Abstract
Rice (Oryza sativa) is the most aluminum (Al)-tolerant crop among small-grain cereals, but the mechanism underlying its high Al resistance is still not well understood. To understand the mechanisms underlying high Al-tolerance, we performed a comparative genome-wide transcriptional analysis by comparing expression profiling between the Al-tolerance cultivar (Koshihikari) and an Al-sensitive mutant star1 (SENSITIVE TO AL RHIZOTOXICITY 1) in both the root tips and the basal roots. Exposure to 20 µM AlCl3 for 6 h resulted in up-regulation (higher than 3-fold) of 213 and 2015 genes including 185 common genes in the root tips of wild-type and the mutant, respectively. On the other hand, in the basal root, genes up-regulated by Al were 126 and 2419 including 76 common genes in the wild-type and the mutant, respectively. These results indicate that Al-response genes are not only restricted to the root tips, but also in the basal root region. Analysis with genes up- or down-regulated only in the wild-type reveals that there are other mechanisms for Al-tolerance except for a known transcription factor ART1-regulated one in rice. These mechanisms are related to nitrogen assimilation, secondary metabolite synthesis, cell-wall synthesis and ethylene synthesis. Although the exact roles of these putative tolerance genes remain to be examined, our data provide a platform for further work on Al-tolerance in rice.
Introduction
Aluminum (Al) toxicity is a major factor limiting crop production on acid soils, which comprise approximately 40% of the world’s arable soils and up to 70% of potentially arable land [1]. At soil pH below 5.0, toxic forms of Al (mainly Al3+) are solubilized into the soil solution, which inhibit root growth and function, consequently reducing crop yields [2], [3]. However, there is a great variation for the ability to withstand Al-toxicity between plant species and cultivars within a species. To survive on acidic soils, some plant species or cultivars have evolved mechanisms to tolerate high levels of toxic Al. Many mechanisms for both Al-tolerance and -toxicity have been proposed [3].
Rice (Oryza sativa) is the most Al-tolerant crop among small-grain cereals [4]. A number of quantitative trait loci (QTLs) for Al-tolerance have been identified in rice by using different populations [5], but responsible QTL genes have not been isolated. Recently, through genome-wide association analysis and QTL mapping, 48 loci associated with Al3+ tolerance have been identified [6] in rice. On the other hand, mutant approaches have revealed an ART1-regualted Al-tolerance mechanism in rice [7]. ART1 (AL3+ RESISTANCE TRANSCRIPTION FACTOR 1) is a Cys2His2-type zinc-finger transcription factor [8]. ART1 is constitutively expressed in the roots and its expression is not induced by Al3+ treatment. ART1 regulates the expression of at least 31 genes with a cis-element [GGN(T/g/a/C)V(C/A/g)S(C/G)] (ART1-binding affinity of nucleotides with small characters is weaker than those with large characters) [9]. Among them, only six genes have been functionally characterized. OsSTAR1 and OsSTAR2 (SENSITIVE TO ALUMINUM RHIZOTOXICITY 1 & 2) encode a ATP-binding domain and a transmembrane domain, respectively, of a bacterial-type ATP binding cassette (ABC) transporter, which transports UDP-glucose [10]. The complex is implicated in cell wall modification [10]. OsFRDL4 (FERRIC REDUCTASE DEFECTIVE3-LIKE 4) encodes a citrate transporter, which secretes citrate from the roots to chelate Al in the rhizosphere [11]. On the other hand, OsNrat1 (NRAMP ALUMINUM TRANSPORTER 1) encodes an Al transporter localized at the plasma membrane, which transports Al into the cells [12], while OsALS1 (ALUMINUM SENSITIVE 1) encodes a tonoplast-localized transporter for Al, which sequestrates Al into the vacuoles [13]. Recently, up-regulation of a Mg transporter, OsMGT1 (MAGNESIUM TRANSPORTER 1), is reported to be required for conferring Al-tolerance in rice [14]. All of these genes are specifically induced by Al and knockout of either gene results in decreased Al-tolerance, indicating their important roles in Al-tolerance. However, the mechanisms underlying high Al-tolerance in rice are not fully understood at the molecular level.
In the present study, we performed a genome-wide transcriptional analysis of Al-responsive genes in rice. By comparing transcriptional profiling between a wild-type rice and an Al-sensitive rice mutant star1, we found that rice possesses novel mechanisms of Al-tolerance in addition to ART1-regulated mechanism in rice.
Materials and Methods
Plant Materials and Growth Conditions
Seeds of wild-type rice (Oryza sativa cv. Koshihikari) and an Al-sensitive mutant, star1 [10], were germinated for 2 days at 30°C. The seedlings were then transferred to a plastic net floating on a 0.5 mM CaCl2 solution in a 1.5 L plastic box. At day 4, the seedlings were exposed to a 0.5 mM CaCl2 solution (pH 4.5) containing 0 or 20 µM AlCl3. Root length was measured with a ruler before and after 6 h treatments. Ten seedlings were used for each treatment.
RNA Isolation, Microarray and Data Analysis
Root tips (0–1 cm) and basal region (1–2 cm) of the roots (20–30 plants per sample) were excised from the seedlings of both wild-type rice and star1 mutant which had been exposed to 20 µM AlCl3 for 6 h and immediately frozen in liquid nitrogen. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, Germany). The RNA quality was assessed on agarose gels and with the Nanodrop ND-1000 (Thermo Fisher Scientific, USA). Microarray analysis was performed according to Agilent Oligo DNA Microarray Hybridization protocols using the Agilent 44 K Rice Oligo DNA Microarray RAP-DB (Agilent Technologies, USA; G2519F#15241) [15] with three biological replicates (Agilent Technologies, USA; G2519F#15241) [15]. The hybridized slides were scanned using a DNA microarray scanner (Agilent Technologies, USA). Signal intensities were extracted by Feature Extraction software (Agilent Technologies, USA). For statistical analysis, we excluded genes we excluded genes with low signal intensities less than 500 (sum of +Al and –Al signal intensity) in all treatments of the wild-type and star1 mutant. This is based on expression level of known Al-tolerance genes (10–14). The average value (arithmetic mean) of fold change (the ratios of Cy3 and Cy5) and standard deviation (SD) of each probe were calculated using three biological replicates. Since the expression of known Al-tolerance genes is usually up-regulated by higher than three folds (10–14), we extracted genes up-regulated or down-regulated by Al more/less than three-fold in the wild-type and star1 mutant. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE40964 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40964).
The gene functions were categorized based on databases including National Center of Biotechnology Information (NCBI) [16], the Rice Annotation Project Database (RAP-DB) build 5.0 [15] by the International Rice Genome Sequencing Project (IRGSP) [17], and the MSU Rice Genome Annotation Database [18]. The majority of Al-responsive transcripts were assigned to one of the following 12 categories by checking one by one using excel (Microsoft); (1) transport; (2) metabolism; (3) protein synthesis and processing; (4) signal transduction; (5) translation initiation or transcription factors; (6) abiotic or biotic stress response; (7) cell-wall, cell cycle, cell growth and cell cytoskeleton modification or metabolism; (8) DNA/RNA binding or metabolism; (9) phytohormone metabolism and response; (10) mitochondria or plastid; (11) other; (12) unknown molecular function.
Quantitative Real-time PCR
To validate microarray data, 12 genes were randomly selected for quantitative real-time PCR (qRT-PCR) (Table S1). Total RNA was prepared from the root tips and basal root regions of wild-type and star1 using RNeasy Plant Mini Kit (Qiagen, Germany) and reversely transcribed using SuperSript™ II Reverse Transcriptase (Invitrogen, USA) and Oligo(dT) primers. The qRT-PCR was performed on an Eppendorf MasterCycler ep realplex real-time PCR (Eppendorf, Germany) using the specific primers described in Table S1.
