Abstract
Fiber cell initiation in the epidermal cells of cotton (Gossypium hirsutum L.) ovules represents a unique example of trichome development in higher plants. Little is known about the molecular and metabolic mechanisms controlling this process. Here we report a comparative analysis of a fiberless seed (fls) mutant (lacking fibers) and a normal (FLS) mutant to better understand the initial cytological events in fiber development and to analyze the metabolic changes that are associated with the loss of a major sink for sucrose during cellulose biosynthesis in the mutant seeds. On the day of anthesis (0 DAA), the mutant ovular epidermal cells lacked the typical bud-like projections that are seen in FLS ovules and are required for commitment to the fiber development pathway. Cell-specific gene expression analyses at 0 DAA showed that sucrose synthase (SuSy) RNA and protein were undetectable in fls ovules but were in abundant, steady-state levels in initiating fiber cells of the FLS ovules. Tissue-level analyses of developing seeds 15 to 35 DAA revealed an altered temporal pattern of SuSy expression in the mutant relative to the normal genotype. Whether the altered programming of SuSy expression is the cause or the result of the mutation is unknown. The developing seeds of the fls mutant have also shown several correlated changes that represent altered carbon partitioning in seed coats and cotyledons as compared with the FLS genotype.
Developing cotton (Gossypium hirsutum L.) seeds are an excellent system with which to study diverse patterns of carbon partitioning, including cellulose, starch, and oil biosynthesis (Ruan et al., 1997). There has been substantial progress in our understanding of cellulose synthesis in developing cotton fibers (Basra and Malik, 1984; Amor et al., 1995; Delmer and Amor, 1995; Ruan et al., 1997). In this regard, the cloning of cellulose synthase genes from cotton (Pear et al., 1996) and Arabidopsis (Arioli et al., 1998) is a remarkable achievement and opens a new era of research in cellulose biosynthesis in plants. By comparison, however, little is known about the early events controlling fiber cell initiation from the outermost layer of ovule epidermis. It has long been recognized that although all epidermal cells are potential fibers, only about 30% of these cells actually differentiate into fibers (Basra and Malik, 1984; Tiwari and Wilkins, 1995). Thus, an understanding of the mechanisms that determine which epidermal cells differentiate would provide the knowledge that is essential for increasing fiber productivity through genetic engineering.
Morphologically, the initiation of each fiber cell is associated with the spherical expansion and protrusion of one epidermal cell above the ovular surface during anthesis (Basra and Malik, 1984; Trelease et al., 1986). The appearance of SuSy (EC 2.4.1.13) marks one of the first signals of the onset of this process (Nolte et al., 1995), and the expression of this protein correlates with cellulose biosynthesis (Amor et al., 1995; Ruan et al., 1997). It should be pointed out, however, that rapid secondary cell wall cellulose biosynthesis in developing cotton fibers starts at about 16 DAA (Basra and Malik, 1984; Ruan et al., 1997). The functional basis for SuSy expression during the fiber cell initiation phase (0 DAA) remains to be elucidated. One approach to a better understanding of this problem would be the use of a mutant with reduced SuSy expression or fiber cell initiation.
Here we present the results of our studies to characterize a fiberless seed (fls) mutant, using a cDNA (SS3) encoding cotton SuSy and a polyclonal antibody raised against this protein (Ruan et al., 1997). There was a dramatic reduction in the expression of both SuSy mRNA and protein in the ovule epidermis of the fls mutant, which correlates with the lack of fiber cell initiation from the ovules. In the wild-type ovules (FLS), only those epidermal cells that show high levels of SuSy expression protrude as fibers. Together, these observations provide strong evidence that SuSy may play a critical role in fiber cell initiation. Finally, the impact of the absence of fibers, a strong sink, on carbon partitioning in the fls seed was analyzed. The results are discussed in the context of feedback regulation of Suc unloading and allocation in developing cotton seed. Overall, characterization of the fls mutation is important for a better understanding of the nature of the controls over the process of fiber cell initiation, an important economic trait, and to elucidate certain basic aspects of a plant developmental process.
MATERIALS AND METHODS
Plant Material
Two lines of cotton (Gossypium hirsutum L.), DH 59–64 as the wild type (FLS) and SL-171 as the mutant (fls), were grown under controlled conditions as previously described (Ruan et al., 1997). Seeds of the mutant stock SL-171 were a gift from Dr. Jim Heitholt (USDA-ARS Cotton Physiology and Genetic Research, Stoneville, MS). Cotton fruit age was determined by tagging the petioles of a flower when it was fully opened. All samples, unless otherwise specified, were frozen in liquid nitrogen and then stored at −80°C until analysis. Frozen, developing seeds were separated into fibers, seed coats, and cotyledons on dry ice before analysis.
