Abstract
Objective
Ribonucleotide reductase (RNR) supplies deoxyribonucleotide diphosphates demanded by cells to repair radiation-induced DNA damage. Here, we investigate the impact of pretherapy RNR M1, M2, and M2b (p53R3) subunit level upon human cervical cancer radiochemosensitivity.
Methods/Materials
Immunohistochemistry was performed on a tissue array comprised of 18 paired benign uterine cervix and stage 1B2 cervical cancers to evaluate the relationship between cytosolic RNR M1, M2, and M2b staining intensity and radiochemotherapy cancer response. Patients underwent surgical hysterectomy (n = 8), or daily radiation (45 Gy), co-administered once weekly cisplatin (40 mg/m2), then low-dose-rate brachytherapy (30 Gy), followed by adjuvant hysterectomy (n = 10). Radiochemotherapy response was determined by RECIST v1.0 criteria at the time of brachytherapy. Cancer relapse rates and disease-free survival were calculated.
Results
M1, M2, and M2b antibody staining intensity was low (0–1+) in benign uterine cervix tissue. M1 and M2b immunoreactivity was 2+ or 3+ in the majority of cervical cancers (13 of 18). M2 immunoreactivity was 3+ in nearly all cervical cancers (16 of 18). Cervical cancers overexpressing M1 and M2b had an increased hazard for incomplete radiochemotherapy response, relapse, and shortened disease-free survival.
Conclusions
RNR subunit levels may predict human cervical cancer radiochemosensitivity and subsequent posttherapy cancer outcome. Further validation testing of RNR subunits as biomarkers for radiochemotherapy response is warranted.
Keywords: ribonucleotide reductase, radiosensitivity, cervical cancer
Introduction
Cancers of the uterine cervix pose a serious health concern worldwide.1 Human papillomaviruses (HPVs) are DNA viruses connected with most invasive cervical cancers, perhaps due to viral hijacking of cell machinery supplying DNA precursors in the form of deoxyribonucleotide diphosphates (dNDPs).1 Coordinated expression of HPV-E6 and HPV-E7 removes restrictions at the G1/S checkpoint and activates ribonucleotide reductase (RNR), increasing the likelihood for developing an oncogenic phenotype.2 Understanding RNR and the ways by which its enzymatic activity can be manipulated 3–7 has taught oncologists why anticancer strategies may or may not be successful in cervical cancer therapy.1
RNR acts in cells as a α6β2 heterotetramer that reduces a 2′-hydroxyl to hydrogen in ribose to generate a 2′-dNDP subsequently used to make the triphosphate form that is incorporated in DNA.8, 9 The large 90 kilodalton (kD) RNR α (M1) subunit contains a catalytic site for ribonucleoside substrates, a specificity site, and an activity site that by allosteric regulation controls specific dNDP output by RNR. The small RNR subunit β (M2 [45 kD] or M2b [p53R2, 39 kD]) houses a tyrosyl free radicals that shuttles to and from the M1 catalytic site.10 The M2 subunit distinguishes itself from M2b by possessing an S-phase specific KEN box that normally directs its timely degradation prior to cell mitosis.11, 12 With G1/S checkpoint restrictions lifted and deregulated RNR dNDP production apparent, cervical cancer cells may have an increased capacity to repair damaged DNA and survive.3–6 This led us to hypothesize that cervical cancers having relatively high RNR subunit levels may not respond to radiochemotherapy.
To further elucidate the role of RNR M1, M2, and M2b in the radiochemotherapy DNA damage response, we interrogated the immunohistochemical expression of RNR proteins and subsequent cancer-related outcomes in women diagnosed with clinical stage IB2 cervical cancers. Our results provide evidence that cancers of the uterine cervix uniformly overexpress RNR M2, which is associated with a poor radiochemotherapy response to treatment. When RNR M1 and M2b are simultaneously overexpressed, cancer relapse rates are high and disease-free intervals are short.
Materials and Methods
Patients and Treatments
Between July 2005, and October 2009, 42 women diagnosed with clinical stage IB2 invasive squamous, adenosquamous, or adenocarcinomas of the uterine cervix were evaluated for suitable archived formalin-fixed paraffin embedded (FFPE) tissue for tissue microarray (TMA) construction (Fig. 1). Women having clinical stage IB2 were selected because treated patients had the highest probability of long-term survival and their hysterectomy specimens were likely to have sufficient tissue for analysis. Paired core punches of benign uterine cervix and of cervical cancer were identified in 19 women. Case Western Reserve University Institutional Review Board (IRB) approval was given for TMA construction.
Figure 1.
