the equilibrium from UgtP•UgtP towards UgtP•FtsZ, reducing the amount of FtsZ available for assembly into the cytokinetic ring, thus delaying division and ultimately increasing cell size (Figure 6C). This model is consistent with affinity data indicating that UgtP is likely to be in complex with either itself or FtsZ at its physiological concentration of ~1.45 μM (Tables 1 and 2), as well as our finding that UgtP oligomerization is sensitive to changes in the concentration of FtsZ both in vivo and in vitro (Figures 2B and 2C) (Weart et al., 2007).
This model rests on two assumptions: 1) that cell division and cell size are dependent on the accumulation of threshold levels of FtsZ, and 2) that the intracellular concentration of FtsZ is constant at all growth rates. With regard to the former, there is significant data supporting the idea that cell division is dependent on the accumulation of FtsZ to threshold levels (Lutkenhaus, 2007, Palacios et al., 1996, Teather et al., 1974). Limited reductions (<20%) in the intracellular concentration of FtsZ increase cell length under steady state conditions by as much as ~50% in both E. coli and B. subtilis (Palacios et al., 1996) (Figure 6A). With regard to the latter, the intracellular concentration of FtsZ is constant at all growth rates in both E. coli and B. subtilis (Weart & Levin, 2003). Thus, nutrient-dependent changes in the equilibrium between UgtP•UgtP and UgtP•FtsZ should have a relatively rapid and predictable impact on the amount of FtsZ available for assembly into the cytokinetic ring.
A dynamic mechanism controlling cell size homeostasis
Consistent with the longstanding observation that cells shifted from a poor nutrient source to a rich one immediately increase their growth rate, but delay division until they have achieved the appropriate size for the given condition (Cooper, 1969), we find that shifting B. subtilis cells from nutrient-poor to nutrient-rich conditions leads to the protein synthesis independent re-localization of UgtP away from the randomly positioned foci to the cytoplasm and cytokinetic ring (Figure 3C). UgtP's high affinity for FtsZ (72 nM in the presence of UDP-glc, approximately equivalent to its affinity for itself under the same conditions) means that increases in UDP-glc availability should shift the equilibrium away from UgtP•UgtP towards UgtP•FtsZ, effectively reducing the pool of FtsZ available for assembly into the cytokinetic ring, and forcing cells to delay division until they have accumulated sufficient additional FtsZ to support cytokinesis at the new, larger size.
Notably, our data do not differentiate between a mechanism in which UgtP inhibits division by delaying the onset of FtsZ ring formation or by delaying the constriction of an already extant ring, or both. The FtsZ ring is present for ~85% of the division cycle in rapidly growing B. subtilis (mass doubling time ~26 minutes) and ~50% of the same cells cultured at a slower growth rate (mass doubling time ~80 minutes) (Weart & Levin, 2003, Weart et al., 2007). Determining the length of the so-called Z-period in single cells following a shift from nutrient-poor to nutrient-rich medium should clarify this issue.
A conformational change coupled to a mutually exclusive interaction?
Precisely how interactions with UDP-glc influence UgtP oligomerization is not clear. Based on data from UgtP homologs whose conformation has been shown to change on substrate binding (Guerin et al., 2009, Vetting et al., 2008), we favor an allosteric mechanism in which binding to UDP-glc at one binding site alters UgtP conformation, reducing its affinity for itself and increasing the pool of free UgtP available to interact with FtsZ. This type of mechanism fits the allosteric model of signal transduction (Changeux & Edelstein, 2005), in which binding to one ligand alters the affinity of an enzyme for another ligand whose binding site is structurally distinct from the first. Allosteric regulation also plays a key role in glycogen synthesis, a process that requires the same glc-1-P precursor as UDP-glc biosynthesis (Wilson et al., 2010).
