Abstract
Four Cladobotryum isolates were collected from four different commercially grown mushroom types infected with cobweb disease in Cheongdo-gun and Chilgok-gun of Gyeongbuk Province, Korea in 2010. The isolates were identified as C. mycophilum from Agaricus bisporus and Pleurotus eryngii, C. varium from Flammulina velutipes and Hypsizygus marmoreus. The cultural characteristics of the four isolates were investigated using potato dextrose agar (PDA) media under nine different temperatures ranging from 5~32℃. Rapid growth of the isolates to colony diameters of 47~82 mm was observed at conditions of 18~22℃. No growth was observed at 32℃. C. mycophilum produced a yellowish red pigment while C. varium produced a cream colored pigment after cultivation for 25 days on PDA. Phylogenetic analysis of the internal transcribed spacer region and partial 28S rDNA from the four isolates confirmed they were C. mycophilum and C. varium. Cross pathogenicity tests revealed that the two isolates of C. mycophilum were highly pathogenic toward three mushroom types, but not toward H. marmoreus. The two isolates of C. varium were less pathogenic than those of C. mycophilum, but were pathogenic toward all mushroom types evaluated.
Keywords: Cobweb disease, Cross pathogenicity, ITS region, Phylogenetic analysis, 28S rDNA
Introduction
Cobweb disease is caused by several species of Cladobotryum and is characterized by the growth of coarse mycelium over the affected mushrooms [1]. The disease is found in all mushroom-growing countries worldwide and causes economic loss in areas it impacts [2-4]. There have been numerous reports of cobweb disease affecting Agaricus bisporus, which is known to be infected by species of Cladobotryum including C. dendroides, C. mycophilum, C. varium, C. multiseptatum, and C. verticillatum [2, 5]. C. mycophilum and C. varium are known to be the dominant pathogens for A. bisporus. In Korea, numerous types of mushrooms including A. bisporus, Pleurotus eryngii, Flammulina velutipes and Hypsizygus marmoreus are commercially cultivated for domestic consumption.
In recent years, two species of cobweb fungi, C. mycophilum on A. bisporus, and C. varium on P. eryngii and F. velutipes, have been reported in Korea [6, 7]. Cladobotryum was identified by the morphological and cultural characteristics of its sporocarp and spores, as well as its internal transcribed spacer (ITS) region and partial 28S rDNA genetic characteristics. However, these two species produce nearly identical symptoms during mushroom cultivation. In order to manage cobweb disease effectively, correct identification of pathogens is important as cobweb disease can be spread through spore distribution. However, the possible infection by Cladobotryum species of different types of mushrooms has yet to be investigated. Therefore, in this study we investigated Cladobotryum isolates from four mushroom types based on their morphological and genetic characteristics, and their cross pathogenic ability.
Materials and Methods
Fungal isolation and identification
Cladobotryum isolates were collected from the fruiting bodies of four different cobweb disease infected mushrooms: A. bisporus, P. eryngii, F. velutipes and H. marmoreus, which were obtained from Cheongdo-gun and Chilgok-gun in Gyeongbuk Province, Korea in 2010. All cultures used in the experiments were derived from a single spore and were grown on potato dextrose agar (PDA) at 20℃ in the dark for 3~4 days. The shape, size and color of 100 conidia and conidiophores of the isolates were microscopically observed. The isolates were then identified based on the morphological characteristics of the conidia and conidiophores according to the descriptions from Gams and Hoozemans [8].
Growth conditions
The isolates were cultured on PDA media at 20℃ for four days under dark conditions. From these cultures, small mycelia plugs (5 mm in diameter) were punched out from the actively growing area using a cork borer and placed at the center of a culture plate (90 mm in diameter) containing PDA media. The influence of temperature on their growth was investigated by incubating the plates at 5, 10, 15, 18, 20, 22, 25, 28, and 32℃, and measuring the resulting colony diameters after four days. Data are presented as the means of three replicates. To investigate the resulting pigmentation on the PDA, the cultures were further kept at 22℃ for 5~25 days.
