Abstract
A number of environmental stresses can lead to enhanced production of superoxide within plant tissues, and plants are believed to rely on the enzyme superoxide dismutase (SOD) to detoxify this reactive oxygen species. We have identified seven cDNAs and genes for SOD in Arabidopsis. These consist of three CuZnSODs (CSD1, CSD2, and CSD3), three FeSODs (FSD1, FSD2, and FSD3), and one MnSOD (MSD1). The chromosomal location of these seven SOD genes has been established. To study this enzyme family, antibodies were generated against five proteins: CSD1, CSD2, CSD3, FSD1, and MSD1. Using these antisera and nondenaturing-polyacrylamide gel electrophoresis enzyme assays, we identified protein and activity for two CuZnSODs and for FeSOD and MnSOD in Arabidopsis rosette tissue. Additionally, subcellular fractionation studies revealed the presence of CSD2 and FeSOD protein within Arabidopsis chloroplasts. The seven SOD mRNAs and the four proteins identified were differentially regulated in response to various light regimes, ozone fumigation, and ultraviolet-B irradiation. To our knowledge, this is the first report of a large-scale analysis of the regulation of multiple SOD proteins in a plant species.
All aerobic organisms are continuously subjected to potentially destructive ROS, including superoxide (O2−), lipid peroxides (ROO·), H2O2, and the highly reactive hydroxyl radical (OH·). These ROS are generated by metabolic processes and their concentrations can be increased by environmental stimuli. To prevent ROS from damaging cellular components, organisms have evolved multiple detoxification mechanisms, including the synthesis of low-Mr antioxidant molecules (e.g. l-ascorbic acid and glutathione) and various enzymes. O2− is an abundant ROS that is formed by univalent electron transfer to O2 and can contribute to the synthesis of OH·, so control of this ROS is essential (Cadenas, 1989; Halliwell and Gutteridge, 1990).
SODs catalyze the conversion of O2− to H2O2. Three classes of SOD activity have been identified that differ by the active site metal cofactors (Fe, Mn, or Cu and Zn). The primary sequences of FeSOD and MnSOD apoproteins are related, whereas CuZnSOD is distinct. Fungi and animals have CuZnSOD and MnSOD, whereas some plants and bacteria have been demonstrated to contain all three forms (Bowler et al., 1992). The plant SOD isoenzymes also differ in their subcellular location. Typically, MnSOD is mitochondrial, FeSOD is plastidic, and CuZnSOD can be plastidic or cytosolic (Bowler et al., 1992). There are also reports of peroxisomal and extracellular SODs (Streller and Wingsle, 1994; Bueno et al., 1995).
The importance of SOD has been demonstrated by analysis of mutants in microbes and animals. SOD mutants in Escherichia coli, Saccharomyces cerevisiae, Neurospora crassa, and Drosophila melanogaster all exhibit increased sensitivity to methyl viologen (paraquat), a redox-active compound that enhances the production of O2− (Carlioz and Touati, 1986; Phillips et al., 1989; Gralla and Valentine, 1991; Chary et al., 1994). SOD is also essential for DNA integrity and normal life span: the E. coli and N. crassa mutations cause an increased spontaneous mutation rate, whereas the D. melanogaster mutant has a significantly shorter life span then the wild type (Carlioz and Touati, 1986; Phillips et al., 1989; Chary et al., 1994). Mutations in human and mouse CuZnSOD have been linked to the disease familial amyotrophic lateral sclerosis, which is characterized by premature neuron death (Rosen et al., 1993). Taken together, this evidence indicates a vital role for SOD in preventing ROS-generated cell damage and death in aerobically growing organisms. SOD is also thought to be important in converting O2− to H2O2 during the pathogen-induced oxidative burst in animal phagocytic immune cells and in plant cells (Desikan et al., 1996; Babior et al., 1997).
In plants exposure to photoinhibitory light, ozone, or other environmental conditions that cause oxidative stress can increase O2− levels (Yruela et al., 1996; Runeckles and Vaartnou, 1997); however, it is not clear whether SOD plays an essential role in attenuating plant oxidative stress in these situations. To date the protective role of SOD in plants has been explored by transgenic approaches, primarily through overexpression or by correlation of SOD expression to the degree of oxidative stress resistance (Bowler et al., 1994; Alscher et al., 1997; Scandalios, 1997). Both approaches have yielded inconclusive and sometimes contradictory results about the role of SOD in plant oxidative stress responses (Bowler et al., 1994; Alscher et al., 1997; Scandalios, 1997).
The existence of three classes of SOD enzymes, each typically encoded by a small gene family, complicates the elucidation of the roles of SOD in plants. This situation is exacerbated by the fact that past work has generally focused on one member of a gene family. To circumvent this problem, we have initiated a thorough analysis of the Arabidopsis SOD genes. The availability of large numbers of cDNA and genomic DNA sequences in Arabidopsis (Newman et al., 1994; Rounsley et al., 1996; Delseny et al., 1997) has made it possible to identify the complete SOD arsenal of this plant. We describe progress toward the detailed understanding of SOD genes and their functions in Arabidopsis, and report on the sequencing of three SOD cDNAs and the map locations of seven SOD structural genes. Information is also provided on the characterization of antisera against five of the seven SOD proteins. Finally, we describe the regulation of SOD activity and mRNA and protein levels in response to ozone, UV-B light, and variations in incident light fluences.
MATERIALS AND METHODS
Strains and Materials
Unless otherwise noted, Arabidopsis ecotype Columbia was planted in a soil mixture (Cornell University, Ithaca, NY) and grown at 70% RH, 21°C, 70 μmol m−2 s−1 PAR from 400 W lamps (Multi-Vapor, General Electric) under a 16-h photoperiod (Landry et al., 1995). Whole rosette tissue was collected for RNA and protein analyses unless otherwise indicated. All treatments were replicated twice with duplicate RNA samples and triplicate protein analysis per replication. Escherichia coli strain XL1 Blue (Stratagene) (F′::Tn10 proA+B+ lacIqΔlacZ M15/recA1 endA1 gyrA96 [NaIr] thi hsdR17 [rk−m+] supE44 relA1 lac) was used as a bacterial host unless otherwise noted.
CuZnSOD2 Microsatellite
Primers CuZnSOD2-Map1F and Map1R (Table I) were designed to amplify a 183-bp fragment from the end sequence of TAMU30D05, which contained a potential microsatellite repeat of (TA)14TG(TA)9. Genomic DNA from Arabidopsis ecotypes Landsberg erecta and Columbia was amplified using the standard microsatellite reaction mixture and program, but with an annealing temperature of 58°C (Bell and Ecker, 1994). The products were separated on a 4% agarose gel, revealing a 4-bp polymorphism between the two ecotypes. This polymorphism was used to map TAMUBAC30D05, and therefore the CSD2 structural gene, using the recombinant inbred lines generated by Lister and Dean (1993).
Table I.
Primers used in this study
| Primer Name | Sequence |
|---|---|
| CuZnSOD1-StuI | 5′-CAAAGGCCTAGTAACAATGGC-3′ |
| CuZnSOD1-HindIII | 5′-ACGTAGAAGCTTTAGCCCTGG-3′ |
| CuZnSOD2-1F | 5′-GTTCCTCTTTCAGCGGCGTCTCT-3′ |
| CuZnSOD2-1R | 5′-CATCGGCATTGGCATTTAT-3′ |
| CuZnSOD2-Map1F | 5′-GCATTACTCCGGTGTCGTC-3′ |
| CuZnSOD2-Map1R | 5′-GAATCTCAATATGTGTCAAC-5′ |
| CuZnSOD3-1F | 5′-TGGATTTCATATTCACTCTTT-3′ |
| CuZnSOD3-1R | 5′-ATTACGTTCTGGTTTACATTAT-3′ |
| FeSOD1-1F | 5′-GGAGCCGCATATGAGCAAACAAA-3′ |
| FeSOD1-1R | 5′-AGCCGAGCACAAGGGGATTCA-3′ |
| FeSOD2-1F | 5′-TGTTATAGTGAAGACGCCCAATG-3′ |
| FeSOD2-1R | 5′-GTTTTAGCCACCCTCAGATACA-3′ |
| MnSOD-1F | 5′-TTACGCTTCCTGATCTTCCTTACG-3′ |
| MnSOD-1R | 5′-CTCCCAAACATCTATACCCACCAG-3′ |
| MnSOD-3A | 5′-GATCCTTTTGGTGGCTCTCC-3′ |
CICYAC Pool PCR
Yeast strains containing a multiplexed CICYAC library were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). Yeast was grown and CICYAC DNA was isolated as described in the provider's handout. Three microliters of the pooled CICYAC DNA was amplified in 25 μL of 2.5 mm MgCl2, 1 mm dNTP, 0.6 μm of each primer, and 3 units of TAQ (Promega) in the follow manner: 2 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 52°C, 90 s at 72°C, and 10 min at 72°C. CICYACs that produced an amplification product were obtained from the Arabidopsis Biological Resource Center and individually tested for amplification.
MSD1 CAPS
The MnSOD structural gene was amplified using the MnSOD 1F and 1R primers (Table I) and the standard CAPS PCR protocol, except that an annealing temperature of 60°C and 1.5 mm MgCl2 were employed (Jarvis et al., 1994). The purified PCR product was sequenced using the MnSOD 1F, 1R, and 3A primers. A Tai1 site was identified in the Columbia sequence that was not in the Landsberg erecta sequence. Digestion of the Landsberg erecta PCR product amplified by MnSOD 1F and 1R with Tai1 yielded DNA fragments of 1 kb and 230 bp, whereas the corresponding Col fragments were 700, 300, and 230 bp. This polymorphism was used to map the MnSOD structural gene using the recombinant inbred lines generated by Lister and Dean (1993).
RNA and Immunoblot Techniques
All RNA isolation and blot hybridizations were as described by Conklin and Last (1995). Five micrograms of RNA was used on blots probed with CAB3, CSD1, CSD2, FSD1, and MSD. RNA blots probed with CSD3 and FSD2 were loaded with 10 μg of total RNA. SDS-PAGE gels and transfers were as described by Zhao and Last (1995). Chemiluminescent detection was as described by Durrant and Fowler (1994). The first and second dimension for two-dimensional SDS-PAGE were as described in the Bio-Rad instructional pamphlet.
