Background: Human 8-oxoguanine DNA glycosylase (OGG1) removes the mutagenic DNA lesion 7,8-dihydro-8-oxoguanine.
Results: OGG1 activity is significantly decreased by an adjacent DNA mismatch.
Conclusion: Tandem lesions can dramatically diminish base excision repair.
Significance: Deamination of 5′-methylcytosine in CpG-rich DNA sequences prone to oxidative DNA damage could be a significant factor in the generation of G to T transversions hot spots.
Keywords: Carcinogenesis, DNA Damage, DNA Repair, Enzyme Kinetics, Mutagenesis, Oxidative Stress, Base Excision Repair, DNA Glycosylase
Abstract
Human 8-oxoguanine DNA glycosylase (OGG1) is a key enzyme involved in removing 7,8-dihydro-8-oxoguanine (8-oxoG), a highly mutagenic DNA lesion generated by oxidative stress. The removal of 8-oxoG by OGG1 is affected by the local DNA sequence, and this feature most likely contributes to observed mutational hot spots in genomic DNA. To elucidate the influence of local DNA sequence on 8-oxoG excision activity of OGG1, we conducted steady-state, pre-steady-state, and single turnover kinetic evaluation of OGG1 in alternate DNA sequence contexts. The sequence context effect was studied for a mutational hot spot at a CpG dinucleotide. Altering either the global DNA sequence or the 5′-flanking unmodified base pair failed to influence the excision of 8-oxoG. Methylation of the cytosine 5′ to 8-oxoG also did not affect 8-oxoG excision. In contrast, a 5′-neighboring mismatch strongly decreased the rate of 8-oxoG base removal. Substituting the 5′-C in the CpG dinucleotide with T, A, or tetrahydrofuran (i.e. T:G, A:G, and tetrahydrofuran:G mispairs) resulted in a 10-, 13-, and 4-fold decrease in the rate constant for 8-oxoG excision, respectively. A greater loss in activity was observed when T:C or A:C was positioned 5′ of 8-oxoG (59- and 108-fold, respectively). These results indicate that neighboring structural abnormalities 5′ to 8-oxoG deter its repair thereby enhancing its mutagenic potential.
Introduction
Cellular DNA is continuously exposed to reactive oxygen species (ROS)2 that are produced by cellular metabolism, exogenous environmental chemicals, and ionizing radiation. 7,8-Dihydro-8-oxoguanine (8-oxoG) is among the most abundant DNA lesions generated by ROS. Cells have developed elaborate multilayered avoidance and repair systems to protect itself from the cytotoxic and mutagenic consequences of oxidative stress. Simple base lesions, such as 8-oxoG, are primarily repaired by base excision repair (BER) (1). In addition, nucleotide excision repair and mismatch repair appear to contribute to oxidized base repair (2–5). In mammalian cells, BER of 8-oxoG is initiated by a DNA glycosylase (OGG1) that is the functional homologue of the Escherichia coli enzyme MutM. OGG1 recognizes 8-oxoG paired with cytosine in double-stranded DNA and efficiently excises the damaged base from nuclear and mitochondrial genomes (6–10). Due to the altered hydrogen bonding pattern of 8-oxoG, unrepaired 8-oxoG can readily base pair with adenine during DNA replication and repair, resulting in a G to T transversion mutation (11). Thus, the persistence of an 8-oxoG lesion in DNA contributes to mutagenesis and has been implicated in cancer development.
OGG1 is a bifunctional glycosylase/apurinic-apyrimidinic lyase that hydrolyzes the N-glycosidic bond and subsequently cleaves the sugar-phosphate backbone 3′ to an apurinic/apyrimidinic (AP) site (12). Although it possesses two activities, previous work suggested that the AP lyase activity of OGG1 is not essential and that OGG1 may act as a monofunctional glycosylase in vivo (13, 14). The OGG1 knock-out mouse exhibits greater levels of genomic 8-oxoG and also shows an increase in G:C to T:A transversions in genomic DNA that is correlated with a predisposition to tumorigenesis (15–17). Human OGG1 suppresses G:C to T:A mutations in vivo (18, 19). Therefore, OGG1 is an important component for the prevention of oxidative damage-induced mutations.
It has long been suggested that mutations in the TP53 tumor suppressor gene contribute to many human carcinomas (20). In this regard, G to T transversions at TP53 mutational hot spots have been detected in several carcinomas (21–23). These mutations may be related to 8-oxoG, because elevated levels of 8-oxoG have been found in several carcinomas (24, 25). Interestingly, formation of 8-oxoG at CpG dinucleotides in the gene promoter region inhibits the binding of methyl-CpG-binding proteins and the SP1 transcription factor (26, 27). Thus, the 8-oxoG lesion could influence epigenetic factors as well as the somatic mutations that are associated with tumorigenesis.
In mammals, 3–6% of cytosines are methylated on the base at the 5-position (5-meC), and 70–80% of CpG dinucleotides in the human genome contain 5-meC (28–30). CpG dinucleotides in the TP53 gene have also been identified as hypermethylation sites and mutational hot spots in carcinoma cells (31). Loci with 5-meC are associated with at least 30% of germ line and somatic point mutations. The hydrolytic deamination of 5-meC to thymine produces a T:G mismatched base pair and initiates a C to T transition mutation. Importantly, 5-meC has been shown to spontaneously deaminate more readily than cytosine in both single- and double-stranded DNA (32, 33). In addition to spontaneous deamination, methyltransferase-catalyzed deamination may also contribute to mutational hot spots (34). The repair of T:G mismatches is less efficient than that of U:G mismatches in TP53 hot spots (35), suggesting that unrepaired T:G may be associated with CpG dinucleotides. It has been reported that CpG to TpT tandem mutations are promoted at methylated CpG dinucleotides under oxidative stress in nucleotide excision repair-deficient cells (36). Similarly, an abasic site adjacent to 8-oxoG enhances the mutagenicity of 8-oxoG and results in tandem mutations in simian kidney cells (37). Several studies suggest that 5-meC may increase the mutagenicity of a neighboring damaged guanine by forming an aberrant base pair adjacent to 8-oxoG creating a tandem DNA lesion (i.e. thymine glycol/8-oxoG or AP-site/8-oxoG), or intra-strand cross-linked DNA damage in the genome (37–39).
The mutagenicity and repair efficiency of 8-oxoG can be strongly affected by the local DNA sequence. For example, E. coli MutM removes 8-oxoG more efficiently in a pyrimidine-rich sequence than a purine-rich sequence (40). Moreover, the stacking interactions of 8-oxoG with its 5′-neighboring base can influence the extrusion of the lesion by E. coli MutM (41). Although the 8-oxoG recognition system of OGG1 is distinct from that of MutM, these enzymes use the same extra-helical base extrusion strategy in performing glycosylase activity (42, 43). Thus, the surrounding sequence contexts are expected to affect OGG1-mediated 8-oxoG repair and potentially contribute to G to T mutagenic hot spot formation. However, the mechanism of OGG1 regulation as a function of surrounding sequence context is poorly understood and is a subject of the current investigation.
In this study, we examined the glycosylase activity of OGG1 on a model substrate to determine the effect of altering the base pair 5′ to the 8-oxoG:C pair in the DNA sequence context of codon 248 of TP53, a CpG hot spot for ROS-induced mutations (22). An earlier qualitative analysis suggested that the repair of 8-oxoG by OGG1 could be diminished by a 5′-flanking 5-meC in the CpG dinucleotide of a transcription factor-binding site (27). Thymine glycol, the product of an oxidation of 5-meC followed by deamination, has also been shown to modulate the OGG1 repair efficiency of a neighboring 8-oxoG when thymine glycol is paired with adenine (44). However, no quantitative analysis of OGG1-mediated excision of 8-oxoG has been conducted regarding the effect of 5-meC or a mismatched base pair, such as T:G that could result from deamination of 5-meC. Here we report that the global DNA sequence and the 5′-flanking unmodified base pairs do not influence the repair efficiency of 8-oxoG. Furthermore, OGG1 can rapidly excise 8-oxoG even in a highly GC-rich DNA substrate. Contrary to the work described above (27), we found that 5′-neighboring 5-meC does not influence the removal of 8-oxoG by OGG1. However, neighboring mismatched base pairs strongly decrease the rate of 8-oxoG excision. These results provide mechanistic insight into OGG1 activity and the mechanism of mutational hot spot formation linked to 8-oxoG.