DNA extraction and PCR amplification
Total genomic DNA was extracted from each fungal isolate using lysis buffer according to the procedure described by Liu et al. [9]. Total genomic DNA was used to amplify the ITS region and partial 28S ribosomal DNA (rDNA). The ITS rDNA regions and the partial 28S rDNA were amplified using the primer pairs ITS1F (5'-CTT GGT CAT TTA GAG GAA GTA A-3')/ITS4 (5'-TCC TCC GCT TAT TGA TAT GC-3') [10] and NL1 (5'-GCA TAT CAA TAA GCG GAG GAA AAG-3')/NL4 (5'-GGT CCG TGT TTC AAG ACG G-3') [11], respectively. PCR amplification was conducted in a 20 µL reaction mixture containing 2 µL of fungal DNA (20 ng), 0.2 µL Taq polymerase (5 units/µL), 2 µL 10 reaction buffer (100 mM Tris-HCl, 400 mM KCl, 15 mM MgCl2, pH 9.0), 0.4 µL dNTPs mixture (10 mM) and 2 µL of each primer (5 pmol/µL) using an Applied Biosystems 2720 thermal cycler (Applied Biosystems, Foster City, CA, USA). The PCR conditions included preheat at 94℃ for 3 min followed by 35 cycles of 94℃ for 30 sec, 52℃ for 30 sec, 72℃ for 1 min and final extension at 72℃ for 7min. The amplified DNA fragments were purified using ExoSAP-IT (GE Healthcare, Buckinghamshire, UK) and then subjected to direct sequencing (Solgent, Daejeon, Korea) using the same primers.
Sequence and phylogenetic analysis
The obtained sequences were aligned using the DNASTAR computer package (DNASTAR Inc., Madison, WI, USA) and phylogenetic trees were constructed using the neighbor-joining method in ClustalW [12]. The phylogenetic trees based on the ITS region and partial sequence of the 28S rDNA were generated using TreeView (Win32 ver. 1.6.1). Bootstrap analysis with 100 replications was performed in order to determine the support the data will provide for various clades.
Cross pathogenicity test
The infection ability of the isolates was studied by inoculating each isolate onto four mushroom types. Inoculums were prepared from 10~15-day-old isolation cultures on PDA media and adjusted to 5 × 105 conidia/mL. Inoculation was conducted by spraying the fruiting bodies of each mushroom thoroughly with the spore suspension (50 mL). After being kept in plastic bags in order to maintain 100% humidity for 24 hr, the inoculated mushrooms were incubated at 20℃. The development of disease symptoms was observed visually one day after inoculation and disease severity was rated based on the following score index: +, 1~30% disease severity; ++, 31~50%; +++, > 51%; and nd, no disease development.
Results and Discussion
Morphological characteristics of the isolates and their identification
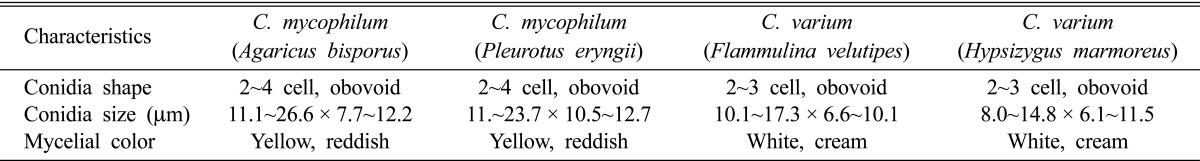
The shape and size of the conidia of the four isolates were observed for 100 conidia (Table 1). The shapes of the four isolates were almost obovoid and consisted of 2~4 cells. The conidia of the four isolates were divided into two groups based on size, one ranging from 11~26 × 7~12 µm and the other from 8~14 × 6~11 µm. These morphological characteristics corresponded to the characteristics of C. mycophilum isolated from A. bisporus and C. varium isolated from F. velutipes [4, 7]. The characteristics of the four isolates also agreed with the description of C. mycophilum and C. varium offered by Gams and Hoozemans [8]. Based on the observed morphological characteristics of the conidia, two isolates from A. bisporus and P. eryngii were identified as C. mycophilum while two isolates from F. velutipes and H. marmoreus were identified as C. varium.
Table 1.