Construction of Fusion Protein Plasmids and Antisera Production
pGEX1 and pGEX3X (Pharmacia Biotech) were digested with SmaI, treated with alkaline phosphatase, and purified using a gel-extraction kit (Qiaex, Qiagen, Inc., Santa Clarita, CA; Smith and Johnson, 1988). MSD1 cDNA (accession no. T44258), CSD2 cDNA (accession no. T21324), and CSD3 cDNA (accession no. T88473) were released from the λPRL2 vector by digestion with SmaI/SnaBI (Newman et al., 1994). MSD1 and CSD2 cDNAs were ligated into the SmaI site of pGEX1 to create pGEX-MSD1 and pGEX-CSD2. The CSD3 cDNA was ligated into the SmaI site of pGEX3X to create pGEX-CSD3. The XmnI/BsaI fragment was digested from pcSODRH (Hindges and Slusarenko, 1992) to isolate the CSD1 cDNA and ligated into XmnI/EcoRI-digested pGEX3X to create pGEX-CSD1. The BsaAI/SspI fragment containing FSD1 was digested from pSOD10 (Van Camp et al., 1990) and ligated into the SmaI site of pGEX3X to create pGEX-FSD1. Synthesis of the GST fusion proteins was induced as described by Smith and Johnson (1988), and fusion protein was isolated by gel chromatography. Antisera were produced as described in Zhao and Last (1995).
The MSD1, FSD1, and CSD2 cDNAs generated above were ligated into pQE32 to create pHIS-MSD1, pHIS-FSD1, and pHIS-CSD2. The CSD3 cDNA isolated above was ligated into pQE31 to create pHIS-CSD3. The PCR primers CuZnSOD1-StuI and CuZnSOD1-HindIII (Table I) were used to amplify the CSD1 ORF, which was cloned into pQE30 to create pHIS-CSD1. The His-tagged fusion proteins were purified by urea solubilization and nickel chromatography, as described by the manufacturer (Qiaexpressionist Handbook, Qiagen). All pHIS strains contained a novel, inducible SOD activity that could be seen by gel analysis, indicating that the expressed fusion proteins were active SOD enzymes (D.J. Kliebenstein and R.L. Last, unpublished results).
SOD Activity Gels
Total protein was isolated by grinding 100 mg of frozen tissue in 300 μL of SOD activity extraction buffer (Van Camp et al., 1994) and centrifuging for 5 min at 15,000 rpm in a Eppendorf microcentrifuge (model no. 5415C, Madison, WI). The protein concentration was quantified using a kit (Bio-Rad). Forty micrograms of total protein was subjected to electrophoresis at 180 V for 2 h in a 12% acrylamide nondenaturing PAGE. The gel was stained for SOD activity as described by Beauchamp and Fridovich (1971). Inhibitor studies were as described by Pan and Yau (1992).
SOD Activity Fractionation
One milligram of total protein from whole Arabidopsis rosettes was loaded onto a 0.75-mm thick, 9- × 11-cm 12% acrylamide PAGE gel and run overnight at 100 V and 4°C. The gel was stained for SOD activity, and slices encompassing either the SOD activities or spaces between the SOD activities were cut out of the gel and ground with a glass pestle in 15-mL plastic conical tubes. The gel fragments were transferred to 2-mL Eppendorf tubes and weighed to equalize the samples, an equal amount (w/v) of SDS loading buffer was added, and the samples were boiled for 15 min and then spun in a microcentrifuge for 5 min at maximum speed. Twenty microliters of each sample was immunodetected.
Plastid Purification
Plastids were isolated from whole Arabidopsis rosettes as described previously (Mishkind et al., 1987). Rubisco large subunit levels in the purified plastid protein and total Arabidopsis leaf protein were determined by Coomassie blue staining of SDS-PAGE gels. Equal Rubisco large subunit protein levels were loaded on SDS-PAGE gels and immunodetected.
Circadian Regulation and Plastid Isolation Tissue Growth Conditions
Plants were grown as described above except they were illuminated with 250 μmol m−2 s−1 PAR from 400-W multivapor lamps (General Electric) for a 9-h photoperiod. Tissue for circadian rhythm experiments was collected every 4 h starting at dawn (8 am) on d 21 postimbibition. We shifted all plants to continuous darkness at dusk by covering them with aluminum foil and turning off all chamber lights. Tissue was collected in the dark from dark-treated plants at d 22 every 4 h starting at 8 am. Tissue for plastid isolations was collected 35 d postimbibition.
Photoinhibition Studies
At 14 d postimbibition, plants were covered with 0.13-mm-thick Mylar and illuminated with 1750 μmol m−2 s−1 PAR (Powerstar HQI-TS 400W/D lamps, Osram, Danvers, MA) at 20°C for 4 h and then allowed to recover overnight. Untreated controls were kept in the same chamber covered with cheesecloth and nylon mesh (Hummerts, St. Louis, MO) to keep the light at 70 μmol m−2 s−1 PAR. For the high-CO2 experiment, CO2 was added to the chamber to bring the concentration to 2250 ppm, as measured by a CO2 detector (model no. 225 MK3, Analytical Development Co., Hertsfordshire, UK) operating in reverse mode. The plants were allowed to acclimate to the high CO2 for 20 min before initiation of the high-light pulse.
Ozone Fumigations
Plants were fumigated 14 d postimbibition with 330 ppb ozone for 8 h. Unfumigated controls were grown with the fumigated plants except during fumigation, when they were kept in an equivalent chamber (Conklin and Last, 1995). Plants were allowed to recover overnight in charcoal-filtered (ozone-free) air.
UV Treatments
After growth for 14 d under continuous 70 μmol m−2 s−1 PAR from cool-white fluorescent lamps (CW1500), plants were exposed to 15 kJ of UV-BBE m−2 d−1 supplemental UV light as described by Landry et al. (1995), except that −UV-B controls were treated under both 3-mm-thick Pyrex glass and 0.13-mm-thick Mylar to remove UV-B and UV-C wavelengths (Landry et al., 1995).
RESULTS
Arabidopsis SOD Gene Family
Single CuZnSODs and FeSODs were previously reported from Arabidopsis (Van Camp et al., 1990; Hindges and Slusarenko, 1992). The EST databases contained cDNAs for two additional CuZnSODs, an additional FeSOD, and a MnSOD (Newman et al., 1994). Genomic sequencing by the Kazusa DNA Research Institute (Chiba, Japan) identified a third FeSOD (http://www.kazusa.or.jp/arabi/). We propose that these genes be named CSD1, CSD2, and CSD3, FSD1, FSD2, and FSD3, and MSD1, respectively, to conform with Arabidopsis nomenclature guidelines (Meinke and Koornneef, 1997). Accession numbers for all sequences utilized in this report are shown in Table II.
Table II.
SOD sequences utilized in evolutionary analysis
| Enzyme | Proposed Location | Species | Reference/Accession No. |
|---|---|---|---|
| CuZnSOD | Extracellular | Pine | Streller and Wingsle (1994) |
| CuZnSOD | Extracellular | Human | J02947 |
| CuZnSOD | Cytosol | Arabidopsis | X60935(CSD1) |
| CuZnSOD | Cytosol | Corn | M54936 |
| CuZnSOD | Cytosol | Human | M13267 |
| CuZnSOD | Cytosol | Ice Plant | U80069 (no. 1) |
| CuZnSOD | Cytosol | Ice Plant | AF034832 (no. 2) |
| CuZnSOD | Cytosol | Pea | M63003 |
| CuZnSOD | Cytosol | Rice | D01000 |
| CuZnSOD | Cytosol | Pine | X58578 |
| CuZnSOD | Peroxisome | Arabidopsis | AF061520(CSD3) |
| CuZnSOD | Peroxisome | Watermelon | Bueno et al. (1995) |
| CuZnSOD | Plastid | Arabidopsis | AF061519(CSD2) |
| CuZnSOD | Plastid | Pea | J04087 |
| CuZnSOD | Plastid | Pine | X58579 |
| CuZnSOD | Plastid | Rice | D85239 |
| CuZnSOD | Plastid | Wheat | U69536 |
| FeSOD | Plastid | Arabidopsis | M55910(FSD1) |
| FeSOD | Plastid | Arabidopsis | Y12641(FSD2) |
| FeSOD | Plastid | Arabidopsis | AF061852(FSD3) |
| FeSOD | Plastid | Chlamydomonas | U22416 |
| FeSOD | E. coli | J03511 | |
| FeSOD | Plastid | Soybean | M64467 |
| FeSOD | Plastid | Tobacco | A09032 |
| MnSOD | Mitochondria | Arabidopsis | AF061518(MSD1) |
| MnSOD | Mitochondria | Corn | M33119 |
| MnSOD | Mitochondria | Human | X65965 |
| MnSOD | Mitochondria | Pea | U30841 |
| MnSOD | Mitochondria | Wheat | U77212 |
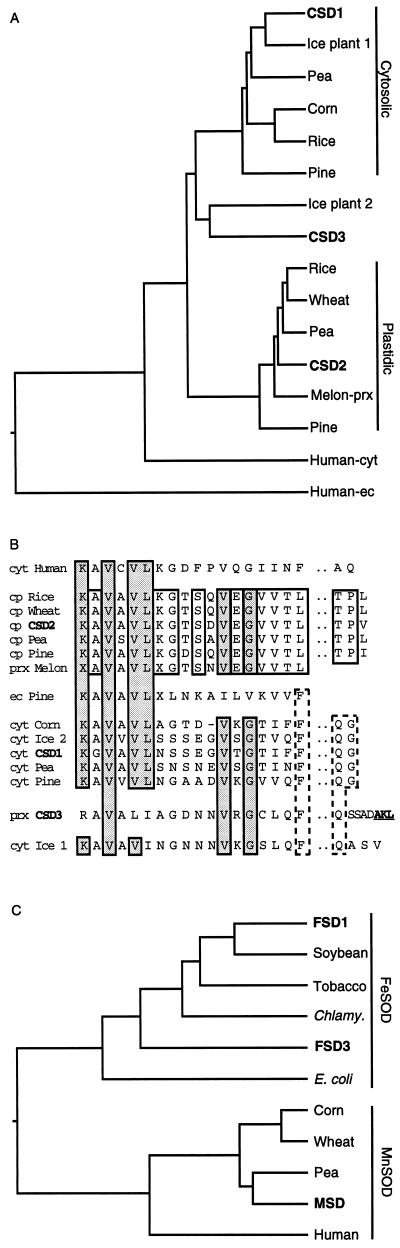
The CSD2 cDNA contains a 651-nucleotide ORF that encodes a predicted 217-amino acid protein containing all of the conserved amino acids for a CuZnSOD protein (Bordo et al., 1994). Two lines of evidence indicate that this protein is plastid targeted. First, phylogenetic analysis indicates that the CSD2 amino acid sequence is most closely related to other known or proposed plastid-targeted CuZnSODs, as shown in Figure 1A. Second, there is an amino-terminal extension that has no homology to nonplant CuZnSOD enzymes. This region has the characteristics of typical plastid-transit peptides in that it is positively charged and rich in hydroxylated amino acids (Keegstra et al., 1989).