In contrast to the somewhat modest impact of interactions with UDP-glc on UgtP oligomerization, our data suggest that UgtP•UgtP and UgtP•FtsZ interactions may be mutually exclusive. FtsZ inhibits UgtP oligomerization in vitro (Weart et al., 2007), and increasing FtsZ levels ~three-fold under nutrient-poor conditions or in cells encoding the putative UDP-glc binding mutant alleles of ugtP, is sufficient to drive UgtP re-localization from punctate foci to the cytokinetic ring (Figures 2C and 5A). Supporting the idea that the same surface of UgtP is required for oligomerization and for interaction with FtsZ, UgtP's affinity for FtsZ is also reduced in the presence of UDP-glc (KD = 72 nM versus 38 nM in the absence of UDP-glc). Alternatively, binding to FtsZ may lead to an allosteric conformational change in UgtP's surface structure that effectively inhibits oligomerization.
A potential role for cellular factors other than UDP-glc in modulating UgtP's interaction with itself and FtsZ
Although our affinity data is consistent with the results of our FtsZ assembly assays (Figure 5B), it is not completely consistent with our in vivo findings. In particular, it is unclear why UgtP is overwhelmingly punctate in vivo in the absence of UDP-glc (Weart et al., 2007), although it still retains a high affinity for FtsZ (Table 2). The ratio of UgtP to FtsZ is ~1:2 during growth in nutrient-rich medium (Weart et al., 2007). Similarly, although we determined that a UgtP mutant defective in both the putative nucleotide- and hexose-binding sites, is a more potent inhibitor of FtsZ assembly in vitro than wild type UgtP, consistent with our affinity data (Figure 5B, Tables 2), a single mutation in UgtP's putative nucleotide or hexose-binding site renders the protein nutrient-blind with regard to both sub-cellular localization and division inhibition (Figure 4B).
There are at least two potential explanations for the apparent discrepancy between our in vitro and in vivo data. First, other cellular factors in addition to UDP-glc almost certainly influence the intricate equilibrium between UgtP•UgtP and UgtP•FtsZ in vivo. Such factors likely include FtsZ binding proteins that compete with UgtP for access to FtsZ as well as proteins and small molecules that interact with UgtP, such as its donor substrate diacylglycerol. In addition, given the facility with which UgtP forms higher order structures in vitro (Weart et al., 2007), it is also possible that cooperative interactions between UgtP monomers favor UgtP oligomerization over interaction with FtsZ in the absence of UDP-glc. Although we did not identify any evidence of cooperativity in our affinity studies, it is possible that cooperative interactions are only apparent at UgtP concentrations below our limit of detection. Future investigations of the interaction between UgtP and FtsZ, both in vitro and in vivo, should clarify this issue.
A new twist on an old model
Although this work represents a significant step forward in our understanding of nutrient-dependent cell size control in bacteria, the model itself has its roots in early work on the eukaryotic cell cycle. In a theoretical paper published in 1975, Fantes et al. suggested four models for the control of cell division (mitosis) and cell size in eukaryotes (Fantes et al., 1975). In the so-called concentration model, an effector protein was proposed to bind to a site within the nucleus. The concentration of this effector at the site increased over the course of the cell cycle, triggering mitosis when it reached critical levels. In subsequent work investigating the impact of changes in nutrient availability on fission yeast cell size, Fantes and Nurse proposed the presence of a signaling molecule that altered the interaction between the effector and receptor by changing the binding kinetics in response to changes in nutrients (Cooper, 1969, Fantes & Nurse, 1977, Schaechter et al., 1958). Although the organisms are different, the models are analogous. In B. subtilis, UDP-glc and UgtP together serve as the signal, FtsZ as the effector, and the cytokinetic ring as the “receptor”.
In concept, this model is also analogous to the mechanism proposed by Marshall and Rosenbaum to govern flagellar length in the photosynthetic protist Chlamydomonas, in which changes in the steady state balance between assembly and disassembly are the primary determinant of flagellar length (Marshall, 2004). In the case of Chlamydomonas, the rate of intraflagellar transport is inherently length-dependent (the longer the flagella the further subunits have to travel), thus the balance between assembly and disassembly is reached only when a specific flagellar length is achieved. Because disassembly is constant regardless of length, changes in subunit availability lead to changes in average flagellar length. Like Chlamydomonas flagella, the FtsZ ring is a dynamic structure with subunits turning over on a regular basis (Anderson et al., 2004). As discussed earlier, while the concentration of FtsZ is growth rate independent, the total amount of FtsZ must reach threshold levels before division can proceed. UDP-glc dependent reductions in the availability of assembly competent FtsZ, thus alter the balance between assembly and disassembly of the FtsZ ring, forcing cells cultured under nutrient-rich conditions to increase size until they have accumulated sufficient FtsZ to support division.
