Abstract
Virulence factor production in Vibrio cholerae is complex, with ToxRS being an important part of the regulatory cascade. Additionally, ToxR is the transcriptional regulator for the genes encoding the major outer membrane porins OmpU and OmpT. ToxR is a transmembrane protein and contains two cysteine residues in the periplasmic domain. This study addresses the influence of the thiol-disulfide oxidoreductase system DsbAB, ToxR cysteine residues and ToxR/ToxS interaction on ToxR activity. The results show that porin production correlates with ToxR intrachain disulfide bond formation, which depends on DsbAB. In contrast, formation of ToxR intrachain or interchain disulfide bonds is dispensable for virulence factor production and in vivo colonization. This study further reveals that in the absence of ToxS, ToxR interchain disulfide bond formation is facilitated, whereat cysteinyl dependent homo- and oligomerization of ToxR is suppressed if ToxS is coexpressed. In summary, new insights into gene regulation by ToxR are presented, demonstrating a mechanism by which ToxR activity is linked to a DsbAB dependent intrachain disulfide bond formation.
Introduction
Vibrio cholerae is a Gram-negative, facultative anaerobic bacterium. It is the causative agent of cholera, which is endemic in India, Bangladesh, Southeast Asia, Africa and South America [1]. Infection starts with the oral ingestion of V. cholerae bacteria from the environment through contaminated food or water supplies [2], [3]. V. cholerae bacteria pass through the gastric acid compartment of the stomach, penetrate the mucus lining of the intestinal epithelia and start colonizing the small intestine. This compartment contains growth inhibitory substances, such as bile salts and organic acids and also factors of the innate immune system, e.g., complement secreted by intestinal epithelial cells [4] and defensins produced by Paneth cells [5]. Therefore, V. cholerae has developed the ability to survive, colonize and produce virulence factors [6] in spite of harsh stress conditions [7], [8].
Extensive studies of cholera pathogenesis revealed that production of the main virulence factors, namely cholera toxin (CT) and toxin-coregulated pili (TCP), is coordinated by a regulatory network [9]. This system is directly controlled by four transcriptional activator complexes identified so far, which act in a regulatory cascade and include AphAB, TcpPH, ToxRS and ToxT [10]–[14]. AphAB act at the beginning of the cascade and regulate transcription of the inner membrane located transcriptional regulator components TcpPH [14] and ToxRS [15]. ToxR is critical for regulation of virulence genes and together with TcpP, it activates transcription of toxT [12], [16]–[21]. Subsequently, the AraC-like transcriptional activator ToxT directly activates transcription of ctx and tcp loci, as well as additional genes [7], [19], [22]. V. cholerae strains lacking ToxT or ToxR do not produce CT or TCP and are avirulent [23]. Moreover, ToxR regulates the transcription of more than 150 additional genes [16], including ompT and ompU, which encode the outer membrane proteins OmpT and OmpU. Both porin genes are inversely regulated [24], [25], i.e., ompU transcription is activated, whereas ompT is repressed by ToxR as determined by OMP analysis and in vivo colonization [25], [26]. For the toxT promoter, ToxR and TcpP binding occurs such that ToxR binds on the distal end and acts as a “scaffold” protein by facilitating TcpP binding adjacent to the RNA polymerase binding site [27], [28]. Recently, it was shown that point mutations in the cytoplasmic domain of ToxR differentially activate ompU and toxT transcription. Based on this observation it was proposed that the orientation of ToxR on its corresponding operators differs for the ompU and toxT promoter regions [29].
The N-terminus of ToxR is located in the cytoplasm and contains the DNA-binding motif, followed by a transmembrane domain and then the periplasmic C-terminus [30]. A long lasting discussion exists about ToxR activity and ToxR dimer- and oligomerization. In Escherichia coli, ToxR-fusion proteins containing defined dimerization signals, were transcriptionally active. However, this activity was not conclusively confirmed in V. cholerae using the same or similar ToxR variants 30–34. As demonstrated by domain analysis, the presence of the ToxR transmembrane domain is essential for its activity [35], [36]. More recent evidence indicates that ToxR transmembrane segment itself possesses some function for ToxR activity and may be involved in bile dependent ToxR activation [37]. The ToxR periplasmic domain has been proposed to act as a sensor for environmental stimuli and contains two cysteine residues at amino acid position 236 and 293, which either can form homodimer or intrachain disulfide bonds [34]. The toxS gene is cotranscribed downstream of toxR [21]. ToxS is an inner membrane protein [7] and evidence suggests that ToxR and ToxS are interacting partners in the periplasm [38] and can form a heterodimer [34]. Additionally, knockout mutants in toxS negatively influence the transcriptional activity of ToxR [21], suggesting that ToxS facilitates the activity of ToxR or has implications for ToxR protein stability [38], [39]. However, major questions remain to be answered. What defines an active ToxR conformation? Is ToxR activity dependent on reduced or oxidized disulfide bonds? Is ToxR/ToxS interaction necessary to yield active ToxR complexes, e.g., homodimer, oligomer or heterodimer? And finally, does ToxR activity respond to environmental stimuli, cellular growth physiology and other input signals?
In this report, the molecular mechanisms that control activity of the membrane bound transcription factor ToxR were addressed using epidemic O1 El Tor and O395 classical strains. The study includes the interplay between ToxR and ToxS and the formation of ToxRS heterodimer. Furthermore, the redox state of ToxR cysteines were characterized in strains encoding knockout mutations in dsbAB and cysteine to serine substitutions in ToxR and ToxR activities were determined for virulence factor and porin expression.
Materials and Methods
Ethics statement
Mice were used for competition colonization experiments in strict accordance to the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, the national “Bundesgesetzblatt fuer die Republik Oesterreich”. Animal protocol (39/158 ex 2000/10), has been approved by the Austrian Federal Ministry of Science and Research Ref. II/10b and the Committee on the Ethics of Animal Experiments of the University of Graz. Housing of mice was conducted with food and water ad libitum and monitored in accordance with the rules of the Institute of Molecular Biosciences at the University of Graz.
Bacterial strains, plasmids and culture conditions
Strains and plasmids used in this study are listed in Table 1. For construction of deletion and chromosomally encoded amino acid substitution mutations in toxR, the suicide vectors pCVD442 and pKEK229 were used. If not stated otherwise, E. coli and V. cholerae strains were transformed by electroporation. E. coli strain SM10λpir was used to introduce plasmids into V. cholerae by conjugation. V. cholerae P27459-S, a spontaneous streptomycin resistant mutant of V. cholerae O1 El Tor clinical isolate P27459 [40] or O395 [41], a spontaneous streptomycin resistant mutant of V. cholerae O1 classical clinical isolate were used as WT strains in all experiments. E. coli strains were grown using LB broth at 37°C. Unless stated otherwise, V. cholerae strains were grown using LB broth or minimal medium M9 supplemented with glycerol (0.4%) as a carbon source at 37°C. For optimal induction of virulence genes, V. cholerae O1 El Tor was grown using AKI conditions [42], whereas the V. cholerae O1 classical strain was grown in LB broth (pH 6.5) at 30°C [25]. If required, streptomycin, ampicillin and kanamycin were used at final concentrations of 100, 100, and 50 µg/ml, respectively. Other supplements were used in the following concentrations: IPTG (0.05 or 0.005 mM), glucose (0.2%), arabinose (0.2%) and sucrose (10%). Strains were stock frozen using LB medium containing glycerol (20%) at −80°C. If not noted otherwise, E. coli refers to XL1-Blue, V. cholerae to O1 El Tor isolate P27459-S and classical to classical O1 isolate O395.
Table 1. Bacteria strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristic | Reference |
| E. coli strains | ||
| XL1-Blue | F′::Tn10 proA + B + lacq Δ(lacZ)M151 recA1 endA1 gyrA46 (Nalr) thi hsdR17 (rK −mK +) supE44 relA1 lac | NEB |
| DH5αλpir | F− Φ80dlacZΔM15Δ(argF lac)U169 deoR recA1 endA1 hsdR17 (rK −mK +) supE44 thi-1 gyrA69 relA1 λ recA::RPA-2-Te::Mu λpirR6K, kmr | [69] |
| SM10λpir | thi thr leu tonA lacY supE recA::RPA-2-Te::Mu λpirR6K, kmr | [25] |
| V. cholerae strains | ||
| O395 | O1 Ogawa, classical, clinical isolate, India 1964, spontaneous smr | [41] |
| P27459-S | O1 Inaba, El Tor, clinical isolate, Bangladesh 1976, spontaneous smr | [40] |
| O395 ΔdsbA::km | dsbA replaced by km cassette, smr, kmr | This study |
| O395 ΔdsbB::km | dsbB replaced by km cassette, smr, kmr | This study |
| O395 ΔtoxR | Deletion in toxR, smr | This study |
| O395 ΔtoxRS | Deletion in toxR and toxS, smr | This study |
| P27459-S ΔdsbA::km | dsbA replaced by km cassette, smr, kmr | This study |
| P27459-S ΔdsbA::km ΔtoxRS | dsbA replaced by km cassette, deletion of toxR and toxS, smr, kmr | This study |
| P27459-S ΔdsbB::km | dsbB replaced by km cassette, smr, kmr | This study |
| P27459-S dsbC::pGP | dsbC inserted by pGP704, amr | This study |
| P27459-S ΔtoxR | Deletion in toxR, smr | This study |
| P27459-S ΔtoxRS | Deletion in toxR and toxS, smr | This study |
| O395 ΔtoxR::FLAGtoxR | toxR replaced by FLAGtoxR, smr | This study |
| O395 ΔtoxR::FLAGtoxRCC | toxR replaced by FLAGtoxRC236SC293S, smr | This study |
| O395 ΔtoxR::FLAGtoxR ΔtoxS | toxR replaced by FLAGtoxR, deletion in toxS, smr | This study |
| P27459-S ΔtoxR::FLAGtoxR | toxR replaced by FLAGtoxR, smr | This study |
| P27459-S ΔtoxR::FLAGtoxRCC | toxR replaced by FLAGtoxRC236SC293S, smr | This study |
| P27459-S ΔtoxR::FLAGtoxR ΔlacZ | toxR replaced by FLAGtoxR, deletion in lacZ, smr | This study |
| P27459-S ΔtoxR::FLAGtoxR ΔtoxS | toxR replaced by FLAGtoxR, deletion in toxS, smr | This study |
| Plasmids | ||
| pKEK229 | OriR6K mobRP4 sacB, apr | [70] |
| pGP704 | OriR6K mobRP4, apr | [25] |
| pCVD442 | OriR6K mobRP4 sacB, apr | [45] |
| pKan π | kmr | [71] |
| pBAD18 | Expression vector, arabinose inducible, apr | [72] |
| pFLAG-MAC™ | Expression vector with N-terminal FLAG-Tag, IPTG inducible, apr | Sigma-Aldrich |
| pGPdsbC | pGP704 carrying internat fragment of dsbC′ apr | This study |
| pKEK229dsbA::km | pCVD442 carrying up and down fragment of dsbA flanking a km cassette, apr | This study |
| pKEK229dsbB::km | pCVD442 carrying up and down fragment of dsbB flanking a km cassette, apr | This study |
| pCVD442lacZ | pCVD442 carrying up and down fragments of lacZ, apr | This study |
| pCVD442toxR | pCVD442 carrying up and down fragment of toxR, apr | This study |
| pCVD442toxRS | pCVD442 carrying up fragment of toxR and down fragments of toxS, apr | This study |
| pdsbA | dsbA of P27459-S in pBAD18, apr | This study |
| pdsbB | dsbB of P27459-S in pBAD18, apr | This study |
| pFLAGtoxR | toxR of P27459-S in pFLAG-MAC™, apr | This study |
| pFLAGtoxRCC | toxRC236SC293S point mutant of P27459-S in pFLAG-MAC™, apr | This study |
| pFLAGtoxRS | toxR and toxS of P27459-P in pFLAG-MAC™, apr | This study |
| pFLAGtoxRCCS | toxRC236SC293S point mutant and toxS of P27459-P in pFLAG-MAC™, apr | This study |
| pFLAGtoxRS(Δ264) | pFLAGtoxRS carrying a 264 bp deletion in toxS generated by two internal AccI sites, apr | This study |
| pFLAGtoxRS_ompU | toxR, toxS and operator region of ompU of P27459-P in pFLAG-MAC™, apr | This study |
| pFLAGtoxRS_toxT | toxR, toxS and operator region of toxT of P27459-P in pFLAG-MAC™, apr | This study |
| pFLAGtoxRS_ompU(Δ264) | pFLAGtoxRS_ompU carrying a 264 bp deletion in toxS generated by two internal AccI sites, apr | This study |
| pFLAGtoxRS_toxT(Δ264) | pFLAGtoxRS_toxT carrying a 264 bp deletion in toxS generated by two internal AccI sites, apr | This study |
| pFLAGtoxRCCS_ompU | toxRC236SC293S point mutant, toxS and operator region of ompU of P27459-P in pFLAG-MAC™, apr | This study |
| pFLAGtoxRCCS_toxT | toxRC236SC293S point mutant, toxS and operator region of toxT of P27459-P in pFLAG-MAC™, apr | |
| pCVD442FLAGtoxR | pCVD442 carrying FLAGtoxR, apr | |
| pCVD442FLAGtoxRCC | pCVD442 carrying FLAGtoxRC236SC293S, apr | |
| pCVD442FLAGtoxRS | pCVD442 carrying FLAGtoxR and toxS, apr |
Recombinant DNA techniques and construction of deletion mutants, point mutants and expression vectors
Oligonucleotide primers used in this study are listed in Table 2 and were purchased from Life Technologies (Life Technologies, Lofer). Chromosomal DNA was prepared as described previously by using ethanol salt precipitation [43]. Purification of DNA fragments from PCR samples and plasmid DNA preparations were performed using QIAgen QIAquick gel extraction, QIAquick PCR purification and QIAprep Spin Miniprep Kits according to the manufacturer's instructions (Quiagen, Hilden). Phusion high-fidelity polymerase (Finnzyme, Espoo) was used in PCR for DNA fragment generation for further subcloning and sequencing. For all other reactions, Taq DNA polymerase, restriction endonucleases and T4 DNA ligase were obtained from New England Biolabs (NEB, Ipswich). DNA sequencing was performed by the dideoxynucleotide chain termination method of Sanger et al. [44] with an automated DNA sequencer, performed at LGC Genomics, Berlin.