One-fifth dilutions of the cDNAs were used as a template for the qRT-PCR in a total volume of 20 µL as follows; 10 µL SYBR Premix Ex Taq™ Perfect Real Time (TaKaRa Biol Inc., Japan), 0.4 µL ROX Reference Dye, 0.8 µL primer mix (50∶50 mix of forward and reverse primers at 10 pmol µL−1 each), 6.8 µL distilled water and 2 µL template. The reaction conditions were: 30 s at 95°C followed by 40 cycles of 30 s at 95°C, 20 s at 60°C and 35 s at 72°C. The rice Histone H3 was used as an internal control. Relative expression levels were calculated by the comparative Ct method. Three independent biological replicates were made for each gene.
Results and Discussion
Tolerance and toxicity of Al stress are a complicated phenomenon, involving many genes and a number of signaling pathways [19]. However, microarray technique has provided a useful tool for investigation of genome-wide changes in transcripts. So far, microarray analysis for Al response has been reported in Arabidopsis [20]–[22], maize [23], [24], Medicago truncatula [25], [26], and wheat [27]. Since the mechanisms for Al-tolerance differ with plant species, in the present study, we performed a microarray analysis with rice, a well-known Al-tolerant species, to understand genes involved in high Al-tolerance at genome-wide scale.
Al-toxicity is characterized by inhibition of root elongation, which occurs within a few hours after exposure to Al [3]. Therefore, to exclude genes associated with Al-toxicity, we sampled the roots exposed to Al solution for 6 h for microarray analysis. Furthermore, to extract genes related to Al-tolerance, we compared the transcriptional profiling between the wild-type rice and an Al-sensitive mutant, star1 [10]. Moreover, we selected a concentration of 20 µM for Al treatment. At this concentration, the root elongation of the wild-type rice was hardly inhibited, whereas that of the mutant was inhibited by 75% (Figure 1), which make possible to extract genes possibly associated with Al-tolerance.
Figure 1. Al-induced inhibition of root elongation.
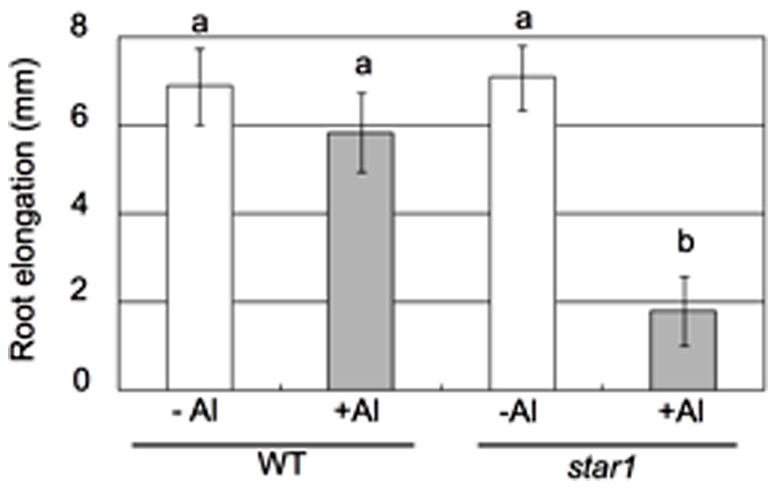
Seedlings (6-d-old) of both wild-type rice (WT) and an Al-sensitive mutant (star1) were exposed to a 0.5 mM CaCl2 solution (pH 4.5) containing 0, 20 µM Al for 6 h. The root length was measured with a ruler before and after Al treatment. Error bars represent ± SD (n = 10). Different letters indicate significant differences at P<0.05 by Tukey’s Honestly Significantly Different test.
Verification of Microarray Results by Quantitative Real-time PCR
To validate the reliability of the microarray data, we randomly selected 12 genes from root tips and basal root regions for the quantitative real-time PCR (qRT-PCR) analysis. There was a good correlation (r = 0.84) between the microarray data and the qRT-PCR results (Figure 2). These results indicated that the microarray data could reflect the transcriptional changes caused by Al stress.
Figure 2. Correlation of gene expression ratio between microarray data and quantatitive RT-PCR data.
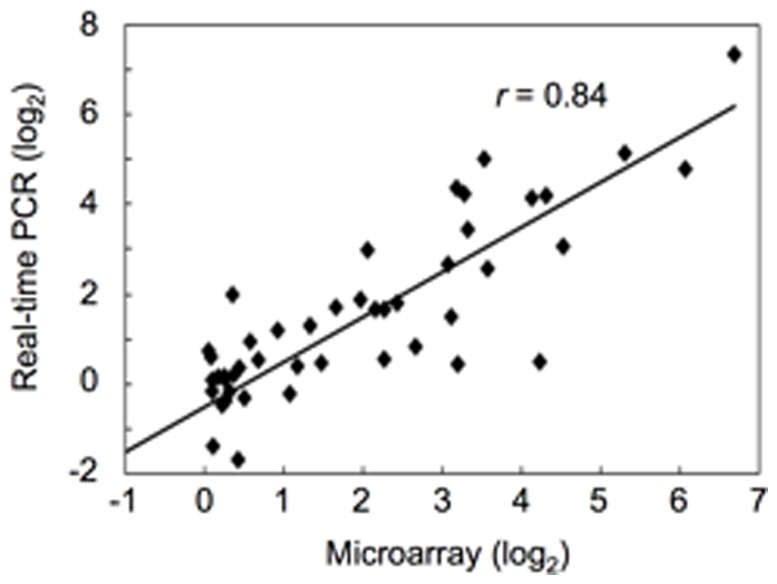
Twelve genes randomly selected were subjected to quantitative real-time PCR analysis. OsHistone H3 was used as an internal standard. Microarray data (fold change of gene expression) were plotted against data (fold change of gene expression) from quantitative real-time PCR. Both x- and y-axes are shown in log2 scale. r indicates correlation coefficient.
Overview of Al-induced Transcriptional Profiling
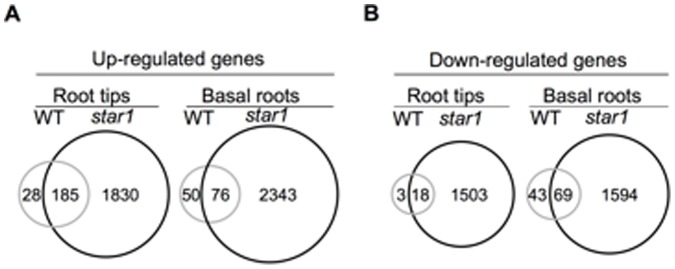
Agilent 44 K Rice Oligo DNA Microarray RAP-DB covers almost genes in rice genome [15]. In the root tips (0–1 cm) of wild-type rice, exposure to 20 µM AlCl3 for 6 h resulted in up-regulation of 213 genes and down-regulation of 21 genes (Figure 3A and 3B). By contrast, much more genes (2015 and 1521) were up- and down-regulated by the same treatment in the star1 mutant (Figure 3A and 3B). In the basal root region (1–2 cm), 126 and 112 genes, respectively, was up- and down-regulated in the wild-type rice, whereas the numbers of up- and down-regulated genes were 2419 and 1663, respectively, in the star1 mutant (Figure 3A and 3B).
Figure 3. Genes up- and down-regulated by Al in the wild-type and star1 mutant roots.
Numbers of Al-responsive genes up-regulated (higher than 3-fold) (A) and down-regulated (lower than 3-fold) (B) are extracted. Wild-type (gray circle) and star1 mutant (black circle) are shown in both the root tips and basal root region.
Functional category analysis showed that 27.0–42.8% of the up- and down-regulated genes are assigned to unknown function (Table 1). Genes related to ‘Metabolism’ and ‘Abiotic or biotic stress response’ were mostly affected by Al stress in both the wild-type rice and star1 roots (Table 1).