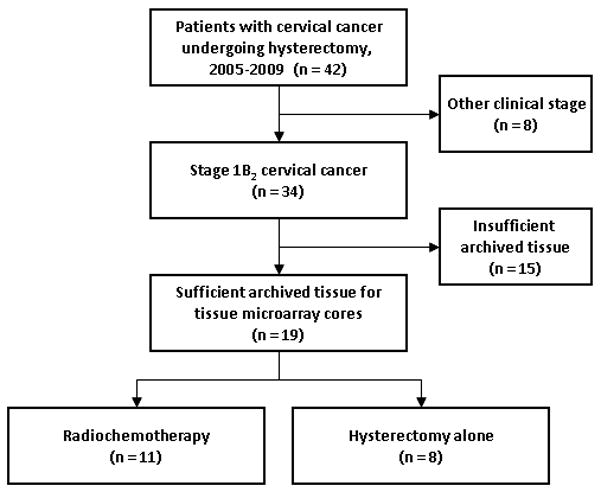
STROBE diagram for progress through stages of analysis. STROBE: Strengthening the Reporting of Observational Studies in Epidemiology.
Demographic, treatment, and tumor characteristics are listed in Table 1. Eight women underwent either an extrafascial (n = 1) or modified radical (n = 7) hysterectomy followed by no adjuvant therapy. Eleven women had pelvic radiation (45 Gy) and co-administered once weekly cisplatin (40 mg/m2) followed by low-dose-rate brachytherapy that preceded an extrafascial (n = 2) or modified radical (n = 9) hysterectomy. Pelvic radiation involved parallel-opposed anteroposterior and lateral external beam portals that administered 25 fractions of 1.8 Gy daily radiation. A parallel opposed anteroposterior parametrial boost with central 4 cm block was used to supplement pelvic radiation dose in eight cases (median 5.4 Gy, range 3.6 Gy – 9 Gy). Intracavitary low-dose-rate brachytherapy consisted of a single tandem and ovoid cesium-137 implant prescribed to point A (median 32 Gy, range 26 Gy – 45 Gy). The median total prescription dose at point A was 77 Gy (range 71 – 90 Gy).
Table 1.
Patient characteristics (n = 19)
| Characteristic | Number patients (%)* | |
|---|---|---|
| Age (years) | ||
| 30–39 | 4 (21) | |
| 40–49 | 10 (53) | |
| 50–59 | 4 (21) | |
| 60–69 | 1 (5) | |
| Stage | ||
| Clinical 1B2 | 19 (100) | |
| Tumor Grade | ||
| Grade 1 | 2 (11) | |
| Grade 2 | 5 (26) | |
| Grade 3 | 12 (63) | |
| Tumor size (mm) | ||
| 40–59 | 9 (47) | |
| 60–79 | 9 (47) | |
| 80 or greater | 1 (5) | |
| Lymphovascular space invasion | ||
| Absent | 8 (42) | |
| Present | 11 (58) | |
| Cervical cancer sub-type | ||
| Squamous cell carcinoma | 14 (74) | |
| Adenocarcinoma | 5 (26) | |
| Adenosquamous carcinoma | 0 (0) | |
| Therapy | ||
| Weekly cisplatin chemotherapy | 10 (53) | |
| Pelvic radiation | 1 (1) | |
| Parametrial boost delivered | 8 (42) | |
| Brachytherapy delivered | 7 (37) | |
| Hysterectomy | 19 (100) | |
Percentages may not add to 100% due to rounding.
Benign and Cervical Cancer Tissue Microarray
From each patient’s FFPE specimen, 1.5 mm circular core punches of their benign cervix and of their cervical cancer were retrieved. Core punches were positioned in a labeled recipient matrix TMA following manufacturer instructions by a semi-automated TMArrayer (Pathology Devices, Inc., Westminster, MD). Single 1.5 mm core punches from human lung, placenta, stomach, and tonsil with lymphocyte proliferation were added to the TMA from de-identified donor tissues banked by the Human Tissue Procurement Core Facility with IRB approval. True negative human tissues for RNR are not available for immunohistochemical negative controls, as a RNR-null phenotype induces renal failure, mitochondrial DNA depletion, and early mortality.13 A lung quality control core was added to the TMA to serve as a low positive staining control for RNR subunits in pneumocytes and moderate positive staining control in lung macrophages.14 Normal stomach and proliferating tonsil cores identified high positive staining for RNR M1, M2, and M2b.15–17 A human placenta core was used as a RNR M1 positive staining control. Five micrometer TMA sections were cut either for hematoxolin and eosin quality control or for RNR subunit immunoreactivity (Fig. 2).