Figure 1.
Evolutionary relationships inferred from SOD sequences. Accession numbers for the sequences utilized are listed in Table II. A, Dendrogram of plant CuZnSOD amino acid sequences. The predicted amino acid sequences were analyzed utilizing a DNASTAR (Madison, WI) program, starting at Lys-Ala-Val-Ala-Val-Leu or a homologous sequence to eliminate any amino-terminal transit sequences. B, Alignment of the first 18 amino acids and conserved carboxy-terminal amino acids of the proteins used for the dendogram in A to illustrate cladistic relationships of the plant CuZnSODs. Underlined amino acids indicate a putative peroxisomal targeting sequence. prx, Peroxisome; cyt, cytosol; ec, extracellular. C, Dendogram of FeSOD and MnSOD amino acid sequences. The amino acid sequences were analyzed from the start of homology to the E. coli FeSOD sequence.
The CSD3 cDNA contains a 465-nucleotide ORF. The deduced protein sequence is 164-amino acids long and contains all of the conserved amino acids for a functional CuZnSOD protein (Bordo et al., 1994). Additionally, the carboxyl terminus has an Ala-Lys-Leu tripeptide that can act as a peroxisomal targeting sequence (Keller et al., 1991), leading us to propose that CSD3 is a putative peroxisomal CuZnSOD. The CSD3 protein does not fall within either the cytosolic or plastidic CuZnSOD clade and forms a separate group with CuZnSOD2 from the ice plant (Fig. 1, A and B). This cladistic divergence is further illustrated by the fact that CSD3 is 70% identical to the ice plant CuZnSOD2, whereas it is at most 65% identical to individuals in either the cytosolic or plastidic groups. In contrast, the plastidic CuZnSODs are 90% identical to each other and the cytosolic CuZnSODs are 80% identical to each other. A previously published watermelon peroxisomal CuZnSOD (melon-prx) appears to be more closely related to plastidic CuZnSODs then to CSD3 (Fig. 1, A and B; Bueno et al., 1995).
As shown in Table II, published studies have identified four FeSOD cDNAs in plants, including the Arabidopsis FSD1. We identified two additional FeSOD cDNAs in the Arabidopsis DNA-sequencing projects. FSD2 is represented by an incomplete cDNA containing an ORF that encodes a protein homologous to the carboxy-terminal half of published plant FeSODs. Due to the truncated nature of the cDNA we did not completely characterize this sequence. The proposed FSD3 cDNA contains a 798-nucleotide ORF encoding a predicted 263-amino acid protein with an amino-terminal peptide extension characteristic of known plastid-transit peptides (Keegstra et al., 1989). Phylogenetic analysis indicates that the inferred FSD3 protein does not evolutionarily cluster with the other plant FeSODs, as it is only 50% identical to the known plant FeSODs, which are 70% to 75% identical to each other (Fig. 1C). However, FSD3 does contain all of the amino acids described by Lah et al. (1995) as conserved between FeSODs, strongly suggesting that it is a plastidic FeSOD protein.
The MSD1 cDNA contains a 691-nucleotide ORF that encodes a 231-amino acid hypothetical protein that is 80% to 90% identical to other plant MnSODs and contains all of the conserved amino acids required for MnSOD activity (Fig. 1C; Lah et al., 1995). Additionally, this protein contains an amino-terminal peptide that is basic and rich in hydroxylated residues, characteristic of mitochondrial transit peptides (Neupert, 1997). Therefore, MSD1 is thought to encode a mitochondrial MnSOD.
Map Positions of the Known SOD Structural Genes
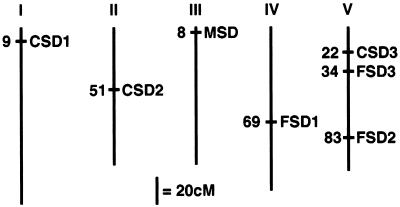
The seven known SOD structural genes were genetically mapped using a variety of methods, and the results are summarized in Figure 2. First, the CuZnSOD cDNAs were used to identify cross-hybridizing Texas A&M University bacterial artificial chromosomes (Choi et al., 1995; K. Dewar and J. Ecker, personal communication). CSD1 was identified on TAMUBAC10I10, 20M22, and 30M24. These BACs are coincident with the CICYAC8C10, placing CSD1 at the top of chromosome 1 between markers PAI1 and PHYA (http://cbil.humgen.upenn.edu/∼atgc/physical-mapping; Creusot et al., 1995). This location agrees with the position identified for the genomic sequence for CSD1 in the Arabidopsis genome project (http://genome-www3. stanford.edu/cgi-bin/Webdriver?MIval=atdb_clone_max &clone_name=T12M4&isprobe=f). CSD2 was identified on TAMUBAC30D05, 03B10, 03B23 and T08I7, which did not correspond to any known genomic location. However, we identified a polymorphic microsatellite in the end sequence from TAMUBAC30D05, and this locus was mapped to chromosome 2, which is 6.5 centimorgans (cM) distal of erecta and 1 cM proximal of m220, using the Lister and Dean recombinant inbred lines (Lister and Dean, 1993; http://nasc.nott.ac.uk/new_ri_map.html). CSD3 was shown to hybridize with TAMUBAC01C09, 01L16, 04D07, 22E21, and 31O14, which had no known genomic location.
Figure 2.
Genetic map positions of known Arabidopsis SOD structural genes. The structural genes for the known SOD proteins were mapped to the Arabidopsis genetic map as described in the text. The numbers given are approximate positions on the February 27, 1998 Lister and Dean Recombinant Inbred map for CSD1, CSD2, CSD3, FSD1, FSD2, FSD3, and MSD1 (http://nasc.nott.ac.uk/new_ri_map.html).
To map CSD3 and the other SOD loci, PCR primers were designed that specifically amplified the SOD genes from pooled CICYAC clones in a multiplex collection of CICYAC DNA (Creusot et al., 1995). These oligonucleotides are listed in Table I. The identified CICYAC address was compared with existing Arabidopsis physical maps to obtain a genomic location. CSD3 is located on CICYAC clones 9H7, 9E10, and 9A8, which are between markers nga151 and GDH on chromosome V (Schmidt et al., 1997). This location was confirmed by genomic sequencing of the P1 element MiRG7 (accession no. AB012246). FSD1 was found on CICYACs 6B12 and 1G9, which are on chromosome IV between markers mi422 and SEP2B (Schmidt et al., 1995). FSD2 was identified on CICYACs 11F9 and 11F10, on chromosome V between nga129 and m435 (Schmidt et al., 1995). MSD1 was amplified from CICYACs 3E3, 3A3, 5D8, and 12H8, but none of these were physically mapped. To map MSD1, a CAPS marker (Konieczny and Ausubel, 1993) was identified between ecotype Columbia and Landsberg erecta within the MSD1 genomic sequence, as described in Methods. This marker was mapped to chromosome III, which is 6.5 cM proximal of GAPC and 2 cM distal of nga126, using the Lister and Dean recombinant inbred lines (Lister and Dean, 1993). FSD3 was identified by genomic sequencing of the P1 clone MiKD15 (accession no. AB007648), which lies on chromosome V between markers m291 and g4556 (Kazusa Institute).
Multiple SOD mRNAs in Arabidopsis Rosette Leaves
To investigate whether the seven SOD genes were expressed during normal plant development, the cDNAs were radiolabeled to similar specific activities and hybridized to RNA blots containing total RNA from Arabidopsis rosette tissue. Under standard 16-h-photoperiod growth conditions, CSD2 was the most abundant mRNA, at 20% that of the CAB3 mRNA level and twice that of the FSD1 mRNA. The CSD1 and MSD1 mRNAs accumulated to approximately 25% the level of CSD2 mRNA, whereas CSD3 and FSD2 mRNAs had a basal accumulation of only about 5% of CSD2 mRNA levels. FSD3 showed the lowest basal mRNA accumulation, at approximately 1% of CSD2 mRNA. Further evidence for the expression of all seven SOD genes was obtained when we observed PCR amplification of each from an Arabidopsis cDNA library using gene-specific primers (D.J. Kliebenstein, S.A. Saracco, and R.L. Last, unpublished data).
Multiple SOD Isoenzymes in Arabidopsis
To promote studies of the subcellular localization and regulation of different SOD isoenzymes, antisera were generated against five different Arabidopsis SOD proteins. E. coli expression plasmids were constructed to produce fusion proteins between GST and CSD1, CSD2, CSD3, FSD1, and MSD1 (Smith and Johnson, 1988). Fusion proteins were not made for FSD2 and FSD3 because cDNA only became available after antiserum production. The GST fusion proteins were largely insoluble, facilitating their partial purification from a lysed cell culture followed by preparative SDS-PAGE gel fractionation. Polyclonal antisera were raised in rabbits against each of the GST-SOD fusion proteins.