Table 1. Functional classification of Al-responsive genes in the roots of the wild-type rice and star1 mutant.
| Up-regulateda | Down-regulatedb | |||||||
| Root tips | Basal roots | Root tips | Basal roots | |||||
| WT (%) | star1 (%) | WT (%) | star1 (%) | WT (%) | star1 (%) | WT (%) | star1 (%) | |
| Transport | 19 (8.8) | 125 (6.2) | 10 (7.9) | 113 (4.7) | 1 (4.8) | 102 (6.7) | 4 (3.6) | 135 (8.1) |
| Metabolism | 31 (14.4) | 251 (12.5) | 18 (14.3) | 340 (14.1) | 4 (19.0) | 119 (7.8) | 19 (17.0) | 204 (12.3) |
| Protein synthesis and processing | 14 (6.5) | 129 (6.4) | 7 (5.6) | 125 (5.2) | 1 (4.8) | 86 (5.7) | 6 (5.4) | 89 (5.4) |
| Signal transduction | 6 (2.8) | 147 (7.3) | 2 (1.6) | 140 (5.8) | 0 (0) | 97 (6.4) | 9 (8.0) | 98 (5.9) |
| Translation initiation or transcription factors | 8 (3.7) | 113 (5.6) | 3 (2.4) | 182 (7.5) | 3 (14.3) | 114 (7.5) | 5 (4.5) | 123 (7.4) |
| Abiotic or biotic stress response | 39 (18.1) | 276 (13.7) | 13 (10.3) | 269 (11.1) | 3 (14.3) | 126 (8.3) | 25 (22.3) | 133 (8.0) |
| Cell-wall, cell cycle, cell growthand cell cytoskeleton modificationor metabolism | 14 (6.5) | 67 (3.3) | 13 (10.3) | 133 (5.5) | 1 (4.8) | 137 (9.0) | 9 (8.0) | 94 (5.7) |
| DNA/RNA binding or metabolism | 1 (0.5) | 26 (1.3) | 1 (0.8) | 60 (2.5) | 0 (0) | 77 (5.1) | 0 (0) | 37 (2.2) |
| Phytohormone metabolism and response | 2 (0.9) | 21 (1.0) | 3 (2.4) | 26 (1.1) | 0 (0) | 22 (1.4) | 1 (0.9) | 17 (1.0) |
| Mitochondria or plastid | 3 (1.4) | 26 (1.3) | 1 (0.8) | 27 (1.1) | 1 (4.8) | 12 (0.8) | 2 (1.8) | 8 (0.5) |
| Other | 0 (0.0) | 19 (0.9) | 2 (1.6) | 31 (1.3) | 0 (0) | 12 (0.8) | 1 (0.9) | 14 (0.8) |
| Unknown molecular function protein | 76 (35.2) | 815 (40.4) | 53 (42.1) | 973 (40.2) | 7 (33.3) | 618 (40.6) | 31(27.7) | 711 (42.8) |
| Total | 213 | 2015 | 126 | 2419 | 21 | 1521 | 112 | 1663 |
Genes which expression was changed higher than 3-fold (fluorescence signal more than 500) in the root tips and the basal roots were categorized.
Genes which expression was changed lower than 3-fold (fluorescence signal more than 500) in the root tips and the basal roots were categorized.
Since the root elongation was hardly inhibited in the wild-type rice, but severely inhibited in the star1 mutant, three different groups for Al-responsive genes could be divided by comparing expression profiling between wild-type rice and star1 mutant. Group 1 includes genes which are up- or down-regulated by Al only in the wild-type rice. These genes are probably involved in Al-tolerance. Twenty eight up-regulated and three down-regulated genes in the root tips, 50 up-regulated and 43 down-regulated genes in the basal root region, belong to this group (Figure 3A and 3B). Group 2 includes genes, which are up- or down-regulated by Al in both the wild-type rice and star1 mutant. These genes are probably involved in Al-tolerance or -toxicity. There are 185 up-regulated and 18 down-regulated genes in this group in the root tip, 76 up-regulated and 69 down-regulated genes in the basal root region (Figure 3A and 3B, Table S2–S5). Genes in Group 3 are those up- or down-regulated only in the mutant. These genes are related to Al-toxicity and included 1830 up-regulated genes and 1503 down-regulated genes in the root tip, 2343 up-regulated and 1594 down-regulated genes in the basal root region (Figure 3A and 3B). Most genes in this group are also response to general stresses and found in microarray data of other plant species such as Arabidopsis [20], maize [23], [24] M. truncatula [25], [26] and wheat [27]. For example, the genes encoding a NADPH oxidase, peroxidase, oxalate oxidase, which are reactive oxygen species (ROS; O2 −, H2O2) generators, were up-regulated (Table S2). Most types of abiotic stresses disrupt the metabolic balance of cells, resulting in enhanced production of ROS [28]. The accumulation of ROS such as 1O2, O2 −, H2O2 and HO•, during abiotic stresses was considered to be a by-product of stress metabolism as well as an overall unwelcome by-product of aerobic metabolism [29]. These findings indicate that these genes are involved in arrest of plant root elongation in response to general stress.
Spatial Profiling of Al-responsive Genes
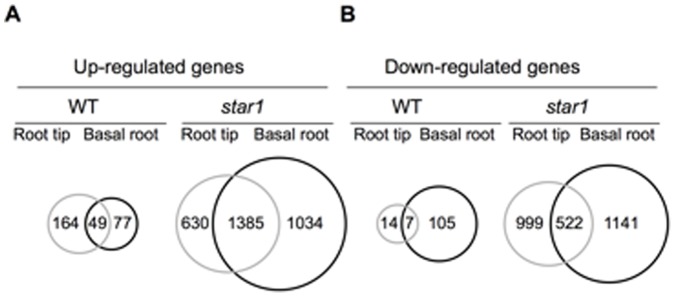
Root tip has been considered as the target of Al-toxicity [3] based on root elongation inhibition, however, surprisingly, similar numbers of genes were up- and down-regulated by Al in the root tips and mature regions of both wild-type rice and star1 mutant (Figure 4A and 4B). This result raises a question on whether the root tip is only the target of Al-toxicity. Among genes affected, 49 up-regulated and 7 down-regulated genes were the same between root tip and basal root region in the wild-type rice (Figure 4A and 4B), but most Al-responsive genes were different between the root tip and basal root region. This was the same in the star1 mutant; 1385 up-regulated and 522 down-regulated genes were the same between the root tip and basal root region, whereas other genes showed root region-dependent (Figure 4A and 4B). These results suggest that basal root region is also a target of Al-toxicity in addition to the root tip. In M. truncatula, Al-induced gene expression is also found not to be restricted to the root tip [25]. In fact, some genes identified from rice were expressed in both the root tips and basal root region. For example, OsFRDL4 was expressed in both the root tip and the mature root zones [11]. The expression of OsSTAR1 and OsSTAR2 was also induced in both regions [10]. These findings suggest that the basal root region is also involved in Al-tolerance and -toxicity.
Figure 4. Genes up- and down-regulated by Al in the root tips and basal roots of the wild-type and star1 mutant.
Number of Al-responsive genes up-regulated (higher than 3-fold) (A) and down-regulated (lower than 3-fold) (B) are extracted. Root tips (gray circle) and basal root regions (black circle) was shown in the wild-type and star1 mutant.
Transcriptional Profiling of ART1-regulated Genes in the Wild-type and the star1 Mutant Roots
ART1-regulated Al-tolerance has been identified as a major mechanism responsible for high Al-tolerance in rice [7], [8]. We compared expression profiling of ART1-reguated downstream genes between wild-type rice and star1 mutant. Among 31 downstream genes, 11 genes showed higher fold changes in the expression in the mutant than in the wild-type rice (Table 2), whereas 13 genes showed higher fold changes in the wild-type rice than in the mutant (Table 2). Seven genes showed similar fold changes in the expression between wild-type rice and mutant (Table 2). Six genes were only up-regulated in the wild-type rice, including genes encoding Expansin (Os04g0583500), Mg2+ transporter/OsMGT1 (Os01g0869200), OsNramp4/OsNrat1 (Os02g0131800), LrgB-like protein family protein (Os10g0578800), Allyl alcohol dehydrogenase (Os12g0227400) and uncharacterized plant-specific domain 01627 containing protein (Os11g0490100, Table 2). Among them, OsNrat1 (Al3+ transporter) and OsMGT1 (Mg2+ transporter) have been demonstrated to be involved in Al-tolerance [12], [14]. Although several ART1-regulated genes were also up-regulated in the mutant, the Al-tolerance was severally decreased, indicating that not a single gene, but multiple genes are required to function together for high Al-tolerance in rice.