In Situ Hybridization
In situ hybridization using paraffin-embedded sections was carried out according to the procedure described by Marrison and Leech (1994). Cotton ovules were collected from fully opened flowers and immediately fixed in formalin-acetic acid. The fixed ovules were dehydrated through a tertiary butyl alcohol series and embedded in paraffin. To eliminate possible RNase contamination, all solutions used prior to and during the hybridization were incubated in 0.1% (v/v) diethylpyrocarbonate overnight at 37°C before autoclaving. Cross-sections (12 μm) were placed on dampened, precharged slides (Probe On Plus, Fisher Scientific) in a water bath at 40°C to 42°C and left to dry on a hot plate overnight at 40°C. For a better comparison, sections from fls and FLS were arranged on the same slides with each genotype section divided into two parts and treated with sense and antisense SuSy RNA probes, respectively.
SS3 cDNA was linearized with ApaI or EcoRI for sense and antisense RNA preparation, respectively. Digoxigenin-labeled sense and antisense RNA probes were synthesized by in vitro transcription reactions using 1 μg of linearized SuSy DNA template, digoxigenin-11-UTP and T3, or T7 RNA polymerases, according to the protocol of the digoxigenin RNA-labeling kit (Boehringer Mannheim). The RNA probes were hydrolyzed to an average size of 150 nucleotides to allow better tissue penetration (Cox and Goldberg, 1988). The hybridized SuSy mRNA was immunologically detected by using anti-digoxigenin antibody conjugated with alkaline phosphatase as described in the digoxigenin nucleic acid detection kit, with modifications as indicated by Marrison and Leech (1994).
Immunolocalization
Immunogold silver staining was conducted according to the procedure recommended by the Histogold kit (Zymed Laboratories, Inc., San Francisco, CA). Paraffin-embedded cross-sections of cotton ovules were cut (12 μm), affixed to slides, deparaffined, rehydrated, and washed with PBS. Thereafter, slides were incubated with serum-blocking solution for 10 min and incubated with 1:1500 diluted SuSy polyclonal antibodies or preimmune serum in a humid environment for 1.5 h. After the slides were washed with PBS, they were incubated for 30 min in a solution of secondary antibody (goat anti-rabbit IgG linked to colloidal gold). Slides were then washed thoroughly with PBS (four times, 3 min each), incubated for 5 min with freshly prepared silver-enhancement reagents, and washed with excess distilled water. Slides were dehydrated in an ethanol series and permanently mounted in Permount (Fisher Scientific) for microscopic examination. Pairs of pre- and immunostained sections from both fls and FLS were treated on the same slide for a better comparison.
RNA and Protein Gel Blots
Total RNA was isolated from developing cotyledons of cotton seeds as previously described (Wadsworth et al., 1988; Ruan et al., 1997). RNA electrophoresis, blotting, and hybridization conditions were as described previously (Ruan et al., 1997). For soluble protein extraction, cotyledons (approximately 0.5 g each) were ground to a fine powder in liquid nitrogen. The grinding continued for 5 min in cold extraction buffer (3:1, v/v) containing 25 mm Hepes/KOH (pH 7.3), 5 mm EDTA, 1 mm DTT, 0.1% soluble PVP (Mr 40,000), 1 mm PMSF, and 0.01 mm leupeptin. The homogenate was centrifuged at 10,000g for 5 min at 4°C. The supernatants were collected and denatured by SDS and boiling treatments. The protein concentrations were determined using the Bio-Rad DC protein assay kit with BSA as a standard. Electrophoresis, blotting, and SuSy antigen detection were carried out as previously described (Ruan et al., 1997). In each case, three independent experiments were conducted and the same results were obtained.
Sugar and Starch Analysis
Soluble sugars and starch were extracted and enzymatically determined as previously described (Ruan et al., 1997).
RESULTS
The fls Trait Is Seed Specific and under Maternal Control
Although the fls mutant SL-171 and the wild-type genotypes are not lineage related, plants of the two genotypes have shown an overall similar pattern in growth rate, development, and timing of flowering. The only exception was the higher levels of red pigmentation in all plant parts of the mutant due to an unknown genetic constitution of its anthocyanin genes. The most striking difference between the two genotypes was seen during seed development. Whereas the developing FLS seeds were covered with young fibers, the fls seeds showed no detectable fiber growth, except for a few rudimentary fibers detectable only under a dissecting microscope. The size and shape of the seeds and fruits were similar in both genotypes. These observations were in agreement with the previous report of this fls mutant (Turley and Ferguson, 1996).