Figure 2.
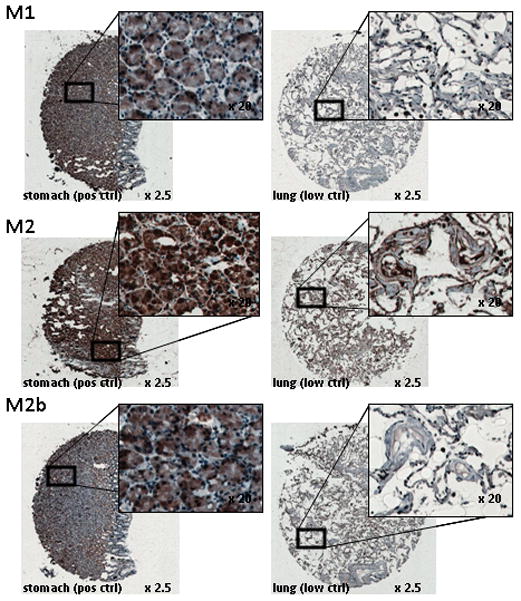
Examples of RNR M1, M2, and M2b antibody staining. IHC staining on FFPE stomach crypt cells for RNR M1, M2, and M2b is intense (3+) in the cytoplasm and nucleus. In contrast, lung pneumocytes show very low (0) stain intensity for RNR M1 and M2b and low (1=) stain intensity for RNR M2.
Immunohistochemistry
TMA slides were baked at 60°C for 75 minutes, deparaffinized in two 7 minute exchanges of xylene, rehydrated in two 2 minute 100% ethanol and 95% ethanol exchanges, rehydrated in a single 70% ethanol exchange, and then finally rinsed in distilled water for 2 minutes. TMA slides were incubated for 20 minutes at 98°C in a 250mL citric acid-based (pH 6.0) antigen unmasking solution (Vector Laboratories, Burlingame, CA). After a 2 minute distilled water wash, slides were immersed for 8 minutes in Peroxidazed1 (BioCare Medical, Concord, CA) and then immersed for 20 minutes in Background Sniper solution (BioCare Medical). Then, a 2 minute wash in 1X Tris-buffered saline (pH 7.4) and 0.1% Tween-20 (TBST) was done. TMA slides were incubated for 60 minutes with primary antibodies and washed in TBST for 2 minutes. TMA slides were incubated for 20 minutes with MACH4 Probe (BioCare Medical), rinsed in TBST for 2 minutes, incubated for 20 minutes with universal horseradish peroxidase-polymer (HRP, BioCare Medical) for 20 minutes, and then rinsed in TBST for 2 minutes. 3, 3′ Diaminobenzidine (DAB, BioCare Medical) applied for 5 minutes in the dark was used as a chromogen. Slides were rinsed with distilled water and hematoxylin counterstaining was applied for 1 minute. Last, alternating washes of TBST and tap water were done, followed by xylene dehydration, and application of coverslips with resin.
Antibodies and dyes
Cell nuclei were stained blue by hematoxylin (BioCare Medical) For primary antibody dilutions, RNR M1 (rabbit polyclonal [ab81085], Abcam, Cambridge, MA) and RNR M2 (mouse monoclonal [ab57653], Abcam) were both used at 1μg (1:200) in 199μL volume of 0.25% casein in phosphate-buffered saline (PBS, Dako North America, Inc., Carpinteria, CA). RNR M2b (rabbit polyclonal [ab8105], Novus Biologicals, Littleton, CO) was used at 1μg (1:250) in 249μL volume of 0.25% casein in PBS. These commercial antibodies were selected for their high relative specificity in recognizing RNR M1, RNR M2, and RNR M2b proteins.18, 19 A biotin-free probe (MACH 4) followed by a HRP (peroxidase) that binds to the MACH 4 probe enabled antibody detection. DAB was used at 32μL chromogen to 1mL DAB substrate buffer concentration to produce a brown precipitate.
To assess quality control of the RNR antibodies from disparate antibody lots, two immunohistochemistry stains of the TMA were done separated in time by nine months. The first lots assessed were: M1 #894101; M2 #3586-1; and M2b #911951. The second lots assessed were: M1 #GR2593-2; M2 #GR30312-1; and M2b #GR1852-4.
