Summary
Neurotensin (NT) is a 13 amino acid peptide that functions as both a neurotransmitter and a hormone through activation of the neurotensin receptor NTS1, a G protein-coupled receptor (GPCR). In the brain, NT modulates activity of dopaminergic systems, opioid-independent analgesia, and the inhibition of food intake, and in the gut NT regulates a range of digestive processes. Here we present the structure at 2.8 Å resolution of NTS1 in an active-like state, bound to NT8-13, the C terminal portion of NT responsible for agonist-induced activation of the receptor. The peptide agonist binds to NTS1 in an extended conformation nearly perpendicular to the membrane plane with the C-terminus oriented towards the receptor core. Our findings provide the first insight into the binding mode of a peptide agonist to a GPCR and may support the development of non-peptide ligands that could be useful in the treatment of neurological disorders, cancer and obesity.
Introduction
Neurotensin (NT) is a short peptide that is found in the nervous system and in peripheral tissues1. NT shows a wide range of biological activities and plays important roles in Parkinsonr’s disease and the pathogenesis of schizophrenia, the modulation of dopamine neurotransmission, hypothermia, antinociception and in promoting the growth of cancer cells2-6. Three neurotensin receptors have been identified. NTS1 (ref.7) and NTS2 (ref.8) belong to the class A GPCR family, whereas NTS3 is a member of the sortilin family with a single transmembrane domain9. Most of the known effects of NT are mediated through NTS1 (ref.5) which signals preferentially via the Gq protein. Many aspects of ligand binding to NTS1 have been addressed by mutagenesis and modeling10-12 but the details of ligand binding remains poorly understood at the molecular level. This has limited the rational design of compounds targeting NTS1 for therapeutic purposes5 in the absence of structural information on NTS1 in complex with agonist or antagonist.
Recent progress in elucidating GPCR structures in complex with either small-molecule antagonists, agonists or in complex with a heterotrimeric G protein has provided detailed molecular insights into GPCR function (reviewed in refs.13-15). However, the majority of receptor structures are of GPCRs belonging to the α group of class A (ref.16), which characteristically bind small endogenous agonists deep within the transmembrane core. NTS1 belongs to a large family of peptide receptors in the β group of class A GPCRs16 (Fig. S1) that includes receptors for substance P, cholecystokinin, neuropeptide Y, endothelin and oxytocin. These peptide receptors bind agonists of a wide range of sizes, from a few amino acid residues to over 60. Current structural information on peptide receptors is limited to representatives of the γ group of class A GPCRs16, namely the opioid receptors17-20 and a chemokine receptor21, all bound to non-peptidic drug-like antagonists. The CXCR4 receptor has also been crystallized in the presence of a cyclic peptide antagonist21. Although the structural understanding of peptide receptors has advanced, structures of peptide GPCRs from the β group have not yet been reported. In addition, no structures have been presented of a GPCR bound to a peptide agonist and only the structures of rhodopsin22,23 and the adenosine A2A receptor (A2AR)24 have been determined bound to endogenous agonists. Here we present the crystal structure of Rattus norvegicus NTS1 from the β GPCR subfamily bound to the C terminal portion (NT8-13) of the endogenous agonist NT.
Crystallization and structure determination of NTS1 in an agonist-bound state
Wild-type NTS1 is unstable in detergent solution and hence a difficult target for structural studies, particularly in the agonist-bound state which is generally thought to be less stable than the antagonist-bound state25. The first step was therefore to stabilize NTS1 using the strategy of conformational thermostabilization26. Previous work had generated a thermostable mutant of NTS1, but the purified receptor was found unsuitable for crystallization, possibly because the receptor contained mutations that were selected in the unliganded state27. NTS1 was therefore thermostabilized in the presence of the agonist NT through the inclusion of 6 stabilizing mutations (mutant NTS1-GW5; A86L1.54, E166A3.49, G215AECL2, L310A6.37, F358A7.42 and V360A7.44; superscripts are the Ballesteros-Weinstein numbers28) (Fig. S2, Table S1). Pharmacological characterization of NTS1-GW5 (Fig. S3, Table S2, Supplementary Discussion) showed that the affinity for the agonist NT was similar to that of the wild-type receptor, but that the apparent affinity for the neutral antagonist SR48692 and the sensitivity to Na+ ions was lower. In G protein coupling assays, NTS1-GW5 did not catalyze nucleotide exchange at Gαq in response to NT (Fig. S3), suggesting that the stabilizing mutations may have limited the ability to propagate the agonist-induced conformational change at the ligand binding pocket to the re-arrangement of the intracellular face of the receptor required for G protein binding and activation.
For crystallization of NTS1-GW5, T4 lysozyme (T4L) was engineered into intracellular loop 3 (ICL3) to improve the probability of obtaining well-diffracting crystals29. NTS1-GW5-T4L was expressed in insect cells using the baculovirus expression system, solubilized and purified in lauryl maltose neopentyl glycol and crystallized using the lipidic cubic phase methodology30. Diffraction data were collected to 2.8 Å resolution from a single crystal and used to determine the structure by molecular replacement (Table S3, Fig. S4, Fig. S5). The final model of NTS1-GW5-T4L, refined to R/Rfree values of 0.23/0.28, includes 454 residues, 2 Hepes molecules, 23 H2O molecules and the agonist ligand NT8-13.
Overall structure of NTS1-GW5-T4L
The receptor core has seven transmembrane helices (TM1-TM7) (Fig. 1, Fig. S6), as expected, but there was no density corresponding to amphipathic helix 8 at the C-terminus of the receptor. Helix 8 has been observed in all GPCR structures except for the chemokine receptor CXCR4 (ref.21), so the absence of helix 8 in the structure of NTS1-GW5-T4L could be because either NTS1 in vivo does not possess H8 or this is the result of a crystallization artifact. On the intracellular side, TM7 is elongated by three helical turns beyond the conserved NP7.50xxY7.53 motif, whilst other GPCRs have only one helical turn. This extension of TM7 is associated with the intracellular end of TM6 through hydrogen bonds between the side chain atoms of N375 and Q378 to the backbone carbonyl oxygen of A3026.29 (Fig. S7).