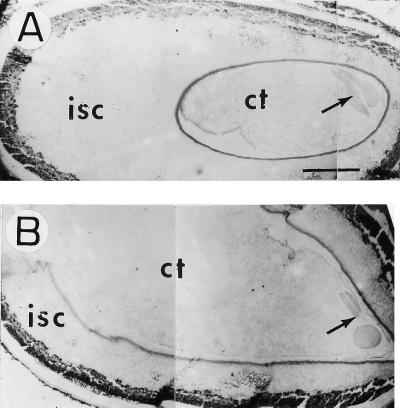
Maternal origin of fiber cells from the ovule epidermis, as early as 16 h preanthesis, is well documented cytologically (Ramsey and Berlin, 1976; Nolte et al., 1995). The fls mutant provided an opportunity to examine this aspect genetically. We emasculated flowers of both genotypes 1 d before anthesis and allowed seed development. Unfertilized seeds at 15 DAA showed massive amounts of fiber in FLS but none in the fls mutant, confirming the maternal control of both traits. It is interesting that seeds at 15 DAA of both genotypes from unpollinated flowers were indistinguishable from the corresponding self-pollinated controls. The only detectable difference was a smaller embryo size in the longitudinal sections of the unpollinated flowers (Fig. 1). The same pattern was also seen in the mutant seed (data not shown). Obviously, normal development of the embryo was dependent on fertilization, but the maternal tissues, including integuments and fiber development, were independent of the pollen parent and pollination.
Figure 1.
Longitudinal sections of FLS cotton seed at 15 DAA from emasculated (A) and pollinated (B) flowers. Note the much reduced size of the “embryo” in A. Bars = 625 μm. isc, Inner seed coat; ct, cotyledon. Arrow indicates location of hypocotyl.
SuSy Expression in the fls Ovule Epidermal Cells Is Reduced
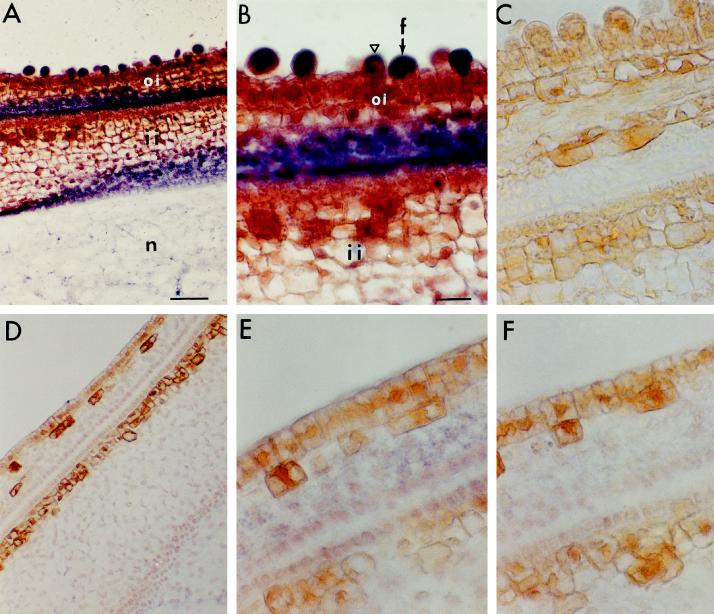
In situ hybridization of ovule sections harvested on the day of anthesis (0 DAA) was done to examine the cellular localization of SuSy mRNA using antisense and sense RNA probes generated from SS3 cDNA. As shown in Figure 2, the appearance of bubble-like structures among the epidermal cells (indicative of the initiation of fiber cells) in the wild type (Fig. 2, A–C) was completely missing in the fls mutant (Fig. 2, D–F). The FLS ovule sections also showed a strong signal for the SuSy mRNA in the initiating cells, whereas a slightly weak signal was seen in the two to three cell layers between the inner and outer integuments inside of the seed (Fig. 2A). Under high magnification the large, spherical “bubbles” showed the strongest signal for the SuSy mRNA (Fig. 2B), whereas the smaller, initiating cells showed weaker signals, and the nondifferentiating cells and those underlying the epidermis did not show any SuSy mRNA (Fig. 2B). In contrast, the mutant fls ovules at 0 DAA showed no signal for SuSy mRNA in the epidermal cells, nor was there any fiber cell initiation (i.e. no bubble formation, Fig. 2, D and E). Significantly, the cell layers between the inner and outer integuments of the fls mutant ovules also showed no SuSy RNA. No signal was seen in the sections of either genotype when treated with the sense control probe (Fig. 2, C and F).
Figure 2.
In situ hybridization of SuSy in cross-sections of FLS (A–C) and fls (D–F) ovules. The purple signals represent SuSy mRNA. A, B, D, and E, Cross-sections were hybridized with an antisense RNA probe generated from SS3 cDNA. Note in B the very strong SuSy mRNA signals in the large and spherically shaped initiating fiber cells (arrow), the weak signals in the small fiber cells (triangle), and the undetectable signals in the nondifferentiating epidermal cells. Also note in E that SuSy mRNA was undetectable in the nondifferentiating epidermis of the fls mutant. C and F, Cross-sections were hybridized with sense RNA probe. Bars in A and B are 50 and 22 μm, respectively (magnifications of A and D, B and C, and E and F). f, Fiber cell; oi, outer integument; ii, inner integument; n, nucellus.