EXPERIMENTAL PROCEDURES
Materials
Ultrapure solutions of dCTP and dGTP were from Sigma. [α-32P]dGTP was from PerkinElmer Life Sciences. Recombinant human OGG1 was overexpressed and purified as described previously (45). Human apurinic/apyrimidinic endonuclease 1 (APE1), DNA polymerase (pol) β, and DNA ligase I (Lig I) were purified as described previously (46–48).
Oligonucleotides
5′-6-Carboxyfluorescein (FAM)-labeled or unlabeled oligonucleotides containing 8-oxoG and unmodified templates were synthesized and polyacrylamide gel electrophoresis purified by Eurofins MWG Operon (Huntsville, AL, USA). OGG1 and BER substrates were constructed by annealing a 5′-6-FAM-labeled or unlabeled oligonucleotide containing 8-oxoG to their template strand at a molar ratio 1:1.2. The sequences of the oligonucleotides are shown in Table 1.
TABLE 1.
Rate constant for 8-oxoG excision by OGG1 under single turnover conditions
| Name | Sequencea | kobs |
|---|---|---|
| s−1 | ||
| std1 | FAM-5′-CTGCAGCTGATGCGCCXTACGGATCCCCGGGTAC-3′ | 0.74 ± 0.015 |
| 3′-GACGTCGACTACGCGGCATGCCTAGGGGCCCATG-5′ | ||
| std2 | FAM-5′-CCCGCCCTGTGGCCCCXCCCGGCCCGCGCTTGCT-3′ | 0.49 ± 0.047 |
| 3′-GGGCGGGACACCGGGGCGGGCCGGGCGCGAACGA-5′ | ||
| std3 | FAM-5′-AGCAAGCGCGGGCCGGGXGGGGCCACAGGGCGGG-3′ | 1.0 ± 0.054 |
| 3′-TCGTTCGCGCCCGGCCCCCCCCGGTGTCCCGCCC-5′ | ||
| C/G | FAM-5′-CATGGGCGGCATGAACCXGAGGCCCATCCTCACC-3′ | 0.71 ± 0.037 |
| 3′-GTACCCGCCGTACTTGGCCTCCGGGTAGGAGTGG-5′ | ||
| A/T | FAM-5′-CATGGGCGGCATGAACAXGAGGCCCATCCTCACC-3′ | 0.76 ± 0.054 |
| 3′-GTACCCGCCGTACTTGTCCTCCGGGTAGGAGTGG-5′ | ||
| G/C | FAM-5′-CATGGGCGGCATGAACGXGAGGCCCATCCTCACC-3′ | 0.91 ± 0.052 |
| 3′-GTACCCGCCGTACTTGCCCTCCGGGTAGGAGTGG-5′ | ||
| T/A | FAM-5′-CATGGGCGGCATGAACTXGAGGCCCATCCTCACC-3′ | 0.77 ± 0.031 |
| 3′-GTACCCGCCGTACTTGACCTCCGGGTAGGAGTGG-5′ | ||
| mC/G | FAM-5′-CATGGGCGGCATGAACCXGAGGCCCATCCTCACC-3′ | 0.61 ± 0.063 |
| 3′-GTACCCGCCGTACTTGGCCTCCGGGTAGGAGTGG-5′ | ||
| mC/GmC | FAM-5′-CATGGGCGGCATGAACCXGAGGCCCATCCTCACC-3′ | 0.92 ± 0.024 |
| 3′-GTACCCGCCGTACTTGGCCTCCGGGTAGGAGTGG-5′ | ||
| T/G-mis | FAM-5′-CATGGGCGGCATGAACTXGAGGCCCATCCTCACC-3′ | 0.068 ± 0.0018 |
| 3′-GTACCCGCCGTACTTGGCCTCCGGGTAGGAGTGG-5′ | ||
| T/G-mis-r | FAM-5′-CATGGGCGGCATGAACCXTAGGCCCATCCTCACC-3′ | 0.78 ± 0.053 |
| 3′-GTACCCGCCGTACTTGGCGTCCGGGTAGGAGTGG-5′ | ||
| A/G-mis | FAM-5′-CATGGGCGGCATGAACAXGAGGCCCATCCTCACC-3′ | 0.055 ± 0.0018 |
| 3′-GTACCCGCCGTACTTGGCCTCCGGGTAGGAGTGG-5′ | ||
| T/C-mis | FAM-5′-CATGGGCGGCATGAACTXGAGGCCCATCCTCACC-3′ | 0.012 ± 0.0025 |
| 3′-GTACCCGCCGTACTTGCCCTCCGGGTAGGAGTGG-5′ | ||
| A/C-mis | FAM-5′-CATGGGCGGCATGAACAXGAGGCCCATCCTCACC-3′ | 0.0066 ± 0.00055 |
| 3′-GTACCCGCCGTACTTGCCCTCCGGGTAGGAGTGG-5′ | ||
| G/T-mis | FAM-5′-CATGGGCGGCATGAACGXGAGGCCCATCCTCACC-3′ | 0.18 ± 0.0079 |
| 3′-GTACCCGCCGTACTTGTCCTCCGGGTAGGAGTGG-5′ | ||
| THF/G | FAM-5′-CATGGGCGGCATGAAC*XGAGGCCCATCCTCACC-3′ | 0.19 ± 0.0010 |
| 3′-GTACCCGCCGTACTTGGCCTCCGGGTAGGAGTGG-5′ |
a X = 8-oxoG. FAM indicates the presence of a fluorescence tag. Mismatched base pairs are underlined. Double-underlined C indicates 5-methylcytosine. Asterisk (*) = tetrahydrofuran.
DNA Cleavage Assays
Glycosylase assays were performed in 50 mm HEPES, pH 7.5, 20 mm KCl, 0.5 mm EDTA, and 0.1% bovine serum albumin at 37 °C. Assays typically included 200 nm 34-mer DNA duplex substrates and 9.6 nm active OGG1. Under single turnover conditions, 50 nm 34-mer DNA substrates and 250 nm active OGG1 were used. To examine pre-steady-state kinetics, 200 nm 34-mer DNA substrates and 40 nm OGG1 were used. A KinTek model RQF-3 rapid quench-flow apparatus (KinTek Corp., Austin, TX) was employed when reactions were too rapid to measure manually. Unless noted otherwise, all concentrations refer to the final concentration after mixing. Reactions were initiated by adding OGG1 to the reaction mixture and terminated with 100 mm NaOH (5 min at 90 °C), to inactivate the enzyme and cleave the resulting AP sites. An equal volume of DNA gel loading buffer (95% formamide, 20 mm EDTA, 0.02% bromphenol blue, and 0.02% xylene cyanol) was added. After incubation at 95 °C for 2 min, the reaction products were separated by electrophoresis in a 15% denaturing polyacrylamide gel containing 8 m urea in 89 mm Tris-HCl, pH 8.8, 89 mm boric acid, and 2 mm EDTA. A Typhoon PhosphorImager was used for gel scanning and imaging, and the data were analyzed with ImageQuant software.
In Vitro BER Reconstitution with Purified Enzymes
The BER assay was performed in a final reaction mixture volume of 20 μl, as described previously (49, 50). The BER reaction mixture contained 50 mm HEPES, pH 7.5, 0.5 mm EDTA, 2 mm dithiothreitol, 20 mm KCl, 5 mm MgCl2, 4 mm ATP, 10 μm [α-32P]dGTP (specific activity, 1 × 106 dpm/pmol), and 250 nm 34-mer DNA substrates. In some cases, the reaction mixtures were supplemented with 10 μm dCTP. The repair reactions were initiated by the addition of OGG1 (40 nm), APE1 (20 nm), and pol β (20 nm) with or without Lig I (200 nm), and incubated at 37 °C. Aliquots (5 μl) were withdrawn at the times indicated in the figures. The reaction was terminated by addition of an equal volume (5 μl) of DNA gel loading buffer (95% formamide, 20 mm EDTA, 0.02% bromphenol blue, and 0.02% xylene cyanol). After incubation at 95 °C for 2 min, the reaction products were separated by electrophoresis in a 15% denaturing polyacrylamide gel and analyzed as described above.