Table 2. Expression changes of ART1-regulated genes in the roots of wild-type rice and star1 mutant.
| Root tips | Basal roots | |||||||||
| WT | star1 | WT | star1 | |||||||
| RAP IDa | Accessionb | Annotationc | Fold change(+Al/−Al)d | ±SDe | Fold change(+Al/−Al) | ±SD | Fold change(+Al/−Al) | ±SD | Fold change(+Al/−Al) | ±SD |
| Cell wall maintenance and Root elongation | ||||||||||
| Os01g0178300 | AK062450 | OsCDT3 | 7.43 | 1.58 | 24.67 | 3.28 | 11.72 | 3.85 | 9.82 | 2.66 |
| Os01g0652100 | AK069291 | Protein of unknown function DUF231 domaincontaining protein | 3.31 | 0.40 | 2.55 | 0.41 | 3.97 | 0.61 | 1.58 | 0.83 |
| Os01g0860500 | AK069860 | Chitinase | 10.32 | 4.51 | 12.09 | 2.34 | 3.67 | 0.84 | 7.41 | 4.20 |
| Os03g0760800 | AK121316 | Gibberellin regulated protein family protein | 4.91 | 1.29 | 21.94 | 6.13 | 7.54 | 2.07 | 9.13 | 5.14 |
| Os04g0583500 | AK062225 | Expansin 4 | 5.28 | 1.23 | 1.36 | 0.42 | 1.01 | 0.19 | 1.47 | 0.84 |
| Os09g0479900 | CI269495 | Peptidase S8 and S53, subtilisin, kexin, sedolisin domaincontaining protein | 3.43 | 0.25 | 6.40 | 0.78 | 1.40 | 0.19 | 9.83 | 5.61 |
| Os10g0524600 | AK069238 | Peptidase S8 and S53, subtilisin, kexin, sedolisin domaincontaining protein | 2.26 | 0.15 | 3.46 | 0.76 | 10.59 | 2.88 | 25.71 | 11.89 |
| Membrane protein | ||||||||||
| Os01g0869200 | AK073453 | Mg2+ transporter/OsMGT1 | 4.43 | 0.77 | 2.12 | 0.42 | 3.24 | 0.59 | 1.78 | 0.48 |
| Os02g0131800 | AK102180 | OsNramp4/OsNrat1 | 7.85 | 0.31 | 0.74 | 0.08 | 5.53 | 0.18 | 0.27 | 0.08 |
| Os02g0755900 | AK104985 | UDP-glucuronosyl/UDP-glucosyltransferase family protein | 5.91 | 0.28 | 23.35 | 1.40 | 1.21 | 0.29 | 44.24 | 6.83 |
| Os03g0755100 | AK066049 | Tonoplast-localized half-size ATP binding cassette (ABC)transporter/OsALS1 | 3.43 | 0.15 | 4.21 | 0.18 | 2.57 | 0.09 | 2.63 | 0.74 |
| Os05g0119000 | AK069359 | Bacterial-type ATP binding cassette (ABC) transporter/OsSTAR2 | 6.75 | 1.86 | 3.62 | 0.85 | 3.68 | 0.90 | 1.63 | 0.47 |
| Os06g0695800 | AK109450 | Bacterial-type ATP binding cassette (ABC) transporter/OsSTAR1 | 4.39 | 0.18 | 4.60 | 0.33 | 3.88 | 0.19 | 2.41 | 0.15 |
| Os09g0426800 | AK060786 | Gl1 protein | 1.97 | 0.38 | 4.67 | 0.53 | 5.34 | 0.71 | 0.99 | 0.22 |
| Os10g0206800 | AK072077 | Multidrug and toxic compound extrusion (MATE) family protein/OsFRDL2 | 5.76 | 0.21 | 3.44 | 0.30 | 5.89 | 0.44 | 1.58 | 0.29 |
| Os10g0578800 | AK065615 | LrgB-like protein family protein | 7.89 | 0.49 | 2.36 | 0.15 | 5.22 | 0.24 | 0.74 | 0.47 |
| Metabolism and Detxification | ||||||||||
| Os01g0716500 | AK101454 | SAM (and some other nucleotide) binding motif domaincontaining protein | 1.90 | 0.10 | 42.65 | 4.84 | 1.99 | 0.30 | 13.98 | 12.53 |
| Os02g0186800 | NM_001052658 | Cytochrome P450 family protein | 12.48 | 4.64 | 5.23 | 0.59 | 5.80 | 0.90 | 1.99 | 0.22 |
| Os02g0770800 | AK102178 | Nitrate reductase | 8.63 | 1.34 | 10.99 | 0.45 | 30.19 | 4.29 | 59.40 | 7.64 |
| Os12g0227400 | CI560939 | Allyl alcohol dehydrogenase | 16.06 | 0.78 | 2.64 | 0.20 | 8.94 | 0.15 | 0.48 | 0.23 |
| Unknown | ||||||||||
| Os01g0731600 | NM_001050684 | Conserved hypothetical protein | 18.18 | 4.39 | 23.14 | 5.08 | 1.68 | 0.31 | 14.15 | 13.58 |
| Os01g0766300 | NM_001050890 | Conserved hypothetical protein | 6.23 | 1.82 | 30.97 | 3.95 | 5.94 | 0.63 | 12.56 | 3.48 |
| Os01g0919200 | AK071325 | Cell division protein FtsZ family protein | 4.24 | 0.80 | 11.58 | 2.03 | 2.24 | 0.51 | 18.69 | 16.31 |
| Os03g0126900 | AK109217 | Conserved hypothetical protein | 7.32 | 0.67 | 4.11 | 0.24 | 7.84 | 0.47 | 2.11 | 0.08 |
| Os03g0304100 | AK111121 | Hypothetical protein | 10.73 | 4.09 | 34.63 | 11.22 | 4.04 | 0.49 | 0.78 | 0.57 |
| Os04g0419100 | AK107777 | Hypothetical protein | 16.41 | 0.44 | 5.38 | 0.39 | 1.09 | 0.23 | 7.96 | 5.25 |
| Os04g0494900 | AK073892 | Protein of unknown function DUF642 family protein | 15.05 | 0.86 | 3.64 | 0.74 | 2.98 | 0.16 | 99.56 | 31.86 |
| Os07g0493100 | AK068708 | Non-protein coding transcript, uncharacterized transcript | 26.08 | 11.10 | 7.18 | 0.87 | 13.41 | 4.30 | 6.97 | 2.12 |
| Os07g0587300 | CI285201 | Hypothetical protein | 6.62 | 1.76 | 208.44 | 71.70 | 6.36 | 1.10 | 104.49 | 46.28 |
| Os11g0488100 | CI197875 | Hypothetical protein | 4.50 | 0.16 | 7.00 | 0.56 | 2.02 | 0.12 | 1.96 | 0.13 |
| Os11g0490100 | AK108872 | Uncharacterized plant-specific domain 01627 containing protein | 5.04 | 0.72 | 1.76 | 0.25 | 9.75 | 1.10 | 21.53 | 10.50 |
RAP-ID based The Rice Annotation Project (RAP) ID numbers.
Accsesion based GenBank locus of the National Center of Biotechnology Information (NCBI).
Annotation based on the Rice Annotation Project Database (RAP-DB) build 3.0 by the International Rice Genome Sequencing Project (IRGSP).
Fold change, ratio of transcript abundance in Al treatement/transcript abundance in control (−Al) treatment.
Standard deviation of the mean.
