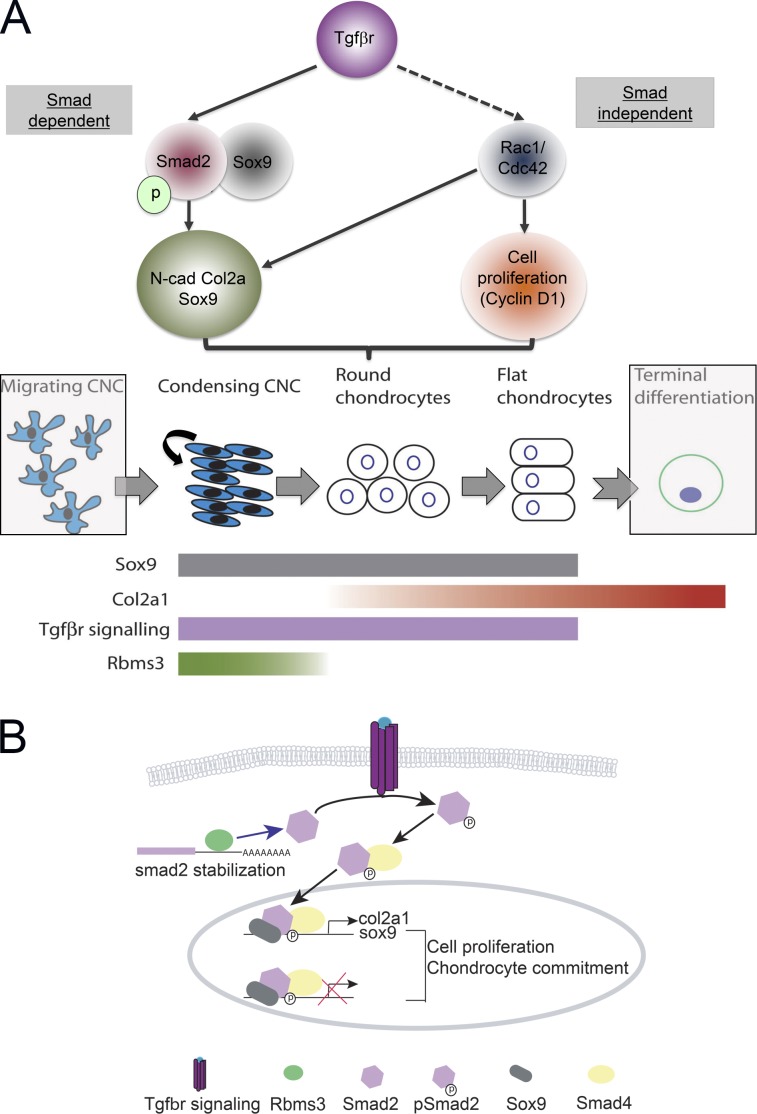
Rbms3 regulates TGF-βr signaling, a critical pathway for chondrogenesis, by binding and stabilizing Smad2 transcripts.
Abstract
Cranial neural crest cells form much of the facial skeleton, and abnormalities in their development lead to severe birth defects. In a novel zebrafish protein trap screen, we identified an RNA-binding protein, Rbms3, that is transiently expressed in the cytoplasm of condensing neural crest cells within the pharyngeal arches. Morphants for rbms3 displayed reduced proliferation of prechondrogenic crest and significantly altered expression for chondrogenic/osteogenic lineage markers. This phenotype strongly resembles cartilage/crest defects observed in Tgf-βr2:Wnt1-Cre mutants, which suggests a possible link with TGF-β signaling. Consistent with this are the findings that: (a) Rbms3 stabilized a reporter transcript with smad2 3′ untranslated region, (b) RNA immunoprecipitation with full-length Rbms3 showed enrichment for smad2/3, and (c) pSmad2 levels were reduced in rbms3 morphants. Overall, these results suggest that Rbms3 posttranscriptionally regulates one of the major pathways that promotes chondrogenesis, the transforming growth factor β receptor (TGF-βr) pathway.
Introduction
RNA processing, localization, and stability from the nucleus to the cytoplasm is coordinated by a multitude of RNA-binding proteins (RBPs) and microRNAs (Keene, 2007). Although the vertebrate genome encodes for hundreds of RBPs, only a small fraction of RBP targets and regulatory roles are known to date (Halbeisen et al., 2008; Hogan et al., 2008).
Neural crest cells are a transient embryonic stem cell population that contributes to important derivatives in vertebrate embryos, including the craniofacial skeleton and peripheral nervous system. Although many critical events in neural crest development are transcriptionally regulated (Sauka-Spengler and Bronner-Fraser, 2008), it is becoming increasingly clear that posttranscriptional modifications, such as SUMOylation (Taylor and LaBonne, 2005) and RNA stabilization/localization, also play important roles (Hogan et al., 2008). However, few studies have examined the role of RNA stabilization in developmental processes.
Interestingly, defects in RNA processing can lead to neural crest-related craniofacial abnormalities. For instance, in Treacher Collins Syndrome, disruption of Tcof1, which is involved in ribosome biosynthesis, leads to craniofacial abnormalities in mice (Dixon et al., 2006). Given that cranial neural crest (CNC) anomalies are among the most common of human birth defects (McKeown and Bronner-Fraser, 2008), it is clearly important to understand the developmental role of RNA stabilization therein.
To this end, we describe the expression and developmental function of an RBP Rbms3 in craniofacial morphogenesis. Isolated in a gene and protein trap screen in zebrafish (Trinh et al., 2011), the Rbms3 RBP belongs to the small family of c-myc single-stranded binding proteins (MSSPs; Penkov et al., 2000). MSSPs contain two ribonucleoprotein (RRM) domains that were originally described in RBPs (Siomi et al., 1993) and required for c-myc promoter binding (Takai et al., 1994). Previously, Rbms3 was shown to exhibit tumor suppressor function via regulation of c-Myc (Li et al., 2011) and to bind/stabilize RNA in vitro (Penkov et al., 2000; Fritz and Stefanovic, 2007; Lu et al., 2012). However, the in vivo function for Rbms3 during development has remained unexplored.
Here, we report that Rbms3 is expressed transiently in condensing CNC in the pharyngeal arches (Penkov et al., 2000). Because our protein trap screen labels endogenous Rbms3, we find that it is predominantly expressed in the cytoplasm of CNC, which suggests that its primary function is in RNA metabolism rather than transcriptional regulation of c-Myc. Embryonic knockdown of rbms3 results in severe craniofacial defects and a reduction in activated Smad2, a downstream effector of TGF-β receptor (TGF-βr) signaling. We present evidence that Rbms3 binds to and stabilizes Smad2 transcripts. Significantly, our results suggest that zebrafish Rbms3 is a novel posttranscriptional regulator of one of the major cartilage differentiation effectors, the TGF-βr pathway. Other novel targets for Rbms3 also include cell cycle regulators (rac1 and cyclin D1) and smad1, which suggests that Rbm3 is a global chondrogenic regulator important for driving CNC down a chondrogenic lineage.
Results
Zebrafish rbms3 is expressed during cartilage formation
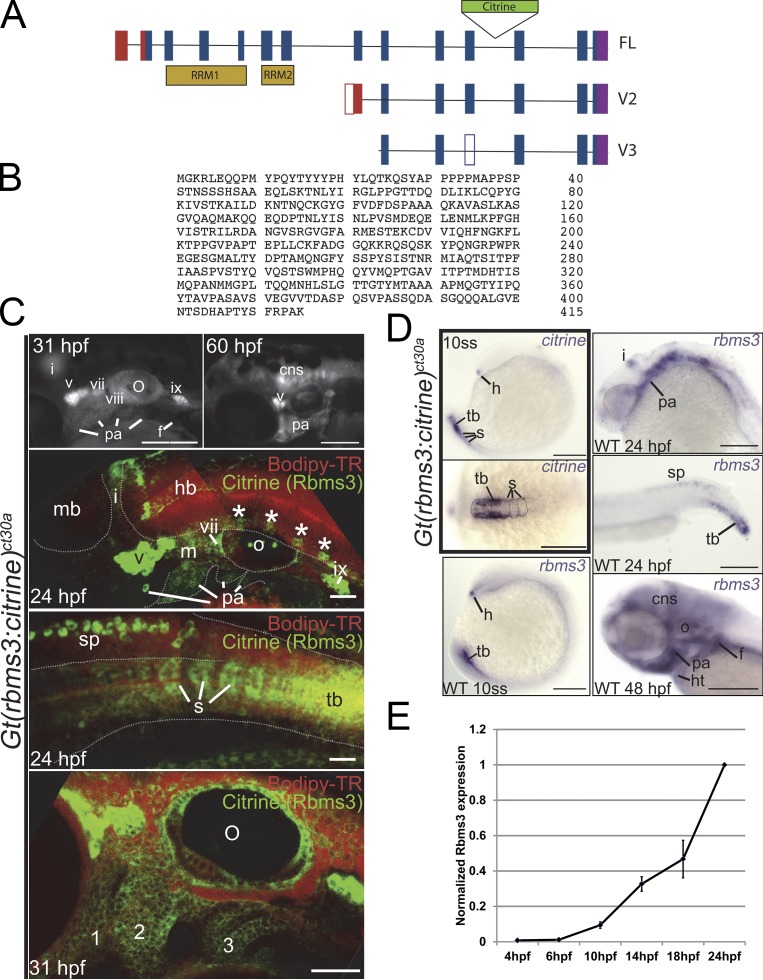
In the Rbms3 trapped line, Gt(rbms3-citrine)ct30a (Trinh et al., 2011), the insertion of the Citrine tag was localized to intron 11. 5′ rapid amplification of cDNA ends (RACE) was performed to retrieve full-length and two C-terminal variants (Fig. 1 A). The full-length rbms3 (exons 2–14) is a 415–amino acid protein; exons 3–7 code for two putative RNA binding domains (RRM).
Figure 1.
Structure and expression of zebrafish rbms3. (A) Full-length and rbms3 variants v2 and v3 were identified using RACE. (B) Full-length zebrafish Rbms3 is 415 amino acids long. (C and D) Endogenous Rbms3 expression as denoted by Citrine expression in the Gt(rbms3-citrine)ct30a line (C); this closely resembles rbms3 transcript expression in wild-type (WT) zebrafish (D). (C) Citrine fluorescence is detected in craniofacial structures from 24 hpf to 60 hpf. 31 hpf and 60 hpf: live whole-mount images. 24 hpf: confocal z-stacks of the head region (top) and tail region (bottom) in live Gt(rbms3-citrine)ct30a homozygotes stained with the vital dye Bodipy-TR (red). Asterisks indicate Rbms3-expressing neurons. cns, central nervous system; cranial ganglia, v, vii, viii, ix, and x; f, fin bud; hb, hindbrain; i, isthmus; m, mesenchyme; mb, midbrain; o, otic vesicle; pa, pharyngeal arches; s, somites; sp, spinal chord; tb, tail bud mesoderm. 31 hpf: confocal section of head region showing expression in the pharyngeal arches (1–3). (D) Citrine transcript is detected at 10 ss (boxed region in the right panel) in the hatching gland (h), tail bud mesoderm (tb), and somites (s, outlined). Lateral (top) and dorsal (bottom) views shown. (E) QPCR analysis of Rbms3 expression during early embryonic development (n = 3). Error bars indicate SD. Bars: (C, top two fluorescent images; and D) 200 µM; (C, bottom three images) 50 µM.