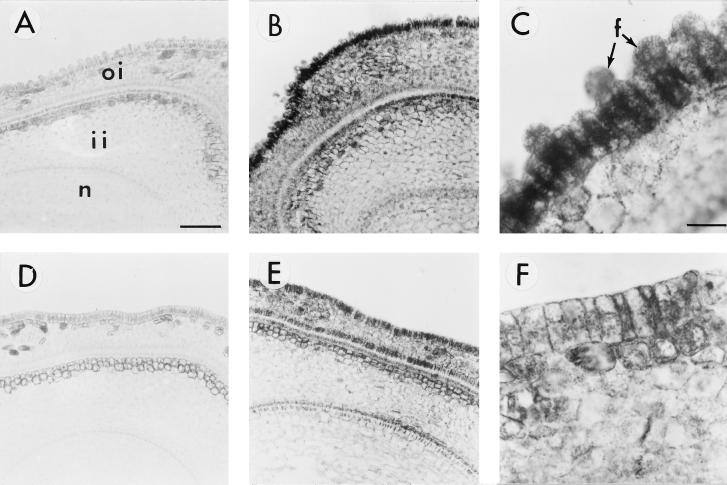
Ovule sections of the two genotypes were also examined at 0 DAA for the SuSy protein by immunolocalization. Figure 3B shows large amounts of SuSy protein in the FLS epidermal cells, but a much-reduced signal was seen in the fls mutant (Fig. 3E). Cell layers inside the seeds of both genotypes were not as sharply contrasted in the levels of SuSy protein as they were for the RNA signal. Under high magnification, however, the FLS epidermal cells showed a strong signal for the protein that was lacking in the mutant cells (Fig. 3, C and F). Preimmune controls for both genotypes are shown in Figure 3, A and D, and, as expected, showed no immunogold reactivity. Similar immunohistological analyses of the serial sections of the same ovules with carrot and maize cell wall invertase antibodies did not detect any signal in either genotype (data not shown).
Figure 3.
Immunogold localization of SuSy protein in cross-sections of FLS (A–C) and fls (D–F) ovules. A and D, Cross-sections were treated with preimmune serum. B, C, E, and F, Cross-sections were treated with polyclonal antibody against SuSy. Note in C the very strong SuSy protein signals in initiating fiber cells (arrows) and weak signals in the nondifferentiating epidermal cells (see the cell between the two initiating fiber cells indicated by arrows). Also note in F that very little SuSy protein can be detected in the ovule epidermis of the fls mutant. Bars in A and C are 77 and 19 μm, respectively (magnifications of A and B, D and E, and C and F). f, Fiber cells; oi, outer integument; ii, inner integument; n, nucellus.
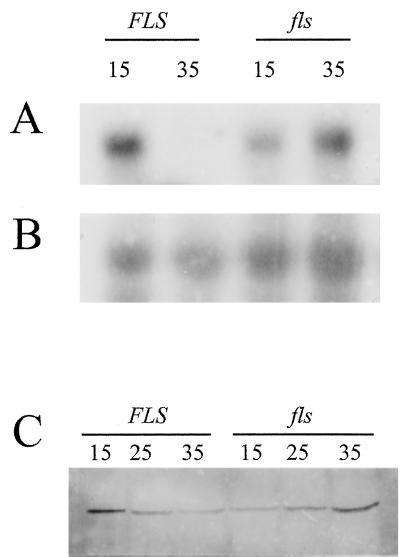
Previously, it was demonstrated that cotyledons but not seed coat cells are enriched in SuSy mRNA and protein in developing cotton seed (Ruan et al., 1997). Thus, SuSy expression patterns were also analyzed by northern- and western-blot analyses in developing cotyledons at 15 DAA and beyond, a stage corresponding to the elongation, secondary cell wall biosynthesis, and maturation phases of fiber development. Steady-state levels of SuSy transcripts in FLS seeds at 35 DAA were markedly reduced to undetectable levels compared with the levels seen at 15 DAA (Fig. 4A). The fls seeds, however, showed much greater abundance of SuSy transcripts at 35 than at 15 DAA. The reversed pattern of temporal expression between the two genotypes was also manifested in the soluble fraction of SuSy protein, as observed by western-blot analyses (Fig. 4C). The highest levels of SuSy polypeptides in the FLS seeds were seen at 15 DAA, followed by a gradual decline, and the least amounts were seen at 35 DAA. In contrast, the fls seeds showed a reverse temporal pattern for the SuSy protein, a gradual increase starting at 15 DAA, and the highest levels at 35 DAA.
Figure 4.