Novel Al-tolerance Mechanism in Rice
Among 28 genes only up-regulated in the root tips of the wild-type (Table 2, Table 3), 6 genes are ART1-regulated, indicating that there are other mechanisms for Al-tolerance except ART1-regulated pathway in rice. Seven genes out of 22 genes belong to unknown function group (Table 3), while other genes are related to transporter (nitrate transporter, iron-regulated transporter), metabolism (nitrate reductase), oxidative stress-responsive genes (germin-like protein), polysaccharide/cell wall metabolism (cell wall invertase, beta-1,3-glucanase precursor) and so on.
Table 3. Genes up- and down-regulated only in the root tips of wild-type rice.
| Functional classificationfa/RAP IDb | Accessionc | Annotationd | Fold change (+Al/−Al)e | ±SDf |
| Up-regulated | ||||
| ART1-regulated genes | ||||
| Os12g0227400 | CI560939 | Allyl alcohol dehydrogenase | 16.06 | 0.78 |
| Os10g0578800 | AK065615 | LrgB-like protein family protein | 7.89 | 0.49 |
| Os02g0131800 | AK102180 | OsNramp4/OsNrat1 | 7.85 | 0.31 |
| Os11g0490100 | AK108872 | Uncharacterized plant-specific domain 01627 containingprotein | 5.04 | 0.72 |
| Os04g0583500 | AF247165 | Expansin 4 | 5.00 | 1.49 |
| Os01g0869200 | AK073453 | Mg2+ transporter/OsMGT1 | 4.43 | 0.77 |
| Other genes | ||||
| Transpot | ||||
| Os05g0410900 | AK119621 | Nitrate transporter/OsNRT1 | 6.91 | 0.93 |
| Os03g0667500 | AY327039 | Iron-regulated transporter 2/OsIRT2 | 3.16 | 0.28 |
| Metabolism | ||||
| Os08g0468100 | AK101662 | Nitrate reductase [NADH] 1/OsNR | 3.02 | 0.37 |
| Protein synthesis and processing | ||||
| Os05g0360400 | AK106046 | Zn-finger, RING domain containing protein | 5.30 | 0.93 |
| Os04g0535200 | AK060585 | Peptidase aspartic family protein | 3.25 | 0.33 |
| Translation initiation or transcription factors | ||||
| Os07g0569100 | AK120160 | Remorin, C-terminal region domain containing protein | 3.17 | 0.52 |
| Abiotic or biotic stress response | ||||
| Os03g0804500 | AF072694 | Germin-like protein subfamily T member 1 precursor/OsGLP | 4.68 | 0.49 |
| Os07g0214900 | NP_001059187 | Chalcone synthase/OsCHS | 4.36 | 0.24 |
| Os04g0456200 | NP_001052967 | TMV induced protein 1–2 | 3.68 | 0.23 |
| Os05g0495900 | AB027431 | Beta-1,3-glucanase precursor | 3.68 | 0.38 |
| Os01g0713200 | AB027429 | Beta-1,3-glucanase precursor | 3.23 | 0.62 |
| Cell-wall, cell cycle, cell growth and cell cytoskeleton modification or metabolism | ||||
| Os04g0664900 | CI550916 | Cell wall invertase | 4.12 | 0.64 |
| Os04g0683700 | AK119512 | 4-coumarate-CoA ligase-like protein | 3.38 | 0.16 |
| Os07g0568700 | AF466357 | Floral organ regulator 1 | 3.35 | 1.09 |
| Hormone metabolism and response | ||||
| Os03g0738600 | AK073529 | Lipoxygenase L-2 | 3.63 | 0.89 |
| Unknown molecular function protein | ||||
| Os10g0137300 | NP_001064130 | Conserved hypothetical protein | 7.55 | 0.91 |
| Os03g0183200 | AK106987 | Conserved hypothetical protein | 4.36 | 0.98 |
| Os01g0915900 | CI543502 | (No Hit) | 3.67 | 1.08 |
| Os11g0211800 | AK059202 | Hypothetical protein | 3.27 | 1.09 |
| Os01g0824800 | AK066200 | Conserved hypothetical protein | 3.13 | 0.62 |
| Os01g0319200 | NP_001042887 | Plant protein of unknown function family protein | 3.10 | 0.22 |
| Os05g0410800 | AK108312 | Conserved hypothetical protein | 2.99 | 0.08 |
| Down-regulated genes | ||||
| Translation initiation or transcription factors | ||||
| Os07g0558100 | Y11415 | Myb protein (similar to ATMYB102) | 0.27 | 0.02 |
| Os03g0279700 | AK111338 | ZPT2-12 | 0.33 | 0.11 |
| Unknown molecular function protein | ||||
| Os10g0391400 | AK107854 | ZIM domain containing protein. (simirlar to JAZ; JA signaling) | 0.28 | 0.06 |
Funcronal classification based on Table 1.
RAP-ID based The Rice Annotation Project (RAP) ID numbers.
Accsesion based GenBank locus of the National Center of Biotechnology Information (NCBI).
Annotation based on the Rice Annotation Project Database (RAP-DB) build 3.0 by the International Rice Genome Sequencing Project (IRGSP).
Fold change, ratio of transcript abundance in Al treatement/transcript abundance in control (−Al) treatment.
Standard deviation of the mean.
Genes encoding nitrate transporter1 (OsNRT1; Os05g0410900) and nitrate reductase (OsNR; Os08g0468100) were up-regulated by 6.9- and 3.0-fold in the root tips of wild-type rice (Table 3). OsNRT1 is a low-affinity transporter for nitrate uptake [30], while OsNR is responsible for the reduction of nitrate to nitrite [31]. Rice takes up nitrogen mainly in the form of ammonium, therefore, it is unlikely that up-regulation of OsNRT1 and OsNR is for enhancing nitrogen uptake. One possibility is that the up-regulation is associated with nitric oxide (NO) production. Nitric oxide is produced from nitrite and a key signal molecule involved in many physiological processes in plants [32]. In fact, addition of exogenous NO enhanced Al-tolerance in rice roots by decreasing the contents of pectin and hemicellulose, increasing the degree of methylation of pectin, and decreasing Al accumulation in root cell walls [32], supporting that up-regulation of OsNRT1 and OsNR is required for Al-tolerance in rice.
Gene encoding iron-regulated transporter 2 (OsIRT2; Os03g0667500) was up-regulated by 3.2-fold (Table 3). Fe uptake is proposed to be mediated through OsIRT1 and OsIRT2 [33]. Interestingly, only IRT2, but not OsIRT1 was up-regulated by Al. Furthermore, this up-regulation seems to be distinct in rice since its homolog is not induced by Al in Arabidopsis, maize, M. truncatula, and wheat roots [20]–[27]. Al inhibits Fe uptake [34], therefore up-regulation of OsIRT2 is necessary for increasing Fe uptake.
Genes related with secondary metabolism were also up-regulated by Al. Chalcone synthase (CHS, EC 2.3.1.74) is a key enzyme of the flavonoid/isoflavonoid biosynthesis pathway. A gene encoding this enzyme was up-regulated by 4.4-fold (Table 3). CHS is quite commonly induced in different plant species under different forms of stress like UV, wounding, herbivory and microbial pathogens, resulting in the production of compounds that have e.g. antimicrobial activity (phytoalexins), insecticidal activity, and antioxidant activity or quench UV light directly or indirectly [35]. CHS expression causes accumulation of flavonoid and isoflavonoid. On the other hand, 4-Coumarate:CoA ligase has a pivotal role in the biosynthesis of plant secondary compounds at the divergence point from general phenylpropanoid metabolism to several major branch pathways [36]. Al is known to induce peroxidation and ROS formation in rice roots [37], [38]. Increased secondary metabolites such as flavonoids may increase anti-oxidative capacity, subsequently alleviating Al-toxicity. In line with this aspect, a gene encoding germin-like protein (OsGLP; Os03g0804500) was also up-regulated (Table 3). Germin-like proteins (GLPs) constitute a diverse family of ubiquitous plant glycoproteins [39]. Many GLPs have manganese-containing superoxide dismutase (SOD) activity [40], [41]. The SOD activities catalyze the dismutation of superoxide into oxygen and hydrogen peroxide. In this study, OsSOD was not up-regulated in rice roots after short exposure to Al stress (Table 3, Table 4, Table S2, S3), indicating OsSOD might not function in rice root after short exposure to Al stress. Thus, OsGLP might function as SOD. Furthermore, the H2O2 produced by OsGLPs is detoxicated by peroxiredoxin (PrxR) and thioredoxin (Trx) because they are only up-regulated antioxidant genes in rice root tips (Table S2). These results suggested that OsGLP, OsPrxR and OsTrx function as major ROS-scavenging enzymes in the rice roots after short exposure to Al stress.