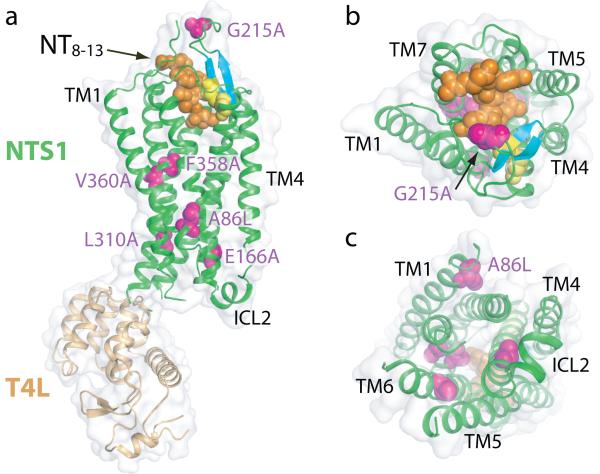
Fig. 1. Overview of the NTS1 structure bound to the peptide agonist NT8-13.
Cartoon representation of NTS1-GW5-T4L; (a) side view, (b) extracellular view, (c) intracellular view. Space filling models are used to depict the agonist NT8-13 (orange), the side chains of thermostabilizing mutations (purple) and the disulphide bond (yellow) between the conserved residues C142 and C225. Also shown are the β-hairpin in extracellular loop 2 (blue-green) and the π-helix in intracellular loop 2 (ICL2). T4L has been omitted from the intracellular view for clarity.
Of the intracellular loops in NTS1, ICL1 was disordered (K93-Q96), ICL3 was disrupted through insertion of T4L and only ICL2 showed density for a structural element, a short π-helix (Fig. 1). ICL2 is thought to be important for G protein coupling and the short helix in NTS1 is similar to those observed in identical positions in both the inactive state31 and in active states32,33 of β-adrenergic receptors (βARs). In the β1-adrenergic receptor (β1AR)31, the helix in ICL2 is stabilized by the interaction of a tyrosine residue in the middle of the helix that forms a hydrogen bond with the aspartate residue of the conserved D/ER3.50Y sequence in TM3. In NTS1, K178 in the middle of the π-helix in ICL2 may perform a similar function, although its side chain is disordered, probably because the stabilizing mutation E166A3.49 prevents the formation of the salt bridge. The extracellular surface of NTS1 is composed of 3 short extracellular loops (ECL1, 2 and 3) and the N-terminal residues N52-D60 that extend across the receptor to form one side of the ligand binding pocket (see below). The longest loop is ECL2, which contains two short antiparallel β-strands that are linked to TM3 by a disulphide bond between the conserved residues C1423.25 and C225ECL2. These β-strands are characteristic of all the peptide GPCR structures determined so far.
NTS1-GW5-T4L contains six thermostabilizing mutations and none of these residues are in the NT binding pocket (Fig. 1). Three of these mutations may directly affect the equilibrium between inactive and active states of NTS1. The mutation F358A7.42 is located at the kink in TM7, with the alanine side chain pointing into the receptor core, and the F358A mutant is known to be constitutively active34. The mutation E166A3.49 is in the conserved D/ER3.50Y motif in TM3 and will prevent the formation of the intrahelical salt bridge with R1673.50, thus facilitating the transition to the active state35,36 by allowing R1673.50 to interact with N2575.58. The R167-N257 interaction may be promoted by the L310A6.37 mutation through decreasing the size of the side chain. The reasons for the thermostabilizing effect of the A86L1.54, V360A7.44 and G215AECL2 mutations are not obvious, although it is interesting that an alanine residue37 is found in human NTS1 (Fig. S8) at the position equivalent to G215 in rat NTS1.
NTS1-GW5 is in an active-like conformation
Upon agonist binding, a GPCR undergoes conformational changes that ultimately allow binding and activation of a G protein at the intracellular surface of the receptor. Agonist-induced conformational changes in the ligand binding pocket are largely receptor specific and thought to be relatively small, leading to larger conformational changes in the intracellular half of the receptor13. There are no reported structures of NTS1 in an inactive conformation, so we are unable to discuss agonist-specific conformational changes in the ligand binding pocket. However, it is possible to compare the NTS1 structure to the inactive and active conformations of rhodopsin and β2AR. This allows an assessment of the overall conformation of NTS1 and, in addition, the rotamer conformations of mechanistically important residues that show characteristic interactions depending on whether the receptor is in an inactive or active state. These comparisons suggest that NTS1 is in an active-like state.
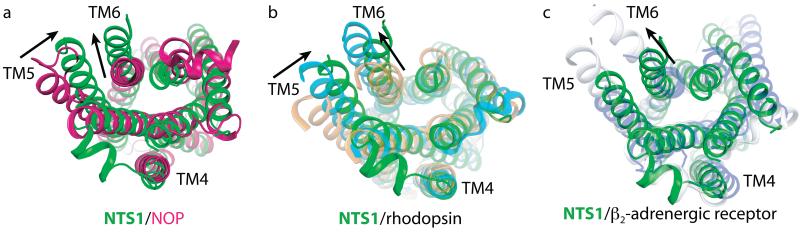
One of the major conformational changes occurring during the transition from the inactive to an active state is an outward tilt of TM6 at the intracellular face of the GPCR. The extent of movement of TM6 upon activation is 6 Å for rhodopsin and 11-14 Å for β2AR32, with TM6 of NTS1 in a position similar to that seen for the active states of rhodopsin22,38 (Fig. 2). The distance between the Cα atoms of R3.50 and E6.30 change upon activation from 9 Å to 15 Å in rhodopsin22,39, and from 11 Å to 19 Å in β2AR32,40. The equivalent distance in NTS1 is 14 Å.
Fig. 2. NTS1-GW5-T4L is in an active-like conformation.
(a) Alignment of NTS1-GW5 (green) with the inactive state of the nociceptin receptor (NOP)19 (red, PDB code 4EA3). NTS1 was most similar to NOP (root mean squared deviation = 2.1 Å for Cα atoms in the TM domains) upon alignment of the TM domains of NTS1 with other peptide receptors17-21. (b) Alignment of the inactive state of rhodopsin39 (light brown, PDB code 1GZM), its active state Meta-II (ref.22) (blue-green, PDB code 3PQR) and NTS1-GW5 (green). (c) Alignment of the inactive state of β2AR40 (pale mauve, PDB code 2RH1) and its active state32 (pale grey, PDB code 3SN6) and NTS1-GW5 (green). All views are from the intracellular surface. The arrows indicate the apparent displacement of TM5 and TM6 of NTS1-GW5 relative to the corresponding helix positions in the inactive receptor structures.