A reversed temporal expression of the SuSy gene in developing cotyledons of the fls mutant compared with that of developing FLS seeds. Numbers indicate DAA. A, RNA gel blot with 16 μg of total RNA in each lane. B, The same blot sequentially hybridized with a maize rRNA probe. C, Protein immunoblot with 50 μg of protein in each lane from crude extracts.
fls Mutant Seeds Display an Altered Mode of Carbon Partitioning
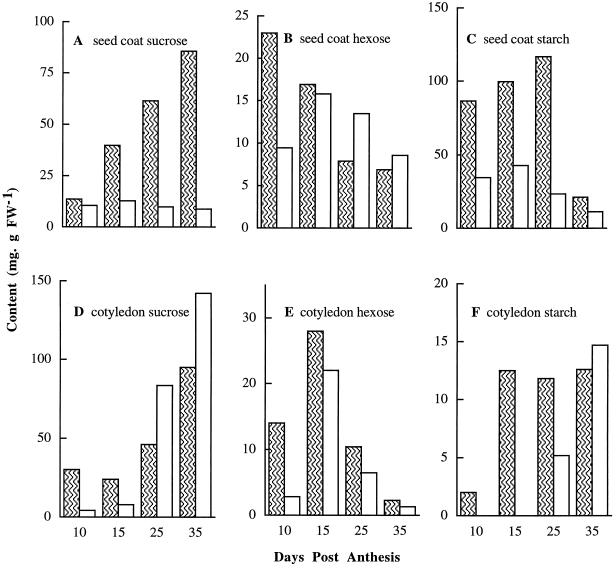
Carbohydrate partitioning, in particular the levels of soluble sugars and starch in developing seeds of the two genotypes, was analyzed to test for possible feedback regulation due to the loss of a major sink site for Suc utilization in the mutant seeds. Figure 5 depicts the results of such a comparative analysis of seed coats and cotyledons at 10 DAA (fiber elongation) through 35 DAA (fiber maturation). Suc levels in the fls seed coat remained relatively low throughout the entire development period, whereas the FLS seed-coat extracts showed a gradual but significant increase in Suc concentration. At 35 DAA, the FLS seed coats displayed levels of Suc that were 8 times higher than the fls genotype (Fig. 5A). The hexose levels in the fls seed coat were lower at 10 DAA, but thereafter, the two genotypes were similar (Fig. 5B). Starch levels in the fls seed coat were reduced to less than one-half that of the wild type during 10 to 25 DAA (Fig. 5C). However, the overall pattern of the developmental changes in starch between the fls and FLS seed coats was similar, with an increase during 10 to 25 DAA and a sharp decrease thereafter (Fig. 5C). These data for the normal genotype are in agreement with those of Hendrix (1990).
Figure 5.
Comparisons of Suc, hexose, and starch levels in developing cotton seed between FLS (hatched bars) and fls (open bars) plants. Each value is the mean of three replicates with se < 11%. FW, Fresh weight.
In the developing cotyledons Suc levels in the mutant were lower than that of the FLS up to 15 DAA but increased thereafter and surpassed the FLS at 25 and 35 DAA (Fig. 5 D). The hexose contents in the fls cotyledons were quite low at 10 to 25 DAA, but the overall pattern of changes in the two genotypes was similar for the later development stages (Fig. 5E). Cotyledonary starch levels in both genotypes at 35 DAA were the highest (Fig. 5F); however, during the early stages, 10 and 15 DAA, the FLS cotyledons showed levels of starch higher than that of the mutant.
DISCUSSION
One of the most important observations of this study is the cytological demonstration that, unlike the FLS genotype, seed epidermal cells of the fls mutant failed to exhibit detectable protrusions or bubbles at 0 DAA. Such changes in the epidermis must precede the elongation phase, leading ultimately to the fiber pathway. We infer from these data that the fls epidermal cells lack the competence to adapt to a specialized mode of growth and development and thus remain fiberless at maturity. To the best of our knowledge, this is the first such description of the fls mutation in cotton. Fiber development in cotton is partitioned into four developmental phases. The first two stages correspond to the initiation and elongation of the epidermal cell, and the last two represent secondary growth and maturation of the fiber marked by massive levels of cellulose biosynthesis.
Developmentally, the fiber growth is in parallel with seed development, which spans a period of about 45 DAA. The fiber cell initiation is analogous to several other systems of tip growth in a single cell, such as trichomes on a leaf or the development of root/shoot hairs. Insertional mutants showing impaired tip-growth functions in Arabidopsis have in general led to genes that encode transcription factors (for review, see Schiefelbein, 1998). The only known exception is the trichome mutation Zwichel (zwi) in Arabidopsis, in which the cloned ZWI gene is shown to encode a member of the kinesin-like family of microtubule proteins (Oppenheimer et al., 1997). In this regard, nothing is known concerning the molecular basis of the initial events that lead seed epidermal cells to the fiber cell pathway in cotton. In a previous study, Turley and Ferguson (1996) made a comparative analysis of the same fls mutant, SL-171, against an unrelated FLS inbred using two-dimensional PAGE analysis of the total ovular proteins. Of the numerous Coomassie blue-stained spots only five polypeptides are unique to the mutant. However, it is difficult to assign any physiological significance to these differences because these proteins are of an unknown nature.