Table 2. Oligonucleotide primers.
| Oligonucleotides | Sequence (5′ - 3′)a |
| SacI_dsbA_1 | TTTGAGCTCCAAGAAGAGATCCCGATCGTCC |
| EcoRI_dsbA_2 | TTTGAATTCCATGACTTTCTCCATTGGATTTATT |
| EcoRI_dsbA_3 | AATGAATTCTAATCTCAACCCATGATTCGGTAT |
| XbaI_dsbA_4 | TTTTCTAGAGATTAAACTGTTGCTGCCGTCAG |
| SacI_dsbB_1 | TTTGAGCTCGTCTTCCTGCCAATGTT |
| EcoRI_dsbB_2 | TTTGAATTCCACAGATAGATCCTTGTTAAAAAGA |
| EcoRI_dsbB_3 | TTTGAATTCTAAGCCAATCGCATCGCTCAAT |
| XbaI_dsabB_4 | TTTTCTAGATAGCATGGAGAGTGAGCCGCCACT |
| EcoRI_dsbC_1 | ATTGAATTCGTGCAAACGTCTGGTGGT |
| XbaI_dsbC_2 | ATTTCTAGAGAGCTCGTGACCCAGCAT |
| SacI_toxRS_1 | TTTGAGCTCATTTGGAAATCACATCGCGCAAAC |
| BamHI_toxRS_2 | TTTGGATCCTCCTAATCCGAACATCTAATGTCC |
| BamHI_toxR_3 | TTTGGATCCAACCCTAACGATGCCATCAAAGT |
| BamHI_toxRS_3 | TTTGGATCCTCCGATGACAATAGTGCAGAAAG |
| XbaI_toxRS_4 | TTTTCTAGAATGACGTTTCCCCGCGGTGAG |
| XbaI_lacZ_1 | TAATCTAGAACACATAACCCTGCAGTA |
| XhoI_lacZ_2 | TTTCTCGAGCTCTACGGCGTACATCCCT |
| XhoI_lacZ_3 | TTTCTCGAGTGCGTGTGGAATGTGACGAT |
| SacI_lacZ_4 | ATGAGCTCTTATTGTGGGGATGACGCTTT |
| SacI_dsbA_5′ | AATGAGCTCGCCACTTTACAAGAACCCCCG |
| XbaI_dsbA_3′ | ATTTCTAGAGATTTACAAAGCCGATTAGCACTG |
| SacI_dsbB_5′ | AATGAGCTCCAATTGAAACTGAAACTAATCCAAG |
| XbaI_dsbB_3′ | AATTCTAGACTTTAAGCGCCTTTTTTATCAACC |
| HindIII_toxR_5′_FLAG | AATAAGCTTATGTTCGGATTAGGACACAACTCA |
| KpnI_toxR_3′_FLAG | AATGGTACCCTACTCACACACTTTGATGGCAT |
| KpnI_toxRC293S_3′_FLAG | AATGGTACCCTACTCAGACACTTTGATGGCATCGTTA b |
| toxRC236S_5′ | GGCTACCGTCAATCGAACTGAGCGTTAAAAAATACAATGA b |
| toxRC236S_3′ | TCATTGTATTTTTTAACGCTCAGTTCGATTGACGGTAGCC b |
| KpnI_toxRS_5′_FLAG | AATGGTACCCATGTTCGGATTAGGACACAACTCA |
| BglII_toxRS_3′_FLAG | TTAAGATCTTTAAGAATTACTGAACAGTACGGT |
| BamHI_ompU_5′ | ATTGGATCCTCCTAAATCGGGTCGGGTT |
| BamHI_ompU_3′ | AATGGATCCGGCTCAGCCATTTTCGTGGC |
| BamHI_toxT_5′ | TTAGGATCCGTATAGCAAAGCATATTCAGAGA |
| BamHI_toxT_3′ | ATTGTCGACTAAATAAACGCAGAGAGCCATC |
| c_FLAGtoxR_3′_F1 | TGTCATCGTCGTCCTTGTAGTCCATCTAATGTCCCAGTATCTCCCTGT |
| c_FLAGtoxR_5′_F2 | GGGACAGGGAGATACTGGGACATTAGATGGACTACAAGGACGACGATGA |
| c_FLAGtoxR_3′_F2 | CTACTCACACACTTTGATGGCAT |
| c_FLAGtoxRC293S_3′_F2 | CTACTCAGACACTTTGATGGCAT b |
| c_FLAGtoxRtoxS_3′_F2 | CTTTCTGCACTATTGTCATCGGTCTACTCACACACTTTGATGGCAT |
| c_FLAGtoxR_5′_F3 | AACCAGTTAACGCTGAATTACATTC |
| c_FLAGtoxRC293S_5′_F3 | GTTGCTAACCCTAACGATGCCATCAAAGTGTCTGAG b |
| c_FLAGtoxRtoxS_5′_F3 | ATGCCATCAAAGTGTGTGAGTAGTCCGATGACAATAG |
| rpoB_fwc | CTGTCTCAAGCCGGTTACAA |
| rpoB_rvc | TTTCTACCAGTGCAGAGATGC |
| VC0633_fw | CTCGCGTACGTCTAAACTTCTTGG |
| VC0633_rv | CGGTTGTCTAGGCTGTTGTTAGAC |
| VC0984_fw | CCTTCATCAGCCACTGTAGTGAAC |
| VC0984_rv | GACCGCTATCAGAATAAGCAGTCG |
| VC1854_fw | ACCCACTAGTGATCGATGAAGACG |
| VC1854_rv | GCCATACTCAGCATATACACGAGC |
| VCr001_fw | AGGGAGGAAGGTGGTTAAGT |
| VCr001_rv | CGCTACACCTGAAATTCTACCC |
Restriction sites are underlined.
Bold letters indicate codons changed to obtain desired amino acid mutations.
Oligonucleotides for rpoB are according to reference [73].
Deletion mutations were generated as described by Donnenberg and Kaper [45] and represent start to stop codon deletions. DNA fragments of approximately 800 bp upstream and downstream of genes of interest were amplified by PCR (oligonucleotides labeled in the format X_Y, in which X stands for the restriction enzyme and Y for the respective gene) and digested with the corresponding restriction endonuclease. After ligation into suicide vector pCVD442 or pKEK229, derivatives were transformed into E. coli SM10λpir and were subsequently conjugated into V. cholerae [45]. Homologous integration of the plasmid into the chromosome, followed by negative selection for loss of plasmid in the presence of sucrose, allowed mutant strains to be constructed that have a deletion in the gene of interest. To improve selection of dsb deletion mutants, a kmr cassette was derived from pKanπ, Table 1, as an EcoRI fragment and ligated in between the dsb up and down flanking DNA fragments. The resulting dsb mutant strains carried chromosomal replacements of the dsb genes by kmr cassettes. The correct deletion for all mutants was confirmed by PCR (BioRad, Wien) (data not shown). Insertion mutants where constructed by using suicide plasmid pGP704 [25]. Therefore an approximately 500 bp internal fragment of the respective gene was amplified by using oligonucleotides labeled in the format X_Y, in which X stands for the restriction enzyme and Y for the respective gene, restricted with the corresponding endonucleases and ligated into similarly digested pGP704. Derivatives were transformed into E. coli SM10λpir and further conjugated into V. cholerae. Correct homologous integration of the plasmid into the chromosome was confirmed by PCR (BioRad, Wien) (data not shown) and maintenance of the plasmid was ensured by culturing the respective strains on media containing ampicillin.
For construction of expression plasmids, using pFLAG-MAC™ or pBAD18, the respective genes were amplified by PCR using oligonucleotides labeled in the format X_Y_5′ and X_Y_3′, in which X stands for the restriction enzyme and Y for the respective genes, Table 2. PCR fragments were digested with the respective restriction enzymes and ligated into similarly digested pFLAG-MAC™ or pBAD18. pFLAGtoxR was constructed by using oligonucleotides HindIII_toxR_5′_FLAG and KpnI_toxR_3′_FLAG. The C293S mutation in toxR was generated by using the oligonucleotide KpnI_toxRC293S_3′_ FLAG, which contains the required amino acid substitution for cysteine to serine residue. The C236S mutation was generated by SOE PCR (splicing by overlap extension), using pFLAGtoxR as a template and oligonucleotide pairs HindIII_toxR_5′_FLAG and toxRC236S_3′ as well as toxRC236S_5′ and KpnI_toxRC293S_3′_FLAG [46]. The two PCR products were used as the template in the second PCR with HindIII_toxR_5′_FLAG and KpnI_toxRC293S_3′_FLAG and the resulting PCR fragments were digested with HindIII and KpnI and ligated into similarly digested pFLAG-MAC™. pFLAGtoxRS and pFLAGtoxRCCS were constructed by using P27459-S ΔtoxR::FLAGtoxR or P27459-S ΔtoxR::FLAGtoxR CC as templates and oligonucleotides KpnI_toxRS_5′_FLAG and BglII_toxRS_3′_FLAG. PCR fragments were digested with KpnI and BglII and ligated into similar digested pFLAG-MAC™. The resulting plasmids were digested with BamHI to integrate ompU or toxT operator sites. ompU and toxT operator fragments were amplified with oligonucleotides BamHI_ompU_5′ and BamHI_ompU_3′ or BamHI_toxT_5′ and BamHI_toxT_3′, respectively, and also digested with BamHI. Similar constructs were digested with AccI to generate a 264 bp deletion of toxS. Constructs were confirmed by PCR and DNA sequencing (data not shown).