Morphological characteristics of Cladobotryum mycophilum and C. varium isolated from four mushrooms on potato dextrose agar media
The color of the mycelia from the two Cladobotryum species differed over time. The mycelia of all isolates were initially white or grayish as grown on PDA media at 22℃. The mycelia of C. mycophilum became yellowish after 5 days of growth and gradually turned reddish over 25 days. The mycelia of the cultures of C. varium remained white or cream color.
Effect of temperature on the cultures
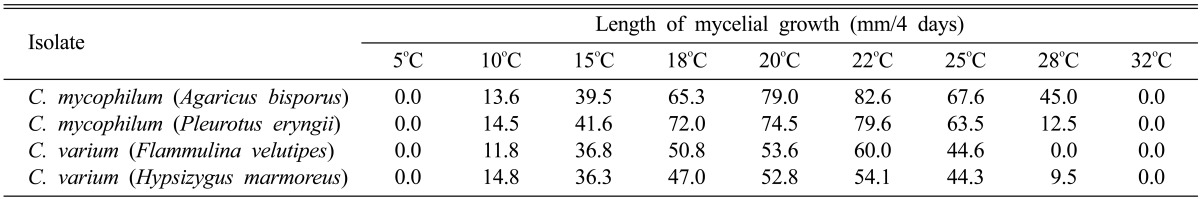
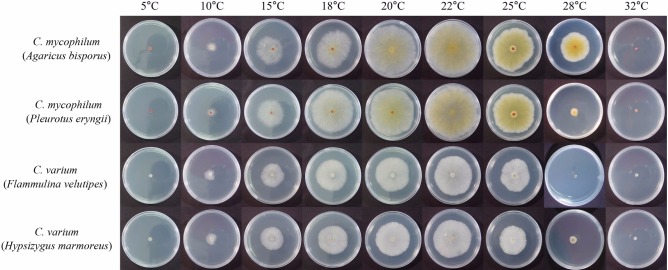
The optimum growth temperature for the four isolates was investigated at temperatures ranging from 5~32℃ (Table 2, Fig. 1). The growth of the isolates was favored at temperatures of 18 to 22℃ with the optimum for all isolates at 22℃. Of the four isolates, only C. mycophilum from A. bisporus grew at 28℃ while the other three types could not grow at this temperature. The growth rate of C. mycophilum (> 79.6 mm at 22℃) was faster than that of C. varium (> 54.1 mm at 22℃). No mycelia grew at less than 10℃, or at a temperature as high as 28℃, except for C. mycophilum from A. bisporus.
Table 2.
Growth of Cladobotryum mycophilum and C. varium isolated from four mushrooms at different temperatures after 4 days incubation on PDA media
Fig. 1.
Effect of temperature on mycelial growth of Cladobotryum mycophilum and C. varium isolated from four mushroom species on potato dextrose agar at 4 days after incubation.
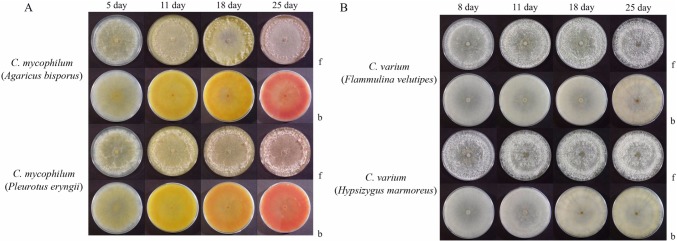
Pigment production
Pigments produced by two Cladobotryum species grown at 22℃ were observed on PDA media (Fig. 2). After five days of growth, two isolates of C. mycophilum produced yellowish pigment in the growth media and gradually turned reddish over 25 days (Fig. 2A). The color of the two isolates of C. varium were cream and white (Fig. 2B). Similarly, red pigment was reportedly produced by C. mycophilum from P. eryngii cultured on PDA [13]. C. varium did not exhibit the pinkish red mycelium coloration such as observed with C. mycophilum from A. bisporus [14]. The red coloration was due to the pigment aurofusarin, a secondary metabolite associated with the Cladobotryum species [15].
Fig. 2.