RESULTS
Determination of the Active Fraction of OGG1
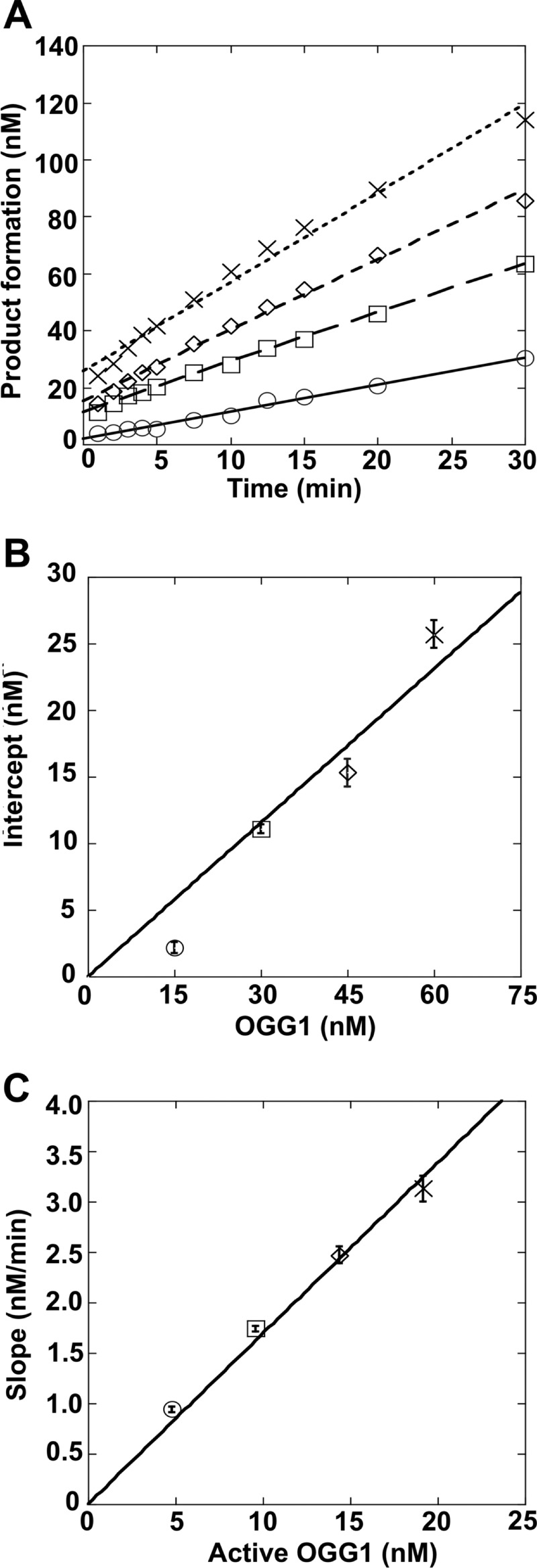
The time course of 8-oxoG removal from duplex DNA is biphasic (Fig. 1A). There is a rapid formation of product that occurs prior to collection of the first time point and a slow linear phase that corresponds to the steady-state rate. The amplitude of the burst phase reflects the fraction of active OGG1 bound to DNA in a productive manner and is dependent on the total concentration of OGG1 added in the reaction mixture (Fig. 1B). This permits a quantitative assessment of the active concentration of enzyme (51). This active site titration was performed using 200 nm 34-mer DNA duplex (Table 1, sequence = std1) and four apparent enzyme concentrations (15, 30, 45, and 60 nm) as determined by a Bradford protein assay. The time courses were fit to a linear equation to determine the intercept (i.e. amplitude) of the burst phase. The estimated active fraction of enzyme was 14, 37, 34, and 43% relative to each protein concentration (average of 32%). The OGG1 concentrations reported below refer to active enzyme concentrations.
FIGURE 1.
Active site titration of purified OGG1. OGG1 (15 nm, ○; 30 nm, □; 45 nm, ♢; or 60 nm, ×) was incubated with 200 nm DNA substrate (std1) at 37 °C for 1–30 min. These enzyme concentrations represent apparent concentrations based on a Bradford protein assay. Product formation was measured as described under “Experimental Procedures.” The data were fit to a linear equation to determine the amplitude of the burst phase (y intercept) and the steady-state rate (slope). The amplitude of the burst phase reflects the true or active concentration of OGG1. A, time courses of product formation. B, plot of the intercepts determined from the linear fits in panel A relative to that determined by the Bradford assay. The slope of the line corresponds to the fraction of active enzyme. C, plot of the slopes determined from the linear fits in panel A relative to the active enzyme concentration. The slope of the line corresponds to the steady-state rate, 0.0028 s−1.
The biphasic nature of the time course indicates that a step after chemistry is rate determining during the slower steady-state phase. This phase corresponds to the dissociation of OGG1 from DNA and limits catalytic cycling. From the active enzyme fraction and the slopes of the linear fits from Fig. 1A, the steady-state rate (kcat = koff,DNA) was determined to be 0.0028 s−1 (Fig. 1C).
Effect of DNA Sequence on OGG1 Excision of 8-OxoG
Because the steady-state rate reflects product dissociation rather than base excision activity, single turnover assays were employed to isolate the excision step. Under single turnover conditions, where the concentration of OGG1 exceeds the DNA substrate concentration, catalytic cycling does not occur so that product release does not influence the observed activity (i.e. N-glycosyl bond hydrolysis). Thus, to directly measure the rate of 8-oxoG excision by OGG1 in alternate DNA sequence contexts, single turnover experiments were conducted with 50 nm DNA duplexes (std1, std2, std3, and alternate 5′-matched base pairs). The sequences of these DNA substrates are shown at Table 1. A 5-fold excess of OGG1 (i.e. 250 nm) was used. Reactions were started by the addition of enzyme, and aliquots were removed at various time periods. These aliquots were heated in the presence of 100 mm NaOH to stop the reaction and cleave the resultant AP site. A minor amount of background cleavage was observed after treatment of the substrate itself with NaOH (Fig. 2A, lane 2). This background was subtracted from the amount of each reaction product. The data were fit to a single exponential equation to determine the first-order rate constant (kobs) as previously described (52).
FIGURE 2.
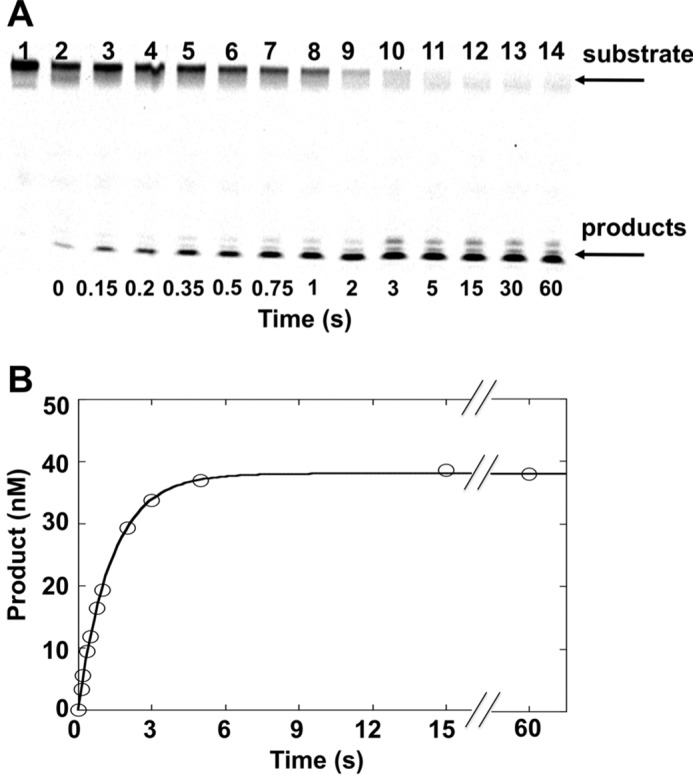
Single turnover analysis to measure base removal by OGG1. OGG1 (250 nm) was incubated with 50 nm substrate DNA (std1) at 37 °C and product formation followed for 60 s as described under “Experimental Procedures.” A, denaturing polyacrylamide gels showing separated substrates/products. Lane 1 is DNA substrate without NaOH treatment. Lanes 2–14 corresponds to time-dependent product formation. B, plot of product formation from panel A. The data were fit to an exponential time course (kobs = 0.74 s−1).