Chromosomal FLAG-tagged toxR and amino acid substitution mutants were constructed by using SOE PCR. For amplification of PCR fragments, pFLAGtoxR and pFLAGtoxRCC were used as templates. Oligonucleotides c_FLAGtoxR_5′_F2 and c_FLAGtoxR_3′_F2 respectively c_FLAGtoxRC293S_3′_F2 were used for generation for P27459-S ΔtoxR::FLAGtoxR and P27459-S ΔtoxR::FLAGtoxRCC. For construction of P27459-S ΔtoxR::FLAGtoxR ΔtoxS oligonucleotides c_FLAGtoxR_5′_F2 and c_FLAGtoxRtoxS_3′_F2 were used. Fragments with about 800 bp each of flanking DNA regions of toxR with one end complementary to the first PCR fragment (see above) were amplified by PCR using oligonucleotides SacI_toxRS_1 and c_FLAGtoxR_3′_F1 and c_FLAGtoxR_5′_F3 or c_FLAGtoxRC293S_5′_F3 and XbaI_toxRS_4 for P27459-S ΔtoxR::FLAGtoxR and P27459-S ΔtoxR::FLAGtoxRCC, respectively. For construction of P27459-S ΔtoxR::FLAGtoxR ΔtoxS oligonucleotides c_FLAGtoxR toxS_5′_F3 and XbaI_toxRS_4 were used. The three PCR products were used as templates in the second PCR with SacI_toxRS_1 and XbaI_toxRS_4 and the resulting PCR fragments were digested with SacI and XbaI and ligated into pCVD442 that had been digested with same restriction enzymes. Resulting ligation products were transformed into E. coli SM10λpir and were further transferred into V. cholerae ΔtoxR or ΔtoxRS by conjugation. V. cholerae cells in which integration of the plasmid occurred by homologous recombination via one of the two fragments and a second homologous recombination step via the other fragment resulted in mutant strains harboring an integration of either FLAG-tagged toxR or FLAG-tagged toxR cysteine to serine substitution mutant in the toxR gene locus. The correct integration of all mutants was confirmed by PCR and DNA sequencing (data not shown).
Membrane protein preparation
Proteins of the membrane and outer membrane of V. cholerae strains were prepared from cells either grown in LB medium or minimal medium M9 glycerol (0.4%). Cells were harvested by centrifugation, washed with HEPES pH 7.5 (10 mM) and lysed by sonification on ice according to standard protocols (Branson Sonifier 250A, Branson Ultrasonics Corp., Danbury). OMP preparations were performed as described previously [47]. For preparation of membrane proteins, lysed cells were centrifuged (13,000 g, 8 min, RT). The supernatants were then transferred to a new tube and centrifuged again (20,000 g, 30 min, RT). The membrane pellets were resuspended in HEPES pH 7.5 (10 mM) supplemented with protease inhibitor (Complete™, Boehringer Mannheim) to obtain proteins of the inner and outer membrane. In order to separate outer membrane proteins, the pellets were resuspended in HEPES pH 7.5 (10 mM) with sarcosyl (1%) and incubated for 30 min at RT. The suspensions were centrifuged (20,000 g, 30 min, RT) and the pellets, containing outer membrane proteins, were first washed and then resuspended in HEPES pH 7.5 (10 mM) supplemented with protease inhibitor. Protein amounts were determined by using the UV absorption 260/280 nm protocol according to Warburg and Christian [48].
SDS-PAGE and immunoblot analyses
For whole cell extracts, E. coli or V. cholerae cultures were either grown in LB, induced with IPTG (0.005 to 0.05 mM) for one to two hours or in M9 glycerol (0.4%) minimal media and induced with IPTG (0.005 to 0.05 mM) for 6.5 h. Equal amounts of cells were harvested by centrifugation in an Eppendorf centrifuge. For immunoblot analyses of whole cell extracts the overall protein contents were assessed to contain similar protein levels by SDS-PAGE coomassie blue staining. Cell pellets were washed with media, resuspended in sample buffer either with or without the reducing agent β-mercaptoethanol and boiled for 10 min. OMP preparations and whole cell extracts were then separated by SDS-PAGE in polyacrylamide (15%) gels, using Mini-PROTEAN Tetra cell (Bio-Rad, Vienna). For detection of membrane and outer membrane proteins, equal protein amounts (60 µg and 4 µg, respectively) were loaded. After SDS-PAGE, proteins were either stained with Coomassie brilliant blue as previously described [49] or transferred for immunoblot analysis to a nitrocellulose membrane (Amersham-Bioscience, Freiburg). Immunoblot analyses were performed as described previously [50]. After transfer and blocking, the membrane was incubated for 2 h at RT with the primary antibody, mouse monoclonal anti-FLAG M2 antibody (Sigma-Aldrich, Taufkirchen) or anti-DDK monoclonal antibody (OriGene Technologies, Inc., Rockville) diluted 1∶2,000 in skim milk (10%) in TBS. The membrane was washed twice in TBS-TT (Tris-HCl pH 7.5, 20 mM, NaCl, 500 mM, Tween 20, 0.05%, Triton X-100, 0.2%) and once in TBS for 10 min each. The membrane was incubated with secondary antibody (horseradish peroxidase-conjugated goat anti-mouse, Dianova GmbH, Hamburg), diluted 1∶10,000 in skim milk (10%) in TBS, for 1 h at RT. Subsequently, the nitrocellulose membrane was washed three times in TBS-TT and once in TBS for 10 min each. Chemiluminescent detection was performed using the Immun-Star™ WesternC™ Kit (Bio-Rad, Vienna) and the result visualized using a Molecular Imager ChemiDoc™ XRS System (Bio-Rad, Vienna).
qRT-PCR analyses
Primers used for quantitative reverse transcriptase PCR (qRT-PCR) are listed in Table 2 (labeled x-fw and x-rv, in which x stands for the respective gene and fw and rv for forward and reverse primers). The efficiencies of fluorescence signaling of the primer pairs amplifying the target and reference genes used in this study are at least 80% compared to a complete duplication per cycle and qRT-PCR was performed as described previously [51]. Briefly, RNA was isolated from six independent cultures grown in M9 glycerol (0.4%) medium at defined time points by using peqGOLD Bacterial RNA Kit (Peqlab, Erlangen). To remove chromosomal DNA, RNA was treated with RQ1 RNase-Free DNase (Promega, Mannheim). By using an iScriptTM Select cDNA Synthesis Kit (Bio-Rad, Vienna) cDNA was synthesized from 200 ng RNA, including controls lacking reverse transcriptase. Quantification of cDNA was performed with SYBR GreenER™ qPCR SuperMix for ABI PRISM® instrument (Invitrogen, Lofer), utilizing a Rotor-GeneTM 600 and Rotor-GeneTM 600 Series Software 1.7 (GenXpress, Wiener-Neudorf). Each reaction mixture contained primers (400 nM) and template (10 ng). Each independent sample was tested in triplicate. For each sample, the mean cycle threshold of the test transcript was normalized to that of reference gene rpoB [51] or 16 s rRNA and to one randomly selected rpoB or 16 s rRNA reference sample of the WT. Values above or below 1 indicate that the transcript is present in higher or lower numbers, respectively, in the mutant compared to the WT strain.
CTX-kmΦ transduction
TCP production was determined by phage transduction frequency, utilizing phage CTX-kmΦ and TCP producing cells [52]. In short, lysogenic strain O395 CTX-kmΦ was grown over night in LB broth. Cells were centrifuged and the supernatant, containing CTX-kmΦ, was sterilized by filtration through a Whatman Klari-Flex filter unit with pore size of 0.22 µm (GE Healthcare, Vienna). Aliquots were stored at 4°C. For induction of TCP production, V. cholerae O1 El Tor was grown in AKI and V. cholerae O1 classical in LB broth (pH 6.5) at 30°C. Cells were incubated with CTX-kmΦ lysate for 30 min and subsequently plated in parallel on LB plates with sm and sm/km to determine transduction frequency.
CT ELISA
CT production in culture supernatants was determined by the ganglioside GM1 ELISA [53]. V. cholerae O1 El Tor was grown in AKI and V. cholerae O1 classical in LB broth (pH 6.5) at 30°C to induce CT production. Cells were removed from CT containing supernatants by centrifugation and supernatants were stored at −20°C. ELISA plates (BD Falcon, Heidelberg) were coated with GM1 ganglioside (10 µg/ml) in Na2CO3 (10 mM) (Sigma-Aldrich, Taufkirchen) for 4 h at 37°C and washed four times with PBS-T pH 7.4 consisting of NaCl (137 mM), KCl (2.7 mM), Na2HPO4×2 H2O (8.1 mM), KH2PO4 (1.76 mM), Tween-20 (0.05%). Free binding sites were blocked with BSA (4 mg/ml) for 1 h at RT. After washing as described above, CT containing supernatants were diluted in PBS and added to the plate. Additionally, purified CT in PBS (Sigma-Aldrich, Taufkirchen) was inoculated in separate wells to generate a standard curve. ELISA plates were incubated with supernatants and purified CT for 1 h at RT and were washed again as described above. After incubation with the primary antibody (anti-CT antibody produced in rabbit, Sigma Aldrich, Taufkirchen), diluted 1∶2,000 in PBS containing BSA (4 mg/ml) for 1 h at RT, ELISA plates were washed four times with PBS-T. After incubation with the secondary antibody (goat anti-rabbit IgG - horseradish peroxidase, Amersham-Biosiences, Freiburg), diluted 1∶5,000 in PBS containing BSA (4 mg/ml) for 1 h at RT and subsequent washing, the ELISA plates were incubated with TMB Substrate Reagent Set (BioLegend, Vienna) for detection of CT. The reaction was stopped, by adding H3PO4 (1 M) and ELISA plates were measured at OD450 by using a microplate reader (FLUOstar Omega, BMG LABtech, Vienna).
In vivo colonization studies
Competition assays for intestinal colonization in infant C57Bl/6 mice (Harlan Laboratories, Inc., Udine) (in vivo) and for growth in LB broth (in vitro) were performed as previously described [54], with a mixture of mutant (LacZ−) and isogenic WT (LacZ+) strains. The competitive index (CI) is the ratio calculated of CFU of mutant to WT recovered at 24 h, normalized to the input ratio.
Statistical analyses
For data analyses Mann-Whitney U test, Kruskal-Wallis followed by Dunns test of selected pairs of columns or unpaired t test were used. Differences were considered significant for P values of <0.05.