The Citrine in Gt(rbms3-citrine)ct30a is flanked by splice acceptor and donor sites and is incorporated as an artificial exon into the Rbms3 protein (Trinh et al., 2011). Homozygous Gt(rbms3-citrine)ct30a fish are viable, suggesting that Citrine incorporation does not interfere with protein function. Citrine (labeling all three rbms3 variants) was observed in the cranial mesenchyme, pharyngeal arches neurons within the neural tube, the isthmus of the midbrain-hindbrain, fin buds, and tail bud/somites (Fig. 1 C). By 48–60 h postfertilization (hpf), Citrine was absent in the tail. At the 10 somite stage (ss; 14 hpf), citrine is expressed in the hatching gland, the mesoderm of the tail bud and somites. Expression for rbms3 was similar to citrine, which confirmed that tagged and endogenous Rbms3 expression patterns overlapped at 24–60 hpf. By 48 hpf, expression in the heart was also apparent (Fig. 1 D). Quantitative PCR (QPCR) analysis showed that onset of rbms3 expression was around 6–10 hpf (gastrulation to 3 ss; Fig. 1 E). By 16–18 hpf, rbms3 was also expressed in the nervous system and cranial mesenchyme (Fig. S1).
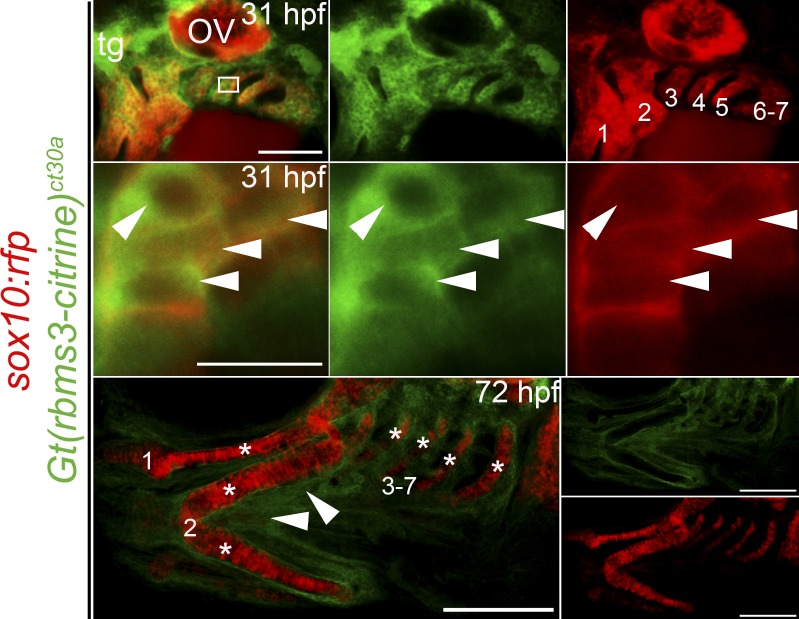
CNC emigrate from the neural tube (5–10 ss/11.5–14 hpf in zebrafish) in a segmental fashion, invade the pharyngeal arches, and form prechondrogenic mesenchyme (Schilling and Kimmel, 1994; Trainor and Krumlauf, 2001; Knecht and Bronner-Fraser, 2002). In Gt(rbms3-citrine)ct30a; Tg(sox10(−7.2):mrfp) fish at 31 hpf, Rbms3 (green) colocalizes with CNC cells (red) in the pharyngeal arches (Fig. 2, top and middle) in the cytoplasm (Fig. 2, middle), which is consistent with a role during RNA metabolism (Penkov et al., 2000; Fritz and Stefanovic, 2007; Lu et al., 2012). By 72 hpf, differentiated cartilage lack Rbms3, but surrounding tissue maintain expression (Fig. 2, bottom, arrowheads). The expression pattern of Rbms3 in prechondrogenic crest (PCC) suggests it may function transiently during cartilage differentiation.
Figure 2.
Rbms3 is expressed by PCC. Citrine-tagged Rbms3 is transiently expressed by PCC (arrowheads, yellow/orange) in the pharyngeal arches (1–7) at 31 hpf (top two rows) and predominantly localized to the cytoplasm (middle row). The boxed region in the top left panel is magnified in the middle row. At 72 hpf, Rbms3 is excluded by cartilage (asterisks) but maintained by surrounding tissue (arrowheads) in the pharyngeal arches (1–7; bottom row). Sox10:rfp (red) marks PCC and cartilage. Bars: (top) 100 µM; (middle) 10 µM; (bottom) 200 µM.
Perturbing rbms3 disrupts craniofacial development
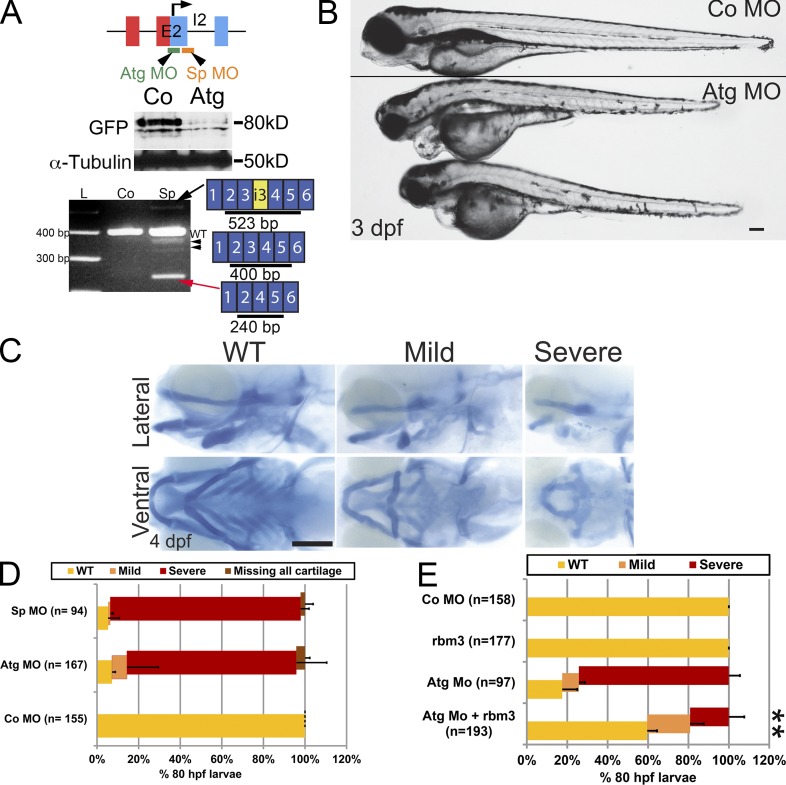
We designed two morpholino oligonucleotides (MOs), a translation blocking translation start site rbms3 morpholino (Atg MO) and a splice site rbms3 morpholino (Sp MO), to perform rbms3 loss-of-function analysis during zebrafish development. Both MOs efficiently blocked Rbms3 production when injected into the Gt(rbms3-citrine)ct30a line, as analyzed by Western blotting using an anti-GFP antibody (Fig. 3 A). The Sp MO was found to skip exon 3 (Fig. 3 B), creating a truncated transcript; sequencing of other minor PCR products also produced other truncated transcripts of rbms3 (Fig. 2 A). Survival of morphant larvae was reduced past 4 d postfertilization (dpf).
Figure 3.
Craniofacial defects in rbms3 morphants. (A) rbms3 MO design (E2, exon 2; I2, intron 2). Atg MO treatment results in knockdown of Citrine-Rbms3 (middle). Sp MO results in skipping of exon 3 (red arrow), and some transcripts maintain a portion of intron 3 (i3, yellow, black arrow), resulting in truncated Rbms3 products. The Sp MO also generates other minor splice variants (black arrowheads). (B) Atg morphants (boxed larvae) show craniofacial defects. (C) Alcian blue cartilage staining in Atg MO–treated fish. Top row, lateral view; bottom row, ventral view. (D) Quantification of cartilage phenotype for Sp and Atg MOs. (E) Full-length rbms3 partially/fully rescues Atg MO severe phenotype (**, P < 0.001, χ2 test). n = total number of treated fish from 3–4 experiments (D and E). Error bars indicate SD. Bars, 200 µM.
Compared with controls, rbms3 morphants displayed cardiac edema, with varying penetrance, smaller body size, a hooked head, and craniofacial defects by 3 dpf (Fig. 3 B).
Rbms3 morphants displayed major disruption to neurocranial and pharyngeal cartilage elements (Fig. 3 C; see Fig. 6 A). About 80–90% of morphants displayed malformed mandibular and hyoid cartilage elements, and with varying penetrance, the cartilage of pharyngeal arches 3–7 was either entirely missing or malformed (Fig. 3, C and D). Co-injecting full-length rbms3 with Atg MO was able to fully or partially rescue the severe phenotype (Fig. 3 E; 74% severe phenotype with Atg MO, n = 97, vs. 20% severe phenotype with Atg MO coinjected with rbms3, n = 193, P < 0.001).
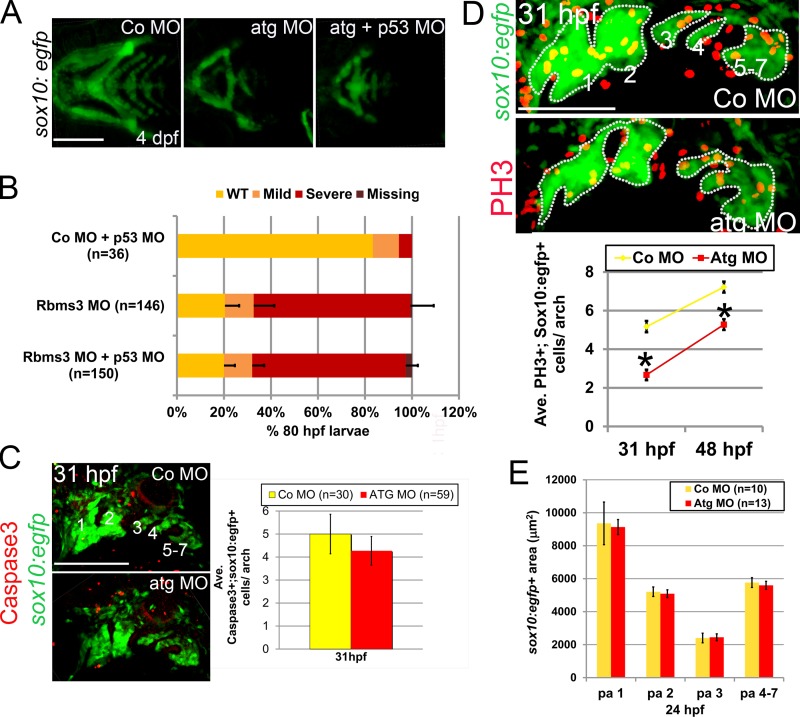
Figure 6.
Cell proliferation is defective in rbms3 morphants. (A and B) Co-injection of Atg MO with p53 MO does not rescue the cartilage phenotype. (A) Ventral views of MO-injected fish. (B) Quantitation of the cartilage phenotype: Atg MO coinjected with p53 MO. P > 0.05 (χ2 test). n = number of larvae pooled from three experiments. Error bars indicate SD. (C) There is no difference in activated Caspase-3 (red) expression in the pharyngeal arches (green; 1–7) in rbms3 morphants. Lateral view of MO-injected sox10:egfp fish at 31 hpf and quantitation of bifluorescent activated Caspase-3+;sox10:egfp+ cells. Error bars indicate SEM. P = 0.5 (Student’s t test). (D) Cell proliferation marker PH3 (red) is reduced in rbms3 morphant pharyngeal arches (green; 1–7). Lateral view of MO-injected sox10:egfp fish at 31 hpf and quantitation of bifluorescent PH3+ and sox10:egfp+ cells in the pharyngeal arches at 31 and 48 hpf. Error bars indicate SEM. *, P < 0.0001 (Student’s t test). (E) Domain of pharyngeal arch (pa) sox10:egfp expression is comparable between MO treatments at 24 hpf. Error bars indicate SD. n = number of fish (B–E). Bars: (A and C) 200 µM; (D) 100 µM.
Rbms3 does not affect early CNC
Tissue interactions and signaling during gastrulation are essential for proper CNC formation in the embryo (Rogers et al., 2012). The cartilage abnormalities observed in rbms3 morphants may be caused by early patterning defects, which subsequently affect early CNC formation and migration (Fig. 4).
Figure 4.