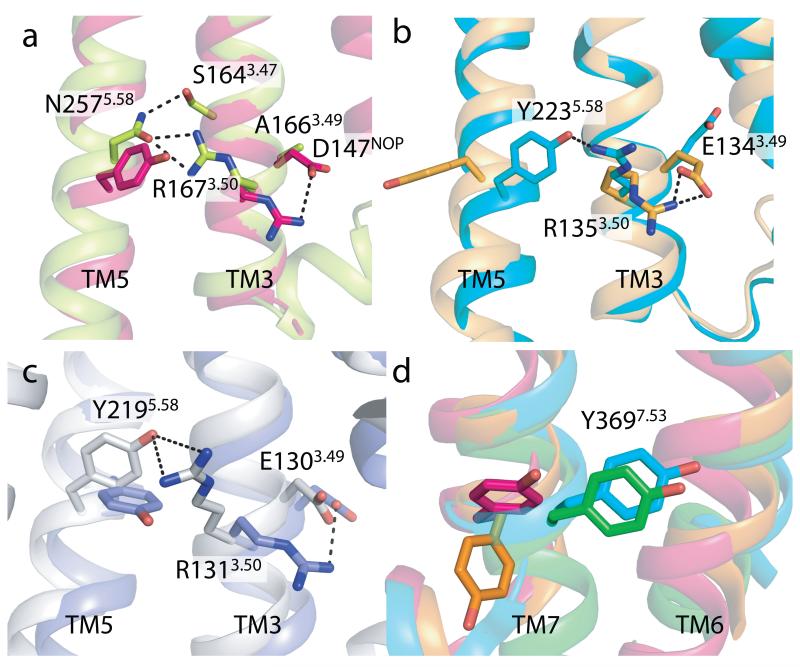
Other features of NTS1 are also remarkably similar to those found in the active conformations of rhodopsin and β2AR. In inactive receptor conformations, R3.50 of the D/ERY motif forms an intrahelical salt bridge with D/E3.49, that is, for example, present in the inactive structure of the nociceptin/orphanin FQ receptor (NOP)19 (Fig. 3), which is the closest structural homologue of NTS1 among peptide receptors. Upon activation, the D/E3.49-R3.50 salt bridge is broken and R3.50 re-orients to form an interhelical hydrogen bond with the conserved tyrosine residue Y5.58 linking TM3 and TM5 (ref.13). In NTS1, the R1673.50 side chain is hydrogen bonded to N2575.58 that interacts with the conserved residue S1643.47 (Fig. 3) stabilizing TM5 in an active-like orientation35. In addition, Y3697.53 of the highly conserved NP7.50xxY7.53 motif occupies space within the helical bundle as seen for Y7.53 in the active states of rhodopsin and β2AR (Fig. 3). In contrast to the active rhodopsin and β2AR, Y3697.53 in NTS1 does not pack against R1673.50 suggesting that N2575.58 has a greater contribution to the active state stabilization than Y3697.53 (ref.35).
Fig. 3. The conserved D/ERY and NPxxY motifs in NTS1-GW5.
(a) Comparison of the side chain orientations of R3.50 and Y/N5.58 of NTS1-GW5 and the inactive nociceptin receptor (NOP)19 (PDB code 4EA3). NTS1-GW5 residues (R167, N257, labeled) in green; corresponding NOP residues (R148, Y235, unlabeled) in red. D1473.49 of NOP and A1663.49 of NTS1 are also shown. (b) In an inactive GPCR, R3.50 interacts with E3.49 as shown here for rhodopsin39 (pale brown, PDB code 1GZM). This salt bridge is broken upon activation allowing R3.50 to contact residue5.58 as depicted for active rhodopsin (ref.22) (blue-green, PDB code 3PQR) (residues E134, R135 and Y223). (c) The same comparison for inactive β2AR40 (pale mauve, PDB code 2RH1) and active β2AR32 (pale grey, PDB code 3SN6) (residues E130, R131 and Y219). (d) Y3697.53 of NTS1 (green) occupies space as seen for Y3067.53 in active rhodopsin (blue-green, PDB code 3PQR). The orientation of Y3067.53 in the inactive rhodopsin (pale brown, PDB code 1GZM) and the inactive NOP (red, PDB code 4EA3) is shown for comparison.
Comparison of the NTS1 structure with rhodopsin and β2AR suggests that it is in an active-like state, which implies that it may couple effectively to G proteins. NTS1-GW5-T4L, as expected, does not couple to G protein (Fig. S3), because T4L replaces most of ICL3 that is essential for G protein binding and sterically blocks access to the intracellular surface of the receptor. Surprisingly, NTS1-GW5 that contains the wild type ICL3 also does not catalyze nucleotide exchange at Gαq in response to NT (Fig. S3). This may be a consequence of the extended region of TM7 on the intracellular surface forming an interhelical contact with TM6 (Fig. S7). This appears to occlude the cavity at the intracellular face of the receptor where the C-terminal portion of the Gα protein binds to fully active receptors22,23,32,38. Thus, although the structure of NTS1 contains many features of an active receptor it is likely that the intracellular surface of the receptor does not represent the structure of fully active NTS1.
The NT8-13 binding site
NTS1-GW5-T4L was co-crystallized with NT8-13, which has higher potency and efficacy than full-length NT10,41, although it does not exist in vivo. NTS1 also binds the NT-related hexapeptide neuromedin N (NN) that is produced along with NT during proteolytic processing of the proNT precursor5. The amino acid sequence of NN (KIPYIL) is similar to NT8-13 (RRPYIL). Potent NT analogues that can cross the blood-brain barrier (e.g. derivatives of RKPWLL) can also be synthesized (see ref.5). The similarity of these agonists to NT8-13 suggests that they all bind in a similar manner. There is charge complementarity between NT8-13 and its binding pocket, with the positive-charged arginine side chains of NT8-13 adjacent to the electronegative rim of the binding site, and the C-terminus of NT8-13 is predicted to form a salt bridge with R3276.54 (Fig. 4, Table S4). There are also extensive van der Waals interactions between NT8-13 and the receptor along with hydrogen bonds and a salt bridge (Table S4, Table S5, Fig. S9). It is striking that only 3 out of 8 hydrogen bonds are made between the side chains of NT8-13 and the receptor, with the bulk of receptor-ligand contacts being van der Waals interactions.