As shown in Fig. 2, product formation followed a single-exponential time course and was nearly complete in 5 s. The observed excision rate constant with the std1 DNA substrate (Table 1) was 0.74 s−1. Using alternate DNA substrates (std1, std2, and std3) or where the 5′-base pair was altered did not significantly influence the rate of excision (0.49–1.0 s−1, Table 1). These results indicate that OGG1 rapidly excised 8-oxoG regardless of the neighboring standard DNA sequence. The magnitude of the excision rates are consistent with the rate of 8-oxoG removal of OGG1 reported recently (53). Notably, the rates were not affected by the nature of the 5′ label utilized on the damaged strand (32P or 6-FAM; data not shown).
Influence of 5′ Modified Nucleotides on OGG1 Excision Activity
To examine the effect of 5′-neighboring modified nucleotides on OGG1-catalyzed excision of 8-oxoG, we examined the single turnover rate where the nucleotide 5′ to 8-oxoG was meC or its deaminated product, thymidine. In both cases, these nucleotides were paired with guanine thereby mimicking codon 248 in the TP53 gene, a CpG hot spot for ROS-induced mutations (22). The excision rate of 8-oxoG with an adjacent meC was similar to that observed for C at this position (kobs = 0.61 s−1, Table 1). In contrast, when T was 5′ of 8-oxoG, creating a T:G mismatch (T/G-mis), the observed rate constant was decreased significantly (Fig. 3; kobs = 0.068 s−1). Interestingly, when a T:G mismatch was positioned 3′ to 8-oxoG (T/G-mis-r), the excision rate of 8-oxoG was not altered (kobs = 0.78 s−1, Table 1).
FIGURE 3.
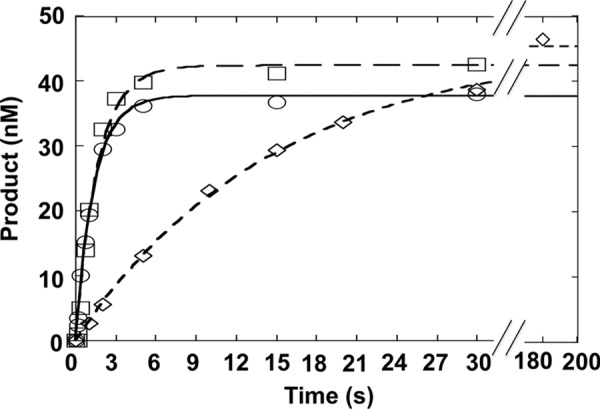
Influence of the 5′-base pair on 8-oxoG excision. Single turnover time course for the removal of 8-oxoG where the 5′-base pair is C:G (○), 5-meC:G (□), or T:G (♢). OGG1 (250 nm) was incubated with 50 nm substrate DNA (C/G, mC/G, or T/G-mis) at 37 °C for up to 180 s. Products were isolated and quantified as described under “Experimental Procedures.”
We further examined how other adjacent mismatches to 8-oxoG, which cause various modifications of the DNA structure, affect the rate of base removal by OGG1. We prepared substrates containing A:G, T:C, A:C, and G:T mismatches, or tetrahydrofuran (THF):G positioned upstream of 8-oxoG (A/G-mis, T/C-mis, A/C-mis, G/T-mis, or THF/G, respectively) and measured the single turnover rate of 8-oxoG excision (Table 1). A significant decrease in the excision activity was observed with all substrates (kobs = 0.0066–0.19 s−1, Table 1). The strongest loss in activity was observed with the A/C-mis, 108-fold lower as compared with C/G, whereas the G/T-mis and THF/G exhibited the least affects, 3.9- and 3.7-fold, respectively. These results indicated that whereas mismatches decreased OGG1 excision activity, the nature of the mismatch also had an affect.
Influence of Neighboring 5-meG:C or T:G on Catalytic Cycling of OGG1
The effect of the neighboring base pair on product release of OGG1 was examined by measuring the steady-state rate. In this situation, excess DNA (200 nm) with C/G, mC/G, or T/G-mis 5′ to 8-oxoG was incubated with 9.6 nm active OGG1 for different periods. Time courses were linear for at least 5–6 turnovers (15 min) and extrapolated to ∼10 nm active enzyme (i.e. y intercept; Fig. 4). The rate of the linear phase that corresponds to the step that limits catalytic cycling can be calculated from the slopes of the linear phase. As illustrated in Fig. 4, the steady-state rates of the alternate substrates do not significantly differ (0.0045–0.0090 s−1). Because MutM excision efficiency was reduced in a purine-rich sequence (40), we also examined the steady-state rate in a purine-rich substrate (i.e. std3) and it was found to be similar to other sequences examined (0.0042 s−1). Likewise, the excision activity of OGG1 is similar in both a pyrimidine- and purine-rich sequence (Table 1, std2 and -3, respectively).
FIGURE 4.
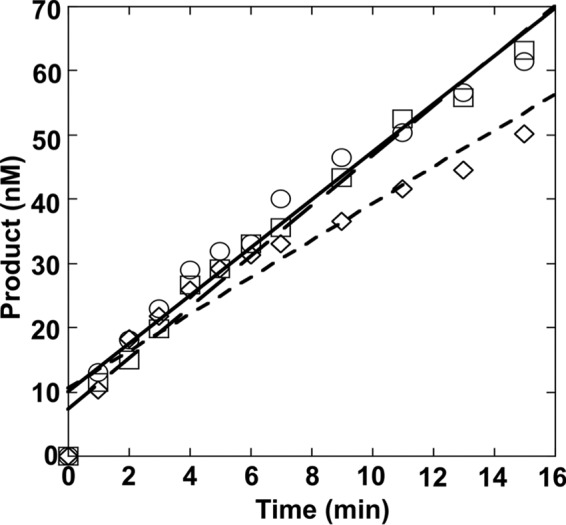
Effect of the 5′-base pair on the steady-state rate of 8-oxoG excision. OGG1 (9.6 nm) was incubated with 200 nm DNA with a C:G (C/G, ○), 5-meC:G (mC/G, □), or T:G (T/G-mis, ♢) base pair 5′ to 8-oxoG at 37 °C for 0–15 min. Products were isolated and quantified as described under “Experimental Procedures.” Data were fit to a straight line with the slopes corresponding to 0.0063, 0.0090, 0.0045 s−1 for the C/G, mC/G, and T/G-mis DNA substrates, respectively.
Effect of the Neighboring Base Pair on the Pre-steady-state Kinetics of 8-OxoG Excision
We confirmed the effect of the adjacent base pair on the excision rate of 8-oxoG and steady-state rate of catalytic cycling by measuring both the pre-steady-state and steady-state phases of a single time course. In this case, 40 nm OGG1 was mixed with 200 nm 34-mer substrates (C/G, mC/G, or T/G) and the reaction followed for 140 s (Fig. 5). These time courses were fit to an equation with rising exponential and linear terms, as shown by Equation 1 (51).
FIGURE 5.