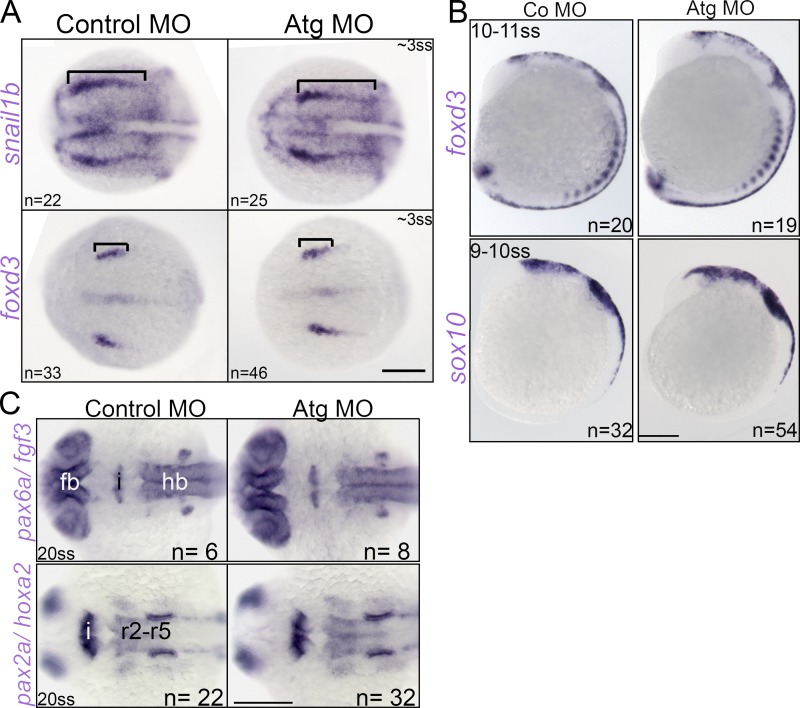
Early CNC and neural development in rbms3 morphants. (A) Dorsal view of neural plate border markers snail1b and foxd3. Bracket: anterior-posterior extent of domain is unchanged. (B) Pre-migratory foxd3 and sox10 CNC expression is unchanged. (C) Dorsal views of neural marker expression. Fb, forebrain (pax6a); hb, hindbrain (pax6a, hox2a); and i, isthmic regions (fgf3, pax2a). r2-r5, rhombomeres 2–5. Bars, 200 µM.
Analysis of the neural plate boarder markers snail1b and foxd3 at 3 ss (10 hpf) showed no differences between controls and rbms3 morphants (Fig. 4 A). No significant changes were also observed in expression of CNC markers sox10 and foxd3 at 10 ss (Fig. 4 A).
Rbms3 is expressed in the isthmus at the midbrain–hindbrain junction. To rule out the possibility that the cartilage defects in rbms3 morphants may be secondary to midbrain–hindbrain patterning defects, we analyzed markers of forebrain (pax6a), isthmus (fgf3/pax2a), and hindbrain (hoxa2) at 20 ss (19 hpf). No significant differences between controls and rbms3 morphants were observed (Fig. 4 C). Collectively, the results suggest that early patterning in rbms3 morphants is normal, and that the cartilage phenotype is not a secondary defect.
Abnormal PCC development in rbms3 morphants
The cartilage phenotype may result from defects in PCC condensation and/or differentiation because early CNC development was normal in rbms3 morphants. Analysis of dlx2a revealed no significant differences in PCC condensation at 24 hpf in the pharyngeal arches (Fig. 5 A). At this stage, Rbms3 is only weakly expressed and later up-regulated in the pharyngeal arches by 31 hpf (Fig. 1 C).
Figure 5.
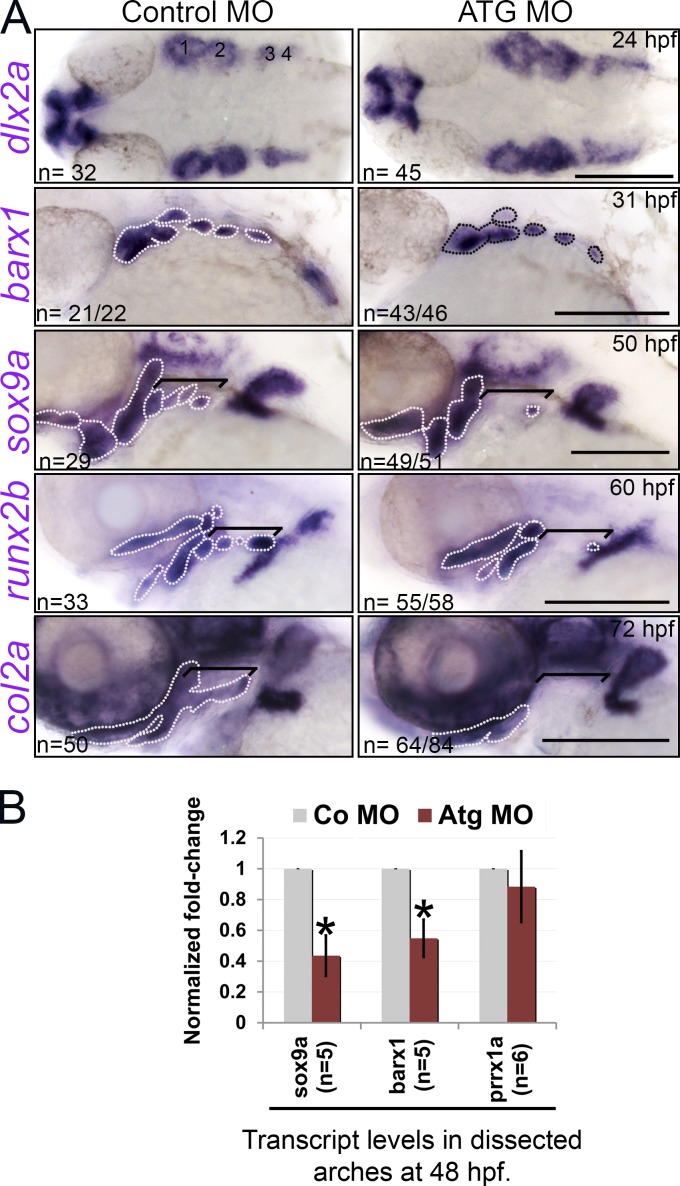
PCC differentiation is affected in rbms3 morphants. (A) dlx2a PCC condensations are normal (1–4) yet PCC/chondrogenic (barx1, sox9a, and col2a1a) and osteogenic (runx2b) markers are severely affected in morphants. Area highlighted by the broken lines and brackets: pharyngeal arch expression. Brackets: posterior arch expression. Bars, 200 µM. (B) QPCR shows reduction in sox9a and barx1 in rbms3 morphants. Error bars indicate SD. *, P < 0.005 (Student’s t test). n = independent pools of RNA.
In contrast, significant effects on PCC and its derivatives were observed in rbms3 morphants from 31 hpf to 72 hpf. Barx1, sox9a (PCC/chondrocytes), and col2a1a (mature chondrocytes) domains of expression were reduced/missing in morphants (Fig. 5 A). QPCR analysis at 48 hpf showed a reduction in sox9a and barx1 levels in rbms3 morphants (P < 0.005), yet no significant difference in prrx1a (cranial mesenchyme) was observed (Fig. 5 B; P = 0.14), which suggests that in rbms3 morphants, CNC-derived mesenchyme in the pharyngeal arches is specifically affected. Examination of runx2b (osteogenic lineage) also showed altered expression in rbms3 morphants at 60 hpf (Fig. 5 A).
Rbms3 controls PCC cell proliferation
Nonspecific MO effects may indirectly cause craniofacial defects through the activation of the p53–apoptotic pathway (Robu et al., 2007). Tg(sox10(−4.7):egfp) embryos coinjected with Atg MO and p53 MO failed to rescue the rbms3 morphant cartilage phenotype, which suggests that the observed craniofacial defects were specifically caused by loss of rbms3 (Fig. 6, A and B). Moreover, no significant difference in the numbers of Caspase-3+; EGFP+ cells were observed in the pharyngeal arches at 31 hpf between controls and rbms3 morphants (Fig. 6 C).
In contrast, cell proliferation was markedly reduced in rbms3 morphants at 31 hpf (Co MO, n = 31; Atg MO, n = 27) and 48 hpf (Co MO, n = 33; Atg MO, n = 34), as assayed by PH3 expression in Tg(sox10(−4.7):egfp) fish (Fig. 6 D; P < 0.001). Comparison of sox10:egfp+ pharyngeal arch domains between controls and rbms3 morphants at 24 hpf showed no significant differences (Fig. 6 E), which suggests that the proliferative differences observed between treatments at 31 hpf are not caused by significant loss of pharyngeal arch cells at early stages. Thus, Rbms3 functions in PCC by regulating cell proliferation.
Tgf-βr signaling is reduced in rbms3 morphants
The CNC phenotype observed in rbms3 morphants is reminiscent of knockdown of mouse TGF-βr2 in Wnt1-Cre mice (Ito et al., 2003; Wurdak et al., 2005). In both cases, the primary deficit is in the capacity of PCC to differentiate along the chondrogenic lineage because of an inability to maintain Sox9 expression. Furthermore, Smad2/3, effectors of the TGF-βr2 pathway, promote Sox9-dependent transcriptional activity on the a1(II) collagen gene (Col2A1) enhancer and induce chondrogenesis in vitro (Furumatsu et al., 2005).
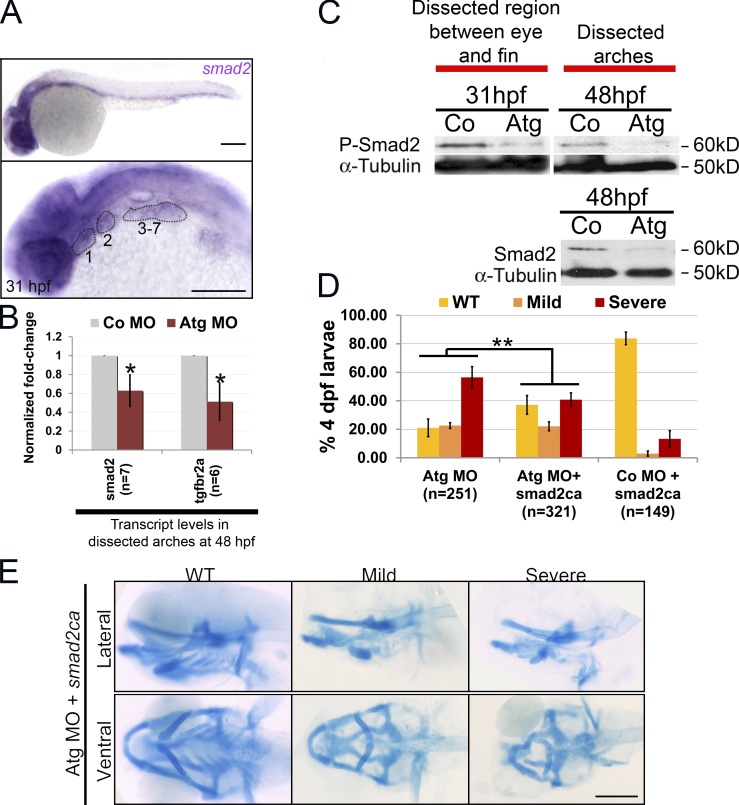
In zebrafish, smad2 is expressed by PCC at 26 hpf (Müller et al., 1999) and 31 hpf (Fig. 7 A). At 48 hpf, there was a reduction in transcription of both smad2 and tgf-βr2a receptor in the pharyngeal arches of rbms3 morphants (Fig. 7 B; P < 0.005). This also corresponded to reduced Smad2 protein levels at 48 hpf in rbms3 morphant arches (Fig. 7 C); quantitation of band intensity revealed a ratio of 1:0.3 ± 0.05 (n = 3) in controls/morphants after normalization to α-tubulin (Fig. 7 C). A reduction in phosphorylated Smad2 (pSmad2, reflecting active Tgf-β signaling) levels in rbms3 morphants at 31 hpf and 48 hpf was also observed in the pharyngeal arches (Fig. 7 C). These results further support the hypothesis that the rbms3 morphant cartilage phenotype may be caused by defects in Tgf-βr signaling and that smad2 is a putative Rbms3 target.