Microscopy
Individual cores of benign and cervical cancer tissues were viewed on an inverted microscope (Olympus, Center Valley, PA) at 2.5X and 20X magnifications. Three independent pathologists, blinded to treatment and outcome, scored the brown color staining intensity of M1, M2, and M2b on a 0 (< 5% of cells), 1 (5% to < 25%), 2 (25% to < 75%), and 3 (≥75%) scale.20 Discrepancies in any individual scored sample were reviewed and resolved to a single score. Digital images of TMA slides were acquired using a D-Metrix DX-40 microscope slide scanner (DMetrix, Inc, Tucson, AZ) at the indicated magnifications of tissue cores. Image processing involved digitalEyepiece software version 1.4.1 (DMetrix, Inc.). One cervical cancer core did not meet quality control standards and was not used for analysis.
Assessment of response and statistics
To measure agreement among inter-raters on qualitative RNR subunit immunohistochemistry, a Kappa statistic was determined (SPSS 18.0; SPSS, Inc., Chicago, IL). Our primary goal was to determine immunohistochemical expressions of RNR proteins and the impact they had on relapse as an outcome. Fisher’s exact test was used to compute P values for such associations. In constructing the TMA, we made an a priori decision to include any clinical stage IB2 cervical cancer patient with suitable hysterectomy material for evaluation of benign uterine cervix and cervical cancer tissue. Although this limits sample size, it permits pairwise analyses of benign versus cancerous tissue and allows exploratory analyses of RNR immunohistochemistry and its impact upon cancer outcome after hysterectomy alone versus radiochemotherapy and hysterectomy.
In this study, we elected to associate RNR subunit expression and cervical cancer radiochemotherapy response at two important clinical time points—at brachytherapy and at adjuvant hysterectomy. For this analysis, responses to radiochemotherapy were recorded following RECIST (version 1.0) criteria. As a second independent measure of response, we calculated a standard uptake value ratio of pretherapy to post-radiation, pre-surgical 18F-deoxyglucose positron emission tomography (18F-FDG PET/CT) to determine metabolic response, as we have done before.21 A ratio of less than 0.33 indicates a complete metabolic response.21
In this study, disease-free survival (DFS) was defined in terms of the time to relapse or any cause of death as measured from the first date of hysterectomy or radiation. Product-limit estimates with 95% confidence intervals (CI) for DFS were computed using the method of Kaplan and Meier. The log-rank test was used to compare pairs of such curves (α = 0.05). A Cox proportional hazards model was used to adjust for prognostic factors and to estimate relative progression-free survival. Factors included in the hazards model included age, pretherapy cervical cancer tumor size and histological grade (1, 2, or 3), lymphovascular space invasion (i.e., present versus absent), treatment (i.e., hysterectomy versus radiochemotherapy followed by hysterectomy), and separate variables of RNR subunit staining intensity (0, 1, 2, or 3+).
Results
RNR M1 expression
RNR M1 is constitutively expressed throughout the cell cycle.16 In the presence of DNA damage from ionizing radiation, RNR M1 transcripts and protein levels are relatively unperturbed in in vitro cervical cancer cells.7 To evaluate whether this observation holds in in situ cervical cancers, we evaluated the pattern of protein expression of RNR M1 in paired benign uterine cervix and cervical cancers. RNR M1 antibody staining intensity in the cytosol was low (0–1+) in all 19 benign uterine cervix cores obtained from hysterectomy specimens (Fig. 3). Cytoplasmic RNR M1 antibody staining intensity was 2+ or 3+ in the majority (6 of 8) of untreated cervix cancers at hysterectomy (Fig. 3). Cytoplasmic RNR M1 was 2+ or 3+ in most (7 of 10) cervix cancers at hysterectomy when pretreated by radiochemotherapy (Fig. 3). Immunohistochemistry staining intensity scores had a strong agreement between observers (kappa value: 0.677, p < 0.001) and between antibody lots (kappa value: 0.808, p < 0.001).
Figure 3.
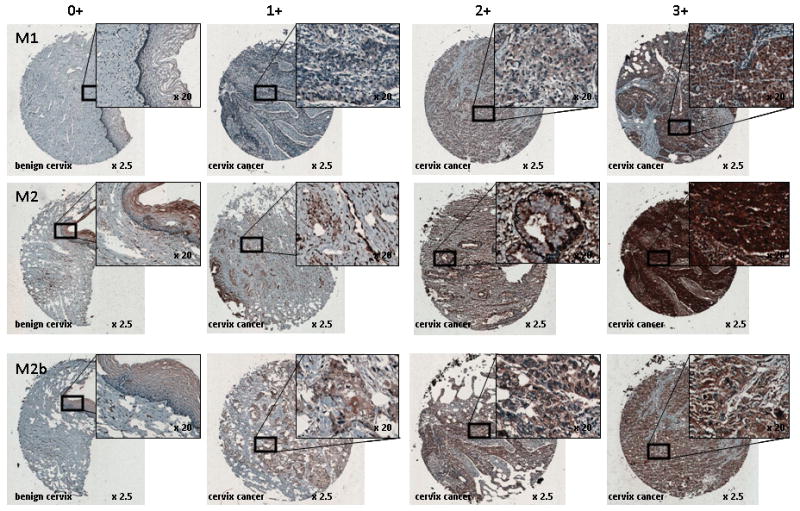
RNR M1, M2, and M2b immunoreactivity in uterine cervix benign and cancer tissues. Columns indicated brown color staining intensity 0 (< 5% of cells), 1 (5% to < 25%), 2 (25% to < 75%), and 3 (≥ 75%) scale. Rows identify RNR subunit. Magnification is indicated.