Our studies at the cellular level have also shown undetectable levels of SuSy RNA and protein at 0 DAA in the fls epidermal cells. In contrast, the FLS cells showed abundant levels of SuSy, and there was much heterogeneity among the FLS cells. Those with the highest levels of SuSy appeared to differentiate into the longest cells, whereas cells with the lower or undetectable levels showed smaller or no initiation out of the epidermis (Figs. 3 and 4; Nolte et al., 1995). The enzyme SuSy catalyzes a reversible conversion of Suc and UDP to UDP-Glc and Fru and plays a major role in energy metabolism by mobilizing Suc into diverse pathways, including metabolic, structural, and storage functions of plant cells. Several lines of evidence from divergent plant sources (Chourey and Nelson, 1979; Geigenberger and Stitt, 1993; Heim et al., 1993) indicate that the enzyme preferentially catalyzes Suc degradation to Fru and UDP-Glc, the immediate precursor for cellulose biosynthesis in cotton fiber (Amor et al., 1995, and refs. therein). However, it is unclear why such high levels of SuSy expression were seen as early as 0 DAA, the stage primarily associated with the initiation phase of the fiber pathway in the FLS epidermal cells. It is significant that cell wall invertase was immunohistologically not detectable in serial sections of the same ovules at 0 DAA using either carrot or maize polyclonal antibodies (W. Cheng and P. Chourey, unpublished data). Thus, SuSy is a key enzyme in metabolizing incoming Suc, presumably both for energy and as a precursor for biosynthetic processes in the initiating fiber cells.
It is also possible that SuSy may contribute to turgor-related functions required for initiation and associated protrusion of the ovular epidermal cells. Such cellular changes are associated with rapid cell expansion driven by high cell turgor (Cosgrove, 1986). Previous in vitro studies have shown that the maintenance of turgor potential is essential for fiber cell expansion and elongation, which is achieved by increasing osmotically active solutes, mainly soluble sugars, K+, and malate (Basra and Malik, 1984). The most recent data of Smart et al. (1998) show high levels of expression of genes encoding plasma membrane H+-ATPase, vacuolar H+-ATPase, proton-translocating pyrophosphatase, PEP carboxylase, and major intrinsic protein, which are putatively engaged in the control of cell turgor in elongating cotton fibers. They also observed high levels of these proteins during the early stages that coincide with fiber initiation. Suc cleavage into Fru and UDP-Glc by SuSy would double the osmotic contribution of Suc. Thus, the enzyme could play a critical role in establishing and maintaining high turgor potential, driving the fiber cell precursor above the ovule epidermis. The high ratio of hexose to Suc in developing fibers (Ruan et al., 1997) concurs with this viewpoint. In the fls mutant the loss or the reduced levels of SuSy in the ovule epidermal cells may lead to low turgor and, consequently, no fiber growth. The role of SuSy in osmotic regulation and maintenance of cell turgor has also been indicated in guard cells of potato leaves in response to water stress (Kopka et al., 1997).
Remarkably, developing seeds of the two genotypes were quite different from each other during later developmental stages, 10 to 35 DAA. Alterations in the temporal patterns of SuSy expression, based on northern- and western-blot analyses, are of particular interest. Overall, the temporal pattern of SuSy expression in the fls mutant was reversed as compared with the normal genotype. In the FLS seeds the highest levels of SuSy RNA and protein were seen at 15 DAA and the lowest at 35 DAA; however, in the fls mutant the highest and the lowest levels were seen at 35 and 15 DAA, respectively. Thus, the mutant seeds have shown a delayed program of SuSy expression at both the RNA and protein levels, i.e. undetectable levels at 0 DAA, followed by a slow but gradual increase such that the highest levels of SuSy protein were seen at 35 DAA.
It should be noted that both genotypes were grown in the same greenhouse and that the samples were harvested at nearly the same time, thus minimizing possible contributions due to environmental factors. In addition, although the two genotypes were not lineage related, there were no obvious developmental differences (such as flowering and maturity) between them. Thus, it is unlikely that an altered temporal program of SuSy expression could be due to differences in either the environment or their inbred backgrounds. Previous reports of alterations of SuSy expression in maize (Chourey and Nelson, 1976; Chourey et al., 1988) and potato (Zrenner et al., 1995) show that the loss or the reduction of tissue-specific gene products are either due to a mutation in a gene that encodes SuSy or to the antisense inhibition. Results concerning fls seeds are unique because they indicate a regulatory type of change, as evidenced by an altered program of gene expression. Whether such a change in SuSy expression is a cause or a feedback effect of a mutation in certain upstream regulatory genes (such as those encoding the transcription factors) remains to be elucidated.