Figure 7.
Smad2 signaling is defective in rbms3 morphants. (A) At 31 hpf, smad2 is expressed in the pharyngeal arches (numbered and highlighted). (B) QPCR shows that smad2 and tgf-βr2a are reduced in rbms3 morphants at 48 hpf. Error bars indicate SD. *, P < 0.005 (two-tailed Student’s t test). n = independent pools of RNA. (C) pSmad2 (top) and total Smad2 (bottom) is reduced in rbms3 morphants. (D and E) Quantitation of phenotype (D) and cartilage staining (E) of fish coinjected with smad2ca and Atg MO. **, P < 0.001 (χ2 test). Bars, 200 µM.
We next tested whether constitutively activated smad2 (smad2ca), which signals independently of TGF-βr, was able to rescue the phenotype. Because injecting smad2ca alone produces a dorsalized phenotype (Müller et al., 1999; Dick et al., 2000), we injected RNA below the threshold of an observable phenotype and excluded any embryos exhibiting axis defects from analysis (Fig. 7, D and E). To improve survival of smad2ca and Atg MO coinjected embryos, we also reduced Atg MO concentration (∼150 µM), which in turn reduced the severe phenotype to ∼60%. Co-injecting smad2ca RNA with Atg MO nearly doubled the number of wild-type larvae (∼40%, n = 321 vs. Atg MO alone, ∼20%, n = 251; Fig. 7 D; P < 0.001). Therefore, the rbms3 morphant cartilage defect is due, at least in part, to reduction in Tgf-βr/Smad2 signaling.
Rbms3 regulates stability of smad2 by binding to smad2 3′ untranslated region (UTR)
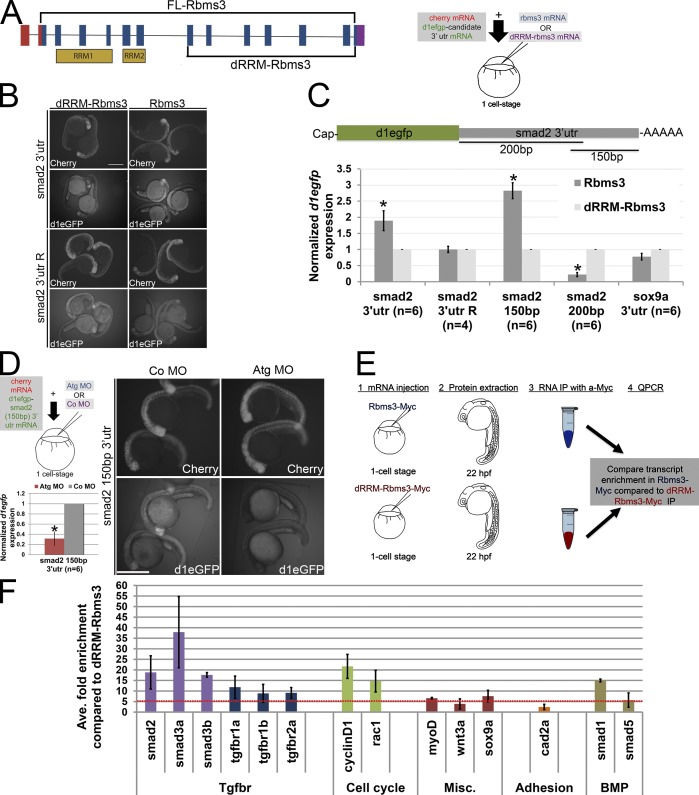
In PCC, Rbms3 may regulate the stability of smad2 transcript and thus affect the pool of protein available for signaling, as evidence suggests that Rbms3 can bind to the 3′ UTRs (Fritz and Stefanovic, 2007; Lu et al., 2012). To test if binding sites were harbored within the 3′ UTR of zebrafish smad2, embryos were coinjected with full-length Rbms3 and destabilized egfp (d1egfp) mRNA fused to smad2 3′ UTR (d1egfp-smad2 3′ UTR; Fig. 8, A–C). Comparing fish at 22–24 hpf injected with a truncated C-terminal version of rbms3 (dRRM-rbms3; Fig. 8 A) versus full-length rbms3 showed higher d1EGFP protein and transcript levels (Fig. 8, B and C), which suggests that the d1egfp-smad2 3′ UTR transcript half-life was extended with full-length Rbms3 (1.7 ± 0.3-fold increase, P = 0.05; Fig. 8 C, asterisk). In contrast, no difference in fluorescence was observed with smad2 3′ UTR in the reverse orientation (smad2 3′ UTR R), since it would not bind directly/indirectly to Rbms3 (P > 0.05) when coinjected with either rbms3 or dRRM-rbms3 (Fig. 8, B and C). The last 150 bp of smad2 3′ UTR was found to harbor Rbms3-mediated stabilization sites (2.8 ± 0.2 fold, P < 0.05; Fig. 8 C, asterisk). However, the first 200 bp region of smad2 3′ UTR was found to destabilize the reporter transcript (Fig. 8 C; *, P < 0.05). Additionally, Rbms3 has no effect on sox9a 3′ UTR (P = 0.7; Fig. 8 C). To further extend these findings, we tested d1egfp-smad2-150bp 3′ UTR stability in the absence of endogenous Rbms3. Co- or Atg MO–treated embryos were coinjected with d1egfp-smad2-150bp 3′ UTR, and Atg MO–treated embryos had significantly less d1egfp transcript (*, P < 0.05) and protein levels compared with controls at ∼22–24 hpf (Fig. 8 D). Collectively, these results suggest that Rbms3 stabilizes smad2 transcripts by acting on the last 150 bp of the smad2 3′ UTR.
Figure 8.
Smad2 transcripts are a target of Rbms3. (A) dRRM-Rbms3 lacks the putative RNA-binding domains (RRM; lower bracket, left). A schematic showing an in vivo stability assay is shown on the right. (B and C) Fluorescent images (B) and QPCR (C; d1egfp normalized to cherry) showing that full-length Rbms3 treatment stabilizes d1egfp-smad2 3′ utr and d1egfp-smad2 (150bp) 3′ utr reporters. (D) Schematic shows the experiment (top left), QPCR (d1egfp normalized to cherry; bottom left), and fluorescent images (right) showing that Atg MO treatment destabilizes d1egfp-smad2 (150bp) 3′ utr. (B and D) Images were taken under same brightness/contrast conditions. (E) Schematic of RIP experiment. (F) QPCR showing transcripts enriched (more than fivefold, red dotted line) for full-length Rbms3 (n = 2–3 experiments). Error bars indicate SEM (C) and SD (D and F). n = RNA from individual embryos (C and D). *, P < 0.05 (Student’s t test). Bars, 500 µM.
Rbms3 binds directly to multiple targets important for chondrogenesis
To test for direct binding, we performed RNA immunoprecipitation (RIP). Either full-length Rbms3-Myc or truncated Rbms3 lacking RNA binding domains (dRRM-Rbms3)-Myc was injected into zebrafish embryos, and c-Myc antibody was used to pull down associated mRNA transcripts (Fig. 8, E and F). Analysis of the Tgf-βr pathway components in the Rbms3-Myc pull-downs revealed higher enrichment for smad2, smad3a, and smad3b compared with tgf-β receptors (compare purple with blue; Fig. 8 F). Other transcripts that were significantly enriched (more than fivefold; Fig. 8 F, broken red line) for full-length Rbms3 were cell cycle regulators (cyclin D1 and rac1) and smad1.
We tested specificity of binding for Rbms3 and found very little enrichment for the following transcripts. These transcripts overlapped with observed domains of expression for endogenous Rbms3 around 10–24 hpf: wnt3a, expressed in the isthmus and other neural tissues (Clements et al., 2009); the adhesion molecule cad2a (n-cad), ubiquitously expressed yet enriched in neural tissues (Harrington et al., 2007); and transcription factors (myod and sox9a), both expressed in somites, with the latter also being expressed in the otic placodes (Fig. 8 F; Weinberg et al., 1996; Yan et al., 2005). These results suggest that posttranscriptional binding and stabilization of smad2 and other transcripts associated with the Tgf-βr signaling pathway may be a direct mechanism by which Rbms3 regulates PCC differentiation into cartilage.
Discussion
From a protein trap screen (Trinh et al., 2011), we isolated zebrafish rbms3, an RBP that belongs to the small family of MSSP (Penkov et al., 2000). For the first time, we have analyzed the in vivo developmental function for Rbms3. Here we report that Rbms3 is predominantly restricted to the cytoplasm and that rbms3 morphants exhibit severe craniofacial defects. Significantly, our results highlight a posttranscriptional role for Rbms3 in regulating one of the major pathways that promote chondrogenesis, the TGF-βr pathway.
Rbms3 cells autonomously affect prechondrogenic transcription factors
Knockdown of rbms3 with two different MOs resulted in severe craniofacial defects (Fig. 3). Early neural crest development proceeded normally (Fig. 4). Although CNC migration to and condensation within the pharyngeal arches appeared unaffected in rbms3 morphants, pharyngeal arch expression domains of barx1, sox9a, col2a1a, and runx2b were severely reduced or affected (Fig. 5), which suggests a major role for Rbms3 during chondrogenesis.
CNC migration and condensation relies on interactions between CNC and their environment (Trainor and Tam, 1995; Trainor and Krumlauf, 2001; García-Castro et al., 2002; Trainor et al., 2002; Crump et al., 2004; Eberhart et al., 2006). Citrine tagged Rbms3 is expressed in the pharyngeal arches and the paraxial cranial mesoderm during CNC condensation in the arches (Figs. 1 and 2). High-resolution analysis in the Tg(sox10(−7.2):mrfp) line revealed that Rbms3 is transiently expressed by PCC and excluded from differentiated cartilage while being maintained by surrounding tissues (Figs. 1 and 2). Collectively, the results suggest that Rbms3 promotes differentiation of PCC to cartilage in a cell-autonomous manner.
Cell proliferation is reduced in rbms3 morphants
A fine balance between cell proliferation and apoptosis is essential for proper cranial cartilage formation (Plaster et al., 2006; Jones et al., 2008). Rbms3 morphants show a reduction in proliferating CNC (Fig. 6). This may be caused by (a) proliferative/apoptotic defects in the neuroepithelium, which influence the number of emigrating CNC; or (b) proliferative/apoptotic defects in the condensing ectomesenchyme. The former seems unlikely because Rbms3 is not expressed in the neuroepithelium during CNC induction. Additionally, condensation of CNC in the pharyngeal arches at 24 hpf is unaffected, and co-injecting rbms3 MO with p53 MO does not rescue the cartilage phenotype (Fig. 6). This differs from the flathead mutants, where decreased proliferation of CNC cells at 48 hpf and activation of the p53 pathway lead to a deficit in zygotic DNA polymerase δ1, thus affecting cartilage morphogenesis. In contrast, in rbms3 morphants, lack of proliferation does not activate the p53 pathway or increase apoptosis (Fig. 6).
Craniofacial defects in rbms3 morphants resemble Tgf-βr pathway defects in mice
We hypothesized that Rbms3 may modulate the TGF-βr pathway. Consistent with this possibility, rbms3 and smad2 (an effector of the TGF-βr pathway) exhibit overlapping expression during CNC condensation in the pharyngeal arches (Figs. 1 and 7). Mice conditionally deficient for the TGF-βr2 pathway exhibit a similar cartilage phenotype to rbms3 morphants (Ito et al., 2003; Wurdak et al., 2005), although previous work in these mutant mice only concentrated on palatogenesis and no comment was made on the more posterior cartilage defects. As observed in Tgf-βr2fl/fl; Wnt1-Cre mice, rbms3 morphants show a reduction in sox9a expression (Fig. 5) and cell proliferation of PCC (Fig. 6), whereas pre-migratory and early migrating CNC appear (Fig. 4) normal, and cell death is unaffected (Fig. 6).