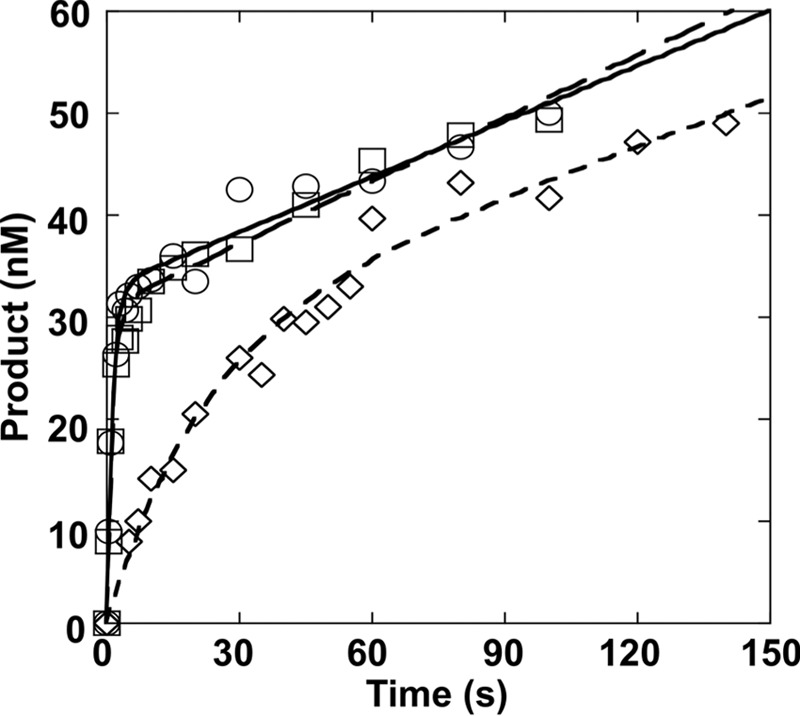
Effect of the 5′-base pair on the pre-steady-state rate of 8-oxoG excision. OGG1 (40 nm) was incubated with 200 nm DNA with a C:G (C/G, ○), 5-meC:G (mC/G, □), or T:G (T/G-mis, ♢) base pair 5′ to 8-oxoG at 37 °C for 0–140 s. Products were isolated and quantified as described under “Experimental Procedures.” Data were fit to Equation 1 as described in the text, and the resulting parameters tabulated in Table 2.
The fits provide a burst rate constant (kobs) and an apparent linear rate (vss). With this analysis, the burst amplitude (A) represents the active concentration of enzyme so that kss = (vss/active enzyme). The results of these fits are summarized in Table 2. Importantly, increasing the substrate concentration (i.e. 300 nm) does not alter the steady-state rate indicating that 200 nm DNA is saturating (data not shown). Consistent with the results of both the single turnover and the multiple turnover experiments, kobs for T/G-mis (0.046 s−1) was 16-fold lower than those for C/G and mC/G (0.75 and 0.72 s−1, respectively), whereas koff for T/G-mis (0.0057 s−1) was similar to those for C/G and mC/G (0.0055 and 0.0068 s−1, respectively). The steady-state turnover numbers (kss = koff,DNA) were 136- and 106-fold lower than the intrinsic 8-oxoG excision rates of C/G and mC/G, respectively. The slower excision rate with the T/G substrate was still 8-fold faster than the steady-state rate.
TABLE 2.
Pre-steady-state kinetic parameters for 8-oxoG excision by OGG1
Data was tabulated from Fig. 5.
| Substrate | kobsa | Ab | vssc | kcatd |
|---|---|---|---|---|
| s−1 | nm | nm s−1 | s−1 | |
| C/G | 0.75 ± 0.083 | 33 ± 0.89 | 0.18 ± 0.018 | 0.0055 ± 0.020 |
| mC/G | 0.72 ± 0.075 | 31 ± 0.80 | 0.21 ± 0.017 | 0.0068 ± 0.021 |
| T/G-mis | 0.046 ± 0.011 | 28 ± 4.4 | 0.16 ± 0.039 | 0.0057 ± 0.00089 |
a Single-exponential rate constants determined from the initial burst phase.
b The amplitude of the burst phase.
c The slope of the linear steady-state phase.
d Steady-state rate corrected for active enzyme fraction; calculated as kcat = (vss/A). The slow catalytic cycling is limited by product dissociation (i.e. koff-).
Neighboring Mismatch Inhibits BER of 8-OxoG:C
To further investigate the effect of an adjacent mismatch on BER efficiency of 8-oxoG, we reconstituted an in vitro BER system for repair of 8-oxoG. The DNA substrates included C/G, mC/G, or T/G-mis adjacent to the 8-oxoG paired with C (Table 1). The DNA was incubated with OGG1, pol β, APE1, and Lig I (Fig. 6). In the absence of Lig I, an unligated repair intermediate increased in a time-dependent manner in the reaction for both C/G and mC/G (Fig. 6, lanes 1–3 and 7–9, respectively). When T/G-mis was used as the substrate, the amount of the intermediate was dramatically decreased (Fig. 6, lanes 13–15). The amount of the ligated product for T/G-mis was also significantly less than that observed for C/G and mC/G (Fig. 6 lanes 4–6, 10–12, and 16–18). Because APE1 has a weak proofreading activity capable of excising the mismatched primer terminus (54), we considered whether the lack of dGMP incorporation might be due to the inability to extend a shortened primer that would require insertion of dCMP. However, addition of both dCTP and dGTP into the reaction mixture for the T/G-mis substrate did not significantly increase the unligated intermediate (lanes 19–21).
FIGURE 6.
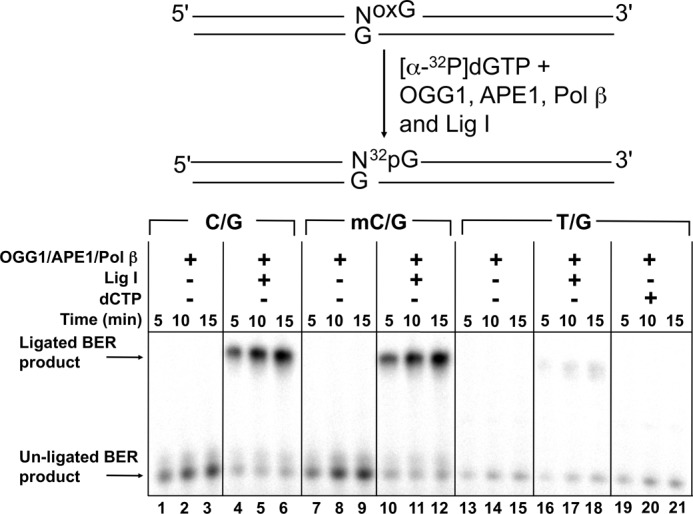
Effect of the 5′-base pair on the BER activity of 8-oxoG excision. Schematic representation of the substrate and the reaction scheme are shown in the top panel. A DNA substrate with C:G (C/G), 5-meC:G (mC/G), or T:G (T/G-mis) positioned 5′ to 8-oxoG (250 nm) was incubated with 40 nm OGG1, 20 nm APE1, 20 nm pol β, with or without 200 nm Lig I as indicated was incubated at 37 °C for 5, 10, or 15 min. In lanes 19–21, 10 μm dCTP was also included in the reaction mixtures. Reaction products were isolated and quantified as described under “Experimental Procedures.”
DISCUSSION
In this study, we analyzed the effect of DNA sequence context on the in vitro excision activity of OGG1 for 8-oxoG using 8-oxoG-modified oligonucleotides and purified human enzyme. The BER system recognizes 8-oxoG and removes it from the genome. Replication of DNA templates with 8-oxoG will often result in G to T transversions due to the efficient insertion of dAMP opposite 8-oxoG. DNA glycosylases initiate BER of 8-oxoG and deficiencies of DNA glycosylases such as OGG1 and adenine DNA glycosylase (MYH; removes adenine misinserted opposite 8-oxoG) result in an increase of G to T mutations contributing to tumorigenesis (55). Previous studies have suggested that the local sequence context might affect the activity of OGG1 (40), and that this may be related to mutational hot spots. In the present study, we set out to investigate this possibility with a focus on the local sequence at a specific CpG dinucleotide in a mutational hot spot of TP53 (i.e. codon 248).
According to our results, alternate DNA sequences of unmodified bases adjacent to 8-oxoG did not affect the excision activity of OGG1 (Table 1). Furthermore, OGG1 rapidly excised 8-oxoG even in a substrate with 3 consecutive purines or pyrimidines 5′ to the lesion (Table 1, std2 and std3). This is in contrast to E. coli MutM, which is strongly affected by the local sequence context (40). Interestingly, an earlier study suggested that OGG1 interacts with the 8-oxoG-modified substrates more avidly than MutM and was not significantly affected by DNA sequence context (56). Thus, a rapid excision of 8-oxoG by OGG1, regardless of the local DNA sequence, may be due to its high specificity for the cognate substrate. Our results indicate that OGG1 can efficiently repair the lesion even in a purine-rich sequence, i.e. CpG island promoter region, suppressing mutations in the human genome.