Table 4. Genes up- and down-regulated only in the basal roots of wild-type rice.
| Functional classificationfa/RAP IDb | Accessionc | Annotationd | Fold change(+Al/−Al)e | ±SDf |
| Up-regulated | ||||
| ART1-regulated genes | ||||
| Os12g0227400 | CI560939 | Allyl alcohol dehydrogenase | 8.94 | 0.15 |
| Os03g0126900 | AK109217 | Conserved hypothetical protein | 7.84 | 0.47 |
| Os10g0206800 | AK072077 | Multidrug and toxic compound extrusion (MATE) family protein/OsFRDL2 | 5.89 | 0.44 |
| Os02g0131800 | AK102180 | OsNramp4/OsNrat1 | 5.53 | 0.18 |
| Os09g0426800 | AK060786 | Gl1 protein | 5.34 | 0.71 |
| Os10g0578800 | AK065615 | LrgB-like protein family protein | 5.22 | 0.24 |
| Os03g0304100 | AK111121 | Hypothetical protein | 4.04 | 0.49 |
| Os06g0695800 | AK064089 | Bacterial-type ATP binding cassette (ABC) transporter/OsSTAR1 | 3.81 | 0.19 |
| Os05g0119000 | AK069359 | Bacterial-type ATP binding cassette (ABC) transporter/OsSTAR2 | 3.68 | 0.90 |
| Os01g0869200 | AK073453 | Mg2+ transporter/OsMGT1 | 3.24 | 0.59 |
| Other genes | ||||
| Transport | ||||
| Os06g0701700 | AB061311 | HKT-type transporter (Sodium ion transporter) | 2.97 | 0.83 |
| Metabolism | ||||
| Os08g0547300 | AK072163 | E-class P450, group I family protein | 7.65 | 3.42 |
| Os04g0405300 | AK110700 | Stem secoisolariciresinol dehydrogenase | 4.04 | 1.05 |
| Os06g0500700 | CI431272 | Cytochrome P450 family protein | 3.48 | 1.21 |
| Os02g0176900 | NP_001046065 | Aldose 1-epimerase family protein | 3.47 | 0.67 |
| Os05g0438600 | AY035554 | Fructose-1,6-bisphosphatase (FBPase) | 3.45 | 0.16 |
| Os11g0487600 | NP_001067918 | Cytochrome P450 family protein | 3.17 | 0.09 |
| Os05g0424300 | AK120987 | Cytochrome P450 family protein | 3.07 | 0.44 |
| Protein synthesis and processing | ||||
| Os12g0108500 | AK122171 | Cyclin-like F-box domain containing protein | 10.16 | 0.26 |
| Os04g0535200 | AK060585 | Peptidase aspartic family protein | 3.10 | 0.35 |
| Translation initiation or transcription factors | ||||
| Os01g0286100 | AK102252 | Basic helix-loop-helix dimerisation region bHLH domain containing protein | 3.34 | 0.45 |
| Abiotic or biotic stress response | ||||
| Os09g0361500 | AK120689 | Isochorismate synthase 1 (ICS1) | 4.08 | 0.76 |
| Os05g0223000 | AK071661 | Calmodulin-related protein 2, touch-induced | 3.32 | 0.77 |
| Os04g0635500 | AK069933 | Wound induced protein | 3.05 | 0.81 |
| Cell-wall, cell cycle, cell growth and cell cytoskeleton modification or metabolism | ||||
| Os04g0506800 | AK070719 | Glycosyl transferase, family 29 protein/OsGT | 5.21 | 0.77 |
| Os11g0444000 | AK099588 | UDP-glucosyltransferase BX8 | 3.50 | 0.37 |
| Os02g0802200 | AK107538 | Glycoside hydrolase, family 79, N-terminal domain containing protein/OsGH | 3.34 | 0.42 |
| Os04g0477500 | AK063950 | Glycosyl transferase, family 17 protein/OsGT | 3.15 | 0.30 |
| Os03g0324700 | AK121618 | Exostosin-like family protein | 3.03 | 0.25 |
| Hormone metabolism and response | ||||
| Os04g0667400 | AK119413 | 2OG-Fe(II) oxygenase domain containing protein | 5.42 | 1.44 |
| Mitochondria or plastid | ||||
| Os07g0469100 | AK120365 | Thylakoid membrane phosphoprotein 14 kDa | 3.06 | 0.34 |
| Unknown molecular function protein | ||||
| Os07g0269000 | CI251879 | (No Hit) | 3.74 | 0.10 |
| Os10g0473200 | AK105229 | Conserved hypothetical protein | 5.05 | 0.76 |
| Os05g0573800 | CI142713 | (No Hit) | 4.49 | 0.56 |
| Os04g0635400 | CI037812 | Conserved hypothetical protein | 4.38 | 1.40 |
| Os04g0603800 | AK063616 | Hypothetical protein | 4.17 | 0.13 |
| Os03g0183200 | AK106987 | Conserved hypothetical protein | 4.10 | 0.75 |
| Os12g0265400 | CI096837 | Hypothetical protein | 4.05 | 0.23 |
| Os09g0459900 | AK063208 | Cyclin-dependent kinase inhibitor family protein | 3.92 | 1.21 |
| Os09g0459500 | AB118006 | Hypothetical protein | 3.87 | 0.49 |
| Os03g0255500 | AK061620 | Phosphoenolpyruvate carboxykinase | 3.69 | 0.36 |
| Os01g0213500 | CI426147 | Conserved hypothetical protein | 3.45 | 0.15 |
| Os11g0259100 | NP_001067644 | Hypothetical protein | 3.43 | 0.60 |
| Os02g0600200 | AK058978 | IQ calmodulin-binding region domain containing protein | 3.23 | 0.70 |
| Os02g0327000 | AK073631 | C2 domain containing protein | 3.15 | 0.28 |
| Os06g0535200 | AK109943 | Zn-finger, RING domain containing protein | 3.10 | 0.17 |
| Os01g0854000 | AK070440 | Conserved hypothetical protein | 3.10 | 0.72 |
| Os04g0520700 | AK065832 | Protein of unknown function DUF584 family protein | 3.08 | 0.81 |
| Os03g0113900 | AK119700 | Protein of unknown function DUF584 family protein | 2.98 | 0.19 |
| Os04g0231800 | AK068417 | Protein of unknown function DUF1165 family protein | 2.95 | 0.03 |
| Down-regulated | ||||
| Transport | ||||
| Os04g0538900 | CI558963 | Glyoxalase/bleomycin resistance protein/dioxygenase domain containing protein | 0.29 | 0.10 |
| Os03g0817200 | AK121940 | Amino acid/polyamine transporter II family protein | 0.31 | 0.03 |
| Os03g0375900 | AK107064 | Amino acid/polyamine transporter I family protein | 0.32 | 0.05 |
| Metabolism | ||||
| Os06g0185500 | C97337 | Transferase family protein | 0.05 | 0.02 |
| Os06g0185300 | – | Transferase family protein | 0.15 | 0.04 |
| Os12g0626400 | AK063967 | Squalene/phytoene synthase family protein | 0.22 | 0.03 |
| Os06g0549900 | AK109673 | FAD linked oxidase, N-terminal domain containing protein | 0.23 | 0.07 |
| Os06g0294600 | AK058424 | Cytochrome P450 family protein | 0.24 | 0.01 |