The phenotypes observed in other TGF-βr pathway mutants are not consistent with the overall rbms3 MO phenotype. In tgf-βr3 morphants, survival of early neural crest is affected, resulting in a secondary cartilage phenotype (Cheah et al., 2010). The requirement for Tgf-βr1 (Dudas et al., 2006) is significantly different from Tgf-βr2 during chondrogenesis, suggesting that Tgf-βr1 may participate in diverse signaling cascades during craniofacial development. Furthermore, signaling via Tgf-βr1 is not necessary for PCC proliferation but is required for survival (Dudas et al., 2006). Although it is true that Tgf-βr1 can activate pSmad2, we suggest that reduction in observed Smad2 signaling is primarily caused by the Tgf-βr2–pSmad2 pathway deficiency.
Early PCC proliferation is likely to be regulated mainly through the TGF-βr receptor pathway in a Smad-independent manner. Although Tgf-βr2 null mice exhibit a cell proliferation defect (Ito et al., 2003), the mechanism by which this occurs has not been established during chondrocyte differentiation (Fig. 9 A, broken line). Previously, the small GTP-binding proteins Cdc42/Rac1 were shown to promote chondrocyte differentiation in mesenchyme condensations by promoting Sox9, Col2A, and N-cadherin expression (Woods et al., 2007). Additionally in a cell-dependent context, Cdc42 can activate cell proliferation in a Smad-independent manner after TGF-βr activation in fibroblasts (Wilkes et al., 2003). Collectively, the decreased proliferation observed in rbms3 morphants is hypothesized to be a direct consequence of down-regulating components of the TGF-βr pathway, likely in a Smad-independent manner. In support of this, we find that the Tgf-β receptor transcripts are enriched after immunoprecipitation for Rbms3 (Fig. 8). The direct interactions between TGF-βr signaling, Cdc42/Rac1, and cell cycle effectors remain unclear, but this study demonstrates that rac1 and cyclin D1 can be independently regulated by Rbms3 (Fig. 8).
Figure 9.
Rbms3 function during cartilage formation. (A) Schematic showing how the TGF-βr signaling (Smad-dependent and -independent pathways) affects chondrocyte differentiation (top) and how chondrocytes differentiate from migrating CNC (bottom). Cells not responding to TGF-βr signaling are in shaded gray boxes. Migrating CNC form PCC condensations in the pharyngeal arches. Here, Sox9+ (gray) CNC proliferate (black arrow) and receive TGF-β signals (purple), which drives them down the chondrogenic lineage. During development, these cells gradually increase production of Col2a1 (graded red) and reach terminal differentiation. Rbms3 (graded green) is transiently expressed as CNC condense and is gradually excluded from committed chondrocytes. (B) Rbms3 functions to stabilize smad2 transcripts by binding to smad2 3′ utr. This increases/maintains the pool of Smad2 available to receive TGF-βr signaling. Upon Tgf-βr signaling, pSmad2–Smad4 complexes enter the nucleus (Liu, 2003) and bind to Sox9 to: (a) activate transcription of cartilage gene col2a1 (Furumatsu et al., 2005), (b) maintain levels of sox9 (Ito et al., 2003; Wurdak et al., 2005), and (c) repress a battery of other transcription factors (Ito et al., 2003). This drives the PCC to proliferate and commit toward the chondrogenic lineage (Ito et al., 2003; Wurdak et al., 2005).
Although the cell proliferation defect is likely to be mainly Smad independent yet TGF-βr signaling dependent, we suggest that an inability to generate cartilage and bone is caused by a Smad2 signaling deficiency in PCC progenitors (Fig. 9). Atg MO treatment resulted in reduced levels of pSmad2 (indicative of active Tgf-βr signaling), probably because of reduced Smad2 transcript and total protein levels in morphants. Importantly, coinjection of capped smad2ca RNA with rbms3 MO significantly rescued the severe cartilage phenotype (Fig. 7), further supporting the notion that Rbms3 modulates the Tgf-βr2 pathway during cartilage formation (Fig. 9).
Rbms3 directly binds to smad2/3 transcripts
The Tgf-βr pathway is highly modulated during development to control the precise timing and duration of signaling (Liu, 2003; Massagué et al., 2005). Steady-state levels of receptor Smads (such as Smad2) are regulated by the ubiquitin pathway in a signal-dependent and -independent manner (Izzi and Attisano, 2004). Another possible mechanism for regulating protein levels is by altering the levels of transcript either by modulating the half-life of mRNA or altering accessibility of mRNA to translational machinery through compartmentalization of transcripts to different regions within the cell. This is facilitated by a multitude of RBPs and microRNAs (Lasko, 2003; Keene, 2007). Evidence from the in vivo stability assay suggests that the smad2 3′ UTR harbors Rbms3-binding sites (Fig. 8), thereby lending further support to the idea that smad2 levels and thus Smad2 steady-state protein levels are regulated by Rbms3. The region localized to the last 150 bp of smad2 3′ UTR is sufficient to stabilize the reporter transcript. However, the first 200 bp of the smad2 3′ UTR caused the reporter transcript to be destabilized, suggesting that there are “functional” domains within the smad2 3′ UTR that both positively and negatively regulated by Rbms3. It may also reflect Rbms3 dependence for RNA secondary structure to facilitate binding of the Rbms3–RNP complex (Hiller et al., 2006; Li et al., 2010). The requirement for endogenous Rbms3 to stabilize smad2 150 bp 3′ UTR and significant enrichment of smad2/3 transcripts after RIP for Rbms3 further provides evidence that Smad2 is a direct target (Fig. 8). Smad3 also participates during chondrocyte differentiation (Ferguson et al., 2000; Yang et al., 2001) and is thus likely to be regulated directly by Rbms3.
The reduction observed in sox9a expression (Fig. 5) is also consistent with the hypothesis that Rbms3 is a modulator of the TGF-βr pathway. Sustained Sox9 expression by condensing prechondrogenic CNC requires active TGF-βr2 signaling (Ito et al., 2003; Wurdak et al., 2005). Analysis of Sox9−/− chimeric mice reveals that loss of Sox9 expression prevents PCC from differentiating into cartilage, which was formed entirely from wild-type cells, whereas Sox9−/− cells, which also lacked Col2a1, were excluded (Bi et al., 1999). Conditional inactivation of Sox9 during CNC condensation maintains these cells in a mesenchymal state, as observed by a lack of Col2A1-positive cartilage extracellular matrix production (Akiyama et al., 2002). This is consistent with the finding that Smad2/3, effectors of the TGF-βr2 pathway, promote Sox9-dependent transcription of Col2A1 in vitro (Furumatsu et al., 2005). Further support of the notion that loss of Smad2 is the primary defect and that sox9a loss in rbms3 morphants is a secondary outcome comes from the finding that the sox9a 3′ UTR is unresponsive to Rbms3 and is not highly enriched during RIP (Fig. 8).
Other pathways are regulated by Rbms3 during chondrogenesis
While TGF-βr signaling is a major pathway that promotes chondrogenesis, other signaling pathways also contribute to this process (Walshe and Mason, 2003a; Wada et al., 2005; Yoon et al., 2005; Eberhart et al., 2006; Sakai et al., 2006; de Frutos et al., 2007; Oka et al., 2008). In support of this, our data suggest that smad2ca alone is not sufficient to fully rescue the rbms3 morphant phenotype (Fig. 7). Mice deficient for Bmpr1a and Bmpr1b exhibit defects in PCC proliferation and differentiation (Yoon et al., 2005). Additionally, a zebrafish transgenic reporter for Smad1/5/8 and expression of smad1 also highlight a requirement for BMP signaling during chondrocyte development (Alexander et al., 2011; Laux et al., 2011). Collectively, we suggest that the cell proliferation defect and the loss in sox9a expression could be partially caused by a loss of BMP–Smad1 signaling and rac1 regulation by Rbms3 (Fig. 8).
A model for Rbms3 function during CNC development
Integrating these data, we propose a model for Rbms3 function during prechondrogenic CNC development in which Rbms3 posttranscriptionally regulates components of the Tgf-βr pathway to influence the proliferative state and initial differentiation of prechondrogenic CNC (Fig. 9). Previously, Prx1 and Ptf1a mRNA was demonstrated to be up-regulated by Rbms3 through direct interaction with the Prx1/Ptf1a 3′ UTR sequence (Fritz and Stefanovic, 2007; Lu et al., 2012). In a similar manner, we predict that Rbms3 influences transcript stability of Tgf-βr pathway components (particularly smad2) and therefore transiently influences the timing and duration of Tgf-βr signaling within CNC cells. Additionally, Rbms3 is likely to also regulate other components of the Smad2-independent pathway, such as cell cycle regulators and the BMP–Smad1 pathway. Once these prechondrogenic cells become committed to form round chondrocytes, rbms3 expression is shut down in these cells. Under normal conditions, PCC are likely to be sensitive to levels of Tgf-βr/BMP signaling and require a certain threshold of signaling to drive them down the chondrogenic lineage. In rbms3 morphants, a reduction in the pool of smad2 transcripts caused by reduced levels of Rbms3 is likely to directly influence the pool of Smad2 protein available in the cytoplasm that is able to respond to external Tgf-β signals, thereby maintaining these cells in a mesenchymal state. Once prechondrogenic cells are driven into the cartilage lineage, we predict that the function of Rmbs3 in the CNC is complete. At this juncture, perhaps other RBPs or other mechanisms help maintain these cells in a committed chondrogenic state.
Materials and methods
Zebrafish husbandry and lines
Wild-type (AB or AB/TL background) and transgenic zebrafish lines (see below) were maintained according to institutional animal care and use protocols. Embryos were collected in egg water, maintained at 28°C, and staged according to Kimmel et al. (1995) based on somite number or landmark structural features.
Gt(rbms3-citrine)ct30a were maintained in a mixed AB/TL background. Previously published Tg(sox10(−7.2):mrfp) (Kucenas et al., 2009) or Tg(sox10(−4.7):egfp) (Dutton et al., 2008) were maintained in the AB background.
Cloning of rbms3
To retrieve the full-length rbms3, wild-type 42–48 hpf cDNA library was used to amplify full length and variants. 5′ RACE was performed using the Generacer kit (Invitrogen) using 5′-AGAGCGTTCCAGCCTCATCACAGGACGC-3′ primer. 3′ RACE was performed as described previously (Jayasena et al., 2011) using a 5′-AGAGCGTTCCAGCCTCATCACAGGACGC-3′ primer. For subcloning of full-length rbms3 containing 5′ UTR and open reading frame into pCS2+ at the Cla1–Xba1 sites are: 5′-GATCCCATCGATACAGCCAGAAGGATTCCAGCTAC-3′ and 5′-TGAGGAGTAGAACCCGTTCTGCATTG-3′.
MO injections
MOs were injected either into AB or AB/TL wild-type background. The translation start site MO (Atg MO) is as follows: 5′-CGTTTGCCCATGTAGCTGGAATCCT-3′. The splice MO (Sp MO) sequence is as follows: 5′-CATCTGGGAGAGACACTTACGGTTG-3′. Atg MO stock injecting solution was at 160–120 µM and Sp MO was at 50 µM (unless otherwise specified). 2.3 nl of the stock solution was injected per embryo at the 1–2-cell stage. For control injections, the standard MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′ (Co MO), was used. p53 MO injection stock solution was at 250 µM (Robu et al., 2007). All MOs were purchased from Gene Tools, LLC. To detect splice variants generated by Sp MO, the following PCR primer pair was used: Exon2F, 5′-AGAACCACAAGCGGAGGCTTT-3′; with Exon5/6R, 5′-GTCCTGCTCTTGTTGCTTTGCCAT-3′.
For rescue experiments with the Atg MO, a kozac sequence (5′-GCCACC-3′) was cloned immediately upstream of a mutagenized full-length rbms3 into pCS2+ at the Cla1–Xba1 sites. The endogenous first two codons (indicated by underlines) following the start site (shown in bold letters; 5′-ATGGGCAAA-3′) of rbms3 were changed to 5′-ATGGGAAAG-3′.