RNR M2 expression
RNR M2 is S-phase specific in the cell cycle.11 Dividing cells may have up to a 20-fold rise in RNR M2 level for replication of the genome.22 In vitro cervical cancer cells show a 17-fold rise in RNR M2 transcripts and protein levels over 12 to 18 hours after irradiation.7 It has been suggested that this rise in RNR M2 assists in DNA damage repair.3–6 In order to distinguish RNR M2 as a marker of cell replication versus a marker of enhanced DNA-damage repair, we set the criteria for RNR M2 overexpression to be 3+ in intensity in the cytoplasm (i.e., ≥75%). For benign uterine cervix, cytosolic RNR M2 intensity was low (0–1+) in half (10 of 19) of the cores (Fig. 3). RNR M2 intensity was 3+ in nearly all (7 of 8) untreated cervix cancers at hysterectomy (Fig. 3). In the radiochemotherapy treated group of cores, RNR M2 in the cytosol was 3+ in the majority (9 of 10) of cervix cancers at hysterectomy (Fig. 3). Immunohistochemical scoring agreed consistently between observers (kappa value: 0.780, p < 0.001) and between antibody lots (kappa value: 0.629, p < 0.001).
RNR M2b expression
RNR M2b has been observed naturally at low levels throughout the cell cycle and may be considered a DNA damage response element.1 Molecularly, up to 90% of cervical cancers contain HPV DNA and proteins.1 Perhaps by disrupting a protein-protein inhibitory interaction of p53 upon RNR M2b (p53R2),23 cervical cancers possessing HPV-E6 may unleash M2b to unite with RNR M1 and may generate dNDPs freely. Benign uterine cervix cytosolic staining intensity for RNR M2b was low (0–1+) in most (17 of 19) of the cores (Fig. 3). As it turns out, RNR M2b intensity was 2+ or 3+ in most (5 of 8) untreated cervix cancers at hysterectomy (Fig. 3). After radiochemotherapy, RNR M2b staining intensity in the cytosol was 2+ or 3+ in the majority (8 of 10) of cervix cancers at hysterectomy (Fig. 3). Agreement was high among observers (kappa value: 0.729, p < 0.001) and between antibody lots (kappa value: 0.749, p < 0.001).
Evaluation of RNR M1, M2, and M2b immunohistochemistry and cancer outcome
Increased RNR M1, M2, or M2b might or might not result in a higher likelihood of cancer relapse. Figure 4 shows immunohistochemical expression of RNR subunits and cancer outcomes. Of the 19 patients who underwent hysterectomy in which we evaluated RNR M1, M2, and M2b expression, eight patients had surgery and no adjuvant cancer therapy. Two of these had cancer relapses, whereas the other six did not. In the two patients with relapse, their tumors showed intense 3+ staining for RNR M1 and RNR M2. Eleven patients had radiochemotherapy preceding hysterectomy; 10 specimens in the TMA could be evaluated by immunohistochemistry. Although our sample size is limited, only two (20%) of 10 patients achieved a complete clinical response by RECIST criteria at the time of brachytherapy. These two responses were confirmed by hysterectomy histopathology (> 90% of cancer cells sterilized, as discussed before).24 As a contrast, 18F-FDG PET complete metabolic response (SUV pre-radiochemotherapy to pre-surgical ratio < 0.33)21 was recorded in most (6 [60%] of 10) patients.
Figure 4.