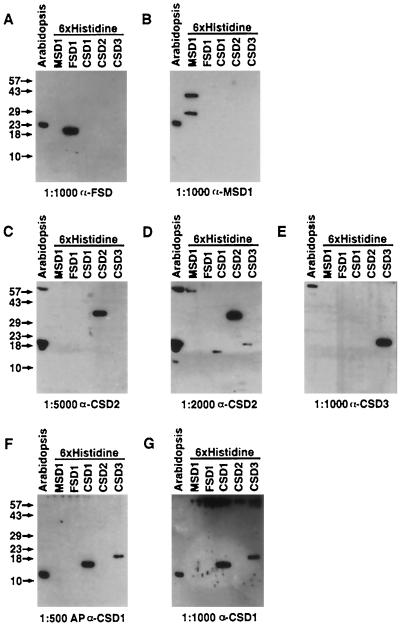
The SOD antisera were tested against Arabidopsis leaf protein extracts. As shown in Figure 3, A and B, line 1, anti-FSD1 and anti-MSD1 antiserum each detected a protein with an apparent molecular mass of 23 kD, close to the expected 22- and 23-kD sizes for FeSOD and MnSOD, respectively, as defined by the start of homology to bacterial proteins (Lah et al., 1995). At a 1:5000 antiserum dilution, nonaffinity-purified anti-CSD2 detected a protein with an apparent molecular mass of 20 kD, which agrees with the 20-kD protein predicted to be encoded by the CSD2 cDNA with the intact transit peptide (Fig. 3C). At a 1:500 antiserum dilution, affinity-purified anti-CSD1 detected a protein of 15-kD apparent molecular mass, the size predicted for the CSD1 protein based on conceptual translation of the cDNA ORF (Fig. 3F). These results suggest that the 15-kD protein is encoded by CSD1, whereas the 20-kD polypeptide corresponds to the CSD2 protein.
Figure 3.
Test of SOD antisera specificity. Twenty-five micrograms of total protein from Arabidopsis rosette leaves and 12 ng of the purified 6× His-tagged MSD1, FSD1, CSD1, CSD2, and CSD3 proteins were run on SDS-PAGE and subjected to immunoblot analysis. Postions of molecular-mass markers are listed on the left of each row of gels. The antisera and dilution (v/v) used are listed at the bottom of each gel. AP, Affinity-purified antiserum.
The specificity and reactivity of these antisera was assessed further using the SOD proteins with His6 attached to their amino termini. Consistent with our having obtained reagents of high reactivity and good specificity, each antiserum recognized the appropriate purified His6-tagged fusion proteins when used at the high dilutions (Fig. 3, A–C, E, and F). At these low serum concentrations, we observed minimal cross-reactivity with the other His6-tagged CuZnSOD fusion proteins (Fig. 3, C, E, and F). However, in one unique case, affinity-purified anti-CSD1 did cross-react with His6-CSD3 at the high 1:500 dilution (Fig. 3F). The experiment was repeated with increased antisera concentrations to investigate potential cross-reactivity of the CSD antisera. As the antiserum concentration increased, all CSD antisera became fairly promiscuous, showing cross-recognition of the other CSD proteins. For example, when the CSD2 antiserum dilution was decreased from 1:5000 to 1:2000, there was cross-recognition of the His6-CSD1 and His6-CSD3 protein (Fig. 3D).
Several lines of evidence support the hypothesis that the 15-kD protein is encoded by CSD1 and the 20-kD polypeptide by CSD2. First, affinity purification of the CSD1 antiserum increased the specificity for the 15-kD CSD1 protein (Fig. 3, compare F and G). The second line of evidence comes from transgenic Arabidopsis plants containing an antisense CSD1 construct driven by the cauliflower mosaic virus 35S promoter, which exhibit reduced accumulation of CSD1 mRNA and the 15-kD protein (D.J. Kliebenstein, L.G. Landry, and R.L. Last, unpublished data). Transgenic Arabidopsis containing a transgene with CSD1 sense expression controlled by the cauliflower mosaic virus 35S promoter have increased levels of CSD1 mRNA and 15-kD protein.
Analysis of 35S-CSD1 sense and antisense lines also yielded circumstantial evidence for the hypothesis that the immunologically detected 20-kD protein is encoded by CSD2 (D.J. Kliebenstein, L.G. Landry, and R.L. Last, unpublished data). The CSD1 antisense lines have increased accumulation of both the CSD2 mRNA and the 20-kD protein. Furthermore, both the CSD2 mRNA and 20-kD protein are decreased in the CSD1 overexpression lines. These results also indicate that there may be a form of compensatory regulation between CSD1 and CSD2.
At a 1:1000 dilution, anti-CSD3 detected several high-molecular-mass proteins from whole leaf extracts, but no protein in the 15- to 20-kD range as predicted by DNA sequence analysis of CSD3 (Fig. 3E). At higher concentrations anti-CSD3 occasionally detected proteins of 20-, 17-, and 15-kD apparent molecular mass, similar to the other CSD antisera (D.J. Kliebenstein and R.L. Last, unpublished results). Two lines of evidence indicate that CSD3 may encode the 17-kD protein. First, anti-CSD3 was highly specific for the His6-CSD3 fusion protein and recognized the 17-kD protein more strongly than either anti-CSD1 or anti-CSD2. Second, affinity purification of the CSD3 antiserum decreased detection of the 15- and 20-kD proteins but not the 17-kD protein. However, even after affinity purification, the 15- and 20-kD signals were still stronger than the faint 17-kD signal. We believe that this faint signal was due to low endogenous levels of the protein, because we were able to easily detect 4 ng of the purified His6-CSD3 protein. This is consistent with the low levels of detectable mRNA for CSD3. Regardless of the reason, we found inconsistent detection of the 17-kD protein in Arabidopsis leaf extracts and did not further characterize the CSD3 antiserum.
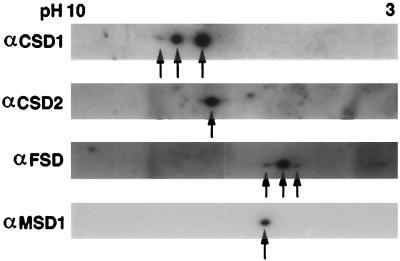
To confirm the specificity of the SOD antisera, total leaf protein extract was separated by two-dimensional PAGE. As shown in Figure 4, anti-CSD1 detects three different isoelectric variants, each with an apparent molecular mass of 15 kD, and anti-CSD2 recognizes one isoelectric variant of the 20-kD protein. Two-dimensional-PAGE analysis with the CSD1 antisense and overexpression transgenic lines were consistent with the 15- and 20-kD variants being derived from the CSD1 and CSD2 genes, respectively (D.J. Kliebenstein and R.L. Last, unpublished results). Anti-MSD1 identified only one 23-kD isoelectric variant in Arabidopsis leaf extracts, whereas the antiserum generated against GST-FSD1 recognizes three isoelectric variants at a 23-kD apparent molecular mass (Fig. 4). The available data do not allow us to assess whether these three isoelectric variants are encoded by distinct genes, so we refer to this antiserum as anti-FSD.
Figure 4.
SOD isoelectric variants in Arabidopsis rosette leaves. Twenty-five micrograms of total protein from Arabidopsis rosette leaves was separated by IEF in the first dimension and by SDS-PAGE in the second dimension prior to immunoblot analysis. Arrowheads indicate the SOD isoelectric variants consistently detected.
Four Major SOD Activities in Arabidopsis Rosettes
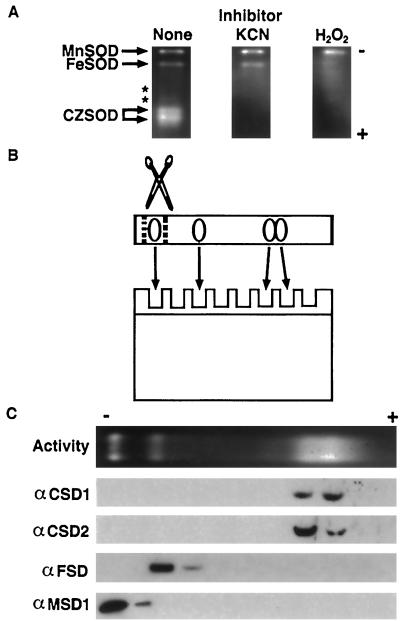
Based upon analysis of SOD mRNA and protein expression results (Figs. 3 and 4), we predicted that Arabidopsis leaves would contain CuZnSOD, FeSOD, and MnSOD activities. The presence of these SOD activities is readily assayed by nondenaturing PAGE enzyme assays (Beauchamp and Fridovich, 1971), and this method identified four major and two minor SOD activities, as shown in Figure 5A. The identities of the major SOD activity bands were tested by preincubating the gels with well-characterized SOD inhibitors: KCN is an inhibitor of CuZnSOD, whereas H2O2 inhibits both CuZnSOD and FeSOD (Pan and Yau, 1992). The results shown in Figure 5A suggest that the activity of slowest mobility is a MnSOD, the next slowest is an FeSOD, and the fastest migrating and diffuse doublet consists of CuZnSOD activities.
Figure 5.
Characterization of the major Arabidopsis SOD activities. A, Forty micrograms of total protein from Arabidopsis rosette tissue was fractionated on a nondenaturing PAGE gel and stained for SOD activity (clear gel regions). Gels were preincubated with KCN (which inhibits CuZnSOD) or H2O2 (which inhibits both CuZnSOD and FeSOD) to facilitate identification of the different activities. Asterisks mark the location of two minor activities that were seen only occasionally. B, Graphic representation of the experiment shown in Figure 5C. SOD activity gels were cut into slices, the slices were boiled in SDS loading buffer to elute and denature proteins, and aliquots were run on SDS-PAGE and immunodetected. C, Immunoblots of SOD activity gel fractions as illustrated in B. Antisera used are listed to the left.