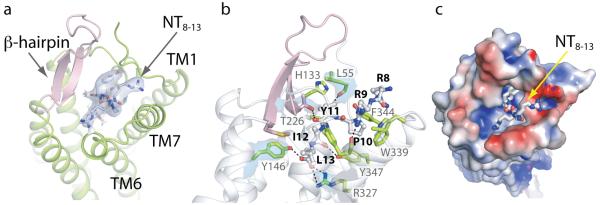
Fig. 4. The NTS1 agonist binding pocket.
All views are from the extracellular side with NT8-13 shown as a stick model. (a) Cartoon representation of the ligand binding pocket with the ECL2 β-hairpin (pink). The quality of the electron density of NT8-13 is depicted as blue isosurface (2Fo-Fc map contoured at 1σ). (b) Key NTS1 residues (green residues with grey labels) in contact with the peptide ligand (grey residues with bold black labels). Hydrogen bonds and salt bridges are indicated by dashed lines (black). The electron density maps of selected NTS1 residues (L55, Y146, T226, R327) are shown as blue isosurface (2Fo-Fc map contoured at 1σ). (c) The charge complementarity between NT8-13 and its binding pocket is depicted with the NTS1 surface colored according to its electrostatic potential (−4 kT/e to +4 kT/e; red, negative; blue, positive). The positively charged arginine side chains of the ligand face the electronegative rim of the binding pocket whereas the negatively charged carboxylate of L13 resides in an electropositive environment. See also Supplementary Information.
The ligand binding pocket for NT8-13 is composed of amino acid residues in the N-terminus, the three extracellular loops (ECL1-ECL3) and six transmembrane α-helices (TM2-TM7), with the greatest number of ligand-receptor contacts from residues in ECL2, ECL3, TM6 and TM7 (Table S5). The binding pocket is wide open on the extracellular surface of NTS1 (Fig. 4, Fig. S10) with density for NT8-13 clearly discernable from a difference electron map (Fig. S11), but we believe that radiation damage during data collection has resulted in reduced density for the C-terminal carboxylate group (Fig. S12). The extended conformation of NT8-13 in the crystal structure is in good agreement with that of NT8-13 bound to wild-type NTS1 analyzed by solid-state NMR42 (Fig. S13). The overall shape of the NTS1 ligand binding pocket is narrower than that observed in the other peptide receptors17-21 due to an inward shift of the extracellular regions of TM2 and TM6 (Fig. S14), probably as a result of thepronounced kink in TM2 at A1202.57 and a change in tilt of TM6. At the extracellular surface, the NT8-13 binding pocket is partially capped by the ECL2 β-hairpin and the proximal end of the receptor N-terminus. NTS1 has been subjected to extensive site-directed mutagenesis studies to define ligand-receptor interactions (see Supplementary Discussion) and there is excellent agreement between those data and the structure of NTS1.
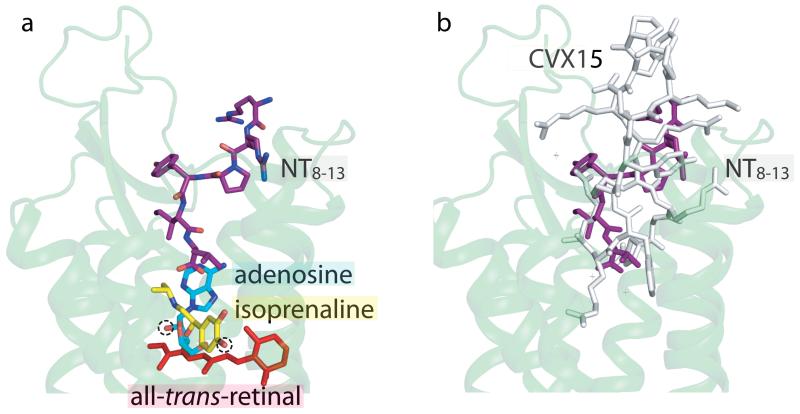
There is a striking difference between the binding mode of NT8-13 in NTS1 with the binding of agonists in rhodopsin, β1AR and A2AR (Fig. 5). The agonist-specific interactions made by isoprenaline in β1AR (ref.43) and adenosine in A2AR (ref.24) all occur at a similar depth in the receptor with respect to the extracellular surface as retinal in rhodopsin22,23. In contrast, NT8-13 does not penetrate the receptor as deeply, with the C-terminus of NT8-13 over 5 Å closer to the extracellular surface than the hydroxyl groups in isoprenaline and adenosine that are characteristic of agonists for βARs and A2AR. This suggests that the mode of activation of NTS1 is subtly different from these receptors. Structures of NTS1 in the inactive state will be necessary to gain a better understanding of how peptide agonists activate GPCRs.
Fig. 5. A new paradigm for peptide agonist binding.
Compared to small endogenous agonists, the NT8-13 binding cavity is located near the receptor surface. (a) The structures of the agonist-bound adenosine A2A receptor24 (PDB ID 2YDO), β1AR43 (PDB ID 2Y03) and meta-rhodopsin II (ref.22) (PDB code 3PQR) were aligned in PyMOL. Only the cartoon representation for NTS1-GW5 is shown (pale green) with NT8-13 depicted in purple. The agonists adenosine (blue-green), isoprenaline (yellow; an isopropyl derivative of the endogenous ligand noradrenaline) and all-trans-retinal (red) are labeled. The chemical groups in adenosine and isoprenaline that make agonist-specific contacts to the receptor are circled (dashed line). (b) The cyclic antagonist peptide CVX15 binds to the CXCR4 receptor21 (grey, PDB ID 3OE0) in a similar fashion to NT8-13 (purple) in NTS1-GW5 (pale green cartoon).