The excision efficiency of 8-oxoG by bacterial MutM is dependent on local sequence context (40); in contrast, the single turnover rates for removal of this lesion in the alternate sequences used in this study by human OGG1 is not. This suggests that the measured rates do not reflect base stacking interactions that must be overcome to expel the lesion from the DNA helix; but probably reflects slow cleavage of the N-glycosidic bond as suggested previously (57). Although these two enzymes are structurally unrelated, they employ a similar strategy for 8-oxoG lesion interrogation and expulsion. Binding of the enzyme, and insertion of an aromatic side chain 3′ to the lesion, induces a dramatic bend in the DNA. This results in unstacking of the estranged cytosine and expulsion of 8-oxoG from the DNA helix. Comparing crystallographic structures of a recognition complex of MutM (43) with that of OGG1 (42) indicates that the interactions with the extra-helical oxidized base for each enzyme is unique. However, it is not obvious how the local DNA sequence would modulate excision activity. In contrast, there are local effects on the base pairs neighboring the site of the lesion. In the case of MutM, both the 3′ and 5′ base pair are buckled away from the site of the lesion. In the case of OGG1, only the 3′ base pair is buckled, whereas the 5′ base pair remains planar. Taken together, the kinetic and structural observations suggest that 5′-base pair buckling might be sensitive to the local DNA sequence context and could alter excision of 8-oxoG presumably by influencing active site geometry.
Cytosines in CpG dinucleotides are highly methylated in the human genome. It has been shown that methylation of cytosine enhances its base-base stacking interaction in DNA duplex due to an increase in polarizability (58, 59). Thus, methylated cytosine could possibly enhance the stacking interaction with neighboring 8-oxoG at CpG dinucleotides. We suspected that the enhanced stacking of 5-meC with the neighboring 8-oxoG might affect the extrusion of 8-oxoG from the DNA helix in the active site of OGG1 and modulate the base removal rate, as previously examined (27). Contrary to our expectation, both the single turnover and steady-state rate of OGG1 were not altered by a 5′-flanking 5-meC (Tables 1 and 2), suggesting that the stacking interaction of 8-oxoG with a neighboring base is not crucial for both the base extrusion and the product release steps for the OGG1 catalytic cycle. Pre-steady-state stopped-flow studies following intrinsic fluorescence changes in OGG1 upon binding an 8-oxoG:C DNA substrate indicate that several rapid enzyme conformational changes occur prior to the slow cleavage of the N-glycosidic bond (57). Thus, our results are consistent with glycosidic bond cleavage as the rate-limiting step during single turnover analysis. The diminished rate with a 5′-mismatch or AP-site indicates that site-specific DNA structural perturbations can interrupt OGG1 active site geometry.
Earlier studies indicated that the binding affinity of OGG1 for a lesion containing substrate was decreased when a T:G mismatched base pair was positioned both 5′ or 3′ of 8-oxoG (56). In the current study, a 5′-flanking T:G mismatched base pair significantly decreased the 8-oxoG base removal of OGG1 in single turnover and pre-steady-state assays (Figs. 3 and 4, Table 1), but did not significantly alter the rate of product release (Figs. 4 and 5, Table 2). Because catalytic cycling is dependent on the DNA dissociation rate constant, the lack of an effect of the T:G mismatch indicates that the DNA binding affinity is not affected by the mismatch positioned adjacent to an AP site. Furthermore, the extent of decrease in the base removal rate was dependent on the identity of the mismatch. As tabulated in Table 1, substitution of the 5′ neighboring C:G base pair with T:G, G:T, or A:G at CpG dinucleotides resulted in a 10-, 3.9-, or 13-fold decrease in excision of 8-oxoG. On the other hand, a much stronger decrease was observed for T:C or A:C (59- or 108-fold, respectively). These differences may reflect a destabilizing effect of the specific mismatch in duplex DNA as previously reported; the introduction of A:C or T:C mismatches resulted in greater destabilization of the helix than T:G or A:G (60, 61). Whereas T:G or A:G base pairs lead to dynamic perturbations of the adjacent base pairs, introduction of A:C or T:C resulted in perturbations that extended several base pairs into the DNA helix (60). The THF:G alteration also decreased the excision rate of 8-oxoG, suggesting that a neighboring AP site leads to a structural alteration in the adjacent 8-oxoG:C in duplex DNA destabilizing the DNA backbone (62). In contrast, a T:G mismatch 3′ to the lesion did not affect N-glycosidic bond cleavage (Table 1) indicating that active site geometry is not disturbed by a structural perturbation at this position.
As shown in Fig. 6, a reconstituted BER system using purified OGG1, pol β, APE1, and Lig I was strongly inhibited by a 5′-neighboring T:G, but not by a 5′-neighboring 5-meC:G. The inhibition of BER by an adjacent T:G mismatch should reflect not only the decreased excision rate of OGG1, but also the poor primer extension activity of pol β with a mismatch terminus (66). The strand joining activity of Lig I could also be decreased by a mismatched base pair upstream of a matched primer terminus. The ability of APE1 to incise the resulting AP site may also be sensitive to a nearby mismatch. Further investigation could clarify the effect of the surrounding sequence for the activity of Lig I and APE1. In summary, the mismatched base pair adjacent to 8-oxoG strongly inhibits successful completion of BER.
Recent studies have shown that thymine glycol (Tg) is formed by oxidation of 5-meC followed by deamination, and that the 5′ neighboring Tg:A base pair perturbed the repair of the adjacent 8-oxoG in the presence of OGG1 and APE1 (38, 44). If Tg is produced at methylated CpG dinucleotides, it could initially form a wobble base pair with guanine as found with a T:G mismatch (67). The structural perturbation in DNA caused by a Tg:G wobble base pair would be expected to be different from that induced by Tg:A (68–70); Tg would be displaced toward the major groove when paired with G, as compared with A, resulting in less steric hindrance between the methyl group of Tg and the 5′-adjacent base. It remains to be seen how an adjacent Tg:G might affect the repair efficiency of a neighboring 8-oxoG in a CpG dinucleotide.
In conclusion, we have shown that an aberrant base pair 5′ to 8-oxoG strongly reduces its excision and repair in a mutational hot spot. Additionally, OGG1 activity in other sequence contexts was not altered significantly. It is noteworthy that several other proteins including APE1, x-ray repair cross-complementing protein 1, and poly(ADP-ribose) polymerase 1 influence the removal of 8-oxoG by OGG1 (63–65). Further studies that examine the influence of other BER proteins/co-factors on the coding potential (polymerase-dependent) and repair of 8-oxoG will provide insights into cooperative protein interactions that modulate the mutagenic potential of an important oxidative DNA lesion in a biologically important sequence context.
Acknowledgments
We thank Dr. Julie Horton for critical reading of the manuscript, David Shock for assistance with the kinetic experiments, Esther Hou for assistance with the purification of human OGG1, and Bonnie Mesmer for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Research Project Grant Z01-ES050158 in the Intramural Research Program, NIEHS.
- ROS
- reactive oxygen species
- 6-FAM
- 6-carboxyfluorescein
- 8-oxoG
- 7,8-dihydro-8-oxoguanine
- OGG1
- 8-oxoguanine DNA glycosylase
- BER
- base excision repair
- AP
- apurinic-apyrimidinic
- 5-meC
- 5-methylcytosine
- Tg
- thymine glycol
- APE1
- apurinic/apyrimidinic endonuclease 1
- pol
- DNA polymerase
- Lig I
- DNA ligase I
- THF
- tetrahydrofuran.