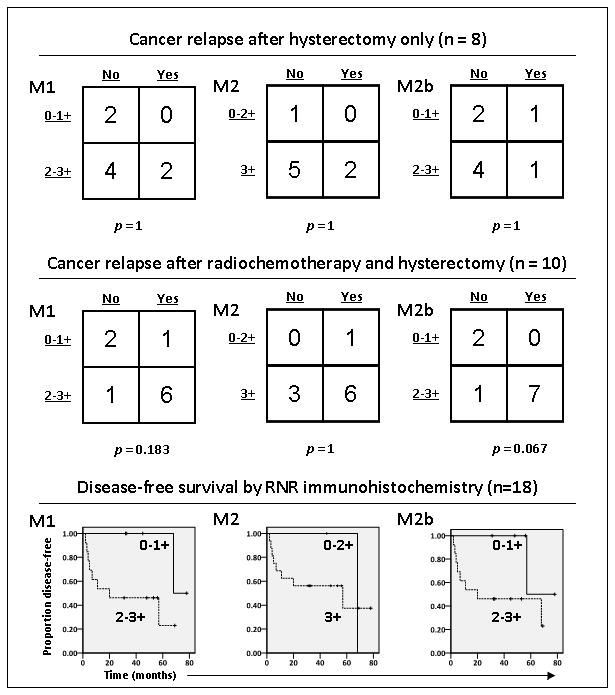
Dichotomized RNR M1, M2, and M2b immunoreactivity in cancer tissues by cancer relapse status. Kaplan-Meier curves for these dichotomies are shown.
Radiation increases at least M2 and possibly M2b expression in cancers.7 When comparing irradiated matched benign uterine cervix and cervical cancer cores, there was an increase in immunohistochemical intensity in the cancers by at least 1+ for RNR M2 in nearly all (9 of 10) cases and for M2b in most (8 of 10) cases. RNR M1 staining intensity rose 1+ in few (3 of 10) cases. Since few (2 of 10) patients achieved a complete clinical response and RNR M2 and M2b expression levels rose in nearly all cases, this data suggests decreased radiochemosensitivity. Additional lines of evidence supporting this claim are revealed in relapse patterns after radiochemotherapy (Fig. 4). Again while the data is limited, a rise in M2b evident during radiochemotherapy is associated strongly (p = 0.067) with cancer relapse. Overexpression of M1 (2–3+), M2 (3+), and M2b (2–3+) associate strongly with shortened DFS (Fig. 4). Among women whose cervical cancers were treated by radiochemotherapy prior to hysterectomy, DFS was estimated to be 33% at 5 years posttherapy. However, in a multivariate analysis, no single RNR subunit expression status increased the hazard beyond entered pathological variables.
Discussion
Substantial evidence supports RNR taking part in the radiochemotherapeutic response of cervical cancers.3–7, 20 Indeed, in vitro cervical cancer cells show increased RNR enzymatic activity, correlating with rapid repair of radiochemotherapy-induced DNA damage.4 Pharmacologic inhibition of RNR, especially when de novo dNDP production is demanded, protracts fixing of damaged DNA and enhances cell death.3–6 RNR blockade in human cervical cancer trials has been met with much success.1 Our findings in this study provide further evidence that cancers of the uterine cervix demonstrate marked immunoreactivity of RNR M2 and M2b (Fig. 3) translating into a diminished radiochemotherapy response and shortened DFS (Fig. 4).
In cancers of the uterine cervix, preclinical data upheld that the regulatory subunit RNR M1 remained level after DNA damage induced by ionizing radiation.7 With the recognized shortcoming of not having a pretherapy RNR M1 status assessment, our data suggest that RNR M1 expression in uterine cervix cancers rises infrequently after radiochemotherapy (Fig. 3).
Conversely, increased expression of RNR M2 and M2b has been identified in cervical cancers.20 When cervical cancer RNR M2 and M2b are pharmacologically inhibited, a high (10 of 10) rate of complete clinical response is observed after radiochemotherapy.20 Here in this study, it seems that overexpression of RNR M2 and M2b lowered radiochemotherapy response and led to earlier than expected cancer relapse (Figs. 3 and 4). To further expand upon this point, posttherapy residual disease after radiochemotherapy in women with bulky clinical stage IB2 cervical cancer patients associates with long-term survival.24 Women whose cancers responded poorly (≥ 10% viable cells in hysterectomy specimens) to radiochemotherapy had an estimated progression-free rate of 48% at 5 years posttherapy. Women whose cancers responded had an estimated progression-free rate of 85% at 10 years posttherapy. Even though our data is very limited, women undergoing radiochemotherapy whose tumor had 3+ M2 or M2b immunoreactivity at hysterectomy had a 33% 5-year estimate for DFS. This is one of a few reports of increased RNR M2 and M2b expression associating with immunoreactivity and cancer outcome, and is the only report of RNR M2 and M2b overexpression linked to shortened DFS. Our study was not designed to detect 18F-FDG PET complete metabolic responses, as in our prior work.21
In biomarker immunohistochemistry of FFPE tissue, consistency can be quite difficult as a result of influential pre-procedural, procedural, and post-procedural factors. Pitfalls include inconsistent time to tissue paraffin embedding and fixation time, uneven antibody epitope access among native in situ versus denatured ex vivo protein forms, polyclonal antibody pools of variable affinity, non-specific and non-reproducible antibody clones from one lot to another, and discordant staining localization such as the case of a predominately cytoplasmic staining when a transcription factor target is known to be restricted to the cell nucleus.25
Some of these factors are difficult to standardize. In our study, we reasoned that a comparison of benign uterine cervix tissue matched to cervical cancer in the same patient would lessen interpretive bias stemming from irregular FFPE tissue retrieval, processing and storage between the 19 patients. By constructing a TMA for batch processing of RNR immunoreactivity, the issue of discordant antibody dilution and intensity variance of peroxidase brown precipitation among single stained slides is avoided.