Results
dsbAB mutants and porin production
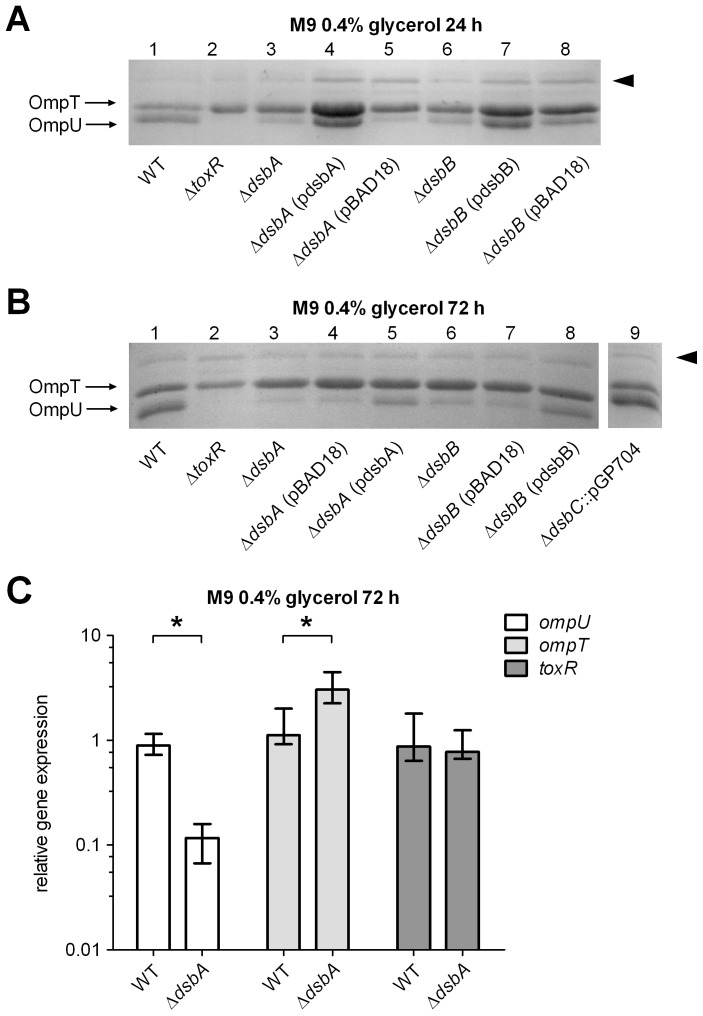
ToxR contains two cysteine residues in its periplasmic domain at amino acid position 236 and 293 [30]. Inter- as well as intrachain disulfide bond formation was found to exist within ToxR, which may alter ToxR activity [34]. To address whether ToxR activity depends on disulfide bond formation via the Dsb system [55], porin production was monitored by comparing a V. cholerae WT strain and corresponding dsbA, dsbB and dsbC deletion mutants. WT and the dsbA mutant showed similar growth kinetics under the conditions tested (Fig. S1). It is essential to monitor dsb defects during growth of cells in minimal media, because in rich LB broth small organic molecules are present that act as oxidizing agents and hence replace Dsb function [56]. Also for monitoring Dsb activity it might be of advantage to use C-sources, which are used for respiration rather than fermentation to establish ubiquinone depending e-transfer activating DsbB. Therefore, cultures were grown in M9 glycerol medium with high respiration to either stationary (24 h) or late growth phase (72 h) and then subjected to OMP preparation. As shown for O1 El Tor (Fig. 1A, B) and classical strain O395 (Fig. S2A), OmpU production was significantly decreased in samples derived from dsbA and dsbB, but not in samples derived from dsbC mutants (Fig. 1A). Complementing activity was observed by expressing dsbA or dsbB from pBAD18 in corresponding dsb knockout mutant strains (Fig. 1A, B). Importantly, if cells were grown in LB broth or M9 supplemented with glucose, no significant change in porin production was observed, neither in dsbA nor dsbB mutant strains (data not shown).
Figure 1. dsb knockout mutations and porin production in V. cholerae P27459-S.
Panel A, B shown are OMP profiles on SDS-PAGE of WT, ΔtoxR, ΔdsbA, ΔdsbA (pBAD18), ΔdsbA (pdsbA), ΔdsbB, ΔdsbB (pBAD18), ΔdsbB (pdsbB) and dsbC::pGP704 (only panel B) strains derived from cells grown for 24 h and 72 h in M9 glycerol, respectively. Arrows mark OmpU and OmpT. As a negative control, ΔtoxR mutant strain showed no production of OmpU and derepressed OmpT protein level. The arrowhead on the right indicates a ToxR independent protein band used as loading control. Panel C, shown are qRT-PCR analyses of WT and ΔdsbA strain for ompU, ompT and toxR transcripts. Fold change ratios were calculated by comparing cDNA levels of genes of interest and the reference gene rpoB, derived from cells grown in M9 glycerol for 72 h. Data are presented as median fold change and the error bars indicate the interquartile range of each data set. Experiments were performed with at least six independent samples, utilizing the Mann-Whitney U test, P<0.05.
Furthermore, qRT-PCR was performed for toxR, ompU and ompT transcription in WT and dsbA mutant strains derived from late stationary grown cells (72 h) in M9 glycerol medium (Fig. 1B). In accordance with the OMP profile, the qRT-PCR data of WT compared with a dsbA mutant showed a 8-fold reduced ompU transcription, whereas ompT transcription was 3-fold upregulated. Notably, no alteration was observed for toxR transcription, indicating that a dsbA deletion has no effect on toxR transcription under the conditions tested.
ToxR disulfide bond formation depends on ToxS and DsbA
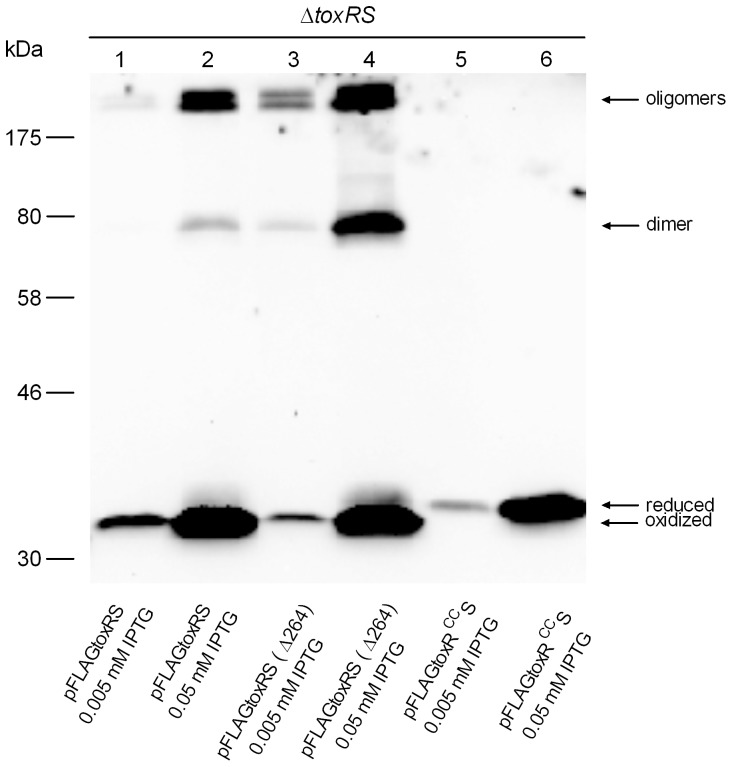
In order to monitor ToxR protein in V. cholerae, toxRS was amplified using chromosomal DNA as template and toxRS encoding reporter plasmids were constructed. They additionally contained a DNA sequence encoding a FLAG tag peptide (N-DYKDDDDK-C) fused in frame to the toxR 5′ end, termed as FLAG-tagged toxRS. Cysteine residue 236 in ToxR was shown to contribute to ToxR intrachain disulfide bond formation [34]. Hence, the two periplasmic cysteine residues 236 and 293 were replaced with serine residues to construct a toxRCCS mutant. Furthermore, to address ToxS function on ToxR disulfide bond formation a control plasmid pFLAGtoxRS(Δ264) was constructed, harboring a 264 bp internal deletion in toxS, yielding an incomplete ToxS protein. Immunoblot analysis was performed using specific FLAG-antibodies to detect ToxR proteins. The latter were produced by plasmid encoded FLAG-tagged toxRS, toxRS(Δ264) and toxRCCS in a V. cholerae ΔtoxRS strain grown in LB medium. Cell extracts were sampled in Laemmli buffer, both with and without the reducing agent ß-mercaptoethanol. In the presence of ß-mercaptoethanol, only a single reduced ToxR or ToxRCC protein band with 35 kDa was observed (data not shown). If cell extracts were not treated with reducing agent, then additional ToxR protein bands became visible. As shown (Fig. 2, lane 4, from bottom to top), ToxR protein bands were visible as an oxidized form at ∼33 kDa and a minor reduced monomer band at 35 kDa, followed by a 70 kDa homodimer and oligomeric forms >170 kDa. In contrast, FLAG-tagged toxRCCS expression (Fig. 2, lane 5 and 6) only yielded one single monomeric ToxR form migrating at 35 kDa, indicating no existing disulfide bond formation. Interestingly, it was observed that by expressing FLAG-tagged toxRS(Δ264), the formation of ToxR disulfide bond dependent homodimer and oligomers was enhanced (Fig. 2, compare lane 1, 2 with lane 3, 4). This observation was also shown in E. coli (see below) and demonstrates that toxRS coexpression negatively influences the formation of cysteinyl dependent ToxR homodimers and oligomers.
Figure 2. toxRS coexpression in V. cholerae P27459-S ΔtoxRS mutant strain acts negatively on ToxR disulfide bond homodimer and oligomers.
Shown is an immunoblot analysis derived from SDS-PAGE analysis performed under non-reducing conditions, utilizing anti-FLAG antibodies and V. cholerae cells harboring various pFLAGtoxRS expressing plasmids, grown in LB medium to mid-log phase and induced with IPTG. Molecular markers are indicated on the left side. Two different IPTG concentrations are indicated, showing different ToxR levels. Immunoblot analysis was performed at least three times, and results were reproducible.
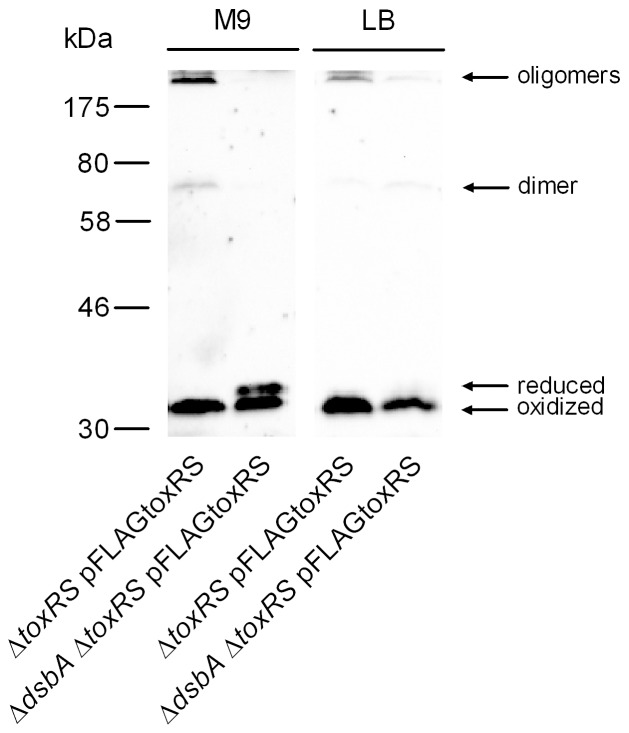
ToxR disulfide bond formation of plasmid encoded FLAG-tagged toxRS was monitored in V. cholerae ΔtoxRS and dsbA+/− strain backgrounds. Cells were grown under two different conditions, M9 glycerol and LB broth. As shown (Fig. 3), if FLAG-tagged toxRS was expressed in a dsbA+ background, mainly oxidized ToxR and oligomeric forms were visible, regardless of the growth medium. In contrast, expression of FLAG-tagged toxRS in a dsbA strain in M9 glycerol medium resulted in both ToxR monomer forms and no observable oligomers, while in LB broth, the oxidized monomer was the dominant expressed ToxR form followed by very minor expressed oligomer forms. Thus, these data demonstrate that disulfide bond formation of ToxR differs in a dsbA dependent manner. Importantly, ToxR intrachain disulfide bond formation existed independently of DsbA under LB broth growth conditions, but responded to DsbA function under M9 glycerol growth conditions (Fig. 3). These results indicate a correlation between the activity of DsbA, ToxR intrachain disulfide bond formation and ToxR activity.
Figure 3. dsbA knockout mutant and ToxR forms.