Imaging and image processing
For live confocal imaging, Gt(pwp2h-citrine)ct30a fish were soaked in BODIPY TR methyl ester vital dye (Invitrogen) for 30–60 min at 28°C to label all cells before mounting. Fish were anesthetized using 0.1% Tricaine, pH 7.2, in egg water and mounted laterally onto imaging molds (1% agarose in egg water).
All confocal imaging was performed at room temperature. Confocal z-stack images ranging from 40 to 100 µM were acquired using a microscope (LSM 510) and 25× LD LCI Plan-Apochromat NA 0.80 DIC 1 mm Korr or LD C-Apochromat 40×/1.1 W Korr UV-VIS-IR objective lenses (all from Carl Zeiss). LSM 510 version 4 software (Carl Zeiss) was used to acquire images.
Imaging of antibody-stained fish in whole-mount was performed as described for live confocal imaging earlier. Details of used fluorochromes are described further on. Imaging for whole-mount in situ hybridization and Alcian blue staining was performed using a microscope (Stemi SV 11; Carl Zeiss) with a Plan-Axiochromatic S 1.6× objective lens. Whole-mount images were acquired using AxioVision 4.6.3 software (Carl Zeiss).
All confocal images were processed using ImageJ 1.38×. Brightness/contrast processing of images was performed using Photoshop 7 (Adobe).
In situ hybridization and immunofluorescent staining
A combination of two probes for rbms3 were used: the first contained 5′ UTR and dRRM domains of rbms3 and was generated using 5′-GTGTGGAGCAACCGGCAG-3′ with 5′-TAATACGACTCACTATAGGGTCTCAGTGGACTCCATTCTGGCAA-3′ containing a T7 promoter (underlined) site. The second probe template spanned from the dRRM domains to the stop codon in pPCRII-TOPO (Invitrogen) and was generated using Not1 and with SP6 RNA polymerase. In situ hybridization was performed as described previously (Jayasena et al., 2008). In brief, embryos or larvae were treated with phenylthiourea (PTU) to prevent pigmentation and fixed in 4% PFA. Fish older than 24 hpf were Proteinase K treated (20–100 µg/ml depending on the stage) at room temperature. RNA probes were generated using SP6, T7, or T3 RNA polymerases, and hybridization was performed at 70°C. The citrine (Jayasena et al., 2011), dlx2a (Sperber et al., 2008), fgf3 (Walshe and Mason, 2003b), sox9a (Yan et al., 2005), sox10 (Dutton et al., 2001), foxd3 (Kelsh et al., 2000), pax2a (Pfeffer et al., 1998), barx1 (Sperber and Dawid, 2008), pax6 (Krauss et al., 1991), hox2a (Prince et al., 1998), runx2b (Flores et al., 2004), col2a1a (Yan et al., 2002), and smad2 (Müller et al., 1999) probes have been described previously. Processed fish were imaged (as described earlier) in either PBS or 70% glycerol.
Immunofluorescent staining on deyolked fish was essentially performed as described previously (Ohyama et al., 2006). In brief, fish were fixed in 4% PFA made in 0.1 M phosphate buffer. Blocking, primary, and secondary antibody incubations were performed in 10% goat serum containing 0.5% Triton X-100 in PBS at either room temperature (blocking step, 2–5 h) or 4°C (antibody incubations, 16–48 h). For confocal imaging, embryos were imaged in PBS. The following antibodies were used: rabbit anti–phospho-histone H3 (PH3; EMD Millipore), rabbit anti-activated Caspase3 (R&D Systems), rabbit anti-GFP (Molecular Probes), goat anti–rabbit Alexa Fluor 488, and goat anti–rabbit Alexa Fluor 594 (Molecular Probes).
Immunoblotting
To detect knockdown with Atg MO, Gt(rbms3-citrine)ct30a embryos treated with either Atg MO or Co MO at 48 hpf were deyolked in cold Ringer’s solution with 0.5 mM DTT and 0.2 mM phenylmethanesulfonylfluoride (PMSF) supplemented with Complete protease inhibitors (EDTA free; Roche). For detecting Smad2 levels, 31-hpf head chunks (the region immediately posterior of the eyes and anterior of the fin buds) were collected, and pharyngeal arches from the same region (without any neural tissue) at 48 hpf were dissected in cold Ringer’s solution. Experiments were performed from at least three independent pools of protein extract. The following antibodies were used: mouse anti–α-tubulin (loading control; Sigma-Aldrich), rabbit anti-phosphorylated Smad2 (pSmad2; Cell Signaling Technology), rabbit anti-GFP (Invitrogen), rabbit anti-Smad2/3 (Cell Signaling Technology), secondary anti-mouse HRP (Promega), and anti–rabbit HRP (Promega). Comparison of band intensity between loading control and Smad2 was performed in ImageJ 1.38×. Loading control intensity was set to 1 and data were expressed as mean ± SD.
Alcian blue cartilage staining
Cartilage staining was performed essentially as described previously (Robu et al., 2007) in 0.1% Alcian blue, 70% ethanol, and 0.37% hydrochloric acid. Larvae were bleached with 3% H2O2 plus 1% KOH in PBS for ∼20 min and treated with Trypsin (1 mg/ml; Sigma-Aldrich) for 30–60 min at 37°C. Experiments were performed at least three times to determine consistency. n values represent total number of larvae analyzed for the cartilage phenotype. Data were expressed as mean ± SD. To determine if there were statistical differences in phenotypes, between treatments, a χ2 test was performed.
Cell proliferation, cell death counts and area measurement
MO was injected into the Tg(sox10(−4.7):egfp) background and imaged confocally. Cells positive for both EGFP and a marker of interest within the pharyngeal arches were counted. Data were expressed as bifluorescent cells/arch. The results were expressed as mean ± SEM, and statistical analysis was performed using a two-tailed Student’s t test. For pharyngeal arch area measurements, 0.1% Tricaine-, pH 7.2, treated embryos were imaged in whole-mount at 24 hpf in egg water at room temperature, as described for confocal imaging (Jayasena et al., 2011). Images were acquired using a microscope (MVX10; Olympus) attached to a 1× objective lens (MV PLAPO; Olympus) and AxioVision version 4.6.3 software (Carl Zeiss). Images were processed using the ImageJ 1.38× measure tool.
RNA extraction and QPCR
Pharyngeal arches removed of contaminating neural tissues/whole embryos were collected. Total RNA was extracted using the RNAqueous-Micro kit (Ambion) and treated with DNase1 (Invitrogen), then cDNA was generated using random hexamers with Superscript III (Invitrogen).
QPCR was performed using SYBR green reagent (Applied Biosystems) on 5–7 independent RNA pools (generated from 2–3 experiments). For each QPCR primer set, amplification was performed in triplicate per pool of cDNA and validated for each MO treatment condition at least three times. The housekeeping gene ef1α (Plaster et al., 2006) was used as an internal control, and the results were analyzed using the comparative cycle threshold method for relative quantitation and normalized to ef1α levels.
For primer sequence details, see Table 1. All primer pairs showed 100% efficiency. Because of high variation between experiments for controls, the fold change was set to 1 to facilitate direct comparison between experiments (different pools of RNA from different injection experiments). All samples were run in triplicate per primer pair. Error bars represent SD from biological replicates (independent pools of RNA) unless otherwise stated. Statistical tests were performed using the two-tailed Student’s t test.
Table 1.
List of QPCR primers
| Gene | Forward primer | Reverse primer |
| barx1 | 5′-AAGTCCCACCGGAGACCTTCTAAA-3′ | 5′-ACAGAGGAGACACCGGAAACTTCA-3′ |
| cad2a | 5′-TGGTCCAGATACCCGTTGCTTTCA-3′ | 5′-AGCCTTGAACGTCAGTCAACCTCT-3′ |
| cherry | 5′-TCAAGACCACCTACAAGGCCAAGA-3′ | 5′-TGTGGGAGGTGATGTCCAACTTGA-3′ |
| cyclin D1 | 5′-AGTATGCACGGATGCCCAAGTGTA-3′ | 5′-TCTTTGATGTCCAGCCATCACCCT-3′ |
| d1egfp | 5′-GACGAGCTGTACAAGAAGCTTAGCCA-3′ | 5′-TTGATCCTAGCAGAAGCACAGGCT-3′ |
| myod | 5′-GCATGAGGGATCTGTCCTGAGTGA-3′ | 5′-AATGGTTTCCTGAGCCTGCTGTTG-3′ |
| prrx1a | 5′-GGGCAGGAGTATGCTGGAGTCT-3′ | 5′-GACTGCTGTTGAACGTGGTTCTGT-3′ |
| rac1 | 5′-ACCTTGAGAGATGAAGAGCGGCAT-3′ | 5′-ACGTGGGCTCACAAAGAAGGAGAA-3′ |
| rbms3 | 5′-AGAACCACAAGCGGAGGCTTT-3′ | 5′-TGTTCCAAGCGTTTGCCCATGT-3′ |
| smad1 | 5′-TCGTGGATGGCTTTACTGACCCAT-3′ | 5′-TCGATGGTTGAGTTCCGGTTGACA-3′ |
| smad2 | 5′-AAGTCTGCGTCAATCCCTACCACT-3′ | 5′-TAGTCGTCCAAAGGTGGCAACTCA-3′ |
| smad3a | 5′-CAAGAGTTTGACATTCTTCTTGACGGGA-3′ | 5′-CTTGCTTCTGCTAAACGCACGAGA-3′ |
| smad3b | 5′-TGATGCACCAGACAAGCCTTCAGA-3′ | 5′-TAGTGTGCAAACAACCGGTTCAGC-3′ |
| smad5 | 5′-TGTCTCGTAATGATGGGCTGACCT-3′ | 5′-TGGTGGCATTCTCCAGGTTGTGTA-3′ |
| sox9a | 5′-AACACTCAGGCCAGTCCCAG-3′ | 5′-TCTCGTTTCAGATCCGCTTTGCCT-3′ |
| Tgf-βr1a | 5′-TCCGTACTGCTGCAACACTCACAT-3′ | 5′-CAGCACAAAGCTCAGCACACAGAT-3′ |
| Tgf-βr1b | 5′-ATCGACATCGCTCCCAATCACAGA-3′ | 5′-TCAGCCCTCTTGAAGGACTCGAAA-3′ |
| Tgf-βr2a | 5′-AGGATTTCAAACAGCGGCCCTTTC-3′ | 5′-AACACCACCTGGTTATTGGTGGGA-3′ |
| wnt3a | 5′-AAAGCTCGCCCAACTTCTGTGAAC-3′ | 5′-TGCGCTTCTCAGTTCGAGTGTTGT-3′ |
Generating capped mRNA and rescues
All DNA coding for desired mRNA were cloned into a pCS2+ vector and linearized with either Not1 or Kpn1 for generating capped mRNA using mMESSAGE mMACHINE SP6 kit (Ambion). For rescue of the MO phenotype, full-length mutagenized rbms3 (described earlier) was injected ∼20–30 pg/embryo (as already described).
Smad2ca (Dick et al., 2000) was injected ∼15–30 pg/embryo, and Atg MO was coinjected at 150 µM. Trace amounts of egfp mRNA (∼5 pg/embryo) were also coinjected as a selection marker.
In vivo stabilization assay
Truncated rbms3 (dRRM-rbms3, lacking the putative RNA binding domains [RRM], amino acids 210–415) was cloned into Cla1–Xba1 site in pCS2+. d1egfp from the O. Pourquie laboratory (Institute of Genetics and Molecular and Cellular Biology, Strasbourg, France) containing the PEST domain of mouse ornithine decarboxylase (d1egfp) was subcloned into pCS2+ with EcoR1–Xba1 after the stop codon. The candidate 3′ UTR sequences were cloned into EcoR1–Xba1 site of pCS2+-d1egfp. pCS2+-cherryh2b was used as an injection control. Each embryo was injected with 70 pg of candidate 3′ UTR, 60 pg rbms3/dRRM-rbms3, and 20 pg cherryh2b. Levels of d1egfp were normalized to levels of cherryh2b. For QPCR, RNA was extracted from 4–6 individual embryos collected from at least two independent experiments per condition. Normalized fold change was graphed as mean ± SEM or SD, and a two-tailed Student’s t test was performed. For 3′ UTR and QPCR primer sequence details, see Tables 1 and 2.