To confirm the provisional identification based upon SOD inhibitor sensitivity the activity gel was fractionated, and the proteins were separated by SDS-PAGE and subjected to immunoblot analysis (the experiment is shown schematically in Fig. 5B). Anti-MSD1 and anti-FSD recognized proteins from the expected regions of the activity gel, lending further support for the identification of the two activities as MnSOD and FeSOD (Fig. 5C). The 20-kD CSD2 protein is enriched in the slower-migrating section of the diffuse CuZnSOD activity doublet, whereas the 15-kD CSD1 protein is more prominent in the faster-migrating area.
Arabidopsis Plastids Contain FeSOD and CuZnSOD2
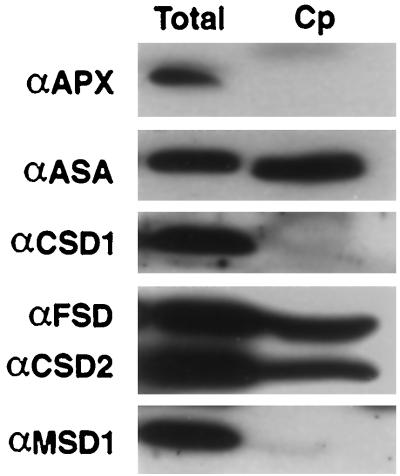
DNA-sequence analysis of CSD2, FSD1, and FSD3 suggested the presence of an amino-terminal transit peptide. These data and published information on the presence of plastidic FeSODs and CuZnSODs in other plant species (Alscher and Hess, 1993; Bowler et al., 1994; Scandalios, 1997) suggested that these Arabidopsis proteins are localized to the chloroplast. To test this hypothesis, extracts of intact Arabidopsis leaf chloroplasts were subjected to immunoblot analysis, and compared with the total cellular protein, as shown in Figure 6. As expected for plastid-localized proteins, the CSD2 and FSD polypeptides copurified with the α-subunit of the plastidic Trp biosynthetic pathway enzyme anthranilate synthase (Zhao and Last, 1995). In contrast, as seen for the cytosolic ascorbate peroxidase, the presumed cytosolic CSD1 and mitochondrial MSD1 proteins were not present in the chloroplast fraction (Fig. 6).
Figure 6.
Copurification of CSD2 and FSD proteins with Arabidopsis chloroplasts. Plastids were purified from rosette leaves, run on SDS-PAGE, and immunodetected with the antisera listed to the left. Cp, Intact chloroplast fraction; Total, total cell extract; APX, ascorbate peroxidase; ASA, anthranilate synthase alpha. Equal protein was loaded based on the level of Rubisco large subunit protein.
FSD1 mRNA Is under Circadian Control
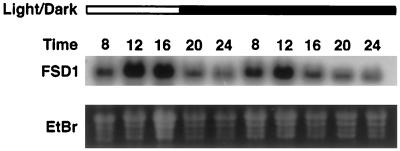
Considering that Arabidopsis expresses at least seven SOD genes encoding proteins targeted to at least three subcellular compartments, it seems likely that these isoenzymes carry out diverse physiological functions. If this is the case, we would expect to observe differences in the regulated expression of these proteins and their mRNAs in response to environmental stimuli. The first environmental stimulus tested was light, which influences expression of a multitude of plant genes involved in diverse biochemical processes (Kreps and Kay, 1997). Three-week-old plants were grown under a 9-h photoperiod that began at 8 am and ended at 5 pm, and tissue was harvested at 4-h intervals from 8 am to 12 am. To evaluate whether any observed diurnal fluctuations reflected circadian regulation, the plants were placed in continuous darkness during d 2, and tissue was collected at the same times as on d 1. Of the seven SOD genes tested (CSD1, CSD2, CSD3, FSD1, FSD2, FSD3, and MSD1), only FSD1 mRNA showed a diurnal oscillation, with maximum expression approximately during midday, as shown in Figure 7. This rhythm persisted in continuous darkness during d 2, indicating that FSD1 mRNA accumulation is under circadian control. In contrast to the mRNA regulation, no changes were seen in protein levels using the CSD1, CSD2, FSD, or MSD1 antisera, nor were changes detected in SOD activities using activity gels (D.J. Kliebenstein and R.L. Last, unpublished results).
Figure 7.
Circadian oscillation of FSD1 mRNA. Total RNA was isolated from tissue collected between 8 am and midnight from plants grown for 21 d in a 9-h photoperiod (lights were on from 8 am to 5 pm) and subjected to RNA blot hybridization analysis conducted using an FSD1 cDNA probe. The open bar at the top of the figure represents samples harvested during the illuminated period; the dark bar represents darkness. The time is presented on a 24-h time scale.
SOD Accumulation in Response to Growth under Differing Light Fluences
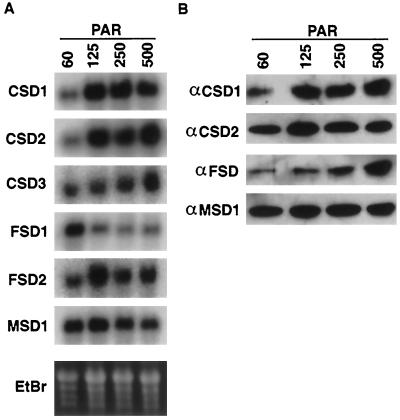
The level of endogenous ROS production can vary depending upon the fluence of light incident upon the plant during growth, with higher light fluences leading to elevated ROS production (Halliwell, 1987). To determine the regulation of the SOD genes in response to growth under different light fluences, plants were grown under 60, 125, 250, or 500 μmol m−2 s−1 PAR of incident light and analyzed for SOD mRNA accumulation. As shown in Figure 8A, CSD1 and CSD2 mRNA steady-state levels increased between 60 and 125 μmol m−2 s−1 PAR, but no further increases were observed at higher fluences. In contrast, FSD1 showed maximal accumulation in the 60 μmol m−2 s−1 PAR sample, and decreased under the other light fluences (Fig. 8A). CSD3 mRNA exhibited an increased mRNA accumulation in only the 500 μmol m−2 s−1 PAR sample (Fig. 8A). The FSD2 and MSD1 mRNAs showed no consistent variation in mRNA accumulation under the light fluences tested (Fig. 8A; D.J. Kliebenstein and R.L. Last, unpublished results).
Figure 8.
Regulation of SOD mRNAs and proteins in response to growth under varied light fluences. Arabidopsis seeds were germinated for 2 d in constant light at 100 μmol m−2 s−1 PAR, and then moved to a different chamber, where the incident light was adjusted to 60, 120, 250, or 500 μmol m−2 s−1 PAR with cheesecloth and nylon mesh. The plants were grown under this fluence for 2 weeks, and tissue was collected for analysis. A, RNA-blot hybridization using the cDNAs listed on the left as probes. B, Immunoblots of protein extracted from plants grown in the same pots that were used for the RNA blots in A. The proteins were immunodetected using the antisera listed.
The same tissue was analyzed for variation in SOD protein accumulation. Levels of CSD1 protein significantly increased between the 60 and 125 μmol m−2 s−1 PAR samples, reflecting the mRNA, but showed no further increase at the higher light levels (Fig. 8B). The CSD2 protein did not mirror the CSD2 mRNA, as the protein showed no striking differences in accumulation between the four fluences (Fig. 8B). The FSD polypeptides increased slightly in the 250 μmol m−2 s−1 PAR sample and to a greater extent at 500 μmol m−2 s−1 PAR (Fig. 8B). As predicted from the mRNA, levels of MSD1 protein did not change throughout the experiment (Fig. 8B). Based upon activity gels, the SOD activities showed no significant variation throughout the experiment (D.J. Kliebenstein and R.L. Last, unpublished results).
Short-Term High-Light Treatment Causes Changes in SOD mRNA Accumulation
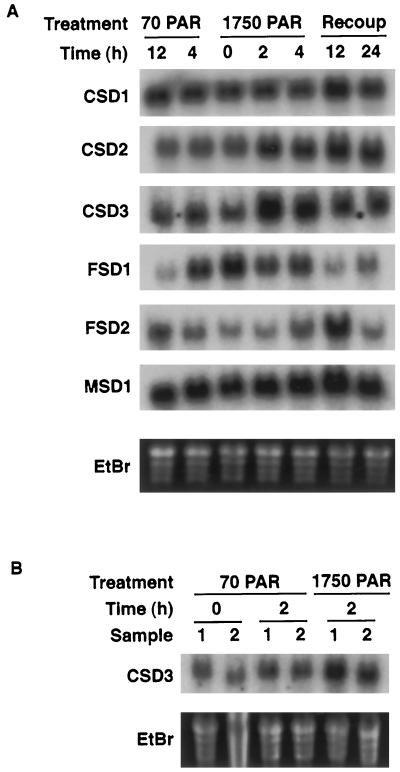
By photoexcitation of Fd, photoinhibitory light pulses can lead to the generation of O2−, which subsequently forms H2O2 in the plastid (Yruela et al., 1996). It is possible that one of the seven SOD genes identified in Arabidopsis is induced in response to a transient burst of high light. To test this, plants were grown under 16 h of 70 μmol m−2 s−1 PAR for 2 weeks, exposed to 4 h of 1750 μmol m−2 s−1 PAR, and the tissue was collected for the mRNA analysis shown in Figure 9. Levels of mRNA encoding the putative peroxisomal CSD3 increased strongly within 2 h of initiation of the high-light pulse (Fig. 9A). These levels then diminished but remained elevated compared with control samples. This induction does not appear to have been due to increased photorespiration, because the same results were obtained on high-light-treated plants grown under 2250 μg mL−1 CO2 (Fig. 9B). This concentration of CO2 should be sufficient to dramatically decrease flux through the photorespiration pathway (Somerville and Ogren, 1981).
Figure 9.