Importantly, our model does not rule out the possibility that something other than UDP-glc also plays a role in UgtP mediated nutrient-dependent cell size control of B. subtilis. Even though our data suggest that UDP-glc influences UgtP's ability as a nutrient-dependent inhibitor of FtsZ assembly and cell division, in vitro the limited impact of UDP-glc on UgtP oligomerization (Figure 1C) and UgtP's inhibition of FtsZ assembly (Figure 5C) raises the possibility that UgtP activity is influenced by something other than UDP-glc. One potential candidate is diacylglycerol (DAG), the acceptor substrate of UgtP's glucosyltransferase activity. It remains to be determined if DAG modulates UgtP localization and UgtP mediated division inhibition together with UDP-glc.
In summary, our data indicate that cell size is coupled to nutrient availability in B. subtilis via a homeostatic mechanism in which UDP-glc dependent changes in UgtP's oligomerization potential alter the steady state balance between FtsZ assembly and disassembly and with it cell size. Significantly, our findings provide a model to explain how cells are able to precisely coordinate size with environmental conditions without the need to invoke a cell size dependent checkpoint or molecular ruler. We are confident this work will serve as a platform to investigate the factors governing cell size in other bacterial systems as well as in eukaryotic model organisms where cell size control remains a largely open question.
EXPERIMENTAL PROCEDURES
General Methods and Bacterial Strains
B. subtilis strains used in this study are described in Table S1. Standard techniques were used for cloning and genetic manipulations. Unless noted, cells were cultured in Luria-Bertani (LB) or S750 minimal media (Jaacks et al., 1989) with appropriate supplements at 30°C or 37°C.
Fluorescence Microscopy
Microscopy was performed using an Olympus BX51 microscope equipped with an OrcaERG camera. Openlab version 4.0 (Improvision) was used for image capture and analysis. Images were processed with Adobe Photoshop CS and Adobe Photoshop Lightroom 3 (Adobe Systems). Live cells were prepared for imaging as described previously (Weart et al., 2007) and stained for membrane using FM4-64 (Invitrogen) or TMA-DPH (Sigma-Aldrich) where necessary. Cell length was calculated as the distance between adjacent septa. Significance of length distributions was established using t-test analysis. For YFP-UgtP localization, cells encoding a xyloseinducible YFP-UgtP fusion were grown at 30°C in the presence of 0.5% xylose for ~3 hours prior to visualization.
Protein Purification
Native B. subtilis FtsZ and Thio-UgtP were expressed and purified as described (Haeusser et al., 2004) (Weart et al., 2007).
Sucrose Density Gradient Centrifugation and Gel Filtration
For sucrose density gradient centrifugation, purified Thio-UgtP and markers were loaded onto 4 ml 20%–50% sucrose gradients in UgtP buffer (100 mM NaCl, 10% glycerol, 20 mM Tris pH 8.0) and centrifuged at 200000 g for 18 hours at 4°C using the 50.1 SW rotor and Optima L-80 XP Ultracentrifuge (Beckman). Ten fractions were collected form the top to the bottom of the gradients and examined by SDS-PAGE. For gel filtration, purified Thio-UgtP and markers were applied to an S300 gel filtration column (GE Healthcare). 2 ml fractions were collected and examined by SDS-PAGE. Sedimentation coefficient, Stokes radius and apparent molecular weight of Thio-UgtP were determined as previously described (Richter, 2005).