Immunoblot analyses are shown using anti-FLAG antibodies to detect FLAG-tagged ToxR produced in V. cholerae P27459-S ΔtoxRS and ΔtoxRS ΔdsbA mutant strain (as indicated in the figure). Bacterial cultures harboring pFLAGtoxRS were grown to mid-log phase in M9 glycerol and in LB broth and induced with IPTG. ToxR mobility in the different samples was monitored and differences for intrachain disulfide bond formation were detected. Immunoblot analysis was performed at least three times, and results were reproducible.
toxRCC mutant affects porin production
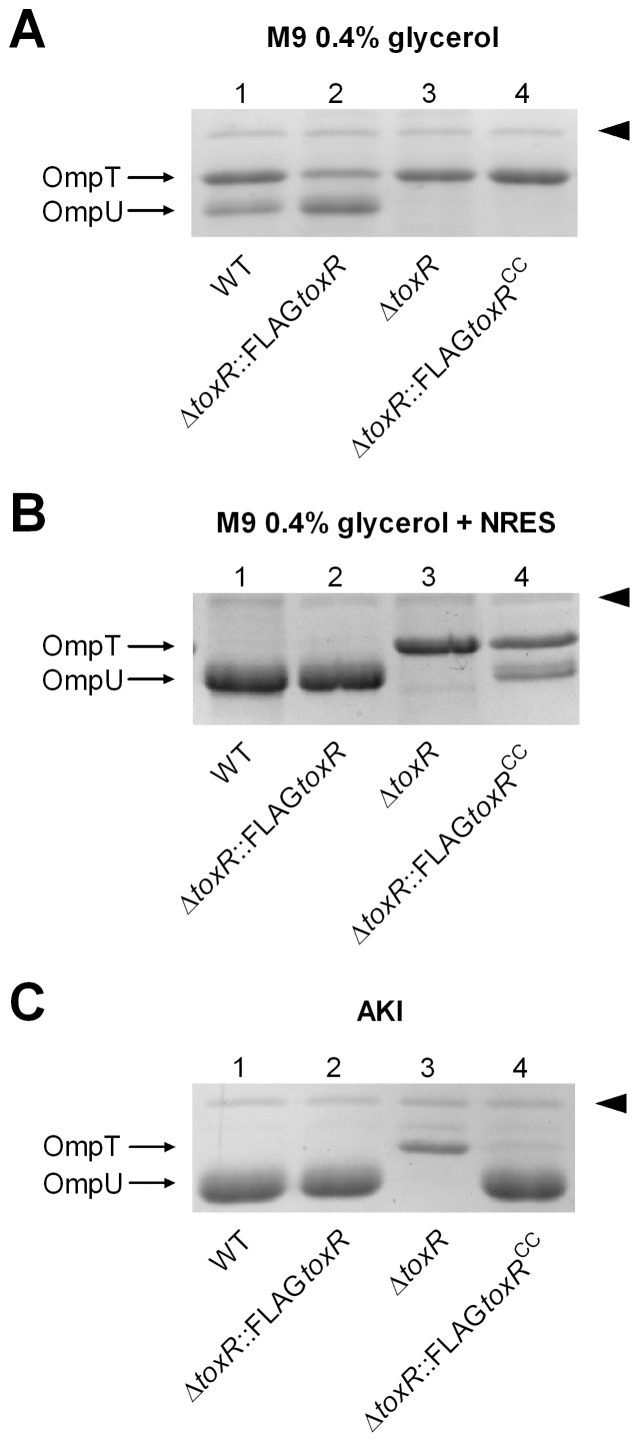
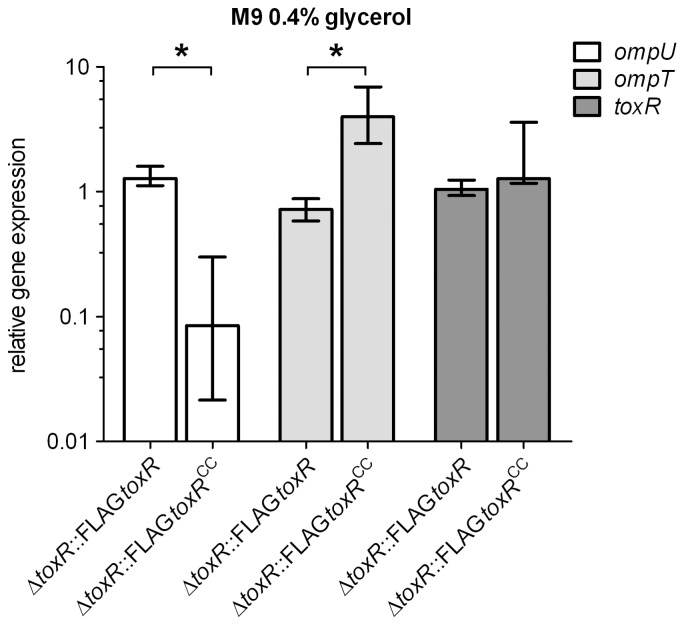
As shown above, a dsbA knockout mutation interfered with disulfide bond formation of ToxR and furthermore resulted in a loss of ToxR activity. Therefore, toxRCC expression should cause a similar defect in ToxR activity. ToxR regulation is considered non-physiological if plasmid encoded toxR is used, because elevated ToxR protein levels counteract ToxR regulation sensitivity [21]. Accordingly, no difference in OmpU and OmpT production was observed by analyzing the OMP profile of ΔtoxRS strains expressing plasmid encoded FLAG-tagged toxRS or toxRCCS (data not shown). Therefore, single copy gene number and chromosomally expressed toxR and toxRCC were tested under different growth conditions. FLAG-tagged toxR and toxRCC gene alleles were transferred into the chromosomal toxR expression locus of a V. cholerae ΔtoxR strain (see Material and Methods). Subsequently, chromosomally produced FLAG-tagged ToxR proteins were collected from membrane extracts derived from cells grown to stationary phase in LB broth and monitored by immunoblot analysis. It is important to note that ToxR proteins could be detected under these conditions, however, the signal intensity was weak such that monitoring ToxR was only possible under high magnification sensitivity (Fig. S3). No signals were detected for samples treated without ß-mercapthoethanol (data not shown). As shown recently, the production of the porins OmpU and OmpT was significantly changed in V. cholerae cells if they were grown in a complex broth medium, such as LB, compared to minimal T medium [57]. It was further shown that the pattern of OMP production observed for cells grown in LB broth could be mimicked by the addition of the amino acids L-asparagine, -arginine, -glutamate and –serine (termed NRES) to minimal growth medium [57]. In order to monitor the effect of the toxRCC mutant on OMP production, standard growth conditions were tested that generally affect porin [57] and virulence factor production, i.e., AKI growth medium [42]. When cells were grown in AKI, M9 glycerol or M9 glycerol NRES medium, the pattern of OmpU and OmpT production in a FLAG-tagged toxR strain was similar to that observed for WT (Fig. 4A, B, C, lane 1 and 2, respectively). In contrast and shown by others [25], no OmpU and derepressed OmpT was observed for the ΔtoxR mutant strain (Fig. 4A, B, C, lane 3). Interestingly, the toxRCC mutant cells, grown in M9 glycerol medium (Fig. 4A, lane 4), showed no OmpU and derepressed OmpT, similarly as observed for the dsbAB mutations (Fig. 1A). Such an effect was also observed in the classical strain O395 (Fig. S2B). Furthermore, production of OmpU was only partially restored and did not reach WT levels in toxRCC mutant cells grown in M9 NRES medium (Fig. 4B, lane 4). Interestingly, if the dsbA mutant strain was grown in M9 glycerol with NRES, OmpU production was similar to that observed for NRES activated WT cells (data not shown). Considering the recently published effects of NRES [57], these data indicate that dsbA mutation does not disrupt NRES activated OmpU production, but toxRCC mutation does. To quantify the effects of toxRCC on transcription, ompU, ompT and toxR mRNA levels were determined by qRT-PCR analysis of cells grown in M9 glycerol medium for 24 h. The results showed that toxR gene transcript levels of FLAG-tagged toxR compared to toxRCC were similar (Fig. 5). In contrast, ompU transcription showed a 15-fold reduction in a FLAG-tagged toxRCC mutant compared to the FLAG-tagged toxR strain, whereas ompT transcription was 6-fold increased. Thereby, the transcriptional pattern corresponds to the observed porin production profile and is related to a similar transcriptional pattern seen in a dsbA mutant (Fig. 1B). It is important to note that in AKI grown WT, FLAG-tagged toxR or FLAG-tagged toxRCC cells, OmpU levels were strongly upregulated, as shown in Fig. 4C. To quantify this observation we monitored toxR and ompU transcription in WT, using qRT-PCR, by comparing cells grown in M9 glycerol and AKI medium. The obtained results showed a 4-fold upregulation for ompU transcription, but no difference in toxR transcription (Fig. S4). Therefore, the increase in OmpU production did not correlate with elevated toxR transcription.
Figure 4. Chromosomal expression of FLAGtoxR and FLAGtoxRCC and porin regulation.
Panel A, B and C, shown are OMP profiles derived from OM preparations, representing WT, ΔtoxR::FLAGtoxR, ΔtoxR and ΔtoxR::FLAGtoxRCC strains, grown to stationary phase in M9 glycerol (A), M9 glycerol NRES (B) and AKI (C) during the anaerobic growth phase, respectively. Arrows mark OmpU and OmpT. Arrowheads on the right indicate a ToxR independent protein band used as loading control.
Figure 5. Transcriptional analysis of toxR and porin genes ompU and ompT in V. cholerae P27459-S.
Using qRT-PCR analysis, transcriptional activity of chromosomal encoding FLAG-tagged toxR and toxRCC strains was monitored for the porin genes ompU and ompT and also for toxR and toxRCC. mRNA levels of rpoB (used as a reference gene) were determined and correlated with the mRNA level of the genes of interest. Data are presented as median fold change and the error bars indicate the interquartile range of each data set. Experiments were performed with six independent samples, the Mann-Whitney U test was used, P<0.05.