Table 2.
Primers used for subcloning candidate 3′ UTRs into pCS2+-d1gefp
| 3′ UTR | Forward primer | Reverse primer |
| smad2 3′ utr | 5′-TGGACAAGGTTCTGACCCAGAT-3′ | 5′-CTACTACTAATACATTTAATATTGAGGGT-3′ |
| smad2 3′ utr R (smad2 3′ utr in the reverse orientation) | 5′-ACCCTCAATATTAAATGTATTAGTAGTAG-3′ | 5′-ATCTGGGTCAGAACCTTGTCCA-3′ |
| smad2 200bp 3′ utr | 5′-TGGACAAGGTTCTGACCCAGAT-3′ | 5′-GGTCCGAACATCAGTATCGAGAGGA-3′ |
| smad2 150bp 3′ utr | 5′-CTCGTGAGCCTTTCTTCTATCCTC-3′ | 5′-CTACTACTAATACATTTAATATTGAGGGTA-3′ |
| sox9a 3′ utr | 5′-CCATCCCTCAATCCAACCACAGT-3′ | 5′-GTGTGTATACTATGGTTACCGATTTCAAAC-3′ |
RIP
RIP with full-length Rbms3-Myc and dRRM-Rbms3-Myc was performed as described previously (Keene et al., 2006), at 4°C unless otherwise stated. Approximately 50 pg of rbms3-Myc or dRRM-rbms3-Myc capped mRNA was injected at the one-cell stage. Embryos (16–24 hpf) were deyolked manually with a pulled glass Pasteur pipette in cold zebrafish Ringer’s solution (116 mM NaCl, 2.9 mM KCl, 5 mM Hepes, pH 7, 20 mM vanadyl ribonucleoside complex [VRC; Sigma-Aldrich], and 1 mM DTT). Per 10 ml of zebrafish Ringer’s solution, the following was added: 1 mini tablet EDTA-free protease inhibitors (Roche) and 10 µl of 0.2 M phenylmethanesulfonylfluoride (PMSF). To isolate total protein (∼0.5–1 mg), an Eppendorf pestle was used. About 100–150 µl of cold polysome lysis buffer (10 mM Hepes, pH 7.9, 100 mM KCl, 5 mM MgCl2, 1% IGEPAL CA-630, 1 mM DTT, 7× complete protease inhibitors [EDTA free; Roche], 100 U/ml SuperaseIn (Ambion), and 250 U/ml RNaseOUT [Invitrogen]) was used, and lysate was stored at −80°C. Approximately 5 µg of mouse anti–c-Myc antibody (clone 9E10; Santa Cruz Biotechnology, Inc.) or mouse IgG (Santa Cruz Biotechnology, Inc.) were incubated with 50 µl of Dynabeads Protein G (Invitrogen) overnight in NT2 (50 mM Tris, pH 7.4, 150 mM NaCl, and 1 mM MgCl2) containing 5% BSA and 0.05% IGEPAL CA-630. The beads were washed several times in NT2 plus 0.05% IGEPAL CA-630. Thawed lysate was centrifuged at 20,000 g for 10 min. The resulting supernatant (0.1× final total volume) was incubated with the beads in NT2 supplemented with 200 U/ml RNaseOut, 1 mM DTT, 0.05% IGEPAL CA-630, and 20 mM EDTA for 2 h. All post-antibody incubation washes were performed in NT2 plus 0.5% IGEPAL CA-630; the final three washes were also supplemented with 1 M urea. 2–3 independent experiments were performed (mean ± SD). For calculations, QPCR results were initially normalized to input samples before calculating fold enrichment. Transcripts enriched greater than fivefold were considered to be significantly enriched. For primer sequence details, see Table 1.
Online supplemental material
Fig. S1 shows rbms3 expression at 16 and 18 hpf. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201204138/DC1.
Supplementary Material
Acknowledgments
We wish to thank the following people: Dr. Le. A. Trinh and Bronner laboratory members for helpful discussions and technical help, Ho-yin Leung for performing 3′ RACE, and Leigh Ann Fletcher and David Mayorga from the Caltech Centers of Excellence in Genomic Science fish facility for fish maintenance.
This work was supported by the US Public Health Service Corps grants P50HG004071, DE017911, and HD037105 to M.E. Bronner.
Author contributions: C.S. Jayasena designed and performed the experiments, executed the data analysis, and wrote and edited the manuscript. M. Bronner performed data analysis and edited the manuscript.
Footnotes
Abbreviations used in this paper:
- Atg MO
- translation start site rbms3 morpholino
- CNC
- cranial neural crest
- dRRM-Rbms3
- truncated Rbms3 lacking RNA binding domains
- d1EGFP
- destabilized EGFP
- hpf
- hours postfertilization
- MO
- morpholino oligonucleotide
- smad2ca
- constitutively activated smad2
- Sp MO
- splice site rbms3 morpholino
- MSSP
- myc single-stranded binding protein
- PCC
- prechondrogenic crest
- QPCR
- quantitative PCR
- RACE
- rapid amplification of cDNA ends
- RBP
- RNA-binding protein
- RIP
- RNA immunoprecipitation
- ss
- somite stage
- TGF-βr
- TGF-β receptor
- UTR
- untranslated region
References
- Akiyama H., Chaboissier M.-C., Martin J.F., Schedl A., de Crombrugghe B. 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16:2813–2828 10.1101/gad.1017802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C., Zuniga E., Blitz I.L., Wada N., Le Pabic P., Javidan Y., Zhang T., Cho K.W., Crump J.G., Schilling T.F. 2011. Combinatorial roles for BMPs and Endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development. 138:5135–5146 10.1242/dev.067801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W., Deng J.M., Zhang Z., Behringer R.R., de Crombrugghe B. 1999. Sox9 is required for cartilage formation. Nat. Genet. 22:85–89 10.1038/8792 [DOI] [PubMed] [Google Scholar]
- Cheah F.S.H., Winkler C., Jabs E.W., Chong S.S. 2010. Tgfbeta3 regulation of chondrogenesis and osteogenesis in zebrafish is mediated through formation and survival of a subpopulation of the cranial neural crest. Mech. Dev. 127:329–344 10.1016/j.mod.2010.04.003 [DOI] [PubMed] [Google Scholar]
- Clements W.K., Ong K.G., Traver D. 2009. Zebrafish wnt3 is expressed in developing neural tissue. Dev. Dyn. 238:1788–1795 10.1002/dvdy.21977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump J.G., Maves L., Lawson N.D., Weinstein B.M., Kimmel C.B. 2004. An essential role for Fgfs in endodermal pouch formation influences later craniofacial skeletal patterning. Development. 131:5703–5716 10.1242/dev.01444 [DOI] [PubMed] [Google Scholar]
- de Frutos C.A., Vega S., Manzanares M., Flores J.M., Huertas H., Martínez-Frías M.L., Nieto M.A. 2007. Snail1 is a transcriptional effector of FGFR3 signaling during chondrogenesis and achondroplasias. Dev. Cell. 13:872–883 10.1016/j.devcel.2007.09.016 [DOI] [PubMed] [Google Scholar]
- Dick A., Mayr T., Bauer H., Meier A., Hammerschmidt M. 2000. Cloning and characterization of zebrafish smad2, smad3 and smad4. Gene. 246:69–80 10.1016/S0378-1119(00)00056-1 [DOI] [PubMed] [Google Scholar]
- Dixon J., Jones N.C., Sandell L.L., Jayasinghe S.M., Crane J., Rey J.-P., Dixon M.J., Trainor P.A. 2006. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc. Natl. Acad. Sci. USA. 103:13403–13408 10.1073/pnas.0603730103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas M., Kim J., Li W.-Y., Nagy A., Larsson J., Karlsson S., Chai Y., Kaartinen V. 2006. Epithelial and ectomesenchymal role of the type I TGF-beta receptor ALK5 during facial morphogenesis and palatal fusion. Dev. Biol. 296:298–314 10.1016/j.ydbio.2006.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton K.A., Pauliny A., Lopes S.S., Elworthy S., Carney T.J., Rauch J., Geisler R., Haffter P., Kelsh R.N. 2001. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 128:4113–4125 [DOI] [PubMed] [Google Scholar]
- Dutton J.R., Antonellis A., Carney T.J., Rodrigues F.S., Pavan W.J., Ward A., Kelsh R.N. 2008. An evolutionarily conserved intronic region controls the spatiotemporal expression of the transcription factor Sox10. BMC Dev. Biol. 8:105 10.1186/1471-213X-8-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart J.K., Swartz M.E., Crump J.G., Kimmel C.B. 2006. Early Hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development. Development. 133:1069–1077 10.1242/dev.02281 [DOI] [PubMed] [Google Scholar]
- Ferguson C.M., Schwarz E.M., Reynolds P.R., Puzas J.E., Rosier R.N., O’Keefe R.J. 2000. Smad2 and 3 mediate transforming growth factor-beta1-induced inhibition of chondrocyte maturation. Endocrinology. 141:4728–4735 10.1210/en.141.12.4728 [DOI] [PubMed] [Google Scholar]
- Flores M.V., Tsang V.W.K., Hu W., Kalev-Zylinska M., Postlethwait J., Crosier P., Crosier K., Fisher S. 2004. Duplicate zebrafish runx2 orthologues are expressed in developing skeletal elements. Gene Expr. Patterns. 4:573–581 10.1016/j.modgep.2004.01.016 [DOI] [PubMed] [Google Scholar]
- Fritz D., Stefanovic B. 2007. RNA-binding protein RBMS3 is expressed in activated hepatic stellate cells and liver fibrosis and increases expression of transcription factor Prx1. J. Mol. Biol. 371:585–595 10.1016/j.jmb.2007.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumatsu T., Tsuda M., Taniguchi N., Tajima Y., Asahara H. 2005. Smad3 induces chondrogenesis through the activation of SOX9 via CREB-binding protein/p300 recruitment. J. Biol. Chem. 280:8343–8350 10.1074/jbc.M413913200 [DOI] [PubMed] [Google Scholar]
- García-Castro M.I., Marcelle C., Bronner-Fraser M. 2002. Ectodermal Wnt function as a neural crest inducer. Science. 297:848–851 [DOI] [PubMed] [Google Scholar]
- Halbeisen R.E., Galgano A., Scherrer T., Gerber A.P. 2008. Post-transcriptional gene regulation: from genome-wide studies to principles. Cell. Mol. Life Sci. 65:798–813 10.1007/s00018-007-7447-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington M.J., Hong E., Fasanmi O., Brewster R. 2007. Cadherin-mediated adhesion regulates posterior body formation. BMC Dev. Biol. 7:130 10.1186/1471-213X-7-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M., Pudimat R., Busch A., Backofen R. 2006. Using RNA secondary structures to guide sequence motif finding towards single-stranded regions. Nucleic Acids Res. 34:e117 10.1093/nar/gkl544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan D.J., Riordan D.P., Gerber A.P., Herschlag D., Brown P.O. 2008. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6:e255 10.1371/journal.pbio.0060255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Yeo J.Y., Chytil A., Han J., Bringas P., Jr, Nakajima A., Shuler C.F., Moses H.L., Chai Y. 2003. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 130:5269–5280 10.1242/dev.00708 [DOI] [PubMed] [Google Scholar]
- Izzi L., Attisano L. 2004. Regulation of the TGFbeta signalling pathway by ubiquitin-mediated degradation. Oncogene. 23:2071–2078 10.1038/sj.onc.1207412 [DOI] [PubMed] [Google Scholar]
- Jayasena C.S., Ohyama T., Segil N., Groves A.K. 2008. Notch signaling augments the canonical Wnt pathway to specify the size of the otic placode. Development. 135:2251–2261 10.1242/dev.017905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena C.S., Trinh A., Bronner M. 2011. Live imaging of endogenous Collapsin response mediator protein-1 expression at subcellular resolution during zebrafish nervous system development. Gene Expr. Patterns. 11:395–400 10.1016/j.gep.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N.C., Lynn M.L., Gaudenz K., Sakai D., Aoto K., Rey J.-P., Glynn E.F., Ellington L., Du C., Dixon J., et al. 2008. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat. Med. 14:125–133 10.1038/nm1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J.D. 2007. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8:533–543 10.1038/nrg2111 [DOI] [PubMed] [Google Scholar]