Single Molecule Studies
A 5 μl droplet of B. subtilis cells suspended in LB was deposited between a manufacturer-cleaned fused silica chip (Hoya) and a glass cover slip. The experimental setup was described previously (DeSantis et al., 2010). In detail, the single molecule experiments were performed using prism-type total internal reflection fluorescence (TIRF) microscopy. The single molecule images were taken by a Nikon Eclipse TE2000-S inverted microscope using a 100 × objective (Nikon). By using a 2 × expansion lens before the EMCCD camera, the resulting pixel size is 79 nm. The samples were excited by a 488 nm laser line (I70C-SPECTRUM Argon/Krypton Laser, Coherent Inc.), which was filtered from the multiline laser emission using a polychromatic acousto-optic filters (48062 PCAOM model, NEOS Technologies). The laser was linearly polarized, and was focused onto a 40 × 20 μm2 region, resulting in a laser excitation intensity of 0.1 kW/cm2. The emitted photons from the samples were collected by an iXon back illuminated electron multiplying charge coupled device camera (DV897ECS-BV, Andor Technology). The images were analyzed using ImageJ. The number of YFP-UgtP molecules per focus was determined using the IPMC method (Wang et al., 2005), in which the ratio between the mean lifetime photon count of YFP-UgtP foci and the mean lifetime photon count of individual YFP molecules is the mean number of YFP-UgtP molecules in a focus. The lifetime photon counts of YFP molecules were obtained by in vitro imaging of YFP adsorbed on fused-silica surfaces and counting their total emission photons before bleaching. The distribution of YFP lifetime photon counts was then fitted to a Gaussian function; the mean of this distribution was the mean YFP lifetime photon count in the study.
Lipid Extraction and Thin Layer Chromatography
Briefly, 1 L of medium was inoculated with 10 ml of an overnight culture and allowed to grow at 37°C until mid-exponential phase. Cultures were chilled on ice and cells were collected by centrifugation. Cells were washed with 0.1 M sodium citrate pH 4.7 and lysed by French press. Lysed cells were centrifuged at 12000 g for 20 minutes at 4°C. Lipids were extracted from cell pellets using a modified Bligh-Dryer method described previously (Grundling & Schneewind, 2007). Prior to separation of extracted lipids, TLC plates were dried for 2 hours at 100°C. Extracted lipids were separated by TLC using Whatman Silica Gel A60 plates in a solvent system containing chloroform-methanol-H2O (70:30:4). TLC plates were then stained with iodine. Diglucosyl-diacylglycerol from B. cereus and plants was used as a chromatographic standard where necessary.
Octet Assay
The assay was performed on the Octet RED96 system (FortéBio) at 30°C. Streptavidin-coated biosensor tips were washed with PBS buffer and equilibrated with UgtP buffer (100 mM NaCl, 10% glycerol, 20 mM Tris pH 8.0) to establish a baseline prior to Thio-UgtP immobilization. 200 μl UgtP buffer containing biotinylated Thio-UgtP was dispensed into 96-well microtiter plates (Millipore) and then coated onto the biosensor tips while agitating for 10 minutes at 1000 rpm. Association between Thio-UgtP at the biosensor tips and various concentrations of Thio-UgtP or FtsZ in the wells was monitored while agitating for 10 minutes at 1000 rpm. Dissociation was monitored for 20 minutes at 1000 rpm when the biosensor tips were dipped into wells containing UgtP buffer alone. Data were generated automatically by the Octet User Software version 3.1 (FortéBio) and were subsequently analyzed by global fitting using Octet Data Analysis Software (FortéBio).
90° Angle Light-Scattering Assay
Light-scattering assays were conducted essentially as described (Weart et al., 2005) using a DM-45 spectrofluorimeter (Olis). Readings were taken four times per second at 30°C, and a baseline was gathered for 1 minute before the addition of 1 mM GTP to the cuvette. Baseline corrections were applied where necessary. Assembly buffer contains 50 mM MES pH 6.5, 5 mM MgCl2, 1 mM EGTA and 75 mM NaCl for Figure 5B and 50 mM MES pH 6.5, 5 mM MgCl2, 1 mM EGTA and 25 mM NaCl for Figure 5D.