ToxR operators negatively influence ToxRS heterodimerization
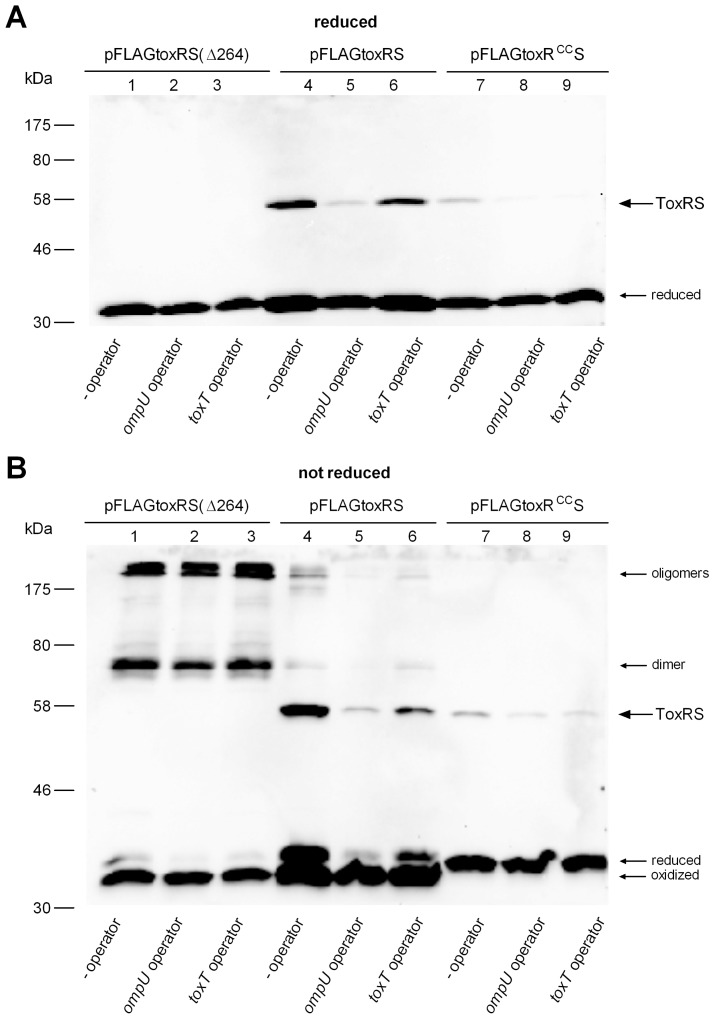
If pFLAGtoxRS was expressed in E. coli cells,grown in LB broth (mid-log phase OD600 of 0.5 and subsequently induced with IPTG for 1 h), a novel and stable SDS-resistant FLAG related protein band of about 55 kDa was observed (Fig. 6A, B, lane 4). This size corresponded well with a reported ToxRS heterodimer [34]. This heterodimer is disulfide bond independent, since ToxS does not have any cysteine residues and the heterodimer was also present in SDS-PAGE analysis using samples treated with ß-mercaptoethanol (Fig. 6A, lane 4). To confirm that this protein band represents a ToxRS heterodimer, again pFLAGtoxRS(Δ264) served as negative control, resulting in the loss of the 55 kDa protein band (Fig. 6A, B, lane 1). Thus, these data indicate that this protein band indeed represented a ToxRS heterodimer. Furthermore, as observed in Fig. 6A, B, lane 7, toxRCCS expression also yielded a heterodimer, which was less pronounced and indicated a decreased interaction of both proteins. Prompted by the observation that ToxRS heterodimer formation only occured in E. coli (Fig. 6, lane 4), but not in V. cholerae (Fig. 2, lane 1, 2), pFLAGtoxRS plasmids were constructed to additionally contain ToxR operator sites, either of ompU [58] or toxT [19] (see Material and Methods). Such plasmids were expressed again in E. coli, showing that in the presence of ToxR binding sites, ToxRS heterodimer formation appeared strongly diminished (Fig. 6B, lane 4–6) and mainly ToxR monomers with the intrachain disulfide bond were detectable. Furthermore, also a slight decrease in ToxRCCS heterodimerization was observed in the presence of ToxR operators (Fig. 6A, B, lane 8, 9). Notably, the presence of ToxR operators had no effect on the appearance of cysteinyl dependent ToxR homodimer or oligomer formation if expressed as toxRS(Δ264) (Fig. 6B, lane 1–3). Thus, it seems that ToxR binding sites negatively influenced ToxRS heterodimerization. These data provide a rational explanation for the failure to observe a heterodimer in V. cholerae, because numerous ToxR regulated genes [16] and therefore multiple ToxR operators are present in V. cholerae.
Figure 6. toxRS coexpression in E. coli XL1-Blue strain.
Shown are immunoblot analyses utilizing anti-FLAG antibodies to monitor FLAG-tagged ToxR production of pFLAGtoxRS constructs, performed under reducing (panel A) and non-reducing conditions (panel B). pFLAGtoxRS was expressed in E. coli cells grown in LB broth to mid-log phase (OD600 of 0.5) and subsequently induced with IPTG for 1 h. From left to right, shown are pFLAGtoxRS(Δ264), pFLAGtoxRS and pFLAGtoxRCCS, either containing no ToxR operator sequence or ompU or toxT operator sequences, respectively. A 55 kDa ToxR cross-reacting protein band, associated with pFLAGtoxRS and pFLAGtoxRCCS, is indicated by an arrow. To note, cysteinyl dependent homodimer and oligomer ToxR bands occurred diminished as observed for pFLAGtoxRS in comparison to pFLAGtoxRS(Δ264). Molecular size markers are indicated on the left. Immunoblot analysis was performed at least three times, and results were reproducible.
Virulence factor production of toxRCC mutants in El Tor and classical strains and in vivo colonization of mice
In order to characterize a toxRCC mutant strain for virulence factor production, the strain encoding chromosomally FLAG-tagged toxRCC was compared with a chromosomally FLAG-tagged toxR strain. The levels of CT and TCP were assessed in both biotypes, O1 V. cholerae El Tor and O395 classical strain, Table 3. As shown, no significant differences were detectable between FLAG-tagged toxRCC and toxR, while slightly enhanced CT production was observed by comparing FLAG-tagged toxR and WT strains. In contrast, a toxR deletion strain was about 82-fold reduced for CT production compared to a FLAG-tagged toxR O1 El Tor strain and >1,700-fold reduced in the classical strain, Table 3. A toxS knockout was about 5- and 3-fold below the CT level compared to the parental strains with FLAG-tagged toxR O1 El Tor and classical, respectively see Table 3. Additionally, the CTX-kmΦ transduction frequency was determined for both biotypes, Table 3. This assay relies on the production level of the type IV bundle forming pili TCP and subsequent CTX-kmΦ transduction utilizing a kanamycin encoding CTX-kmΦ [52]. CTX-kmΦ transduction frequencies of WT, FLAG-tagged toxR and toxRCC strains did not show significant differences and for ΔtoxR mutants, no detectable CTX-kmΦ transductants were observed. ΔtoxS mutants of O1 El Tor and classical strains compared to a corresponding FLAG-tagged toxR strain showed 16- and 7-fold lower frequencies, respectively. In addition, in vivo and in vitro competition assays were performed to further elucidate a putative loss of function for the toxRCC mutant. A 1∶1 mixture of chromosomally encoded FLAG-tagged toxR (LacZ−) and FLAG-tagged toxRCC (LacZ+) strains was administered orally to infant mice (in vivo). As a control, LB broth was inoculated with this mixture and incubated for 24 h at 37°C (in vitro). Competitive indices from at least four independent competition experiments were obtained, Table 4, demonstrating no significant difference in colonization fitness between both strains tested. Finally, and as shown above, OMP analysis of the FLAG-tagged toxRCC mutant strain grown in AKI medium showed a similar porin pattern as obtained for a WT strain (Fig. 4C). Thus, under AKI growth conditions, FLAG-tagged toxRCC did not disrupt the porin production pattern. In summary, expression of toxRCC has no effect on virulence gene transcription under the conditions tested. Consequently, these data suggest that other ToxR activation mechanisms exist that do not require ToxR disulfide bond formation.
Table 3. Virulence factor production of chromosomal encoded FLAG-tagged toxR and FLAG-tagged toxRCC mutants.
| Strain | CTX-kmΦ transduction rate [cfu×ml−1]a | CT production [ng×ml−1]b |
| El Tor P27459-S | ||
| ΔtoxR::FLAGtoxR | 4.76×10−5 (1.5×10−5–1.03×10−4) | 7502 (4341–15734) |
| ΔtoxR | <LOD | 92 * (77–127) |
| ΔtoxR::FLAGtoxRCC | 3.83×10−5 (2.48×10−5–6.84×10−5) | 5012 (2433–11578) |
| ΔtoxR::FLAGtoxR ΔtoxS | 2.96×10−6 (8.15×10−7–2.76×10−5) | 1543 * (988–2570) |
| WT | 4.54×10−5 (2.85×10−5–6.09×10−5) | 4654 (2605–7806) |
| Classical O395 | ||
| ΔtoxR::FLAGtoxR | 1.55×10−4 (2.78×10−5–1.98×10−4) | 17178.5 (10372–40044) |
| ΔtoxR | <LOD | 10 * (10–10) |
| ΔtoxR::FLAGtoxRCC | 5.71×10−5 (1.63×10−5–3.13×10−4) | 12725.5 (3961–48650) |
| ΔtoxR::FLAGtoxR ΔtoxS | 2.22×10−5 (8.87×10−6–1.01×10−4) | 5490 (3042–15710) |
| WT | 1.34×10−4 (2.02×10−5–5.60×10−4) | 11354.5 (9978–25108) |
median and interquartile range of at least 7 independent experiments.
median and interquartile range of 9 independent experiments.
significant by Kruskal-Wallis test followed by Dunn's test of selected pairs of columns with P<0.05.
<LOD below limit of detection of 5×10−8.
Table 4. In vitro and in vivo competition of chromosomal encoded FLAG-tagged toxR versus FLAG-tagged toxRCC mutant in El Tor P27459-S.
median and interquartile range of 12 independent experiments.
median and interquartile range of 5 independent experiments.
Discussion
A previous study [34] reported that ToxR formed a heterodimer with ToxS and also existed as a homodimer and monomer based on inter- and intrachain disulfide bonds of cysteine 236 and 293. Moreover, in vitro analysis using the purified periplasmic domain of ToxR showed that ToxR homodimers exist and rely on cysteine 293 by forming an intermolecular disulfide bond [59]. We revisited these earlier characterizations with our work, because we observed that dsbAB mutants affect porin production of OmpU and OmpT. Therefore, we focused our studies on the influence of cysteinyl dependent ToxR forms, ToxR activity, ToxRS interaction and response to the Dsb system. As identified in this study, dsbA, dsbB deletions and toxRCC mutants showed altered activity for porin regulation. In V. cholerae, dsbAB and dsbCD homologues exist, encoding functions for disulfide bond formation [60] and correction of protein folding [61] that are located in the periplasm. In E. coli, it is known that DsbAB activity is important for disulfide bond formation if cells are grown in minimal medium. This is because in full broth media, such as LB, small organic molecules are present, which act as oxidizing agents on cysteine residues and will therefore also lead to disulfide bond formation [56]. As observed in this study, dsbA or dsbB, but not dsbC deletion mutants affected ToxR dependent porin regulation. As shown for dsbA or dsbB mutant strains grown under M9 glycerol growth conditions, low OmpU and derepressed OmpT levels were observed, which corresponded to altered ompU and ompT transcription if compared to the WT strain. Interestingly, in a dsbA mutant a statistic significant increase of ompT transcription was observed, also the OMP profile exposed a slightly higher expressed OmpT band. However, the latter observation also indicated a higher OmpT expression in dsb mutants if compared with toxR knockout mutant, thereby it can be excluded that dsbA deficient disulfide bond formation of ToxR is the cause of increased OmpT expression. Further investigation is necessary to provide an explanation for this observation. No alteration in porin production was observed in the dsbC mutant, which suggests that the DsbCD system does not participate in regulating ToxR activity. However, its participation cannot be entirely excluded since DsbCD activity may have an influence on porin production under conditions not tested in this study. Since the dsbAB as well as toxRCC phenotypes were also detectable in classical strain O395, we conclude that the herein characterized ToxR cysteine requirement is significant for V. cholerae strains in general.
To identify whether DsbA disulfide oxidoreductase activity per se is influencing ToxR forms, FLAG-tagged toxRS expression was analyzed in a dsbA toxRS knockout strain. Interestingly, it was demonstrated that by expression of toxRS in the absence of dsbA ToxR showed intrachain disufide bond and reduced monomer forms in M9 glycerol, but only the intrachain disulfide bond form in LB broth. The latter is explainable because small organic molecules are present in LB and can catalyze intrachain disulfide bond formation supporting ToxR activity. Taken together, these data imply that ToxR activity is stimulated by DsbA or alternative oxidizing mechanisms, producing thiol-dependent intrachain disulfide bond formation.
To confirm dsbA dependent ToxR phenotypes, a toxRCC mutant was characterized, which is defined by amino acid substitutions of cysteines to serines. The appearance and electrophoretic mobility of ToxRCC tested without reducing agents in SDS-PAGE was as expected. ToxRCC showed no intrachain disulfide bonded monomer, homodimer or oligomeric forms. Instead, ToxRCC only produceed a single protein band corresponding to the size of a reduced ToxR monomer. Therefore, the activity of ToxRCC was associated with the reduced monomeric form. By using a chromosomally encoded toxRCC mutant, it was demonstrated that porin production is influenced profoundly, quite similar to that observed for the dsbA or dsbB deletion mutants. Additionally, we tested a double mutant comprising dsbA and toxRCC and observed no differences in the OMP profile compared to a toxRCC mutant (data not shown). Furthermore, no difference was observed between toxR or toxRCC transcription levels, hence we argue that toxRCC represents a mutation affecting ToxR activity. Therefore, we provide evidence that DsbA activity is targeting cysteine residues of ToxR and this influences ToxR activation.