| Os11g0644800 | CI019806 | Tyrosine/nicotianamine aminotransferase family protein | 0.25 | 0.04 |
| Os07g0643400 | AK061012 | Esterase/lipase/thioesterase domain containing protein | 0.31 | 0.02 |
| Protein synthesis and processing | ||||
| Os01g0124100 | AK062394 | Proteinase inhibitor I12, Bowman-Birk family protein | 0.13 | 0.01 |
| Os10g0537800 | AK061277 | Peptidase A1, pepsin family protein | 0.21 | 0.06 |
| Os03g0318400 | AK106440 | Peptidase A1, pepsin family protein | 0.21 | 0.07 |
| Signal transduction | ||||
| Os07g0186200 | NP_001059070 | Protein kinase family protein | 0.32 | 0.03 |
| Os04g0618700 | AK120799 | Protein kinase domain containing protein | 0.34 | 0.02 |
| Os01g0699600 | AK105196 | Protein kinase domain containing protein | 0.34 | 0.23 |
| Translation initiation or transcription factors | ||||
| Os02g0624300 | AK112056 | MYB1 protein | 0.11 | 0.09 |
| Os11g0702400 | AK105226 | Zn-finger, C2H2 type domain containing protein | 0.26 | 0.03 |
| Abiotic or biotic stress response | ||||
| Os07g0129300 | AF306651 | Pathogenesis-related protein 1 precursor | 0.07 | 0.02 |
| Os06g0546500 | AK073833 | Peroxidase | 0.16 | 0.03 |
| Os05g0427400 | CI551987 | Phenylalanine ammonia-lyase | 0.19 | 0.01 |
| Os02g0627100 | AK068993 | Phenylalanine ammonia-lyase | 0.22 | 0.04 |
| Os09g0417800 | AK067834 | DNA-binding WRKY domain containing protein | 0.24 | 0.07 |
| Os09g0417600 | AF467736 | DNA-binding WRKY domain containing protein | 0.26 | 0.05 |
| Os10g0542900 | AB016497 | Chitinase | 0.28 | 0.06 |
| Os05g0135400 | AK063587 | Plant peroxidase family protein | 0.29 | 0.06 |
| Os05g0149400 | AK061064 | 1-aminocyclopropane-1-carboxylate oxidase/OsACC | 0.30 | 0.10 |
| Os01g0687400 | AB110201 | Chitinase | 0.31 | 0.04 |
| Os11g0592000 | AK121059 | Barwin | 0.33 | 0.04 |
| Os01g0933900 | AF309383 | Glutathione transferase III(B) | 0.35 | 0.02 |
| Cell-wall, cell cycle, cell growth and cell cytoskeleton modification or metabolism | ||||
| Os02g0267200 | CI377660 | Alpha-expansin OsEXPA13 | 0.32 | 0.04 |
| Unknown molecular function protein | ||||
| Os04g0368000 | CI447876 | (No Hit) | 0.15 | 0.04 |
| Os06g0587300 | AK121885 | Conserved hypothetical protein | 0.05 | 0.03 |
| Os06g0586000 | AK063903 | Conserved hypothetical protein | 0.07 | 0.03 |
| Os12g0437800 | AK063833 | CI2E | 0.08 | 0.02 |
| Os01g0796000 | CI508923 | (No Hit) | 0.12 | 0.04 |
| Os10g0391400 | AK107854 | ZIM domain containing protein | 0.18 | 0.15 |
| Os05g0368000 | NP_001055341 | Conserved hypothetical protein | 0.20 | 0.03 |
| Os06g0282000 | CI563293 | (No Hit) | 0.21 | 0.03 |
| Os06g0292400 | CI409636 | Embryogenesis transmembrane protein | 0.27 | 0.02 |
| Os02g0520100 | AK072610 | NUDIX hydrolase domain containing protein | 0.28 | 0.01 |
| Os03g0187800 | AK105352 | Protein of unknown function DUF250 domain containing protein | 0.30 | 0.05 |
| Os06g0155400 | NP_001056850 | Hypothetical protein | 0.33 | 0.06 |
RAP-ID based The Rice Annotation Project (RAP) ID numbers.
Accsesion based GenBank locus of the National Center of Biotechnology Information (NCBI).
Annotation based on the Rice Annotation Project Database (RAP-DB) build 3.0 by the International Rice Genome Sequencing Project (IRGSP).
Fold change, ratio of transcript abundance in Al treatement/transcript abundance in control (−Al) treatment.
Standard deviation of the mean.
Among genes up-regulated by Al only in the basal region of wild-type rice, some are related to polysaccharide/cell wall metabolism, including genes encoding glycoside hydrolase (GH; Os02g0802200) and glycosyl transferases (GTs; Os04g0506800, Os04g0477500) (Table 4). Glycoside hydrolases (GHs) catalyze the hydrolysis of the glycosidic linkage to release smaller sugars [42]. Glycosyl transferases (GTs) catalyze the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, thereby forming glycosidic bonds [43]. Al causes the thickening and rigidification of cell walls [44]. Increased expression of OsGH and OsGTs may contribute to the cell wall synthesis, hence alleviating the Al-induced inhibition of longitudinal cell expansion.
Gene encoding 1-aminocyclopropane-1-carboxylate oxidase (OsACC; Os05g0149400) was down-regulated in the basal root (Table 4). OsACC is related to biosynthesis of ethylene. Ethylene production is associated with inhibition of root elongation in Lotus japonicus and M. truncatula [45]. Down-regulation of OsACC may prevent further inhibition of root growth caused by Al. The association between other genes and Al-tolerance remain to be examined in future.
As a conclusion, our comparative genome-wide transcriptional analysis reveals that there are other mechanisms for Al-tolerance except for ART1-regulated one in rice including those related to nitrogen assimilation, secondary metabolite synthesis, cell-wall synthesis and ethylene synthesis. Although the exact roles of these putative tolerance genes remain to be examined, our data provide a platform for further work on Al-tolerance in rice.
Supporting Information
Primer sequences used for quantitative real-time PCR.
(XLS)
Genes up-regulated in the root tips of both the wild-type and star1 mutant.
(XLS)
Genes up-regulated in the basal root regions of both the wild-type and star1 mutant.
(XLS)
Genes down-regulated in the root tips of both the wild-type and star1 mutant.
(XLS)
Genes down-regulated in the basal roots of both the wild-type and star1 mutant.