REFERENCES
- 1. Nilsen H., Krokan H. E. (2001) Base excision repair in a network of defence and tolerance. Carcinogenesis 22, 987–998 [DOI] [PubMed] [Google Scholar]
- 2. Colussi C., Parlanti E., Degan P., Aquilina G., Barnes D., Macpherson P., Karran P., Crescenzi M., Dogliotti E., Bignami M. (2002) The mammalian mismatch repair pathway removes DNA 8-oxo-dGMP incorporated from the oxidized dNTP pool. Curr. Biol. 12, 912–918 [DOI] [PubMed] [Google Scholar]
- 3. Larson E. D., Iams K., Drummond J. T. (2003) Strand-specific processing of 8-oxoguanine by the human mismatch repair pathway. Inefficient removal of 8-oxoguanine paired with adenine or cytosine. DNA Repair 2, 1199–1210 [DOI] [PubMed] [Google Scholar]
- 4. Reardon J. T., Bessho T., Kung H. C., Bolton P. H., Sancar A. (1997) In vitro repair of oxidative DNA damage by human nucleotide excision repair system. Possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc. Natl. Acad. Sci. U.S.A. 94, 9463–9468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scott A. D., Neishabury M., Jones D. H., Reed S. H., Boiteux S., Waters R. (1999) Spontaneous mutation, oxidative DNA damage, and the roles of base and nucleotide excision repair in the yeast Saccharomyces cerevisiae. Yeast 15, 205–218 [DOI] [PubMed] [Google Scholar]
- 6. Rosenquist T. A., Zharkov D. O., Grollman A. P. (1997) Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 94, 7429–7434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zharkov D. O., Rosenquist T. A., Gerchman S. E., Grollman A. P. (2000) Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J. Biol. Chem. 275, 28607–28617 [DOI] [PubMed] [Google Scholar]
- 8. Boiteux S., Radicella J. P. (2000) The human OGG1 gene. Structure, functions, and its implication in the process of carcinogenesis. Arch. Biochem. Biophys. 377, 1–8 [DOI] [PubMed] [Google Scholar]
- 9. Takao M., Aburatani H., Kobayashi K., Yasui A. (1998) Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res. 26, 2917–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nishioka K., Ohtsubo T., Oda H., Fujiwara T., Kang D., Sugimachi K., Nakabeppu Y. (1999) Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol. Biol. Cell 10, 1637–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shibutani S., Takeshita M., Grollman A. P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349, 431–434 [DOI] [PubMed] [Google Scholar]
- 12. Radicella J. P., Dherin C., Desmaze C., Fox M. S., Boiteux S. (1997) Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 94, 8010–8015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dalhus B., Forsbring M., Helle I. H., Vik E. S., Forstrøm R. J., Backe P. H., Alseth I., Bjørås M. (2011) Separation-of-function mutants unravel the dual-reaction mode of human 8-oxoguanine DNA glycosylase. Structure 19, 117–127 [DOI] [PubMed] [Google Scholar]
- 14. Morland I., Luna L., Gustad E., Seeberg E., Bjørås M. (2005) Product inhibition and magnesium modulate the dual reaction mode of hOgg1. DNA Repair 4, 381–387 [DOI] [PubMed] [Google Scholar]
- 15. Minowa O., Arai T., Hirano M., Monden Y., Nakai S., Fukuda M., Itoh M., Takano H., Hippou Y., Aburatani H., Masumura K., Nohmi T., Nishimura S., Noda T. (2000) Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl. Acad. Sci. U.S.A. 97, 4156–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sakumi K., Tominaga Y., Furuichi M., Xu P., Tsuzuki T., Sekiguchi M., Nakabeppu Y. (2003) Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Cancer Res. 63, 902–905 [PubMed] [Google Scholar]
- 17. Kohno T., Shinmura K., Tosaka M., Tani M., Kim S. R., Sugimura H., Nohmi T., Kasai H., Yokota J. (1998) Genetic polymorphisms and alternative splicing of the hOGG1 gene that is involved in the repair of 8-hydroxyguanine in damaged DNA. Oncogene 16, 3219–3225 [DOI] [PubMed] [Google Scholar]
- 18. Sunaga N., Kohno T., Shinmura K., Saitoh T., Matsuda T., Saito R., Yokota J. (2001) OGG1 protein suppresses G:C → T:A mutation in a shuttle vector containing 8-hydroxyguanine in human cells. Carcinogenesis 22, 1355–1362 [DOI] [PubMed] [Google Scholar]
- 19. Yamane A., Shinmura K., Sunaga N., Saitoh T., Yamaguchi S., Shinmura Y., Yoshimura K., Murakami H., Nojima Y., Kohno T., Yokota J. (2003) Suppressive activities of OGG1 and MYH proteins against G:C to T:A mutations caused by 8-hydroxyguanine but not by benzo[a]pyrene diol epoxide in human cells in vivo. Carcinogenesis 24, 1031–1037 [DOI] [PubMed] [Google Scholar]
- 20. Levine A. J., Momand J., Finlay C. A. (1991) The p53 tumor suppressor gene. Nature 351, 453–456 [DOI] [PubMed] [Google Scholar]
- 21. Oliva M. R., Ripoll F., Muñiz P., Iradi A., Trullenque R., Valls V., Drehmer E., Sáez G. T. (1997) Genetic alterations and oxidative metabolism in sporadic colorectal tumors from a Spanish community. Mol. Carcinog. 18, 232–243 [PubMed] [Google Scholar]
- 22. Agar N. S., Halliday G. M., Barnetson R. S., Ananthaswamy H. N., Wheeler M., Jones A. M. (2004) The basal layer in human squamous tumors harbors more UVA than UVB fingerprint mutations. A role for UVA in human skin carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 4954–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hussain S. P., Raja K., Amstad P. A., Sawyer M., Trudel L. J., Wogan G. N., Hofseth L. J., Shields P. G., Billiar T. R., Trautwein C., Hohler T., Galle P. R., Phillips D. H., Markin R., Marrogi A. J., Harris C. C. (2000) Increased p53 mutation load in nontumorous human liver of Wilson disease and hemochromatosis. Oxyradical overload diseases. Proc. Natl. Acad. Sci. U.S.A. 97, 12770–12775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Olinski R., Zastawny T., Budzbon J., Skokowski J., Zegarski W., Dizdaroglu M. (1992) DNA base modifications in chromatin of human cancerous tissues. FEBS Lett. 309, 193–198 [DOI] [PubMed] [Google Scholar]
- 25. Jaruga P., Zastawny T. H., Skokowski J., Dizdaroglu M., Olinski R. (1994) Oxidative DNA base damage and antioxidant enzyme activities in human lung cancer. FEBS Lett. 341, 59–64 [DOI] [PubMed] [Google Scholar]
- 26. Valinluck V., Tsai H. H., Rogstad D. K., Burdzy A., Bird A., Sowers L. C. (2004) Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG-binding protein 2 (MeCP2). Nucleic Acids Res. 32, 4100–4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zawia N. H., Lahiri D. K., Cardozo-Pelaez F. (2009) Epigenetics, oxidative stress, and Alzheimer disease. Free Radic. Biol. Med. 46, 1241–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vanyushin B. F., Tkacheva S. G., Belozersky A. N. (1970) Rare bases in animal DNA. Nature 225, 948–949 [DOI] [PubMed] [Google Scholar]
- 29. Antequera F., Bird A. (1993) Number of CpG islands and genes in human and mouse. Proc. Natl. Acad. Sci. U.S.A. 90, 11995–11999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bird A. P. (1995) Gene number, noise reduction and biological complexity. Trends Genet. 11, 94–100 [DOI] [PubMed] [Google Scholar]
- 31. Tornaletti S., Pfeifer G. P. (1995) Complete and tissue-independent methylation of CpG sites in the p53 gene. Implications for mutations in human cancers. Oncogene 10, 1493–1499 [PubMed] [Google Scholar]
- 32. Ehrlich M., Norris K. F., Wang R. Y., Kuo K. C., Gehrke C. W. (1986) DNA cytosine methylation and heat-induced deamination. Biosci. Rep. 6, 387–393 [DOI] [PubMed] [Google Scholar]
- 33. Shen J. C., Rideout W. M., 3rd, Jones P. A. (1994) The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 22, 972–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shen J. C., Rideout W. M., 3rd, Jones P. A. (1992) High frequency mutagenesis by a DNA methyltransferase. Cell 71, 1073–1080 [DOI] [PubMed] [Google Scholar]
- 35. Schmutte C., Yang A. S., Beart R. W., Jones P. A. (1995) Base excision repair of U:G mismatches at a mutational hot spot in the p53 gene is more efficient than base excision repair of T:G mismatches in extracts of human colon tumors. Cancer Res. 55, 3742–3746 [PubMed] [Google Scholar]