GTPase Assay
GTPase activity was measured as previously described (Buske & Levin, 2012, Ingerman & Nunnari, 2005) using a SPECTRAmax Plus spectrophotometer (Molecular Devices). Reaction buffer contains 1 mM GTP, 1 mM phosphoenolpyruvate, 250 μM NADH, 80 units ml−1 lactose dehydrogenase, 80 units ml−1 pyruvate kinase and various concentrations of FtsZ and UgtP where necessary. Absorbance at 340 nm for NADH was recorded at 30°C for 3 minutes in a quartz cuvette (1 cm pathlength). The data set was exported and analyzed in Microsoft Excel.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Ernst Heinz for the gift of lipid markers. Bradley Ford provided assistance with the Octet assay, Cindy Richard-Fogal offered her expertise on protein purification, and Aaron Collins helped with sucrose density gradient centrifugation and thin layer chromatography. We are also indebted to Harold Erickson, Heidi Arjes, P.J. Buske, Norbert Hill, Suckjoon Jun and Sattar Taheri for their insightful comments on the manuscript. We are thankful to all members of the Levin lab for help, support and discussions. AC was a Lee Foundation Fellow of the McDonnell International Scholars Academy. This work was supported by NIH Public Health Service grant GM64671 to P.A.L.
REFERENCES
- Anderson DE, Gueiros-Filho FJ, Erickson HP. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol. 2004;186:5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botte C, Jeanneau C, Snajdrova L, Bastien O, Imberty A, Breton C, Marechal E. Molecular modeling and site-directed mutagenesis of plant chloroplast monogalactosyldiacylglycerol synthase reveal critical residues for activity. J Biol Chem. 2005;280:34691–34701. doi: 10.1074/jbc.M505622200. [DOI] [PubMed] [Google Scholar]
- Buske PJ, Levin PA. The extreme C-terminus of the bacterial cytoskeletal protein FtsZ plays a fundamental role in assembly independent of modulatory proteins. J Biol Chem. 2012 doi: 10.1074/jbc.M111.330324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- Cooper S. Cell division and DNA replication following a shift to a richer medium. J Mol Biol. 1969;43:1–11. doi: 10.1016/0022-2836(69)90074-6. [DOI] [PubMed] [Google Scholar]
- Dajkovic A, Mukherjee A, Lutkenhaus J. Investigation of regulation of FtsZ assembly by SulA and development of a model for FtsZ polymerization. J Bacteriol. 2008;190:2513–2526. doi: 10.1128/JB.01612-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis MC, DeCenzo SH, Li JL, Wang YM. Precision analysis for standard deviation measurements of immobile single fluorescent molecule images. Opt Express. 2010;18:6563–6576. doi: 10.1364/OE.18.006563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donachie WD, Begg KJ. Cell length, nucleoid separation, and cell division of rod-shaped and spherical cells of Escherichia coli. J Bacteriol. 1989;171:4633–4639. doi: 10.1128/jb.171.9.4633-4639.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes P, Nurse P. Control of cell size at division in fission yeast by a growth-modulated size control over nuclear division. Exp Cell Res. 1977;107:377–386. doi: 10.1016/0014-4827(77)90359-7. [DOI] [PubMed] [Google Scholar]
- Fantes PA, Grant WD, Pritchard RH, Sudbery PE, Wheals AE. The regulation of cell size and the control of mitosis. J Theor Biol. 1975;50:213–244. doi: 10.1016/0022-5193(75)90034-x. [DOI] [PubMed] [Google Scholar]
- Graumann PL. Cytoskeletal elements in bacteria. Annu Rev Microbiol. 2007;61:589–618. doi: 10.1146/annurev.micro.61.080706.093236. [DOI] [PubMed] [Google Scholar]
- Grundling A, Schneewind O. Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus. J Bacteriol. 2007;189:2521–2530. doi: 10.1128/JB.01683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin ME, Schaeffer F, Chaffotte A, Gest P, Giganti D, Kordulakova J, van der Woerd M, Jackson M, Alzari PM. Substrate-induced conformational changes in the essential peripheral membrane-associated mannosyltransferase PimA from mycobacteria: implications for catalysis. J Biol Chem. 2009;284:21613–21625. doi: 10.1074/jbc.M109.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusser DP, Schwartz RL, Smith AM, Oates ME, Levin PA. EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol Microbiol. 2004;52:801–814. doi: 10.1111/j.1365-2958.2004.04016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldal M, Norland S, Tumyr O. X-ray microanalytic method for measurement of dry matter and elemental content of individual bacteria. Appl Environ Microbiol. 1985;50:1251–1257. doi: 10.1128/aem.50.5.1251-1257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerman E, Nunnari J. A continuous, regenerative coupled GTPase assay for dynamin-related proteins. Methods Enzymol. 2005;404:611–619. doi: 10.1016/S0076-6879(05)04053-X. [DOI] [PubMed] [Google Scholar]