- Keene J.D., Komisarow J.M., Friedersdorf M.B. 2006. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 1:302–307 10.1038/nprot.2006.47 [DOI] [PubMed] [Google Scholar]
- Kelsh R.N., Dutton K., Medlin J., Eisen J.S. 2000. Expression of zebrafish fkd6 in neural crest-derived glia. Mech. Dev. 93:161–164 10.1016/S0925-4773(00)00250-1 [DOI] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203:253–310 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Knecht A.K., Bronner-Fraser M. 2002. Induction of the neural crest: a multigene process. Nat. Rev. Genet. 3:453–461 10.1038/nrg819 [DOI] [PubMed] [Google Scholar]
- Krauss S., Johansen T., Korzh V., Fjose A. 1991. Expression pattern of zebrafish pax genes suggests a role in early brain regionalization. Nature. 353:267–270 10.1038/353267a0 [DOI] [PubMed] [Google Scholar]
- Kucenas S., Wang W.-D., Knapik E.W., Appel B. 2009. A selective glial barrier at motor axon exit points prevents oligodendrocyte migration from the spinal cord. J. Neurosci. 29:15187–15194 10.1523/JNEUROSCI.4193-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko P. 2003. Gene regulation at the RNA layer: RNA binding proteins in intercellular signaling networks. Sci. STKE. 2003:RE6 10.1126/stke.2003.179.re6 [DOI] [PubMed] [Google Scholar]
- Laux D.W., Febbo J.A., Roman B.L. 2011. Dynamic analysis of BMP-responsive smad activity in live zebrafish embryos. Dev. Dyn. 240:682–694 10.1002/dvdy.22558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Quon G., Lipshitz H.D., Morris Q. 2010. Predicting in vivo binding sites of RNA-binding proteins using mRNA secondary structure. RNA. 16:1096–1107 10.1261/rna.2017210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chen L., Nie C.J., Zeng T.T., Liu H., Mao X., Qin Y., Zhu Y.H., Fu L., Guan X.Y. 2011. Downregulation of RBMS3 is associated with poor prognosis in esophageal squamous cell carcinoma. Cancer Res. 71:6106–6115 10.1158/0008-5472.CAN-10-4291 [DOI] [PubMed] [Google Scholar]
- Liu F. 2003. Receptor-regulated Smads in TGF-beta signaling. Front. Biosci. 8:s1280–s1303 10.2741/1149 [DOI] [PubMed] [Google Scholar]
- Lu C.K., Lai Y.C., Chen H.R., Chiang M.K. 2012. Rbms3, an RNA-binding protein, mediates the expression of Ptf1a by binding to its 3’UTR during mouse pancreas development. DNA Cell Biol. 31:1245–1251 10.1089/dna.2012.1619 [DOI] [PubMed] [Google Scholar]
- Massagué J., Seoane J., Wotton D. 2005. Smad transcription factors. Genes Dev. 19:2783–2810 10.1101/gad.1350705 [DOI] [PubMed] [Google Scholar]
- McKeown S.J., Bronner-Fraser M. 2008. Saving face: rescuing a craniofacial birth defect. Nat. Med. 14:115–116 10.1038/nm0208-115 [DOI] [PubMed] [Google Scholar]
- Müller F., Blader P., Rastegar S., Fischer N., Knöchel W., Strähle U. 1999. Characterization of zebrafish smad1, smad2 and smad5: the amino-terminus of smad1 and smad5 is required for specific function in the embryo. Mech. Dev. 88:73–88 10.1016/S0925-4773(99)00173-2 [DOI] [PubMed] [Google Scholar]
- Ohyama T., Mohamed O.A., Taketo M.M., Dufort D., Groves A.K. 2006. Wnt signals mediate a fate decision between otic placode and epidermis. Development. 133:865–875 10.1242/dev.02271 [DOI] [PubMed] [Google Scholar]
- Oka K., Oka S., Hosokawa R., Bringas P., Jr, Brockhoff H.C., II, Nonaka K., Chai Y. 2008. TGF-beta mediated Dlx5 signaling plays a crucial role in osteo-chondroprogenitor cell lineage determination during mandible development. Dev. Biol. 321:303–309 10.1016/j.ydbio.2008.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkov D., Ni R., Else C., Piñol-Roma S., Ramirez F., Tanaka S. 2000. Cloning of a human gene closely related to the genes coding for the c-myc single-strand binding proteins. Gene. 243:27–36 10.1016/S0378-1119(99)00515-6 [DOI] [PubMed] [Google Scholar]
- Pfeffer P.L., Gerster T., Lun K., Brand M., Busslinger M. 1998. Characterization of three novel members of the zebrafish Pax2/5/8 family: dependency of Pax5 and Pax8 expression on the Pax2.1 (noi) function. Development. 125:3063–3074 [DOI] [PubMed] [Google Scholar]
- Plaster N., Sonntag C., Busse C.E., Hammerschmidt M. 2006. p53 deficiency rescues apoptosis and differentiation of multiple cell types in zebrafish flathead mutants deficient for zygotic DNA polymerase delta1. Cell Death Differ. 13:223–235 10.1038/sj.cdd.4401747 [DOI] [PubMed] [Google Scholar]
- Prince V.E., Moens C.B., Kimmel C.B., Ho R.K. 1998. Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development. 125:393–406 [DOI] [PubMed] [Google Scholar]
- Robu M.E., Larson J.D., Nasevicius A., Beiraghi S., Brenner C., Farber S.A., Ekker S.C. 2007. p53 activation by knockdown technologies. PLoS Genet. 3:e78 10.1371/journal.pgen.0030078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C.D., Jayasena C.S., Nie S., Bronner M.E. 2012. Neural crest specification: tissues, signals, and transcription factors. Wiley Interdisciplinary Reviews: Developmental Biology. 1:52–68 10.1002/wdev.8 [DOI] [PubMed] [Google Scholar]
- Sakai D., Suzuki T., Osumi N., Wakamatsu Y. 2006. Cooperative action of Sox9, Snail2 and PKA signaling in early neural crest development. Development. 133:1323–1333 10.1242/dev.02297 [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T., Bronner-Fraser M. 2008. A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 9:557–568 10.1038/nrm2428 [DOI] [PubMed] [Google Scholar]
- Schilling T.F., Kimmel C.B. 1994. Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development. 120:483–494 [DOI] [PubMed] [Google Scholar]
- Siomi H., Siomi M.C., Nussbaum R.L., Dreyfuss G. 1993. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 74:291–298 10.1016/0092-8674(93)90420-U [DOI] [PubMed] [Google Scholar]
- Sperber S.M., Dawid I.B. 2008. barx1 is necessary for ectomesenchyme proliferation and osteochondroprogenitor condensation in the zebrafish pharyngeal arches. Dev. Biol. 321:101–110 10.1016/j.ydbio.2008.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber S.M., Saxena V., Hatch G., Ekker M. 2008. Zebrafish dlx2a contributes to hindbrain neural crest survival, is necessary for differentiation of sensory ganglia and functions with dlx1a in maturation of the arch cartilage elements. Dev. Biol. 314:59–70 10.1016/j.ydbio.2007.11.005 [DOI] [PubMed] [Google Scholar]
- Takai T., Nishita Y., Iguchi-Ariga S.M.M., Ariga H. 1994. Molecular cloning of MSSP-2, a c-myc gene single-strand binding protein: characterization of binding specificity and DNA replication activity. Nucleic Acids Res. 22:5576–5581 10.1093/nar/22.25.5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K.M., LaBonne C. 2005. SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev. Cell. 9:593–603 10.1016/j.devcel.2005.09.016 [DOI] [PubMed] [Google Scholar]
- Trainor P.A., Krumlauf R. 2001. Hox genes, neural crest cells and branchial arch patterning. Curr. Opin. Cell Biol. 13:698–705 10.1016/S0955-0674(00)00273-8 [DOI] [PubMed] [Google Scholar]
- Trainor P.A., Tam P.P. 1995. Cranial paraxial mesoderm and neural crest cells of the mouse embryo: co-distribution in the craniofacial mesenchyme but distinct segregation in branchial arches. Development. 121:2569–2582 [DOI] [PubMed] [Google Scholar]
- Trainor P.A., Ariza-McNaughton L., Krumlauf R. 2002. Role of the isthmus and FGFs in resolving the paradox of neural crest plasticity and prepatterning. Science. 295:1288–1291 10.1126/science.1064540 [DOI] [PubMed] [Google Scholar]
- Trinh A., Hochgreb T., Graham M., Wu D., Ruf-Zamojski F., Jayasena C.S., Saxena A., Hawk R., Gonzalez-Serricchio A., Dixson A., et al. 2011. A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes Dev. 25:2306–2320 10.1101/gad.174037.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada N., Javidan Y., Nelson S., Carney T.J., Kelsh R.N., Schilling T.F. 2005. Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development. 132:3977–3988 10.1242/dev.01943 [DOI] [PubMed] [Google Scholar]
- Walshe J., Mason I. 2003a. Fgf signalling is required for formation of cartilage in the head. Dev. Biol. 264:522–536 10.1016/j.ydbio.2003.08.010 [DOI] [PubMed] [Google Scholar]
- Walshe J., Mason I. 2003b. Unique and combinatorial functions of Fgf3 and Fgf8 during zebrafish forebrain development. Development. 130:4337–4349 10.1242/dev.00660 [DOI] [PubMed] [Google Scholar]
- Weinberg E.S., Allende M.L., Kelly C.S., Abdelhamid A., Murakami T., Andermann P., Doerre O.G., Grunwald D.J., Riggleman B. 1996. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 122:271–280 [DOI] [PubMed] [Google Scholar]
- Wilkes M.C., Murphy S.J., Garamszegi N., Leof E.B. 2003. Cell-type-specific activation of PAK2 by transforming growth factor beta independent of Smad2 and Smad3. Mol. Cell. Biol. 23:8878–8889 10.1128/MCB.23.23.8878-8889.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A., Wang G., Dupuis H., Shao Z., Beier F. 2007. Rac1 signaling stimulates N-cadherin expression, mesenchymal condensation, and chondrogenesis. J. Biol. Chem. 282:23500–23508 10.1074/jbc.M700680200 [DOI] [PubMed] [Google Scholar]
- Wurdak H., Ittner L.M., Lang K.S., Leveen P., Suter U., Fischer J.A., Karlsson S., Born W., Sommer L. 2005. Inactivation of TGFbeta signaling in neural crest stem cells leads to multiple defects reminiscent of DiGeorge syndrome. Genes Dev. 19:530–535 10.1101/gad.317405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.-L., Miller C.T., Nissen R.M., Singer A., Liu D., Kirn A., Draper B., Willoughby J., Morcos P.A., Amsterdam A., et al. 2002. A zebrafish sox9 gene required for cartilage morphogenesis. Development. 129:5065–5079 (published erratum appears in Development 2002. 129:5551) [DOI] [PubMed] [Google Scholar]
- Yan Y.-L., Willoughby J., Liu D., Crump J.G., Wilson C., Miller C.T., Singer A., Kimmel C., Westerfield M., Postlethwait J.H. 2005. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 132:1069–1083 10.1242/dev.01674 [DOI] [PubMed] [Google Scholar]
- Yang X., Chen L., Xu X., Li C., Huang C., Deng C.X. 2001. TGF-β/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J. Cell Biol. 153:35–46 10.1083/jcb.153.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon B.S., Ovchinnikov D.A., Yoshii I., Mishina Y., Behringer R.R., Lyons K.M. 2005. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc. Natl. Acad. Sci. USA. 102:5062–5067 10.1073/pnas.0500031102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.