Methods summary
The stabilized NTS1 mutant with T4 lysozyme replacing most of intracellular loop 3 was expressed in insect cells and purified by cobalt affinity chromatography. It was crystallized in lipidic cubic phase. Diffraction data were collected at the GM/CA-CAT beamline of the Advanced Photon Source at the Argonne National Laboratory. The structure was solved by molecular replacement using data from a single crystal.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Methods
Materials
The tritiated agonist [3H]NT ([3,11-tyrosyl-3,5-3H(N)]-pyroGlu-Leu-Tyr-Glu-Asn-Lys-Pro-Arg-Arg-Pro-Tyr-Ile-Leu) was purchased from Perkin Elmer. Unlabeled NT and NT8-13 (Arg-Arg-Pro-Tyr-Ile-Leu) were synthesized by the Center for Biologics Evaluation and Research (Food and Drug Administration) or purchased from AnaSpec. The detergents n-dodecyl-β-D-maltopyranoside (DDM), n-decyl-β-D-maltopyranoside (DM), lauryl maltose neopentyl glycol (2,2-didecylpropane-1,3-bis-β-D-maltopyranoside) (LMNG)44, 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonic acid (CHAPS) and cholesteryl hemisuccinate Tris salt (CHS) were obtained from Anatrace. TCEP was also from Anatrace. Monoolein (M-7765), cholesterol (C-75209), lysozyme (L-6876) and DNaseI (D-4527) were purchased from Sigma. Talon resin was from Clontech. BioSpin-30 Tris columns were obtained from BioRad. PD10 columns were purchased from GE Healthcare.
Expression of NTS1 in insect cells
The baculovirus construct NTS1-GW5-T4L consisted of the hemagglutinin signal peptide and the Flag tag45, followed by the thermostabilized rat NTS1 (T43-K396 containing the mutations A86L, E166A, G215A, L310A, F358A, V360A) with the ICL3 residues H269-R299 replaced by the cysteine-free bacteriophage T4 lysozyme (N2-Y161 with the mutations C54T and C97A) and a GSGS linker. A deca-histidine tag was placed at the C-terminus. NTS1-GW5 contained the wild-type ICL3 sequence. Wild-type NTS1 (Met-T43-Y424) started at T43 but did not contain any other modifications. Recombinant baculoviruses were generated using a modified pFastBac1 transfer plasmid (Invitrogen). Trichoplusia ni cells were infected at a cell density of 0.8-1 million cells/ml with recombinant virus at a multiplicity of infection (MOI) of 5, and the temperature was lowered from 28°C to 21°C. Cells were harvested by centrifugation 48 hours post infection, resuspended in hypotonic buffer (10 mM Hepes pH 7.5, 10 mM MgCl2, 20 mM KCl), flash-frozen in liquid nitrogen and stored at −80°C until use.
Preparation of urea-washed P2 insect cell membranes
NTS1-enriched membranes were obtained as a P2 fraction from insect cells46,47. Prior to G protein coupling assays and agonist binding experiments, the P2 membranes were treated with urea to remove peripherally bound membrane proteins48,49. The receptor density in urea-washed P2 membranes was determined by [3H]NT saturation binding analysis47.
Stability tests in detergent solution
Cell pellets from 10 ml of insect cell cultures (NTS1-GW5-T4L; cell density at harvest ~1.5-2.5 million cells/ml with ~10 million receptors/cell) or P2 membranes (wild-type NTS1) were resuspended in 2 ml buffer containing DM or LMNG/CHS to give a final buffer composition of 50 mM TrisHCl pH 7.4, 200 mM NaCl, 1% DM or 1% LMNG / 0.1% CHS. The samples were placed on a rotating mixer at 4°C for 1 hour. Cell debris and non-solubilized material were removed by ultracentrifugation (TL120.2 rotor, 60k rpm, 4°C, 30 min in Optima Max bench-top ultracentrifuge, Beckman), and the supernatants containing detergent-solubilized NTS1 were used to test for thermal stability in the +NT format47. For thermal denaturation curves, the supernatants were diluted 6.67-fold into assay buffer (50 mM TrisHCl pH 7.4, 200 mM NaCl) containing 10 nM [3H]NT and incubated for 1-2.5 hrs on ice to allow [3H]NT binding to NTS1. Samples (120 μl aliquots) were exposed to different temperatures between 0°C and 70°C for 30 min and placed on ice. Separation of receptor-ligand complex from free ligand (100 μl) was achieved by centrifugation-assisted gel filtration (spin assay) using Bio-Spin 30 Tris columns, equilibrated with RDB buffer (50 mM TrisHCl pH 7.4, 1 mM EDTA, 0.1% DDM, 0.2% CHAPS, 0.04% CHS). Control reactions on ice were recorded at the start and at the end of each denaturation experiment. The percentage of activity remaining after heat exposure was determined with respect to the unheated control. Data were analyzed by nonlinear regression using a Boltzmann sigmoidal equation in the Prism software (GraphPad).
Ligand binding experiments
All radio-ligand binding assays were conducted with urea-washed P2 insect cell membranes containing wild-type NTS1, NTS1-GW5-T4L or NTS1-GW5.
For agonist [3H]NT saturation binding experiments, receptors were incubated on ice for 1 hour in 250 μl TEBB buffer (50 mM Tris-HCl pH 7.4, 1 mM EDTA, 0.1% BSA, 0.004% bacitracin) containing [3H]NT at a concentration of 0.6-20 nM. Nonspecific [3H]NT binding was determined in the presence of 50 μM unlabeled NT. Separation of bound from free ligand was achieved by rapid filtration through GF/B glass fiber filters (Whatman) pretreated with polyethylenimine. The amount of radioactivity was quantified by liquid scintillation counting (Beckman LS6500). Data were analyzed by nonlinear regression using the GraphPad Prism software and best fit to a one-site binding equation to determine the equilibrium dissociation constant (Kd) for wild-type NTS1 / “apparent” dissociation constants for NTS1-GW5-T4L and NTS1-GW5. Note that the saturation binding experiments using wild-type NTS1 were conducted at equilibrium. In contrast, binding of [3H]NT to NTS1-GW5-T4L and NTS1-GW5 did not reach equilibrium within the incubation time because of the very slow agonist off-rates.
Competition assays with the non-peptide antagonist SR48692 (ref.50) were performed in the presence of [3H]NT (TEBB buffer, 5 nM [3H]NT, NTS1 concentration < 0.5 nM, incubation for 2 hours on ice, 250 μl assay volume). Data were fit to a sigmoidal dose-response equation with standard slope using the concentrations of total SR48692 added versus bound [3H]NT.
The effect of Na+ ions on [3H]NT binding was measured with NaCl concentrations ranging from 0 to 1 M (TEBB buffer, 8 nM [3H]NT, NTS1 concentration < 0.5 nM, incubation for 1.5 hours on ice, 300 μl assay volume). Data were analyzed by nonlinear regression using the GraphPad Prism four-parameter dose-response equation (variable slope) with the top and bottom plateaux constrained from 100%-15% (wild-type NTS1) and 100%-50% (NTS1-GW5-T4L, NTS1-GW5), respectively.