- 36. Lee D. H., O'Connor T. R., Pfeifer G. P. (2002) Oxidative DNA damage induced by copper and hydrogen peroxide promotes CG → TT tandem mutations at methylated CpG dinucleotides in nucleotide excision repair-deficient cells. Nucleic Acids Res. 30, 3566–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kalam M. A., Basu A. K. (2005) Mutagenesis of 8-oxoguanine adjacent to an abasic site in simian kidney cells. Tandem mutations and enhancement of G → T transversions. Chem. Res. Toxicol. 18, 1187–1192 [DOI] [PubMed] [Google Scholar]
- 38. Yuan B., Jiang Y., Wang Y. (2010) Efficient formation of the tandem thymine glycol/8-oxo-7,8-dihydroguanine lesion in isolated DNA and the mutagenic and cytotoxic properties of the tandem lesions in Escherichia coli cells. Chem. Res. Toxicol. 23, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cao H., Wang Y. (2007) Quantification of oxidative single-base and intrastrand cross-link lesions in unmethylated and CpG-methylated DNA induced by Fenton-type reagents. Nucleic Acids Res. 35, 4833–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hatahet Z., Zhou M., Reha-Krantz L. J., Morrical S. W., Wallace S. S. (1998) In search of a mutational hot spot. Proc. Natl. Acad. Sci. U.S.A. 95, 8556–8561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sung R. J., Zhang M., Qi Y., Verdine G. L. (2012) Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM. J. Biol. Chem. 287, 18044–18054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bruner S. D., Norman D. P., Verdine G. L. (2000) Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 403, 859–866 [DOI] [PubMed] [Google Scholar]
- 43. Fromme J. C., Verdine G. L. (2003) DNA lesion recognition by the bacterial repair enzyme MutM. J. Biol. Chem. 278, 51543–51548 [DOI] [PubMed] [Google Scholar]
- 44. Jiang Y., Wang Y. (2009) In vitro replication and repair studies of tandem lesions containing neighboring thymidine glycol and 8-oxo-7,8-dihydro-2′-deoxyguanosine. Chem. Res. Toxicol. 22, 574–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kovtun I. V., Liu Y., Bjoras M., Klungland A., Wilson S. H., McMurray C. T. (2007) OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 447, 447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Beard W. A., Wilson S. H. (1995) Purification and domain-mapping of mammalian DNA polymerase β. Methods Enzymol. 262, 98–107 [DOI] [PubMed] [Google Scholar]
- 47. Strauss P. R., Beard W. A., Patterson T. A., Wilson S. H. (1997) Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs-Haldane mechanism. J. Biol. Chem. 272, 1302–1307 [DOI] [PubMed] [Google Scholar]
- 48. Chen X., Pascal J., Vijayakumar S., Wilson G. M., Ellenberger T., Tomkinson A. E. (2006) Human DNA ligases I, III, and IV purification and new specific assays for these enzymes. Methods Enzymol. 409, 39–52 [DOI] [PubMed] [Google Scholar]
- 49. Singhal R. K., Prasad R., Wilson S. H. (1995) DNA polymerase β conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J. Biol. Chem. 270, 949–957 [DOI] [PubMed] [Google Scholar]
- 50. Prasad R., Singhal R. K., Srivastava D. K., Molina J. T., Tomkinson A. E., Wilson S. H. (1996) Specific interaction of DNA polymerase β and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J. Biol. Chem. 271, 16000–16007 [DOI] [PubMed] [Google Scholar]
- 51. Vande Berg B. J., Beard W. A., Wilson S. H. (2001) DNA structure and aspartate 276 influence nucleotide binding to human DNA polymerase β. Implication for the identity of the rate-limiting conformational change. J. Biol. Chem. 276, 3408–3416 [DOI] [PubMed] [Google Scholar]
- 52. Porello S. L., Leyes A. E., David S. S. (1998) Single turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates. Biochemistry 37, 14756–14764 [DOI] [PubMed] [Google Scholar]
- 53. McKibbin P. L., Kobori A., Taniguchi Y., Kool E. T., David S. S. (2012) Surprising repair activities of nonpolar analogs of 8-oxoG expose features of recognition and catalysis by base excision repair glycosylases. J. Am. Chem. Soc. 134, 1653–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cistulli C., Lavrik O. I., Prasad R., Hou E., Wilson S. H. (2004) AP endonuclease and poly(ADP-ribose) polymerase-1 interact with the same base excision repair intermediate. DNA Repair 3, 581–591 [DOI] [PubMed] [Google Scholar]
- 55. Xie Y., Yang H., Cunanan C., Okamoto K., Shibata D., Pan J., Barnes D. E., Lindahl T., McIlhatton M., Fishel R., Miller J. H. (2004) Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 64, 3096–3102 [DOI] [PubMed] [Google Scholar]
- 56. Kirpota O. O., Endutkin A. V., Ponomarenko M. P., Ponomarenko P. M., Zharkov D. O., Nevinsky G. A. (2011) Thermodynamic and kinetic basis for recognition and repair of 8-oxoguanine in DNA by human 8-oxoguanine-DNA glycosylase. Nucleic Acids Res. 39, 4836–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kuznetsov N. A., Koval V. V., Zharkov D. O., Nevinsky G. A., Douglas K. T., Fedorova O. S. (2005) Kinetics of substrate recognition and cleavage by human 8-oxoguanine-DNA glycosylase. Nucleic Acids Res. 33, 3919–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Acosta-Silva C., Branchadell V., Bertran J., Oliva A. (2010) Mutual relationship between stacking and hydrogen bonding in DNA. Theoretical study of guanine-cytosine, guanine-5-methylcytosine, and their dimers. J. Phys. Chem. B 114, 10217–10227 [DOI] [PubMed] [Google Scholar]
- 59. Sowers L. C., Shaw B. R., Sedwick W. D. (1987) Base stacking and molecular polarizability. Effect of a methyl group in the 5-position of pyrimidines. Biochem. Biophys. Res. Commun. 148, 790–794 [DOI] [PubMed] [Google Scholar]
- 60. Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. (1984) Dynamics of DNA duplexes containing internal G:T, G:A, A:C, and T:C pairs. Hydrogen exchange at and adjacent to mismatch sites. Fed. Proc. 43, 2663–2670 [PubMed] [Google Scholar]
- 61. Modrich P. (1987) DNA mismatch correction. Annu. Rev. Biochem. 56, 435–466 [DOI] [PubMed] [Google Scholar]
- 62. Miller J. H., Aceves-Gaona A., Ernst M. B., Haranczyk M., Gutowski M., Vorpagel E. R., Dupuis M. (2005) Structure and energetics of clustered damage sites. Radiat. Res. 164, 582–585 [DOI] [PubMed] [Google Scholar]
- 63. Hill J. W., Hazra T. K., Izumi T., Mitra S. (2001) Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease. Potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 29, 430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Noren Hooten N., Kompaniez K., Barnes J., Lohani A., Evans M. K. (2011) Poly(ADP-ribose) polymerase 1 (PARP-1) binds to 8-oxoguanine-DNA glycosylase (OGG1). J. Biol. Chem. 286, 44679–44690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Marsin S., Vidal A. E., Sossou M., Ménissier-de Murcia J., Le Page F., Boiteux S., de Murcia G., Radicella J. P. (2003) Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J. Biol. Chem. 278, 44068–44074 [DOI] [PubMed] [Google Scholar]
- 66. Beard W. A., Shock D. D., Wilson S. H. (2004) Influence of DNA structure on DNA polymerase β active site function. Extension of mutagenic DNA intermediates. J. Biol. Chem. 279, 31921–31929 [DOI] [PubMed] [Google Scholar]
- 67. Brown K. L., Basu A. K., Stone M. P. (2009) The cis-(5R,6S)-thymine glycol lesion occupies the wobble position when mismatched with deoxyguanosine in DNA. Biochemistry 48, 9722–9733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Brown K. L., Adams T., Jasti V. P., Basu A. K., Stone M. P. (2008) Interconversion of the cis-5R,6S- and trans-5R,6R-thymine glycol lesions in duplex DNA. J. Am. Chem. Soc. 130, 11701–11710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kao J. Y., Goljer I., Phan T. A., Bolton P. H. (1993) Characterization of the effects of a thymine glycol residue on the structure, dynamics, and stability of duplex DNA by NMR. J. Biol. Chem. 268, 17787–17793 [PubMed] [Google Scholar]
- 70. Kung H. C., Bolton P. H. (1997) Structure of a duplex DNA containing a thymine glycol residue in solution. J. Biol. Chem. 272, 9227–9236 [DOI] [PubMed] [Google Scholar]