Pigments produced by Cladobotryum mycophilum (A) and C. varium (B) on PDA media kept in darkness at 22℃ for 5, 8, 11, 18 and 25 days (f, front view; b, back view of plates).
Molecular analysis
Sequences of the ITS region (596~600 bp) and partial 28S rDNA (~563 bp) from the four isolates were compared using the GENETYX program. Two isolates from C. mycophilum exhibited 100% homology in their ITS regions and partial 28S rDNA sequences, as did the two isolates from C. varium. The sequence from the C. mycophilum ITS region and partial sequence of 28S rDNA from A. bisporus and P. eryngii showed high homology (100%) with those from C. mycophilum (FN859436 and FN859434, respectively). The sequences from the ITS region and partial 28S rDNA of C. varium from F. velutipes and H. marmoreus revealed 98.5% and 100% homology with Hypomyces aurantius and the anamorphs of C. varium (AF055297, AF160230, respectively).
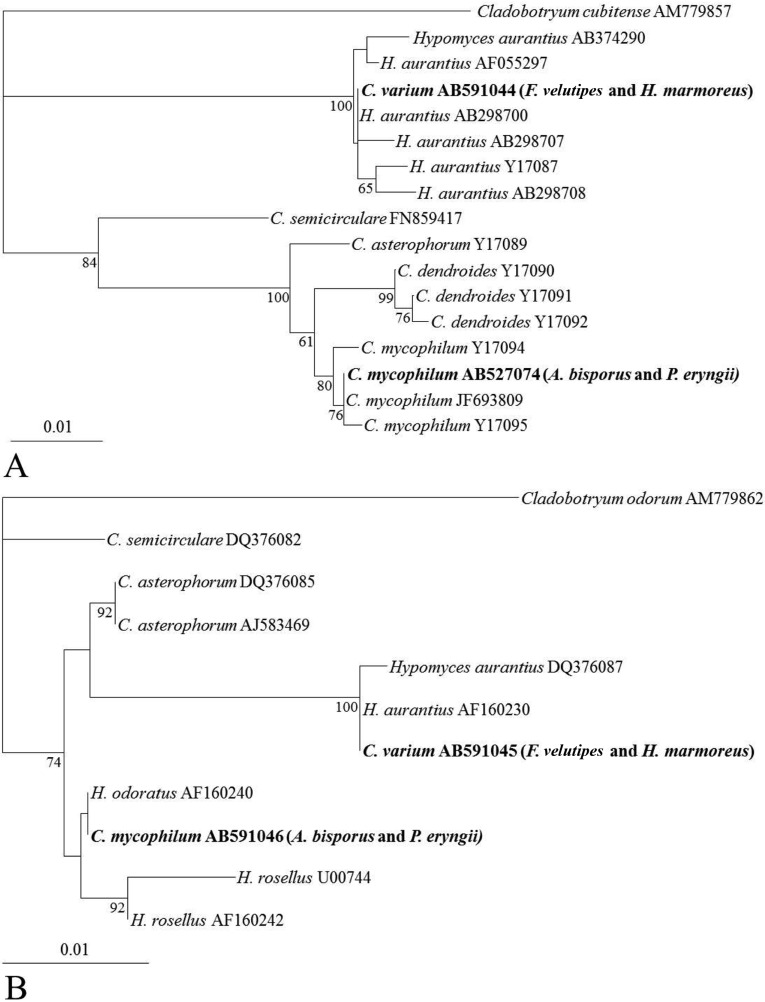
The phylogenetic relationship between the two Cladobotryum species was analyzed based on comparison of their ITS regions (Fig. 3A) and partial 28S rDNA sequences (Fig. 3B) with those of other Cladobotryum sequences obtained from Genbank. The ITS regions of the two isolates of C. mycophilum (AB527074) from this study were clustered with that of C. mycophilum (JF693809), while C. varium (AB591044) clustered with H. aurantius (AB298700, anamorph of C. varium). Similarly, the sequence of the partial 28S rDNA of the two isolates of C. mycophilum clustered with H. odoratus (AF160240, anamorph of C. mycophilum) while that of C. varium clustered with H. aurantius (AF160230, anamorph of C. varium).
Fig. 3.