- Jaacks KJ, Healy J, Losick R, Grossman AD. Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis. J Bacteriol. 1989;171:4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loferer-Krossbacher M, Klima J, Psenner R. Determination of bacterial cell dry mass by transmission electron microscopy and densitometric image analysis. Appl Environ Microbiol. 1998;64:688–694. doi: 10.1128/aem.64.2.688-694.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- Marshall WF. Cellular length control systems. Annu Rev Cell Dev Biol. 2004;20:677–693. doi: 10.1146/annurev.cellbio.20.012103.094437. [DOI] [PubMed] [Google Scholar]
- Mijakovic I, Poncet S, Boel G, Maze A, Gillet S, Jamet E, Decottignies P, Grangeasse C, Doublet P, Le Marechal P, Deutscher J. Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J. 2003;22:4709–4718. doi: 10.1093/emboj/cdg458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibori A, Kusaka J, Hara H, Umeda M, Matsumoto K. Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes. J Bacteriol. 2005;187:2163–2174. doi: 10.1128/JB.187.6.2163-2174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios P, Vicente M, Sanchez M. Dependency of Escherichia coli cell-division size, and independency of nucleoid segregation on the mode and level of ftsZ expression. Mol Microbiol. 1996;20:1093–1098. doi: 10.1111/j.1365-2958.1996.tb02549.x. [DOI] [PubMed] [Google Scholar]
- Richter W. Determining the subunit structure of phosphodiesterases using gel filtration and sucrose density gradient centrifugation. Methods Mol Biol. 2005;307:167–180. doi: 10.1385/1-59259-839-0:167. [DOI] [PubMed] [Google Scholar]
- Sargent MG. Control of cell length in Bacillus subtilis. J Bacteriol. 1975;123:7–19. doi: 10.1128/jb.123.1.7-19.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaechter M, Maaloe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- Teather RM, Collins JF, Donachie WD. Quantal behavior of a diffusible factor which initiates septum formation at potential division sites in Escherichia coli. J Bacteriol. 1974;118:407–413. doi: 10.1128/jb.118.2.407-413.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetting MW, Frantom PA, Blanchard JS. Structural and enzymatic analysis of MshA from Corynebacterium glutamicum: substrate-assisted catalysis. J Biol Chem. 2008;283:15834–15844. doi: 10.1074/jbc.M801017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YM, Tegenfeldt JO, Reisner W, Riehn R, Guan XJ, Guo L, Golding I, Cox EC, Sturm J, Austin RH. Single-molecule studies of repressor-DNA interactions show long-range interactions. Proc Natl Acad Sci U S A. 2005;102:9796–9801. doi: 10.1073/pnas.0502917102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wass MN, Kelley LA, Sternberg MJ. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010;38:W469–473. doi: 10.1093/nar/gkq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weart RB, Lee AH, Chien AC, Haeusser DP, Hill NS, Levin PA. A metabolic sensor governing cell size in bacteria. Cell. 2007;130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weart RB, Levin PA. Growth rate-dependent regulation of medial FtsZ ring formation. J Bacteriol. 2003;185:2826–2834. doi: 10.1128/JB.185.9.2826-2834.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weart RB, Nakano S, Lane BE, Zuber P, Levin PA. The ClpX chaperone modulates assembly of the tubulin-like protein FtsZ. Mol Microbiol. 2005;57:238–249. doi: 10.1111/j.1365-2958.2005.04673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WA, Roach PJ, Montero M, Baroja-Fernandez E, Munoz FJ, Eydallin G, Viale AM, Pozueta-Romero J. Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol Rev. 2010;34:952–985. doi: 10.1111/j.1574-6976.2010.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.