Regulation of SODs in response to photoinhibitory light. Plants were grown for 2 weeks in 70 μmol m−2 s−1 PAR and exposed to 1750 μmol m−2 s−1 PAR for 4 h. Controls were placed in the same chamber covered with filters to maintain the incident light at 70 μmol m−2 s−1 PAR. All plants were returned to the original chamber for the recovery period. Shown are RNA-blot hybridizations using the cDNAs listed on the left as probes. A, Atmospheric conditions (CO2 at 335–360 μg mL−1); B, CO2 at 2250 μg mL−1.
In contrast to CSD3, mRNA for the plastidic CSD2 only gradually increased following the high-light pulse (Fig. 9A). Of the two plastidic FeSODs tested, FSD2 mRNA displayed a strong increase only at the 12-h time point, whereas the diurnal rhythm of FSD1 mRNA was unaltered by the high-light pulse (Fig. 9A). MSD1 and CSD1 mRNA showed no significant variation throughout the experiment (Fig. 9A). Again, the SOD proteins and activities did not alter throughout this treatment (D.J. Kliebenstein and R.L. Last, unpublished results).
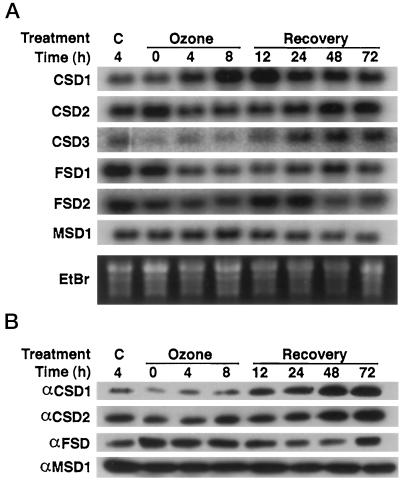
SOD Regulation in Response to Ozone-Mediated Oxidative Stress.
Ozone can directly degrade to generate apoplastic ROS in plants (Runeckles and Vaartnou, 1997). To test the regulation of SOD isoenzymes by ozone-mediated oxidative stress, 2-week-old Arabidopsis plants that had been grown in ozone-depleted (charcoal-filtered) air were treated with 330 ppb ozone for 8 h, followed by recovery in charcoal-filtered (ozone-free) air. Tissue was collected during the fumigation and recovery periods, and the levels of CSD1, CSD2, CSD3, FSD1, FSD2, and MSD1 mRNAs were assayed by RNA-blot hybridization, with the results shown in Figure 10A. As previously described (Sharma and Davis, 1994; Conklin and Last, 1995), CSD1 mRNA accumulation was induced by ozone fumigation and continued to increase during the beginning of the recovery period (Fig. 10A). The mRNA had decreased 24 h after the start of the fumigation to a steady-state level that was elevated compared with unfumigated controls, similar to the results seen by Sharma and Davis (1994). Although stimulated by ozone exposure, mRNA for the putative peroxisomal CSD3 showed a delayed induction that started 24 h after the start of the fumigation and increased through the end of the experiment (Fig. 10A).
Figure 10.
Regulation of SOD isoenzyme expression in response to ozone fumigation. Plants were fumigated with 330 ppb ozone for 8 h. C represents unfumigated control plants maintained in charcoal-filtered air. The ozone-treated plants were returned to charcoal-filtered air during the recovery period. A, RNA-blot hybridization using the cDNAs listed on the left as probes. B, Immunoblots of protein extracted from plants grown in the same pots as used for the RNA blots in A. The proteins were immunodetected using the antisera listed on the left.
In contrast, mRNAs for two plastid-targeted SODs decreased in response to ozone. FSD1 mRNA decreased within 4 h after the start of the fumigation and recovered to nearly prefumigation levels between 48 and 72 h after the fumigation. This decrease is even more dramatic compared with the 4-h control, because this should be near the maximum of the circadian oscillation in untreated plants. The CSD2 gene, which also encodes a plastidic protein, showed a pattern similar to FSD1 (Fig. 10A). In contrast, the mRNAs detected with the FSD2 and MSD1 probes did not show any reproducible changes in response to ozone exposure (Fig. 10A).
The same tissue was used to analyze the regulation of the SOD proteins and enzyme activities in response to ozone fumigation. CSD1 protein began to increase during the fumigation, reaching a plateau 48 h after the start of the ozone treatment (Fig. 10B). In contrast, CSD2 protein showed no response until an increase in the protein level at the 48- and 72-h time points, whereas the FSD protein showed a slight decrease in protein accumulation 24 to 48 h after the onset of the ozone fumigation (Fig. 10B). MSD1 protein did not vary throughout the experiment, as predicted from the mRNA results (Fig. 10B). None of the SOD activities showed any significant changes throughout the experiment (D.J. Kliebenstein and R.L. Last, unpublished results).
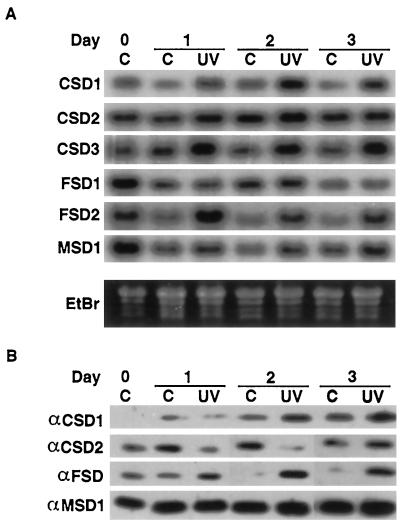
Differential Response of SODs to UV-B Exposure
UV-B causes oxidative stress in plants (Kramer et al., 1991; Landry et al., 1995; Conklin et al., 1996) by leading to ROS production through activation of UV-B chromophores, rather then by direct chemical transformation, as occurs with ozone (Caldwell, 1993; Heath, 1994). This difference in mechanisms of oxidative stress production prompted a test of the response of the various SOD genes to acute UV-B exposure. Two-week-old Arabidopsis plants were treated for 3 d with 15 kJ of UV-BBE m−2 d−1, and treated and untreated tissue was harvested on each day of exposure. As seen in Figure 11A, CSD1 mRNA increased within 1 d of UV-B exposure, to a maximum at d 2. FSD2 mRNA was also up-regulated within 24 h, but decreased slightly on d 2 and 3 (Fig. 11A). UV-B treatment increased CSD3 mRNA accumulation within 24 h, and the levels remained constant throughout d 2 and 3 (Fig. 11A). CSD2, FSD1, and MSD1 mRNA levels showed no significant differences between the treated plants and untreated controls (Fig. 11A).
Figure 11.
Regulation of SOD isoenzyme expression in response to UV-B. UV-B-treated plants were exposed to 15 kJ of UV-B light. Controls were grown under Mylar filters. Both the controls and the UV-B-treated plants were protected from UV-C by Pyrex glass filters placed over the pots. Samples were harvested directly before starting the UV-B treatment and after 1, 2, and 3 d of treatment. C, −UV-B control samples; UV, UV-B-treated samples. A, RNA-blot hybridization using the cDNAs listed on the left as probes. B, Immunoblots of protein extracted from plants grown in the same pots as used for the RNA blots in A. The proteins were immunodetected using the antisera listed on the left.
The same tissue was tested for levels of SOD protein and activities (Fig. 11B; D.J. Kliebenstein and R.L. Last, unpublished results). CSD1 protein showed a gradual increase in the control samples throughout the experiment, presumably due to environmental changes caused by covering the pots with Pyrex glass filters. However, the UV-B treatment caused increases in CSD1 protein levels above the controls on d 2 and 3 (Fig. 11B). CSD2 showed the opposite response, with decreased protein accumulation on d 1 and 2 in response to the UV-B treatment. Protein levels recovered to control levels by d 3 (Fig. 11B). FSD protein was higher in the UV-B-treated samples then in the controls on all days of the experiment. Additionally, the FSD and CSD1 protein in UV-B-treated samples steadily increased from d 1 to 3 (Fig. 11B). Consistent with the mRNA analysis, MSD1 protein showed no variation in response to UV-B (Fig. 11B). The MnSOD and CuZnSOD activities showed no significant variation throughout this experiment (D.J. Kliebenstein and R.L. Last, unpublished results). FeSOD activity was extremely low throughout the experiment and did not measurably increase.
DISCUSSION
Multiple SOD Genes in Arabidopsis
Arabidopsis contains multiple SOD genes encoding at least three CuZnSODs, three FeSODs, and one MnSOD. The seven SOD structural genes were mapped onto the Arabidopsis genome by identifying BAC or YAC clones that contain these genes by hybridization or multiplex PCR analysis, as well as by using recombinant inbred genetic mapping (Fig. 2). To our knowledge, this is the first mapping of multiple SOD genes in a single plant species. Six of the genes have representatives in the EST databases and all seven genes identify transcripts on RNA blots, indicating that they are expressed genes. Given that FSD3 was identified by genomic sequencing, and was not in the public domain EST databases, it is likely that there are other SOD genes yet to be discovered in Arabidopsis.
Analysis of the DNA sequences for CSD2, FSD1, FSD3, and MSD1 revealed the presence of amino-terminal transit peptides. CSD2 clusters within the plastidic CuZnSOD clade (Fig. 1, A and B), suggesting that it is plastidic. Other studies have typically identified plant FeSODs as plastidic and MnSODs as mitochondrial, suggesting that FSD1 and FSD3 are plastidic and MSD1 is mitochondrial (Bowler et al., 1994; Alscher et al., 1997; Scandalios, 1997). Our cellular-fractionation studies showed that Arabidopsis chloroplasts contain CSD2 and FeSOD protein, consistent with the analysis of FSD1, FSD3, and CSD2 cDNA sequences.
An Ala-Lys-Leu tripeptide was identified within the CSD3-coding region, which is sufficient for peroxisomal targeting in yeast (Keller et al., 1991). Such carboxyl-terminal peroxisomal targeting sequences have been shown to be functionally conserved between plants and yeast (Gould et al., 1990). The peroxisomal CuZnSOD from watermelon groups phylogenetically with plastidic CuZnSODs and not CSD3 (Bueno et al., 1995; Fig. 1, A and B), suggesting that there could be several classes of peroxisomal CuZnSODs in plants.