In developing fibers after initiation, although reducing sugar levels are high (Hendrix, 1990; Ruan et al., 1997), the predominant sugar nucleotide is UDP-Glc during both primary and secondary cell wall formation (Carpita and Delmer, 1981), and SuSy is the major enzyme in degrading Suc. Based on enzyme activity and immunohistological assays, invertase appears to play a minor role in developing fibers (Hendrix, 1990) or in ovular cells initiating to fibers (Y. Ruan and P. Chourey, unpublished data). Similarly, we have also observed only low to undetectable levels of cell wall invertase and Suc phosphate synthase RNAs in fibers at 15 DAA by northern-blot analyses using carrot and spinach cDNA clones, respectively (however, the same clone detected invertase RNA in sink leaves and seedlings, and the spinach clone detected Suc phosphate synthase RNA in source leaves; data not shown).
Seed weights at maturity for FLS (without the fibers) and fls genotypes were similar (data not shown); however, the two genotypes showed significant differences in both the relative distributions and the amounts of soluble sugars and starch in developing seeds during 10 to 35 DAA (Fig. 5). In developing cotton seeds, phloem-unloaded Suc is differentially partitioned into three major sink tissues: fiber, seed coat, and cotyledons (Ruan et al., 1997). Obviously, the loss of Suc utilization in fiber development in the fls mutant has led to several secondary alterations in carbon partitioning. Among these, the most striking changes were in the reduced levels of Suc and starch in the fls seed coat throughout the period of 10 to 35 DAA (Fig. 5, A and C). In addition, the mutant cotyledons had undetectable levels of starch during the very early (10–15 DAA) phases of development (Fig. 5F). A steady increase in the levels of Suc in the FLS seed coat would steepen the gradient of Suc concentration between seed coat and fibers, thus facilitating symplastic Suc transport to the growing fibers (Ryser, 1992; Ruan et al., 1997). In contrast, the absence of fibers and the loss of the associated major utilization sink in the mutant may cause a reduction in the levels of incoming Suc to the seed coat. Reduced levels of starch in the mutant seed coat and cotyledon, particularly during the early developmental phases, were most likely due to the reduced levels of SuSy protein, as has been previously demonstrated in SuSy-deficient mutants of maize and potato (Chourey and Nelson, 1976; Zrenner et al., 1995).
ACKNOWLEDGMENTS
We thank Drs. Jim Heitholt and Rickey Turley (USDA-ARS, Stoneville, MS) for many consultations, Deborah P. Delmer (University of California, Davis) and Arnd Sturm (Friedrich Miescher Institute, Basel, Switzerland) for cotton SuSy and carrot cell wall invertase cDNA clones and antibodies, Michael Salvucci for the spinach Suc phosphate synthase cDNA clone, and Earl W. Taliercio and Susan Carlson for critical reading of the manuscript. We also thank Dr. W.-H. Cheng for assistance with immunolocalization analyses for invertases.
Abbreviations:
- DAA
days after anthesis
- SuSy
Suc synthase
Footnotes
This work was supported in part by the U.S. Department of Agriculture (USDA)-Agricultural Research Service (ARS) and by the United States-Israel Binational Agricultural Research and Development Fund (grant no. IS-2282-93). It was a cooperative investigation between the USDA-ARS and the Institute of Food and Agricultural Sciences, University of Florida. This paper is Florida Agricultural Experiment Station journal series no. R-06228.