The association of [3H]NT was assessed at a concentration of 10 nM (TEBB buffer, NTS1 concentration < 0.5 nM). At the indicated time points, 250 μl aliquots were filtered over glass fiber filters. After 2 hours, [3H]NT dissociation was initiated by adding 41.7 μM unlabeled NT or by addition of 41.7 μM NT and 833 mM NaCl; this step reduced the concentration of [3H]NT to 8 nM. Samples were subjected to filtration after the indicated time points. No attempt was made to quantitatively compare the observed rates of association and the dissociation rate constants between NTS1 constructs because of the very fast association of agonist to wild-type NTS1, the very fast dissociation of agonist from wild-type NTS1 in the presence of NaCl, and the very slow dissociation of [3H]NT from NTS1-GW5-T4L and NTS1-GW5.
GTPγS assays
Prior to G protein coupling assays, the P2 membranes were treated with urea to remove peripherally bound membrane proteins48,49. GDP/[35S]GTPγS exchange assays were performed as previously described47 with 1 nM receptor, 100 nM Gαq (purified from dark-adapted Sepia officinalis retina48), 500 nM Gβ1γ1 (purified from bovine retina51), and 10 μM NT, 40 μM nonpeptide antagonist SR48692 (ref.50) or no ligand in the reaction (5 min at 30°C).
Large-scale purification of NTS1-GW5-T4L from insect cell membranes
Cells from 3L of insect cell culture were thawed and the volume was brought to approximately 240 ml with hypotonic buffer (10 mM Hepes pH 7.5, 10 mM MgCl2, 20 mM KCl). The cells were then resuspended using a Turrax T-25 (IKA) homogenizer at 8,200 rpm for 2 min. After centrifugation (45Ti rotor, 40,000 rpm, 45 min, 4°C, Optima L90K, Beckman), the membranes were resuspended (Turrax T-25) in approx. 240 ml of high-salt buffer (10 mM Hepes pH 7.5, 1 M NaCl, 10 mM MgCl2, 20 mM KCl) supplemented with DNaseI (final concentration 10 μg/ml) and AEBSF (100 μM), and centrifuged again. All subsequent steps were performed at 4°C or on ice, and AEBSF (100 μM final concentration) was repeatedly added throughout the procedure. The washed membranes were resuspended in 60 ml of buffer (100 mM TrisHCl pH 7.4, 60% glycerol) containing 20 μM NT8-13, and stirred for 45 min to allow agonist binding to membrane-inserted NTS1-GW5-T4L. The receptor was extracted by drop-wise addition of 40 ml of a 3% LMNG / 0.3% CHS solution. After 1 hour, NaCl was added, and the solution was gently stirred for an additional hour. The final volume was 120 ml containing 50 mM TrisHCl pH 7.4, 30% glycerol, 500 mM NaCl, 1% LMNG / 0.1% CHS and 10 μM NT8-13. The sample was clarified by centrifugation (45Ti rotor, 40,000 rpm, 1 hour, Optima L90K, Beckman), adjusted with imidazole to a final concentration of 20 mM, and then passed through a 0.2 μm filter (Stericup). Next, the sample was loaded at a flow rate of 0.2 ml/min onto 2.5 ml Talon resin packed into an XK16 column (GE Healthcare) equilibrated with Talon-A+ buffer (50 mM TrisHCl pH 7.4, 30% glycerol, 500 mM NaCl, 20 mM imidazole, 1 μM NT8-13, 0.1% LMNG / 0.01% CHS). After washing with 29 column volumes of buffer Talon-A+, NTS1-GW5-T4L was eluted with Talon-B+ buffer (50 mM TrisHCl pH 7.4, 30% glycerol, 500 mM NaCl, 200 mM imidazole, 5 μM NT8-13, 0.1% LMNG / 0.01% CHS). Peak fractions were collected (5 ml) and desalted using PD10 columns equilibrated in PD10 buffer (20 mM TrisHCl pH 7.4, 100 mM NaCl, 0.003% LMNG / 0.0003% CHS). NT8-13 was then added to a concentration of 10 μM, and the sample was used for crystallization. Three liters of insect cell culture yielded ~1.7 mg of purified NTS1-GW5-T4L.
Crystallization
Purified desalted NTS1-GW5-T4L was diluted 4-fold into buffer (25 mM TrisHCl pH 7.4, 100 mM NaCl) containing 100 μM TCEP and then concentrated to an estimated 20-25 mg/ml using a 100,000 MWCO concentrator (Amicon Ultra, Millipore). After addition of NT8-13 to 1 mM and centrifugation (TLA 120.1 rotor, 60,000 rpm, 15 min, 8°C, Beckman), the sample was mixed with 1.5 parts by weight of a mix of monoolein with cholesterol (10:1) using the two-syringe method52. The resulting lipidic cubic phase53 mix was dispensed in 65 nl drops onto Laminex plates (Molecular Dimensions) and overlaid with 750 nl precipitant solution. Crystals grew after one week in precipitant solution consisting of 20.8% PEG 400, 80 mM Hepes pH 7.0, 2 mM TCEP and 43 mM NaK tartrate. Crystals were harvested directly from LCP using 50 μm micro-loops (M5-L18SP-50, MiTeGen) and immediately flash frozen in liquid nitrogen without adding extra cryoprotectant.
Data collection and structure determination
Data collection was performed with the JBluIce-EPICS data acquisition software at the GM/CA-CAT (23-ID-B) beamline at the Advanced Photon Source of Argonne National Laboratory. Crystals within the loop were located by diffraction using the automated rastering module of JBluIce-EPICS54,55. Most crystals showed diffraction spots to ~4-5 Å; a few crystals diffracted to ~3-3.5 Å with a single crystal showing diffraction to ~2.6 Å. A single dataset was collected from this crystal containing 150 frames with an oscillation of 1 degree/frame and was processed in space group P21 to 2.80 Å with an overall completeness of 93.1%.