Phylogenetic trees constructed by the neighbor-joining method based on comparison of the internal transcribed spacer region (A) and partial 28S rDNA (B) sequences of Cladobotryum sp. with those of other Cladobotryum species from GenBank. C. cubitense and C. odorum were used as the outgroup. Cladobotryum species observed in this study are shown in bold. Numbers on branches are the confidence values obtained for 100 replicates (only values above 80% are shown). The bar represents a phylogenetic distance of 1%.
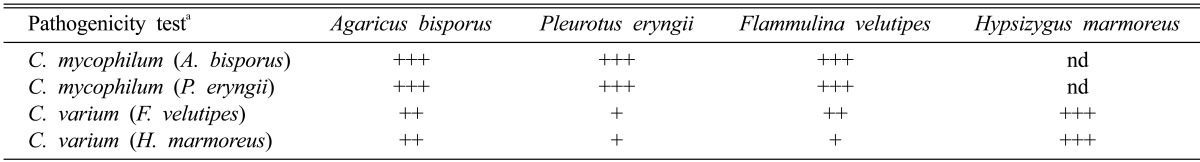
Cross pathogenicity test
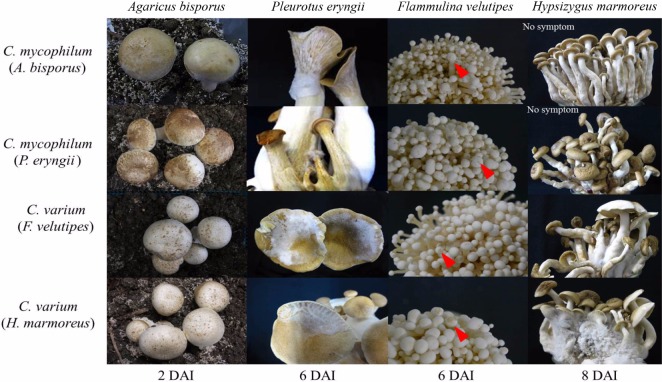
The cross pathogenicity of the isolates was tested using four types of mushrooms. The two isolates of C. mycophilum were pathogenic to three of the mushroom types, but not H. marmoreus (Table 3, Fig. 3). The two isolates of C. varium were pathogenic to all mushroom types. The severity of disease caused by C. mycophilum was more severe than that of C. varium. In the period after inoculation, disease severity was observed to be highest against the original host, e.g., the isolate obtained from H. marmoreus caused the most severe infection in H. marmoreus (Fig. 4). Typical cobweb symptoms including small brown spots were observed 2~3 days after inoculation (DAI). White mycelia were observed at 3 DAI, and after 5 DAI, the fruiting bodies were rotten and covered with massive spores. In this study, C. mycophilum isolates from A. bisporus and P. eryngii were unable to directly infect H. marmoreus. Conversely, C. varium could infect all four mushroom types even though they were less pathogenic. These findings suggest that F. velutipes was a potential host for C. mycophilium and that A. bisporus and P. eryngii were potential hosts for C. varium.
Table 3.
Pathogenicity of two isolates of Cladobotryum mycophilum and C. varium inoculated on four mushrooms
aDisease rate: +, 1~30% disease severity; ++, 31~50%; +++, > 50%; nd, no disease development.
Fig. 4.
Cross pathogenicity of two isolates of Cladobotryum mycophilum and two isolates of C. varium on four mushrooms at 20℃. Arrows indicate black spots on cap. DAI, days after inoculation.
In this study, the four isolates of Cladobotryum were identified as C. mycophilum or C. varium based on their morphological and genetic characteristics. Previously, cobweb fungi on mushrooms were reported as C. mycophilum on A. bisporus based on morphological and genetic characteristics [7], as C. mycophilum on P. eryngii, and C. varium on F. velutips based on the morphological characteristics of their spores and conidiophores [4, 6]. Based on the results of the morphological assessments and phylogenetic analyses conducted in this study, we confirmed that the Cladobotryum isolates from the four mushroom types belonged to C. mycophilum and C. varium. The growth of both species was favored at 18~22℃ and C. mycophilum grew faster than C. varium.