The matter of RNR epitope specificity is more complicated. Here, we used a commercial polyclonal antibody to M1, a monoclonal antibody to M2, and a polyclonal antibody to M2b. Rigorous RNR antibody validation has been done for M2 and M2b by others.7, 18, 19 It has been recommended that antibodies should be validated in Western immunoblots to show single band immunoreactivity in isogenic cell lines overproducing the protein of interest and the absence of bands in null or siRNA or shRNA knockdown cell lines.25 As RNR is not devoid in human tissues, we rationally decided upon benign lung tissue as a “negative” control and benign stomach, proliferating tonsil, and placenta as “positive” controls for our RNR M1, M2, and M2b proteins of interest. While cytosolic versus nuclear compartmentalization of RNR subunits remains under close scrutiny, we rationally scored intensity of cytoplasmic immunoreactivity because RNR M1-M2 or RNR M1-M2b isoforms are expected to generate de novo dNDPs demanded by the nucleus and by the mitochondria. Benign lung and cervix cells showed low levels of RNR subunits, while replicating cervical cancer cells had high levels of RNR subunits. This reduces some of the concerns of epitope specificity and interpretive bias relating to the possible heterogeneous expression of RNR epitopes within a tumor. Lastly, immunohistochemical epitope noise raises issues of antibody nonspecificity. Variance of immunoreactivity intensity by antibody lot can be observed. In our study, the uterine cervix TMA was stained twice by different antibody lots. A high concordance between the first and second lots was observed in our study, as indicated by the kappa statistic. While we advocate more robust scrutiny of antibody lot validation using objective image scoring techniques, at the current time we do not have this capability.
In summary, we investigated the immunohistochemical expression of RNR M1, M2, and M2b proteins in benign tissue and cancers of the uterine cervix. We found that cancers of the uterine cervix uniformly overexpress RNR M2. If RNR M1 and M2b are simultaneously overexpressed, cancer relapse rates are high and disease-free intervals are short. As commercial antibodies raised against RNR M1, M2, and M2b are reasonably informative and specific, studies are warranted of banked tissue from clinical trials conducted in cervical cancer patients.
Acknowledgments
SOURCES OF SUPPORT: Supported in part by NIH grant P30CA43703 for use of the Human Tissue Procurement and Pathology Core Facilities, Case Western Reserve University and the CASE Comprehensive Cancer Center (Cleveland, Ohio).
Footnotes
CONFLICT OF INTEREST: There are no potential conflicts of interest among the authors and this manuscript. This manuscript has been seen, read, and agreed upon in its content by all designated authors. This manuscript has not been submitted or published elsewhere.