Structure determination was performed by molecular replacement using the Phaser-MR module of PHENIX56. Two search models were created using the structure of β2AR57 (PDB code 3NY9) with one containing the T4 lysozyme domain and one containing the receptor seven-helix bundle without side chains. One copy of each search model was found, producing a single solution. Subsequent refinement was performed by PHENIX56 and model building with COOT58. Strong difference density was observed within the ligand binding cavity which we were able to model unambiguously as the six residue neurotensin peptide RRPYIL (NT8-13). The structure was refined with final R/Rfree values of 0.23/0.28. A summary of data collection and refinement statistics is reported in Supplementary Table S3.
We noted during the later stages of structure determination that no electron density was observed for the C-terminal carboxyl group of NT8-13 (using an Fo-Fc map contoured at ~0.1 e−/Å3, ~2 σ) despite clear electron density for the rest of the agonist peptide. However, relatively strong density was found in close proximity to the modeled C-terminal carboxyl group of L13 (Fig. S12). We interpreted the lack of density for the modeled C-terminal carboxyl group of NT8-13 and the presence of electron density close by as a likely result of decarboxylation caused by radiation damage59,60 (estimated absorbed dose over 150 frames ~314 MGy, 10x experimental dose limit, RADDOSE61) with the cleaved entity being stabilized by a potential salt-bridge like interaction with R327. However, for the purpose of understanding the biology and the interactions involved in agonist binding by NTS1 in vivo i.e. in the absence of radiation damage, and to minimize any confusion for those scientists who may not be familiar with x-ray crystallography yet who may view the NTS1-GW5-T4L structure, we have chosen to model NT8-13 with a carboxyl group at its C-terminus in the final NTS1-GW5-T4L model.
Figures were prepared in PyMOL (Schrödinger). Structural alignments were done with the ‘align’ command of PyMOL.
Supplementary Material
Acknowledgements
This research was supported by the Intramural Research Program of the National Institutes of Health (JFW, JG-J, PS, RG: National Institute of Neurological Disorders and Stroke; NN, JS: National Institute of Diabetes and Digestive and Kidney Diseases) and a joint grant from Pfizer Global Research and Development and the MRCT Development Gap Fund in addition to core funding from the UK Medical Research Council MRC U105197215 (YS, CGT). The Protein Production Facility of the New York Consortium on Membrane Protein Structure was supported by the National Institutes of Health grant U54GM075026 (JL, BK). We thank the staff at the General Medicine and Cancer Institute’s Collaborative Access Team (GM/CA-CAT) beamline at the Advanced Photon Source, Argonne National Laboratory for their assistance during data collection. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357.
Abbreviations
- AEBSF
4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]propanesulfonic acid
- CHS
cholesteryl hemisuccinate Tris salt
- DDM
n-dodecyl-β-D-maltopyranoside
- DM
n-decyl-β-D-maltopyranoside
- EDTA
ethylenediaminetetraacetic acid
- ICL
intracellular loop
- LMNG
lauryl maltose neopentyl glycol (2,2-didecylpropane-1,3-bis-β-D-maltopyranoside)
- T4L
cysteine-free bacteriophage T4 lysozyme (C54T, C97A)
- TCEP
Tris (2-carboxyethyl) phosphine hydrochloride
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions
JFW characterized various NTS1 constructs by ligand binding and G-protein assays, tested NTS1 mutants for stability, and purified NTS1 for crystallization. NN collected diffraction data and solved the structure. YS performed alanine scanning mutagenesis and tested NTS1 mutants for stability, and CGT was responsible for the mutagenesis strategy. JL and BK explored and performed the automation of alanine scanning mutagenesis. FX performed crystallization experiments and stability tests. JG-J and PS did alanine scanning and molecular biology on NTS1. JS performed large-scale fermentation. RG performed crystallization experiments, assisted with data collection and was responsible for the overall project strategy. The manuscript was written by RG and CGT.
Author information
Coordinates and structure factors for NTS1-GW5-T4L are deposited in the Protein Data Bank (accession code 4GRV). Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Carraway R, Leeman SE. The isolation of a new hypotensive peptide, neurotensin, from bovine hypothalami. J. Biol. Chem. 1973;248:6854–6861. [PubMed] [Google Scholar]
- 2.Bissette G, Nemeroff CB, Loosen PT, Prange AJ, Jr., Lipton MA. Hypothermia and intolerance to cold induced by intracisternal administration of the hypothalamic peptide neurotensin. Nature. 1976;262:607–609. doi: 10.1038/262607a0. [DOI] [PubMed] [Google Scholar]
- 3.Carraway RE, Plona AM. Involvement of neurotensin in cancer growth: evidence, mechanisms and development of diagnostic tools. Peptides. 2006;27:2445–2460. doi: 10.1016/j.peptides.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 4.Griebel G, Holsboer F. Neuropeptide receptor ligands as drugs for psychiatric diseases: the end of the beginning? Nat. Rev. Drug Discov. 2012;11:462–478. doi: 10.1038/nrd3702. [DOI] [PubMed] [Google Scholar]
- 5.Kitabgi P. Targeting neurotensin receptors with agonists and antagonists for therapeutic purposes. Curr. Opin. Drug Discov. Devel. 2002;5:764–776. [PubMed] [Google Scholar]
- 6.Schimpff R-M, et al. Increased plasma neurotensin concentrations in patients with Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry. 2001;70:784–786. doi: 10.1136/jnnp.70.6.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka K, Masu M, Nakanishi S. Structure and functional expression of the cloned rat neurotensin receptor. Neuron. 1990;4:847–854. doi: 10.1016/0896-6273(90)90137-5. [DOI] [PubMed] [Google Scholar]
- 8.Chalon P, et al. Molecular cloning of a levocabastine-sensitive neurotensin binding site. FEBS Lett. 1996;386:91–94. doi: 10.1016/0014-5793(96)00397-3. [DOI] [PubMed] [Google Scholar]
- 9.Mazella J. Sortilin/neurotensin receptor-3: a new tool to investigate neurotensin signaling and cellular trafficking? Cell. Signal. 2001;13:1–6. doi: 10.1016/s0898-6568(00)00130-3. [DOI] [PubMed] [Google Scholar]
- 10.Barroso S, et al. Identification of residues involved in neurotensin binding and modeling of the agonist binding site in neurotensin receptor 1. J. Biol. Chem. 2000;275:328–336. doi: 10.1074/jbc.275.1.328. [DOI] [PubMed] [Google Scholar]
- 11.Härterich S, Koschatzky S, Einsiedel J, Gmeiner P. Novel insights into GPCR-peptide interactions: mutations in extracellular loop 1, ligand backbone methylations and molecular modeling of neurotensin receptor 1. Bioorg. Med. Chem. 2008;16:9359–9368. doi: 10.1016/j.bmc.2008.08.051. [DOI] [PubMed] [Google Scholar]