As shown recently [57], when an O1 El Tor V. cholerae strain was grown in minimal medium OmpT was expressed as a major porin and no OmpU protein was observed. In LB broth, the porin production pattern was reversed. If NRES amino acids were added to the minimal medium, the porin production profile appeared similar to that observed for cells cultured in LB broth. Mey et al. further showed that NRES amino acids added to the minimal medium led to elevated toxR transcription, which they concluded, is the cause for the switched porin production. As shown in here, we also confirmed the NRES effect in showing that addition of NRES to M9 glycerol medium enhances OmpU production for WT and FLAG-tagged toxR strains. Also FLAG-tagged toxRCC mutant cells responded to NRES but to a lower extend. However, toxRCC mutant cells grown in AKI showed maximum OmpU production, similar as observed for FLAG-tagged toxR or WT strains. Hence, we assumed that under AKI growth conditions, elevated toxR and toxRCC transcription would occur, explaining the increase in OmpU production. This assumption was not confirmed, since qRT-PCR analysis of WT cells cultured in M9 glycerol compared with AKI cultures showed no significant difference in toxR transcription. These data indicate, that other mechanisms exist, which influence ToxR activity. For example, we cannot rule out, that DsbAB activity influences ompU post-transcriptional, -translational or secretion pathways, neither can we exclude that dsbA or toxRCC mutants are solely responsible for the observed decreased ompU transcription. Therefore, other yet unknown factors may contribute or facilitate OmpU expression, especially under growth conditions such as AKI or LB broth media. Also to mention is that if toxRCC is expressed from multi-copy plasmid, then we observed that cells were producing high OmpU levels. Similar behavior of ToxR activity was observed earlier [21], however such conditions were regarded as non-physiological, hence we will not address this for further discussions. In summary, our data consistently show that under minimal growth conditions, porin regulation solely depends on ToxR intrachain disulfide bond formation if toxR is expressed from its chromosomal loci.
If toxS was coexpressed along with toxR, cysteinyl dependent ToxR homodimer and oligomer formation was decreased as shown in our results. The latter finding is supported by earlier published data [34], showing suppression of the ToxR homodimer in the presence of ToxS. According to these findings, it can be assumed that under physiological expression conditions, ToxRS counteract the formation of homodimers. We further suggest, that oligomer ToxR forms most likely represent artifacts, resulting from toxR overexpression, since this form was not observed earlier, if chromosomal expressed toxR was monitored [34]. As found earlier by others, e.g., [21], [39] the combined action of ToxR and ToxS contributed to full ToxR operational activity and both proteins were shown to form a heterodimer [34]. With the expression system we used, heterodimer formation was observed only in E. coli, it was SDS- and heat-resistant and was stably detectable without any cross-linking chemistry. This finding is not exceptional, since other SDS-resistant protein-protein interactions are known to occur [62]–[64]. Such interactions were explained by the failure of SDS to access tightly formed contact sites between the proteins. In contrast, a ToxRS heterodimer could not be visualized if both genes were coexpressed in V. cholerae. To further characterize this, defined ToxR binding sites, located upstream of ompU [58] and toxT [19], were subcloned onto the toxRS expression plasmids and analyzed in E. coli. Interestingly, if ToxR binding sites were present, heterodimer signals were strongly diminished, indicating that the presence of operator sites negatively interfered with ToxRS heterodimerization. Earlier data also described ToxRS heterodimerization in V. cholerae, using cross-linking chemistry [34]. There, detection of the heterodimer was very weak. Since V. cholerae contained multiple ToxR regulated genes [16], the presented data may support the view whereby only weak ToxRS heterodimerization might be observable in the presence of ToxR operator sites. Interestingly, in another report [39] it was shown that ToxS was not necessary for ToxR binding to DNA in vitro. Therefore, it seems questionable whether ToxRS heterodimers can be found in a DNA bound state. So far it can only be speculated whether disaggregation of heterodimer is a consequence of ToxR binding and this needs further characterization.
Finally, we tested virulence factor production in O1 El Tor or classical strains and neither the level of CT, nor CTX-kmΦ transduction frequencies showed any significant differences between FLAG-tagged toxRCC and toxR strains. This indicated that under virulence factor inducing conditions, ToxRCC can participate in the regulation cascade of the virulence factor production system. Moreover, the toxRCC mutant in O1 El Tor V. cholerae strain did also not display a phenotype for in vivo colonization. In summary, this suggests that cysteine associated ToxR forms are dispensable for ToxR activity under virulence factor inducing conditions. Thereby, we argue that multiple ToxRS activation conditions may exist, which do not rely on thiol-dependent disulfide bond forms. So far, we cannot explain why porin gene regulation seems to respond sensitive to ToxR disulfide bond formation, whereas virulence gene expression is not. Recently, a possible hint was provided by Morgan and colleagues [29]. They published data addressing the isolation of toxR point mutations, which differentially target toxT and ompU transcription. All of their toxR mutations were found in the cytoplasmic part of ToxR. For example, for amino acid residues V71, F69 and E39 it was proposed that they interfere with RNAP engagement on the ompU promoter, rather then with DNA binding. Interestingly, the same point mutants had much less effect on toxT activation. In contrast, amino acids R65 and D73 affected more severely ompU activation than toxT. Based on these results, the authors concluded that the facing of ToxR upon operator binding seems differently oriented for the promoter regions of ompU and toxT. Thereby, it can only be speculated that ToxRCC protein configuration is sufficient to serve for toxT activation by correctly facing to the toxT promoter, but becomes conditionally insufficient for ompU activation for yet unknown reasons.
Finally, the question arises whether ToxR may represent a thiol-based redox switch regulator. Although, several periplasmic proteins depend on DsbAB folding activity to obtain function, e.g. PhoA [65], while no examples to our knowledge exist for DsbAB depending thiol-based redox switches for periplasmic proteins. However, the latter does not exclude the possibility that defined environmental conditions exist, e.g., in the aquatic environment, that modulate thiol-dependent intrachain disulfide bond formation in ToxR, hence leading to changes in ToxR activity. For example, such influences could be derived from stress responses, as dsb gene transcription is under the control of the σE-membrane stress pathway in V. cholerae and additionally of the Cxp regulon as shown in E. coli [66]–[68]. ToxR activity may also be linked to the cellular metabolism status of the cells. The status may be reflected by electron transfer activity, which is known to influence DsbAB activity whereby menaquinone or ubiquinone act as recipients for e− derived from disulfide bond formations [55].
Supporting Information
Growth and cell survival for P27459-S and ΔdsbA mutant strains. Shown are growth curves (OD600 left Y axis) and colony forming units (cfu/ml right Y axis) of WT strain P27459-S and corresponding ΔdsbA strains over 72 h in M9 minimal media supplemented with glycerol (0.4%).
(TIF)
OMP profiles of V. cholerae O1 classical strain O395. Arrows indicate OmpU and OmpT. Panel A, shown are WT O395, ΔtoxR, ΔdsbA and ΔdsbB strains grown to stationary phase in M9 glycerol medium. Panel B, shown are WT O395, ΔtoxR, ΔtoxR::FLAGtoxR and ΔtoxR::FLAGtoxRCC strains. Cells were grown to stationary phase in LB broth medium.. Arrowheads on the right indicate a ToxR independent protein band used as loading control.
(TIF)
Detection of chromosomal encoded FLAG-tagged ToxR expressed fusion proteins. Immunoblot analysis is shown, using anti-FLAG antibodies to detect chromosomal expression of FLAG-tagged toxR and toxRCC in V. cholerae P27459-S and mutant strains ΔtoxR and ΔtoxRS of isolated membrane fractions. Cross-reacting background bands are marked with asterisks and ToxR is indicated by an arrow. Molecular size markers are indicated on the left. Immunoblot analysis was performed at least two times, and results were reproducible.
(TIF)
Transcriptional analysis of toxR and porin gene ompU in V. cholerae P27459-S grown in M9 glycerol compared to AKI conditions. The WT strain was cultured in M9 glycerol medium to mid log growth phase and shifted to fresh M9 glycerol or AKI medium for 45 min. Subsequently mRNA was prepared and qRT-PCR was performed for the ompU porin gene and also for toxR. mRNA level of 16S rRNA was determined as a reference and correlated with the mRNA level of the genes of interest. Experiments were performed with three independent samples and data represent means and standard deviations. The unpaired t test was used, P<0.05.
(TIF)
Acknowledgments
For helpful discussion and critical reading of the manuscript we thank Michelle Dziejman.