(XLS)
Funding Statement
The research was supported by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 22119002 to JFM) and by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation, IPG-0006 to JFM). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. von Uexkull HR, Mutert E (1995) Global extent, development and economic impact of acid soils. Plant Soil 171: 1–15. [Google Scholar]
- 2. Kochian LV, Hoekenga OA, Piñeros MA (2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol 55: 459–493. [DOI] [PubMed] [Google Scholar]
- 3. Ma JF (2007) Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int Rev Cytol 264: 225–253. [DOI] [PubMed] [Google Scholar]
- 4. Ma JF, Shen R, Zhao Z, Wissuwa M, Takeuchi Y, et al. (2002) Response of rice to Al stress and identification of quantitative trait loci for Al tolerance. Plant Cell Physiol 43: 652–659. [DOI] [PubMed] [Google Scholar]
- 5. Ma JF, Furukawa J (2003) Recent progress in the research of external Al detoxification in higher plants. J Inorg Biochem 97: 46–51. [DOI] [PubMed] [Google Scholar]
- 6. Famoso AN, Zhao K, Clark RT, Tung CW, Wright MH, et al. (2011) Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet 7: e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Delhaize E, Ma JF, Ryan PR (2012) Transcriptional regulation of aluminium tolerance genes. Trends Plant Sci 6: 341–348. [DOI] [PubMed] [Google Scholar]
- 8. Yamaji N, Huang CF, Nagao S, Yano M, Sato Y, et al. (2009) A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21: 3339–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsutsui T, Yamaji N, Ma JF (2011) Identification of a cis-acting element of ART1, a C2H2-type zinc finger transcription factor for aluminum tolerance in rice. Plant Physiol 156: 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang CF, Yamaji N, Mitani N, Yano M, Nagamura Y, et al. (2009) A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21: 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yokosho K, Yamaji N, Ma JF (2011) An Al-inducible MATE gene is involved in external detoxification of Al in rice. Plant J 69: 1061–1069. [DOI] [PubMed] [Google Scholar]
- 12. Xia J, Yamaji N, Kasai T, Ma JF (2010) Plasma membrane-localized transporter for aluminum in rice. Proc Natl Acad Sci USA 107: 18381–18385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang CF, Yamaji N, Chen Z, Ma JF (2011) A tonoplast-localized half-size ABC transporter is required for internal detoxification of Al in rice. Plant J 69: 857–867. [DOI] [PubMed] [Google Scholar]
- 14. Chen ZC, Yamaji N, Motoyama R, Nagamura Y, Ma JF (2012) Up-regulation of a magnesium transporter gene OsMGT1 is required for conferring aluminum tolerance in rice. Plant Physiol 159: 1624–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tanaka T, Antonio BA, Kikuchi S, Matsumoto T, Nagamura Y, et al. (2008) The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res 36: D1028–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, et al. (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36: W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- 18.Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, et al.. (2007) The TIGR Rice Genome Annotation Resource: improvements and new features. NAR 35 Database Issue: D846–851. [DOI] [PMC free article] [PubMed]
- 19. Hoekenga OA, Vision TJ, Shaff JE, Monforte AJ, Lee GP, et al. (2003) Identification and characterization of aluminum tolerance loci in Arabidopsis (Landsberg erecta x Columbia) by quantitative trait locus mapping. A physiologically simple but genetically complex trait. Plant Physiol 132: 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kumari M, Taylor GJ, Deyholos MK (2008) Transcriptomic responses to aluminum stress in roots of Arabidopsis thaliana . Mol Genet Genomics 279: 339–357. [DOI] [PubMed] [Google Scholar]
- 21. Goodwin SB, Sutter TR (2009) Microarray analysis of Arabidopsis genome response to aluminum stress. Biol Plant 53: 85–99. [Google Scholar]
- 22. Zhao CR, Ikka T, Sawaki Y, Kobayashi Y, Suzuki Y, et al. (2009) Comparative transcriptomic characterization of aluminum, sodium chloride, cadmium and copper rhizotoxicities in Arabidopsis thaliana . BMC Plant Biol 9: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maron LG, Kirst M, Mao C, Milner MJ, Menossi M, et al. (2008) Transcriptional profiling of aluminum toxicity and tolerance responses in maize roots. New Phytol 179: 116–128. [DOI] [PubMed] [Google Scholar]
- 24. Mattiello L, Kirst M, da Silva FR, Jorge RA, Menossi M (2010) Transcriptional profile of maize roots under acid soil growth. BMC Plant Biol 10: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chandran D, Sharopova N, Ivashuta S, Gantt JS, Vandenbosch KA, et al. (2008) Transcriptome profiling identified novel genes associated with aluminum toxicity, resistance and tolerance in Medicago truncatula . Planta 228: 151–166. [DOI] [PubMed] [Google Scholar]
- 26. Chandran D, Sharopova N, VandenBosch KA, Garvin DF, Samac DA (2008) Physiological and molecular characterization of aluminum resistance in Medicago truncatula . BMC Plant Biol 8: 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Houde M, Diallo AO (2008) Identification of genes and pathways associated with aluminum stress and tolerance using transcriptome profiling of wheat near-isogenic lines. BMC Genomics 9: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410. [DOI] [PubMed] [Google Scholar]
- 29. Miller G, Shulaev V, Mittler R (2008) Reactive oxygen signaling and abiotic stress. Physiol Plant 133: 481–489. [DOI] [PubMed] [Google Scholar]
- 30. Lin CM, Koh S, Stacey G, Yu SM, Lin TY, et al. (2000) Cloning and functional characterization of a constitutively expressed nitrate transporter gene, OsNRT1, from rice. Plant Physiol 122: 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xiong J, Fu G, Yang Y, Zhu C, Tao L (2012) Tungstate: is it really a specific nitrate reductase inhibitor in plant nitric oxide research?. J Exp Bot 63: 33–41. [DOI] [PubMed] [Google Scholar]
- 32. Zhang Z, Wang H, Wang X, Bi Y (2011) Nitric oxide enhances aluminum tolerance by affecting cell wall polysaccharides in rice roots. Plant Cell Rep 30: 1701–1711. [DOI] [PubMed] [Google Scholar]
- 33. Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, et al. (2006) Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+ . Plant J 45: 335–346. [DOI] [PubMed] [Google Scholar]
- 34. Chen RF, Shen RF, Gu P, Dong XY, DU CW, et al. (2006) Response of rice (Oryza sativa) with root surface iron plaque under aluminium stress. Ann Bot 98: 389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dao TT, Linthorst HJ, Verpoorte R (2011) Chalcone synthase and its functions in plant resistance. Phytochem Rev 10: 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hamberger B, Hahlbrock K (2004) The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc Natl Acad Sci USA 101: 2209–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meriga B, Reddy BK, Rao KR, Reddy LA, Kishor PB (2004) Aluminium-induced production of oxygen radicals, lipid peroxidation and DNA damage in seedlings of rice (Oryza sativa). J Plant Physiol 161: 63–68. [DOI] [PubMed] [Google Scholar]
- 38. Sharma P, Dubey RS (2007) Involvement of oxidative stress and role of antioxidative defense system in growing rice seedlings exposed to toxic concentrations of aluminum. Plant Cell Rep 26: 2027–2038. [DOI] [PubMed] [Google Scholar]
- 39. Dunwell JM, Gibbings JG, Mahmood T, Naqvi SMS (2008) Germin and germin-like proteins: evolution, structure, and function. Crit Rev Plant Sci 27: 342–375. [Google Scholar]
- 40. Carter C, Thornburg RW (2000) Tobacco nectarin I. Purification and characterization as a germin-like, manganese superoxide dismutase implicated in the defense of floral reproductive tissues. J Biol Chem 275: 36726–36733. [DOI] [PubMed] [Google Scholar]
- 41. Gucciardo S, Wisniewski JP, Brewin NJ, Bornemann S (2007) A germin-like protein with superoxide dismutase activity in pea nodules with high protein sequence identity to a putative rhicadhesin receptor. J Exp Bot 58: 1161–1171. [DOI] [PubMed] [Google Scholar]
- 42.Coutinho PM, Henrissat B (1999) Carbohydrate-active enzymes: an integrated database approach. In: Gilbert HJ, Davies G, Henrissat B, Svensson B, editors. The Royal Society of Chemistry. 3–12.
- 43. Coutinho PM, Deleury E, Davies GJ, Henrissat B (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328: 307–317. [DOI] [PubMed] [Google Scholar]
- 44. Jones DL, Blancaflor EB, Kochian LV, Gilroy S (2006) Spatial coordination of aluminium uptake, production of reactive oxygen species, callose production and wall rigidification in maize roots. Plant Cell and Environ 29: 1309–1318. [DOI] [PubMed] [Google Scholar]
- 45. Sun P, Tian QY, Zhao MG, Dai XY, Huang JH, et al. (2007) Aluminum-induced ethylene production is associated with inhibition of root elongation in Lotus japonicus L. Plant Cell Physiol. 48: 1229–1235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences used for quantitative real-time PCR.
(XLS)
Genes up-regulated in the root tips of both the wild-type and star1 mutant.
(XLS)
Genes up-regulated in the basal root regions of both the wild-type and star1 mutant.
(XLS)
Genes down-regulated in the root tips of both the wild-type and star1 mutant.
(XLS)
Genes down-regulated in the basal roots of both the wild-type and star1 mutant.
(XLS)