- 12.Pang YP, Cusack B, Groshan K, Richelson E. Proposed ligand binding site of the transmembrane receptor for neurotensin(8-13) J. Biol. Chem. 1996;271:15060–15068. doi: 10.1074/jbc.271.25.15060. [DOI] [PubMed] [Google Scholar]
- 13.Deupi X, Standfuss J. Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr. Opin. Struct. Biol. 2011;21:541–551. doi: 10.1016/j.sbi.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol. Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebon G, Warne T, Tate CG. Agonist-bound structures of G protein-coupled receptors. Curr. Opin. Struct. Biol. 2012;22:1–9. doi: 10.1016/j.sbi.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 17.Granier S, et al. Structure of the delta-opioid receptor bound to naltrindole. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manglik A, et al. Crystal structure of the mu-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson AA, et al. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature. 2012;485:395–399. doi: 10.1038/nature11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H, et al. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012;485:327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choe HW, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 23.Standfuss J, et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lebon G, et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521–525. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gether U, et al. Structural instability of a constitutively active G protein-coupled receptor. Agonist-independent activation due to conformational flexibility. J. Biol. Chem. 1997;272:2587–2590. doi: 10.1074/jbc.272.5.2587. [DOI] [PubMed] [Google Scholar]
- 26.Tate CG. A crystal clear solution for determining GPCR structures. Trends Biochem. Sci. 2012;37:343–352. doi: 10.1016/j.tibs.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Shibata Y, et al. Thermostabilization of the neurotensin receptor NTS1. J. Mol. Biol. 2009;390:262–277. doi: 10.1016/j.jmb.2009.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 29.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 30.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warne T, et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barroso S, Richard F, Nicolas-Etheve D, Kitabgi P, Labbe-Jullie C. Constitutive activation of the neurotensin receptor 1 by mutation of Phe(358) in helix seven. Br. J. Pharmacol. 2002;135:997–1002. doi: 10.1038/sj.bjp.0704546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goncalves JA, et al. Highly conserved tyrosine stabilizes the active state of rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 2010;107:19861–19866. doi: 10.1073/pnas.1009405107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogel R, et al. Functional role of the “ionic lock”--an interhelical hydrogen-bond network in family A heptahelical receptors. J. Mol. Biol. 2008;380:648–655. doi: 10.1016/j.jmb.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Vita N, et al. Cloning and expression of a complementary DNA encoding a high affinity human neurotensin receptor. FEBS Lett. 1993;317:139–142. doi: 10.1016/0014-5793(93)81509-x. [DOI] [PubMed] [Google Scholar]
- 38.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J. Mol. Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 40.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henry JA, Horwell DC, Meecham KG, Rees DC. A structure-affinity study of the amino acid side-chains in neurotensin: N and C terminal deletions and Ala-scan. Bioorg. Med. Chem. Lett. 1993;3:949–952. [Google Scholar]
- 42.Luca S, et al. The conformation of neurotensin bound to its G protein-coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10706–10711. doi: 10.1073/pnas.1834523100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warne T, et al. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chae PS, et al. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat. Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobilka BK. Amino and carboxyl terminal modifications to facilitate the production and purification of a G protein-coupled receptor. Anal. Biochem. 1995;231:269–271. doi: 10.1006/abio.1995.1533. [DOI] [PubMed] [Google Scholar]
- 46.Hellmich MR, Battey JF, Northup JK. Selective reconstitution of gastrin-releasing peptide receptor with G alpha q. Proc. Natl. Acad. Sci. U.S.A. 1997;94:751–756. doi: 10.1073/pnas.94.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shibata Y, et al. Thermostabilization of the neurotensin receptor NTS1. J. Mol. Biol. 2009;390:262–277. doi: 10.1016/j.jmb.2009.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hartman JL, Northup JK. Functional reconstitution in situ of 5-hydroxytryptamine 2c (5HT2c) receptors with alphaq and inverse agonism of 5HT2c receptor antagonists. J. Biol. Chem. 1996;271:22591–22597. doi: 10.1074/jbc.271.37.22591. [DOI] [PubMed] [Google Scholar]
- 49.Jian X, et al. The bombesin receptor subtypes have distinct G protein specificities. J. Biol. Chem. 1999;274:11573–11581. doi: 10.1074/jbc.274.17.11573. [DOI] [PubMed] [Google Scholar]
- 50.Gully D, et al. Biochemical and pharmacological profile of a potent and selective nonpeptide antagonist of the neurotensin receptor. Proc. Natl. Acad. Sci. U.S.A. 1993;90:65–69. doi: 10.1073/pnas.90.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inagaki S, et al. Modulation of the interaction between neurotensin receptor NTS1 and Gq protein by lipid. J. Mol. Biol. 2012;417:95–111. doi: 10.1016/j.jmb.2012.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Landau EM, Rosenbusch JP. Lipidic cubic phases: A novel concept for the crystallization of membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cherezov V, et al. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 microm size X-ray synchrotron beam. J. R. Soc. Interface. 2009;6(Suppl 5):S587–S597. doi: 10.1098/rsif.2009.0142.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hilgart MC, et al. Automated sample-scanning methods for radiation damage mitigation and diffraction-based centering of macromolecular crystals. J. Synchrotron Radiat. 2011;18:717–722. doi: 10.1107/S0909049511029918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wacker D, et al. Conserved binding mode of human beta2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. J. Am. Chem. Soc. 2010;132:11443–11445. doi: 10.1021/ja105108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burmeister WP. Structural changes in a cryo-cooled protein crystal owing to radiation damage. Acta Crystallogr. D Biol. Crystallogr. 2000;56:328–341. doi: 10.1107/s0907444999016261. [DOI] [PubMed] [Google Scholar]
- 60.Weik M, et al. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc. Natl. Acad. Sci. U.S.A. 2000;97:623–628. doi: 10.1073/pnas.97.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paithankar KS, Owen RL, Garman EF. Absorbed dose calculations for macromolecular crystals: improvements to RADDOSE. J. Synchrotron Radiat. 2009;16:152–162. doi: 10.1107/S0909049508040430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.