Analysis of Multiple SOD Isoenzymes in Arabidopsis
The presence of multiple SOD genes raises the possibility that each protein may protect against a subset of oxidative stresses and that a variety of SODs are deployed to fully combat environmental stresses. To study the significance of multiple Arabidopsis SODs, polyclonal antisera were generated against CSD1, CSD2, CSD3, FSD1, and MSD1. When tested against Arabidopsis rosette tissue, anti-CSD1, CSD2, FSD1, and MSD1 reproducibly detected protein (Figs. 3 and 4). The CSD2 protein had an apparent molecular mass of 20 kD, which is the expected molecular mass for a protein with an intact transit peptide. This could be because the mature protein migrates slower then expected on a PAGE gel, because the transit peptide is not cleaved during transport into the plastid, or because there are 5 kD of posttranslational modifications to the protein. In addition to the immunodetected SOD proteins, we identified two CuZnSOD activities, one activity each for FeSOD and MnSOD, and other faint, unclassified SOD activities in Arabidopsis leaf tissue (Fig. 6). We confirmed the identity of the four major activities using immunoblot analysis of the SOD activity gel.
Published reports have also identified multiple SOD activities in Arabidopsis. Pan and Yau (1992) cataloged three CuZnSOD activities, one FeSOD activity, and one MnSOD activity, and Ormrod et al. (1995) found that Arabidopsis contained four CuZnSOD activities, no FeSOD activity, and one MnSOD activity. Our identification of the slowest-migrating activity as MnSOD and the fastest-migrating activities as CuZnSODs agrees with these reports. However, Pan and Yau (1992) identified no SOD activity in a similar location to the activity we identified as FeSOD, whereas Ormrod et al. (1995) identified a CuZnSOD activity in this region. It is possible that these inconsistencies are caused by physiological variations due to different growing conditions or genetic backgrounds. The four major activities we detected are present in all organs analyzed (juvenile, adult and cauline leaves, siliques, stems, and roots) throughout the life cycle of the plant, and the minor activities do not significantly increase in any organ or developmental stage (D.J. Kliebenstein and R.L. Last, unpublished results).
Differential Regulation of Multiple mRNAs for Plastidic SODs
Our analysis has suggested that Arabidopsis plastids could contain protein from at least four different genes, CSD2, FSD1, FSD2, and FSD3. These mRNAs are differentially regulated in response to a number of environmental stimuli. For example, only FSD1 is under circadian regulation in response to a diurnal light rhythm (Fig. 7). However, FSD1 is not necessarily coordinated with the level of photosynthesis. In fact, FSD1 mRNA actually decreases when Arabidopsis is grown under increasing incident light fluences, whereas CSD2 mRNA increases and FSD2 mRNA does not significantly change (Fig. 8). These results indicate that FSD1 responds to diurnal-light-mediated ROS fluctuations, whereas CSD2 reacts to long-term changes in photoproduced ROS, and FSD2 may be used to adapt to transient, photoproduced ROS increases.
Ozone fumigation and UV-B illumination also differentially affect the mRNAs encoding plastidic SODs. Ozone fumigation caused a transient decrease in the accumulation of the CSD2 and FSD1 mRNAs, although no reproducible effect was seen on the FSD2 mRNA. In contrast, UV-B led to a dramatic increase in accumulation of the FSD2 mRNA but had no effect on the accumulation of CSD2 or FSD1 mRNA. These results indicate that CSD2, FSD1, and FSD2 are independently regulated and raises the possibility that they are responsible for protecting Arabidopsis plastids against different oxidative stresses.
Although the mRNAs encoding plastidic SODs showed significant regulation in response to the various stresses, the corresponding activities and proteins typically did not. SOD activity analysis is complicated by the possibility that more then one isoenzyme could be present in each activity band. Therefore, any change in the level of activity from one isoenzyme could be masked by the other isoenzymes present in the activity band. Additionally, comparing the anti-FSD results with any specific FSD mRNA is not valid, because we do not know whether the antiserum specifically detects only one of the three known FeSOD isoenzymes. However, we believe that the anti-CSD2 is a specific antiserum. The fact that the CSD2 protein does not mirror the pattern of CSD2 mRNA regulation suggests that there is a translational or posttranslational level of control for CSD2 protein. Madmanchi and coworkers (1994) also found evidence for posttranscriptional regulation of plant SODs.
Regulation of mRNA for the Putative Peroxisomal CSD3
Photoinhibitory light pulses lead to the generation of ROS in plastids through the excitation of Fd and subsequent reaction with oxygen (Halliwell, 1987). Oddly, none of the mRNAs for the plastidic SODs were rapidly induced in response to a high-light pulse. In contrast, mRNA for the putative peroxisomal CSD3 was induced within 2 h of initiation of the high-light pulse (Fig. 10). If CSD3 is peroxisomal, this result suggests that while high light primarily leads to the generation of ROS within the chloroplast, there is also a signal that leads to the induction of mRNA for a CuZnSOD located within the peroxisome. However, this signal does not result simply from an increase in the rate of photorespiration, because CSD3 was induced by a high-light pulse even under levels of CO2 that should dramatically diminish photorespiration. This hints that the high-light pulse could more directly cause increasing ROS in the peroxisome, leading to increased CSD3 mRNA accumulation. In addition to a high-light pulse, growth under increasing light fluences, ozone fumigation, and UV-B irradiation also caused the induction of CSD3, suggesting that these treatments may also lead to increased peroxisomal ROS.
Regulation of Cytosolic CSD1 by Oxidative Stress
Previous research showed that mRNA for cytosolic CSD1 is induced in response to ozone fumigation (Sharma and Davis, 1994; Conklin and Last, 1995). We confirmed this observation and extended it to show that the CSD1 protein is also rapidly induced in response to ozone (Fig. 10). Additionally, we showed that the mRNA and protein for CSD1 are induced in response to UV-B illumination and to growth under increasing light fluences (Figs. 9 and 11). These treatments are thought to generate plant oxidative stress by very different mechanisms. This general responsiveness to different mechanisms that cause oxidative stress suggests that cytosolic CSD1 could be a general stress-response enzyme.
MnSOD Is Unresponsive to the Environmental Stimuli Tested
In contrast to the general inducibility of CSD1, the mRNA, protein, and activity of MSD1 appear to be good candidates for experimental loading controls in plant stress analysis. mRNA, protein, and activity showed minimal changes in response to any treatment tested in this paper (Figs. 7–11). We also found that MSD1 protein levels were not altered in response to virulent or avirulent Pseudomonas syringae pv maculicola, Rose Bengal, or dark-induced senescence (D.J. Kliebenstein, E.H. Williams, and R.L. Last, unpublished data). This lack of change in MSD1 expression in response to the oxidative stresses tested suggests that: (a) the mitochondria may have not been subjected to significantly increased ROS levels during the treatments tested; (b) MSD is not a primary mechanism responsible for protecting mitochondria against increased ROS; or (c) MSD1 activity is not rate limiting for ROS detoxification in the mitochondria.
We are currently using a genetic approach to further our understanding of why plants contain multiple SODs within the same subcellular compartments. Our goal is to identify Arabidopsis mutants deficient in specific SODs to test the role of individual SODs in ROS protection. We are also screening for mutants that alter the accumulation of multiple SODs as a way to understand the molecular basis for their regulation. Through a combination of biochemical, molecular, and mutant analysis, we hope to obtain a better understanding of the roles of SOD in plants.
ACKNOWLEDGMENTS
The authors thank Drs. Katherine Denby and Patricia Conklin (Boyce Thompson Institute for Plant Research) for helpful comments concerning the manuscript; Dr. Paul King for help with the high-CO2 experiment; the Arabidopsis Biological Resource Center for all cDNAs used in this research; and Drs. Kenneth Dewar and Joseph Ecker for performing the BAC library hybridization analysis.
Abbreviations:
- CAPS
cleaved amplified polymorphic sequence
- CICYAC
CEPH/Institut National de la Recherche Agronomique/Centre National de la Recherche Scientifique yeast artificial chromosome
- EST
expressed sequence tag
- GST
glutathione-S-transferase tag
- His6
six-His residue tag
- ORF
open reading frame
- ROS
reactive oxygen species
- SOD
superoxide dismutase
- TAMUBAC
Texas A&M University bacterial artificial chromosome
Footnotes
This work was supported by grant no. 96-35100-3212 from the Plant Responses to the Envrionment Program of the National Research Initiative Competitive Grants Program, U.S. Department of Agriculture. D.J.K. received support from a National Institutes of Health predoctoral training grant fellowship through the Section of Genetics and Development, Cornell University.