C. mycophilum exhibited greater pathogenicity than C. varium against the mushroom types evaluated in this study. The observed cross pathogenic ability of the two Cladobotryum species should aid in designing control measure for cobweb disease as mushrooms are grown year round in Korea. This disease can spread rapidly because Cladobotryum sp. produce masses of spores. Moreover, some species of Cladobotryum from A. bisporus were observed to be fungicide resistant [16] and fungicide application is restricted for edible mushrooms due to its residual toxicity. Disinfection using near-UV irradiation has been described as an effective method for reducing pathogenic fungi and bacteria in mushroom growing spaces [17]. However, further studies are needed in order to ensure prevention of outbreaks of cobweb disease during mushroom cultivation.
Acknowledgements
This research was supported by Kyungpook National University Research Fund, 2012.
References
- 1.Fletcher JT, White PF, Gaze RH. Mushrooms: pest and disease control. Andover: Athenaeum Press; 1989. pp. 58–63. [Google Scholar]
- 2.Adie B, Grogan H, Archer S, Mills P. Temporal and spatial dispersal of Cladobotryum conidia in the controlled environment of a mushroom growing room. Appl Environ Microbiol. 2006;72:7212–7217. doi: 10.1128/AEM.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fletcher JT, Gaze RH. Mushrooms pest and disease control: a color handbook. San Diego: Academic Press; 2008. pp. 76–79. [Google Scholar]
- 4.Kim HK, Seok SJ, Kim GP, Moon BJ, Terashita T. Occurrence of disease caused by Cladobotryum varium on Flammulina velutipes in Korea. Korean J Mycol. 1999;27:415–419. [Google Scholar]
- 5.McKay GJ, Egan D, Morris E, Scott C, Brown AE. Genetic and morphological characterization of Cladobotryum species causing cobweb disease of mushrooms. Appl Environ Microbiol. 1999;65:606–610. doi: 10.1128/aem.65.2.606-610.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim TS, Lee HW, Song GW, Shin WG. King oyster mushroom (Pleurotus eryngii) white mold disease caused by Cladobotryum varium. KSM Newsl. 1998;11:46. [Google Scholar]
- 7.Back CG, Kim YH, Jo WS, Chung H, Jung HY. Cobweb disease on Agaricus bisporus caused by Cladobotryum mycophilum in Korea. J Gen Plant Pathol. 2010;76:232–235. [Google Scholar]
- 8.Gams W, Hoozemans AC. Cladobotryum-konidien formen von hypomyces-Arten. Persoonia. 1970;6:95–110. [Google Scholar]
- 9.Liu D, Coloe S, Baird R, Pedersen J. Rapid mini-preparation of fungal DNA for PCR. J Clin Microbiol. 2000;38:471. doi: 10.1128/jcm.38.1.471-471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 11.O'Donnell K. Fusarium and its near relatives. In: Reynolds DR, Taylor JW, editors. The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. Wallingford: CABI; 1993. pp. 225–233. [Google Scholar]
- 12.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gea FJ, Navarro MJ, Suz LM. First report of Cladobotryum mycophilum causing cobweb on cultivated king oyster mushroom in Spain. Plant Dis. 2011;95:1030. doi: 10.1094/PDIS-03-11-0255. [DOI] [PubMed] [Google Scholar]
- 14.Eicker A, Van Greuning M. Fungi in the cultivation of Agaricus bisporus: an updated list of species; Proceedings of the First International Seminar on Mushroom Science; 1991 May 14-17; Horst. Wageningen: PUDOC; 1991. pp. 89–96. [Google Scholar]
- 15.Rogerson CT, Samuels GJ. Boleticolous species of Hypomyces. Mycologia. 1989;81:413–432. [Google Scholar]
- 16.Grogan HM, Gaze RH. Fungicide resistance among Cladobotryum spp.: causal agents of cobweb disease of the edible mushroom Agaricus bisporus. Mycol Res. 2000;104:357–364. [Google Scholar]
- 17.Sawada D, Ohmasa M, Fukuda M, Masuno K, Koide H, Tsunoda S, Nakamura K. Disinfection of some pathogens of mushroom cultivation by photocatalytic treatment. Mycoscience. 2005;46:54–60. [Google Scholar]