Funding Statement
This work was supported by the Austrian Science Fund (FWF) W901 (DK Molecular Enzymology) to VHIF, AS, SR, SS and JR and by the German Research Foundation (DFG) Re1561/4-1. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Faruque SM, Albert MJ, Mekalanos JJ (1998) Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae . Microbiol and Mol Biol Rev 62: 1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pruzzo C, Vezzulli L, Colwell RR (2008) Global impact of Vibrio cholerae interactions with chitin. Environ Microbiol 10: 1400–1410. [DOI] [PubMed] [Google Scholar]
- 3. Reidl J, Klose KE (2002) Vibrio cholerae and cholera: Out of the water and into the host. FEMS-Microbiol Rev 26: 125–139. [DOI] [PubMed] [Google Scholar]
- 4. Andoh A, Fujiyama Y, Sakumoto H, Uchihara H, Kimura T, et al. (1998) Detection of complement C3 and factor B gene expression in normal colorectal mucosa, adenomas and carcinomas. Clin Exp Immunol 111: 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mallow EB, Harris A, Salzman N, Russell JP, DeBerardinis RJ, et al. (1996) Human enteric defensins. J Biol Chem 271: 4038–4045. [DOI] [PubMed] [Google Scholar]
- 6. Herrington DA, Hall RH, Losonsky G, Mekalanos JJ, Taylor RK, et al. (1988) Toxin, toxin-coregulated pili and the toxR regulation are essential for Vibrio cholerae pathogenesis in humans. J Exp Med 168: 1487–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DiRita VJ, Parsot C, Jander G, Mekalanos JJ (1991) Regulatory cascade controls virulence in Vibrio cholerae . Proc Natl Acad Sci 88: 5403–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Faruque SM, Nair GB, Mekalanos JJ (2004) Genetics of stress adaptation and virulence in toxigenic Vibrio cholerae . DNA Cell Biol 23: 723–741. [DOI] [PubMed] [Google Scholar]
- 9. Matson JS, Withey JH, DiRita VJ (2007) Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect Immun 5542–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karaolis DK, Johnson JA, Bailey CC, Boedeker EC, Kaper JB, et al. (1998) A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA 95: 3134–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kovach ME, Shaffer MD, Peterson KM (1996) A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae . Microbiology 142: 2165–2174. [DOI] [PubMed] [Google Scholar]
- 12. Miller VL, Mekalanos JJ (1984) Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR . Proc Nat Acad Sci USA 81: 3471–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Osorio CR, Klose KE (2000) A region of the transmembrane regulatory protein ToxR That tethers the transcriptional activation domain to the cytoplasmic membrane displays wide divergence among Vibrio species. J Bacteriol 182: 526–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Skorupski K, Taylor RK (1999) A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol Microbiol 31: 763–771. [DOI] [PubMed] [Google Scholar]
- 15. Xu X, Stern AM, Liu Z, Kan B, Zhu J (2010) Virulence regulator AphB enhances toxR transcription in Vibrio cholerae . BMC Microbiology doi:101186/1471-2180-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bina J, Zhu J, Dziejman M, Faruque S, Calderwood S, et al. (2003) ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc Natl Acad Sci USA 100: 2801–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Childers BM, Klose KE (2007) Regulation of virulence in Vibrio cholerae: the ToxR regulon. Future Microbiol 2: 335–344. [DOI] [PubMed] [Google Scholar]
- 18. Hase CC, Mekalanos JJ (1998) TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae . Proc Natl Acad Sci USA 95: 730–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Higgins DE, DiRita VJ (1994) Transcriptional control of toxT, a regulatory gene in the ToxR regulon of Vibrio cholerae . Mol Microbiol 14: 17–29. [DOI] [PubMed] [Google Scholar]
- 20. DiRita VJ (1994) Multiple regulatory systems in Vibrio cholerae pathogenesis. Trends Microbiol 2: 37–38. [DOI] [PubMed] [Google Scholar]
- 21. Miller V, DiRita VJ, Mekalanos JJ (1989) Identification of toxS, a regulatory gene whose product enhances ToxR-mediated activation of the cholera toxin promoter. J Bacteriol 171: 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Withey JH, DiRita VJ (2006) The toxbox: specific DNA sequence requirements for activation of Vibrio cholerae virulence genes by ToxT. Mol Microbiol 59: 1779–1789. [DOI] [PubMed] [Google Scholar]
- 23. Champion GA, Neely MN, Brennan MA, DiRita VJ (1997) A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol Microbiol 23: 323–331. [DOI] [PubMed] [Google Scholar]
- 24. Li CC, Merrell DS, Camilli A, Kaper JB (2002) ToxR interferes with CRP-dependent transcriptional activation of ompT in Vibrio cholerae . Mol Microbiol 43: 1577–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miller VL, Mekalanos JJ (1988) A novel suicide vector and its use in construction of insertion mutations: Osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR . J Bacteriol 170: 2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Provenzano D, Klose KE (2000) Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc Natl Acad Sci USA 97: 10220–10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krukonis ES, Yu RR, DiRita VJ (2000) The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol Microbiol 38: 67–84. [DOI] [PubMed] [Google Scholar]
- 28. Krukonis ES, DiRita VJ (2003) DNA binding and ToxR responsiveness by the wing domain of TcpP, an activator of virulence gene expression in Vibrio cholerae . Mol Cell 12: 157–165. [DOI] [PubMed] [Google Scholar]
- 29. Morgan SJ, Felek S, Gadwal S, Koropatkin NM, Perry JW, et al. (2011) The two faces of ToxR: activator of ompU, co-regulator of toxT in Vibrio cholerae . Mol Microbiol 81: 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller VL, Taylor RK, Mekalanos JJ (1987) Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell 48: 271–279. [DOI] [PubMed] [Google Scholar]
- 31. Dziejman M, Mekalanos JJ (1994) Analysis of membrane protein interaction: ToxR can dimerize the amino terminus of phage lambda repressor. Mol Microbiol 13: 485–494. [DOI] [PubMed] [Google Scholar]
- 32. Kolmar H, Hennecke F, Götz K, Janzer B, Vogt B, et al. (1995) Membrane insertion of the bacterial signal transduction protein ToxR and requirements of transcriptional activation studied by modular replacement of different protein substructures. EMBO J 14: 3895–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ottemann KM, Mekalanos JJ (1995) Analysis of Vibrio cholerae ToxR function by construction of novel fusion proteins. Mol Microbiol 15: 719–731. [DOI] [PubMed] [Google Scholar]
- 34. Ottemann KM, Mekalanos JJ (1996) The ToxR protein of Vibrio cholerae forms homodimers and heterodimers. J Bacteriol 178: 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crawford JA, Krukonis ES, DiRita VJ (2003) Membrane localization of the ToxR winged-helix domain is required for TcpP-mediated virulence gene activation in Vibrio cholerae . Mol Microbiol 47: 1459–1473. [DOI] [PubMed] [Google Scholar]
- 36. Dziejman M, Kolmar H, Fritz HJ, Mekalanos JJ (1999) ToxR co-operative interactions are not modulated by environmental conditions or periplasmic domain conformation. Mol Microbiol 31: 305–317. [DOI] [PubMed] [Google Scholar]
- 37. Hung DT, Mekalanos JJ (2005) Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc Natl Acad Sci, USA 22: 3028–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. DiRita VJ, Mekalanos JJ (1991) Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell 64: 29–37. [DOI] [PubMed] [Google Scholar]
- 39. Pfau JD, Taylor RK (1998) Mutations in toxR and toxS that separate transcriptional activation from DNA binding at the cholera toxin gene promoter. J Bacteriol 180: 4724–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pearson GDN, Woods A, Chiang SL, Mekalanos JJ (1993) CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc Natl Acad Sci USA 90: 3750–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mekalanos JJ (1983) Duplication and amplification of toxin genes in Vibrio cholerae . Cell 35: 253–263. [DOI] [PubMed] [Google Scholar]
- 42. Iwanaga M, Yamamoto K (1985) New medium for the production of cholera toxin by Vibrio cholerae O1 Biotype El Tor. J Clin Microbiol 22: 405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grimberg J, Maguire S, Belluscio L (1989) A simple method for the preparation of plasmid and chromosomal E. coli DNA. Nucleic Acids Res 17: 8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Donnenberg MS, Kaper JB (1991) Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun 59: 4310–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77: 61–68. [DOI] [PubMed] [Google Scholar]
- 47. Carlone ACY, Myrtle LT, Rumschlag HS, Sottner FO (1986) Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus-species. J Clin Microbiol 24: 330–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Warburg O, Christian W (1941) Isolierung und Kristallisation des Gärungsferments Enolase. Biochem Z 310: 384–423. [Google Scholar]
- 49. Kang D, Gho YS, Suh M, Kang C (2002) Highly sensitive and fast protein detection with Coomassie Brilliant Blue in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. BKCS 23: 11. [Google Scholar]
- 50. Schild S, Nelson EJ, Camilli A (2008) Immunization with Vibrio cholerae outer membrane vesicles induces protective immunity in mice. Infect Immun 76: 4554–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schild S, Tamayo R, Nelson EJ, Qadri F, Calderwood SB, et al. (2007) Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe 2: 264–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Waldor KW, Mekalanos JJ (1996) Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272: 1910–1914. [DOI] [PubMed] [Google Scholar]
- 53. Svennerholm AM, Holmgren J (1978) Identification of Escherichia coli heat-labile enterotoxin by means of a ganglioside immunosorbant assay (GM1-ELISA) procedure. Curr Microbiol 1: 19–23. [Google Scholar]
- 54. Moisi M, Jenul C, Butler SM, New A, Tutz S, et al. (2009) A novel regulatory protein involved in motility of Vibrio cholerae . J Bacteriol 191: 7027–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kadokura H, Beckwith J (2010) Mechanisms of oxidative protein folding in the bacterial cell envelope. Antioxid Redox Signal 13: 1231–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bardwell JC, Lee JO, Jander G, Martin N, Belin D, et al. (1993) A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci USA 90: 1038–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mey AR, Craig SA, Payne SM (2012) Effects of amino acid supplementation on porin expression and ToxR levels in Vibrio cholerae. Infect Immun 80: 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Crawford JA, Kaper JB, DiRita VJ (1998) Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae . Mol Microbiol 29: 235–246. [DOI] [PubMed] [Google Scholar]
- 59. Chatterjee A, Dutta PK, Chowdhury R (2007) Effect of fatty acids and cholesterol present in bile on expression of virulence factors and motility of Vibrio cholerae . Infect Immun 75: 1946–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yu J, Webb H, Hirst TR (1992) A homologue of the Escherichia coli DsbA protein involved in disulphide bond formation is required for enterotoxin biogenesis in Vibrio cholerae . Mol Microbiol 6: 1949–1958. [DOI] [PubMed] [Google Scholar]
- 61. Segatori L, Murphy L, Arredondo S, Kadokura H, Gilbert H, et al. (2006) Conserved role of the linker alpha-helix of the bacterial disulfide isomerase DsbC in the avoidance of misoxidation by DsbB. J Biol Chem 281: 4911–4919. [DOI] [PubMed] [Google Scholar]
- 62. Salahpour A, Angers S, Bouvier M (2000) Functional significance of oligomerization of G-protein-coupled receptors. TEM 11: 163–168. [DOI] [PubMed] [Google Scholar]
- 63. Gentile F, Amodeo P, Febbraio F, Picaro F, Motta A, et al. (2002) SDS-resistant active and thermostable dimers are obtained from the dissociation of homotetrameric beta-glycosidase from hyperthermophilic Sulfolobus solfataricus in SDS. Stabilizing role of the A–C intermonomeric interface. J Biol Chem 277: 44050–44060. [DOI] [PubMed] [Google Scholar]
- 64. Lueders J, Pyrowolakis G, Jentsch S (2003) The ubiquitin-like protein HUB1 forms SDS-resistant complexes with cellular proteins in the absence of ATP. EMBO Rep 4: 1169–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Akiyama Y, Ito K (1993) Folding and assembly of bacterial alkaline phosphatase in vitro and in vivo. J Biol Chem 268: 8146–8150. [PubMed] [Google Scholar]
- 66. Ding Y, Davis BM, Waldor MK (2004) Hfq is essential for Vibrio cholerae virulence and downregulates SigmaE expression. Mol Microbiol 53: 345–354. [DOI] [PubMed] [Google Scholar]
- 67. Danese PN, Silhavy TJ (1997) The sigma(E) and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli . Genes Dev 11: 1183–1193. [DOI] [PubMed] [Google Scholar]
- 68. Pogliano J, Lynch AS, Belin D, Lin EC, Beckwith J (1997) Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev 11: 1169–1182. [DOI] [PubMed] [Google Scholar]
- 69. Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166: 557–580. [DOI] [PubMed] [Google Scholar]
- 70. Correa NE, Lauriano CM, McGee R, Klose KE (2000) Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol Microbiol 35: 743–755. [DOI] [PubMed] [Google Scholar]
- 71. Oka A, Sugisaki H, Takanami M (1981) Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol 147: 217–226. [DOI] [PubMed] [Google Scholar]
- 72. Guzman LM, Belin D, Carson MJ, Beckwith J (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Quinones M, Kimsey HH, Waldor MK (2005) LexA cleavage is required for CTX prophage induction. Mol Cell 17: 291–300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth and cell survival for P27459-S and ΔdsbA mutant strains. Shown are growth curves (OD600 left Y axis) and colony forming units (cfu/ml right Y axis) of WT strain P27459-S and corresponding ΔdsbA strains over 72 h in M9 minimal media supplemented with glycerol (0.4%).
(TIF)
OMP profiles of V. cholerae O1 classical strain O395. Arrows indicate OmpU and OmpT. Panel A, shown are WT O395, ΔtoxR, ΔdsbA and ΔdsbB strains grown to stationary phase in M9 glycerol medium. Panel B, shown are WT O395, ΔtoxR, ΔtoxR::FLAGtoxR and ΔtoxR::FLAGtoxRCC strains. Cells were grown to stationary phase in LB broth medium.. Arrowheads on the right indicate a ToxR independent protein band used as loading control.
(TIF)
Detection of chromosomal encoded FLAG-tagged ToxR expressed fusion proteins. Immunoblot analysis is shown, using anti-FLAG antibodies to detect chromosomal expression of FLAG-tagged toxR and toxRCC in V. cholerae P27459-S and mutant strains ΔtoxR and ΔtoxRS of isolated membrane fractions. Cross-reacting background bands are marked with asterisks and ToxR is indicated by an arrow. Molecular size markers are indicated on the left. Immunoblot analysis was performed at least two times, and results were reproducible.
(TIF)
Transcriptional analysis of toxR and porin gene ompU in V. cholerae P27459-S grown in M9 glycerol compared to AKI conditions. The WT strain was cultured in M9 glycerol medium to mid log growth phase and shifted to fresh M9 glycerol or AKI medium for 45 min. Subsequently mRNA was prepared and qRT-PCR was performed for the ompU porin gene and also for toxR. mRNA level of 16S rRNA was determined as a reference and correlated with the mRNA level of the genes of interest. Experiments were performed with three independent samples and data represent means and standard deviations. The unpaired t test was used, P<0.05.
(TIF)