LITERATURE CITED
- Alscher RG, Donahue JL, Cramer CL. Reactive oxygen species and antioxidants: relationships in green cells. Physiol Plant. 1997;100:224–233. [Google Scholar]
- Alscher RG, Hess JL. Antioxidants in Higher Plants. Boca Raton, FL: CRC Press; 1993. [Google Scholar]
- Babior BM, El Benna J, Chanock SJ, Smith RM. The NADPH oxidase of leukocytes: The respiratory burst oxidase. In: Scandalios JG, editor. Oxidative Stress and the Molecular Biology of Antioxidant Defenses, Vol 34. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 737–783. [Google Scholar]
- Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bordo D, Djinovic K, Bolognesi M. Conserved patterns in the Cu, Zn superoxide dismutase family. J Mol Biol. 1994;238:366–386. doi: 10.1006/jmbi.1994.1298. [DOI] [PubMed] [Google Scholar]
- Bowler C, Van Camp W, Van Montagu M, Inzé D. Superoxide dismutase in plants. Crit Rev Plant Sci. 1994;13:199–218. [Google Scholar]
- Bowler C, Van Montagu M, Inzé D. Superoxide dismutase and stress tolerance. Annu Rev Plant Physiol Mol Biol. 1992;43:83–116. [Google Scholar]
- Bueno P, Varela J, Gimenez GG, Del Rio LA. Peroxisomal copper, zinc superoxide dismutase: characterization of the isoenzyme from watermelon cotyledons. Plant Physiol. 1995;108:1151–1160. doi: 10.1104/pp.108.3.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenas E. Biochemistry of oxygen toxicity. Annu Rev Biochem. 1989;58:79–110. doi: 10.1146/annurev.bi.58.070189.000455. [DOI] [PubMed] [Google Scholar]
- Caldwell CR. Ultraviolet-induced photodegradation of cucumber (Cucumis sativus L.) microsomal and soluble protein tryptophanyl residues in vitro. Plant Physiol. 1993;101:947–953. doi: 10.1104/pp.101.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chary P, Dillon D, Schroeder AL, Natvig DO. Superoxide dismutase (sod-1) null mutants of Neurospora crassa: oxidative stress sensitivity, spontaneous mutation rate and response to mutagens. Genetics. 1994;137:723–730. doi: 10.1093/genetics/137.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Creelman RA, Mullet JE, Wing RA. Construction and characterization of a bacterial artificial chromosome library in Arabidopsis thaliana. Weeds World. 1995;2:17–20. [Google Scholar]
- Conklin PL, Last RL. Differential accumulation of antioxidant mRNAs in Arabidopsis thaliana exposed to ozone. Plant Physiol. 1995;109:203–212. doi: 10.1104/pp.109.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Williams EH, Last RL. Environmental stress sensitivity of an ascorbic acid-deficient Arabidopsis mutant. Proc Natl Acad Sci USA. 1996;93:9970–9974. doi: 10.1073/pnas.93.18.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creusot F, Fouilloux E, Dron M, Lafleuriel J, Picard G, Billault A, Le P-D, Cohen D, Chaboute ME, Durr A and others. The CIC library: a large insert YAC library for genome mapping in Arabidopsis thaliana. Plant J. 1995;8:763–770. doi: 10.1046/j.1365-313x.1995.08050763.x. [DOI] [PubMed] [Google Scholar]
- Delseny M, Cooke R, Raynal M, Grellet F. The Arabidopsis thaliana cDNA sequencing projects. FEBS Lett. 1997;405:129–132. doi: 10.1016/s0014-5793(97)00184-1. [DOI] [PubMed] [Google Scholar]
- Desikan R, Hancock JT, Coffey MJ, Neill SJ. Generation of active oxygen in elicited cells of Arabidopsis thaliana is mediated by a NADPH oxidase-like enzyme. FEBS Lett. 1996;382:213–217. doi: 10.1016/0014-5793(96)00177-9. [DOI] [PubMed] [Google Scholar]
- Durrant I, Fowler S. Chemiluminescent detection systems for protein blotting. In: Dunbar BS, editor. Protein Blotting, Vol 140. Oxford, UK: Oxford University Press; 1994. pp. 141–152. [Google Scholar]
- Gould S, Keller JGA, Schneider M, Howell SH, Garrard LJ, Goodman JM, Distel B, Tabak H, Subramani S. Peroxisomal protein import is conserved between yeast, plants, insects and mammals. EMBO J. 1990;9:85–90. doi: 10.1002/j.1460-2075.1990.tb08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla EB, Valentine JS. Null mutants of Saccharomyces cerevisiae Cu, Zn superoxide dismutase: characterization and spontaneous mutation rates. J Bacteriol. 1991;173:5918–5920. doi: 10.1128/jb.173.18.5918-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B. Oxidative damage, lipid peroxidation and antioxidant protection in chloroplasts. Chem Phys Lipids. 1987;44:327–340. [Google Scholar]
- Halliwell B, Gutteridge J. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- Heath RL. Possible mechanisms for the inhibition of photosynthesis by ozone. Photosynth Res. 1994;39:439–451. doi: 10.1007/BF00014597. [DOI] [PubMed] [Google Scholar]
- Hindges R, Slusarenko A. cDNA and derived amino acid sequence of a cytosolic Cu, Zn superoxide dismutase from Arabidopsis thaliana (L.) Heyhn. Plant Mol Biol. 1992;18:123–125. doi: 10.1007/BF00018463. [DOI] [PubMed] [Google Scholar]
- Jarvis P, Lister C, Szabo V, Dean C. Integration of CAPS markers into the RFLP map generated using recombinant inbred lines of Arabidopsis thaliana. Plant Mol Biol. 1994;24:685–687. doi: 10.1007/BF00023565. [DOI] [PubMed] [Google Scholar]
- Keegstra K, Olsen LJ, Theg SM. Chloroplastic precursors and their transport across the envelope membranes. Annu Rev Plant Physiol Mol Biol. 1989;40:471–502. [Google Scholar]
- Keller GA, Krisans S, Gould SJ, Sommer JM, Wang CC, Schliebs W, Kunau W, Brody S, Subramani S. Evolutionary conservation of a microbody targeting signal that targets proteins to peroxisomes glyoxysomes and glycosomes. Cell Biol. 1991;114:893–904. doi: 10.1083/jcb.114.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Kramer GF, Norman HA, Krizek DT, Mirecki RM. Influence of UV-B radiation on polyamines, lipid peroxidation and membrane lipids in cucumber. Phytochemistry. 1991;30:2101–2108. [Google Scholar]
- Kreps JA, Kay SA. Coordination of plant metabolism and development by the circadian clock. Plant Cell. 1997;9:1235–1244. doi: 10.1105/tpc.9.7.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lah MS, Dixon MM, Pattridge KA, Stallings WC, Fee JA, Ludwig ML. Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus. Biochemistry. 1995;34:1646–1660. doi: 10.1021/bi00005a021. [DOI] [PubMed] [Google Scholar]
- Landry LG, Chapple CCS, Last RL. Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 1995;109:1159–1166. doi: 10.1104/pp.109.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Madamanchi NR, Donahue JL, Cramer CL, Alscher RG, Pedersen K. Differential response of Cu, Zn superoxide dismutases in two pea cultivars during a short-term exposure to sulfur dioxide. Plant Mol Biol. 1994;26:95–103. doi: 10.1007/BF00039523. [DOI] [PubMed] [Google Scholar]
- Meinke D, Koornneef M. Community standards for Arabidopsis genetics. Plant J. 1997;12:247–253. [Google Scholar]
- Mishkind ML, Greer KL, Schmidt GW. Cell-free reconstitution of protein transport into chloroplasts. Methods Enzymol. 1987;148:274–294. [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M and others. Genes galore: a summary of methods for accessing results from large-scale partial sequence of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormrod DP, Landry LG, Conklin PL. Short-term UV-B radiation and ozone exposure effects on aromatic secondary metabolite accumulation and shoot growth of flavonoid-deficient Arabidopsis mutants. Physiol Plant. 1995;93:602–610. [Google Scholar]
- Pan S-M, Yau Y-Y. Characterization of superoxide dismutase in Arabidopsis. Plant Cell Physiol. 1992;37:58–66. [Google Scholar]
- Phillips JP, Campbell SD, Michaud D, Charbonneau M. Null mutation of copper/zinc superoxide dismutase in Drosphila confers hypersensitivity to paraquat and reduced longevity. Proc Natl Acad Sci USA. 1989;86:2761–2765. doi: 10.1073/pnas.86.8.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng H-X and others. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Rounsley SD, Glodek A, Sutton G, Adams MD, Somerville CR, Venter JC, Kerlavage AR. The construction of Arabidopsis expressed sequence tag assemblies. Plant Physiol. 1996;112:1177–1183. doi: 10.1104/pp.112.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runeckles VC, Vaartnou M. EPR evidence for superoxide anion formation in leaves during exposure to low levels of ozone. Plant Cell Environ. 1997;20:306–314. [Google Scholar]
- Scandalios JG. Molecular genetics of superoxide dismutases in plants. In: Scandalios JG, editor. Oxidative Stress and the Molecular Biology of Antioxidant Defenses, Vol 34. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 527–568. [Google Scholar]
- Schmidt R, Love K, West J, Lenehan Z, Dean C. Description of 31 YAC contigs spanning the majority of Arabidopsis thaliana chromosome 5. Plant J. 1997;11:563–572. doi: 10.1046/j.1365-313x.1997.11030563.x. [DOI] [PubMed] [Google Scholar]
- Schmidt R, West J, Love K, Lenehan Z, Lister C, Thompson H, Bouchez D, Dean C. Physical map and organization of Arabidopsis thaliana chromosome 4. Science. 1995;270:480–483. doi: 10.1126/science.270.5235.480. [DOI] [PubMed] [Google Scholar]
- Sharma YK, Davis KR. Ozone-induced expression of stress-related genes in Arabidopsis thaliana. Plant Physiol. 1994;105:1089–1096. doi: 10.1104/pp.105.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL. Photorespiration-deficient mutants of Arabidopsis thaliana lacking mitochondrial serine transhydroxymethylase activity. Plant Physiol. 1981;67:666–671. doi: 10.1104/pp.67.4.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streller S, Wingsle G. Pinus sylvestris L. needles contain extracellular CuZn superoxide dismutase. Planta. 1994;192:195–201. [PubMed] [Google Scholar]
- Van Camp W, Bowler C, Villarroel R, Tsang EWT, Van Montagu M, Inzé D. Characterization of iron superoxide dismutase cDNAs from plants obtained by genetic complementation in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:9903–9907. doi: 10.1073/pnas.87.24.9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Camp W, Willekens H, Bowler C, Van Montagu M, Inzé D, Reupold-Popp P, Sandermann H, Jr, Langebartels C. Elevated levels of superoxide dismutase protect transgenic plants against ozone damage. Biotechniques. 1994;12:165–168. [Google Scholar]
- Yruela I, Pueyo JJ, Alonso PJ, Picorel R. Photoinhibition of photosystem II from higher plants: effect of copper inhibition. J Biol Chem. 1996;271:27408–27415. doi: 10.1074/jbc.271.44.27408. [DOI] [PubMed] [Google Scholar]
- Zhao J, Last RL. Immunological characterization and chloroplast localization of the tryptophan biosynthetic enzymes of the flowering plant Arabidopsis thaliana. J Biol Chem. 1995;270:6081–6087. doi: 10.1074/jbc.270.11.6081. [DOI] [PubMed] [Google Scholar]