LITERATURE CITED
- Amor Y, Haigler CH, Johnson S, Wainscott M, Delmer DP. A membrane-associated form of SuSy and its potential role in synthesis of cellulose and callose in plants. Proc Natl Acad Sci USA. 1995;92:9353–9357. doi: 10.1073/pnas.92.20.9353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R and others. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- Basra A, Malik CP. Development of the cotton fiber. Int Rev Cytol. 1984;89:65–113. [Google Scholar]
- Carpita NC, Delmer DP. Concentration and metabolic turn-over of UDP-glucose in developing cotton fibers. J Biol Chem. 1981;256:308–315. [PubMed] [Google Scholar]
- Chourey PS, DeRobertis GA, Still PE. Altered tissue specificity of the revertant shrunken allele upon dissociation (Ds) excision is associated with loss of expression and molecular rearrangement at the corresponding nonallelic isozyme locus in maize. Mol Gen Genet. 1988;214:300–306. [Google Scholar]
- Chourey PS, Nelson OE. The enzymatic deficiency conditioned by the shrunken 1 mutation in maize. Biochem Genet. 1976;14:1041–1055. doi: 10.1007/BF00485135. [DOI] [PubMed] [Google Scholar]
- Chourey PS, Nelson OE. Interallelic complementation at the shrunken locus of maize at the enzyme level. Genetics. 1979;91:317–325. doi: 10.1093/genetics/91.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D. Biophysical control of plant cell growth. Annu Rev Plant Physiol. 1986;37:377–405. doi: 10.1146/annurev.pp.37.060186.002113. [DOI] [PubMed] [Google Scholar]
- Cox KH, Goldberg RB (1988) Analysis of gene expression. In CH Shaw, ed, Plant Molecular Biology: A Practical Approach. Oxford IRL Press, Oxford, UK, pp 1–35
- Delmer DP, Amor Y. Cellulose biosynthesis. Plant Cell. 1995;7:987–1000. doi: 10.1105/tpc.7.7.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M. Sucrose synthase catalyses a readily reversible reaction in vivo in developing potato tubers and other plant tissues. Planta. 1993;189:329–393. doi: 10.1007/BF00194429. [DOI] [PubMed] [Google Scholar]
- Heim U, Weber H, Baumlein H, Wobus U. A sucrose synthase gene of Vicia faba L.: expression pattern in developing seeds in relation to starch synthesis and metabolic regulation. Planta. 1993;191:394–401. doi: 10.1007/BF00195698. [DOI] [PubMed] [Google Scholar]
- Hendrix DL. Carbohydrate and carbohydrate enzymes in developing cotton ovules. Physiol Plant. 1990;78:85–92. [Google Scholar]
- Kopka J, Provart NJ, Müller-Röber B. Potato guard cells respond to drying soil by a complex change in the expression of genes related to carbon metabolism and turgor regulation. Plant J. 1997;11:871–882. doi: 10.1046/j.1365-313x.1997.11040871.x. [DOI] [PubMed] [Google Scholar]
- Marrison JL, Leech RM. The subcellular and intra-organelle recognition of nuclear and chloroplast transcripts in developing leaf cells. Plant J. 1994;6:605–614. [Google Scholar]
- Nolte KD, Hendrix DL, Radin JW, Koch KE. Sucrose synthase localization during initiation of seed development and trichome differentiation in cotton ovules. Plant Physiol. 1995;109:1285–1293. doi: 10.1104/pp.109.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer DG, Pollock MA, Vacik J, Szymanski DB, Erickson B, Feldman K, Marks DM. Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc Natl Acad Sci USA. 1997;94:6261–6266. doi: 10.1073/pnas.94.12.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey JC, Berlin JD. Ultrastructure of early stages of cotton fiber differentiation. Bot Gaz. 1976;137:11–19. [Google Scholar]
- Ruan YL, Chourey PS, Delmer DP, Perez-Grau L. The differential expression of sucrose synthase in relation to diverse patterns of carbon partitioning in developing cotton seed. Plant Physiol. 1997;115:375–385. doi: 10.1104/pp.115.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser U. Ultrastructure of the epidermis of developing cotton (Gossypium) seeds: suberin, pits, plasmodesmata, and their implication for assimilate transport into cotton fibers. Am J Bot. 1992;79:14–22. [Google Scholar]
- Schiefelbein J. Cell fate specificities in the root epidermis. Trends Plant Sci. 1998;3:3–4. [Google Scholar]
- Smart LB, Vojdani F, Maeshima M, Wilkins TA. Genes involved in osmoregulation during turgor-driven cell expansion of developing cotton fibers are differentially regulated. Plant Physiol. 1998;116:1539–1549. doi: 10.1104/pp.116.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SC, Wilkins TA. Cotton (Gossypium hirsutum) seed trichomes expand via diffuse growing mechanism. Can J Bot. 1995;73:746–757. [Google Scholar]
- Trelease RN, Miernyk JA, Choinski JS Jr, Bortman SJ (1986) Synthesis and compartmentalization of enzymes during cotton seed maturation. In JR Mauney, JMD Steward, eds, Cotton Physiology. The Cotton Foundation, Memphis, TN, pp 441–462
- Turley RB, Ferguson DL. Changes of ovule proteins during early fiber development in a normal and fiberless line of cotton (Gossypium hirsutum L.) J Plant Physiol. 1996;149:695–702. [Google Scholar]
- Wadsworth GJ, Redinbaugh MG, Scandlios JG. A procedure for the small-scale isolation of plant RNA suitable for RNA blot analysis. Anal Biochem. 1988;172:279–283. doi: 10.1016/0003-2697(88)90443-5. [DOI] [PubMed] [Google Scholar]
- Zrenner R, Salanoubat M, Willimitzer L, Sonnewald U. Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.) Plant J. 1995;7:97–107. doi: 10.1046/j.1365-313x.1995.07010097.x. [DOI] [PubMed] [Google Scholar]