References
- 1.Kunos C. Therapeutic mechanisms of treatment in cervical and vaginal cancer. Oncology & Hematology Review. 2012;8:55–60. [PMC free article] [PubMed] [Google Scholar]
- 2.Hebner C, Laimins L. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev Med Virol. 2006;16:83–97. doi: 10.1002/rmv.488. [DOI] [PubMed] [Google Scholar]
- 3.Kunos C, Chiu S, Pink J, Kinsella T. Modulating radiation resistance by inhibiting ribonucleotide reductase in cancers with virally or mutationally silenced p53 protein. Radiation Res. 2009;172:666–76. doi: 10.1667/RR1858.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kunos C, Radivoyevitch T, Pink J, et al. Ribonucleotide reductase inhibition enhances chemoradiosensitivity of human cervical cancers. Radiation Res. 2010;174:574–81. doi: 10.1667/RR2273.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunos C, Colussi V, Pink J, et al. Radiosensitization of human cervical cancer cells by inhibiting ribonucleotide reductase: enhanced radiation response at low dose rates. Int J Radiat Oncol Biol Phys. 2011;80:1198–204. doi: 10.1016/j.ijrobp.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunos C, Ferris G, Pyatka N, et al. Deoxynucleoside salvage facilitates DNA repair during ribonucleotide reductase blockade in human cervical cancers. Radiat Res. 2011;176:425–33. doi: 10.1667/rr2556.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuo M-L, Kinsella T. Expression of ribonucleotide reductase after ionizing radiation in human cervical carcinoma cells. Cancer Res. 1998;58:2245–52. [PubMed] [Google Scholar]
- 8.Rofougaran R, Vodnala M, Hofer A. Enzymatically active mammalian ribonucleotide reductase exists primarily as an alpha6beta2 octamer. J Biol Chem. 2006;281:27705–11. doi: 10.1074/jbc.M605573200. [DOI] [PubMed] [Google Scholar]
- 9.Fairman J, Wijerathna S, Ahmad M, et al. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat Struct Mol Biol. 2011;18:316–22. doi: 10.1038/nsmb.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reece S, Hodgkiss J, Stubbe J, Nocera D. Proton-coupled electron transfer: the mechanistic underpinning for radical transport and catalysis in biology. Phil Trans R Soc B. 2006;361:1351–64. doi: 10.1098/rstb.2006.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chabes A, Thelander L. Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J Biol Chem. 2000;275:17747–53. doi: 10.1074/jbc.M000799200. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka H, Arakawa H, Yamaguchi T, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–9. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 13.Kimura T, Takeda S, Sagiya Y, et al. Impaired function of p53R2 in Rrm2b-null mice causes severe renal failure through attenuation of dNTP pools. Nat Genet. 2003;34:440–5. doi: 10.1038/ng1212. [DOI] [PubMed] [Google Scholar]
- 14.Zheng Z, Chen T, Li X, et al. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356:800–8. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 15.Morikawa T, Hino R, Uozaki H, et al. Expression of ribonucleotide reductase M2 subunit in gastric cancer and effects of RRM2 inhibition in vitro. Hum Pathol. 2010;41:1742–8. doi: 10.1016/j.humpath.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Mann G, Musgrove E, Fox R, Thelander L. Ribonucleotide reductase M1 subunit in cellular proliferation, quiescence, and differentiation. Cancer Res. 1988;48:5151–6. [PubMed] [Google Scholar]
- 17.Feder J, Guidos C, Kusler B, et al. A cell cycle analysis of growth-related genes expressed during T lymphocyte maturation. J Cell Biol. 1990;111:2693–701. doi: 10.1083/jcb.111.6.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pontarin G, Ferraro P, Rampazzo C, et al. Deoxyribonucleotide metabolism in cycling and resting human fibroblasts with a missense mutation in p53R2, a subunit of ribonucleotide reductase. J Biol Chem. 2011;286:11132–40. doi: 10.1074/jbc.M110.202283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pontarin G, Fijolek A, Pizzo P, et al. Ribonucleotide reduction is a cytosolic process in mammalian cells independently of DNA damage. Proc Natl Acad Sci USA. 2008;105:17801–6. doi: 10.1073/pnas.0808198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunos C, Waggoner S, Von Gruenigen V, et al. Phase I trial of intravenous 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP, NSC #663249) in combination with pelvic radiation therapy and weekly cisplatin chemotherapy for locally advanced cervical cancer. Clin Cancer Res. 2010;16:1298–306. doi: 10.1158/1078-0432.CCR-09-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunos C, Radivoyevitch T, Abdul-Karim F, Faulhaber P. 18F-fluoro-2-deoxy-d-glucose positron emission tomography standard uptake value as an indicator of cervical cancer chemoradiation therapeutic response. Int J Gynecol Cancer. 2011;21:1117–23. doi: 10.1097/IGC.0b013e31821dc8b5. [DOI] [PubMed] [Google Scholar]
- 22.Hakansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281:7834–41. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 23.Xue L, Zhou B, Liu X, et al. Wild-type p53 regulates human ribonucleotide reductase by protein-protein interaction with p53R2 as well as hRRM2 subunits. Cancer Res. 2003;63:980–6. [PubMed] [Google Scholar]
- 24.Kunos C, Ali S, Abdul-Karim F, et al. Posttherapy residual disease associates with long-term survival after chemoradiation for bulky stage 1B cervical carcinoma: a Gynecologic Oncology Group Study. Am J Obstet Gynecol. 2010;203:351.e1–8. doi: 10.1016/j.ajog.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bordeaux J, Welsh A, Agarwal S, et al. Antibody validation. BioTechniques. 2010;48:197–209. doi: 10.2144/000113382. [DOI] [PMC free article] [PubMed] [Google Scholar]


