Abstract
Nuclear factor erythroid 2-related factor 2 (Nrf2) is the master transcription factor of the antioxidant response element (ARE) pathway, coordinating the induction of detoxifying and antioxidant enzymes. Nrf2 is normally sequestered in the cytoplasm by Kelch-like ECH associating protein 1 (Keap1). To identify novel small molecules that will disturb Nrf2:Keap1 binding and promote activation of the Nrf2-ARE pathway, we generated a quantum model based on the structures of known Nrf2-ARE activators. We used the quantum model to perform in silico screening on over 18 million commercially available chemicals to identify the structures predicted to activate the Nrf2-ARE pathway based on the quantum model. The top hits were tested in vitro and half of the predicted hits activated the Nrf2-ARE pathway significantly in primary cell culture. In addition, we identified a new family of Nrf2-ARE activating structures that all have comparable activity to tBHQ and protect against oxidative stress and dopaminergic toxins in vitro. The improved ability to identify potent activators of Nrf2 through the combination of in silico and in vitro screening described here improves the speed and cost associated with screening Nrf2-ARE activating compounds for drug development.
Introduction
Nuclear factor erythroid 2-related factor 2 (Nrf2) is a basic leucine zipper transcription factor that binds the cis-acting antioxidant response element (ARE) regulatory sequence(1-3). Binding of Nrf2 to the ARE results in a coordinated upregulation of various phase II detoxifying and antioxidant enzymes, including those involved in glutathione synthesis (glutamine cysteine ligase), redox regulation (catalase and peroxiredoxin), and quinone recycling [NAD(P)H dehydrogenase (quionone 1) (NQO1)](4, 5).
Activation of the Nrf2-ARE pathway has been shown to be protective in a variety of disorders, including in mouse models of neurodegenerative disorders (6-8). This is of special interest in the development of therapeutics for Parkinson’s disease (PD), which is characterized by high levels of oxidative stress (9-11). Unfortunately, current PD therapeutics target only the symptoms and not the mechanisms responsible for disease progression. Identification of novel molecules that could cross the blood-brain-barrier and activate Nrf2 may have a valuable role in neuroprotection in Parkinson’s disease.
Under basal conditions, Nrf2 is quarantined in the cytoplasm through binding to kelch-like ECH associated protein 1 (Keap1), with binds both Nrf2 and cytoskeletal elements in the cell, restricting Nrf2 in the cytoplasm (12), and also promoting the ubiquitination and degradation of Nrf2 through its role as a E3 ubiquitin ligase adaptor protein (13). The specific features of the Nrf2:Keap1 interaction, as well as the necessary structural features for small molecules to disrupt Nrf2:Keap1 binding to allow nuclear translocation of Nrf2, are still being discovered. However, it is clear that Keap1 acts as a redox sensor for the complex: the human Keap1 protein has 27 cysteine residues as opposed to only 6 in the human Nrf2 protein. Of the cysteines in Keap1, research has confirmed that the primary electrophilic sensors are C273, C288 and C151 (14-16).
Novel small molecule activators are continually being developed and tested for Nrf2-ARE activation. Two are currently being tested in clinical trials: an oral formulation of dimethyl fumarate marketed by Biogen Idec, Inc. is in clinical trials for treatment of multiple sclerosis, while an oral formulation of 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid-methyl amide (CDDO-MA) marketed by Reata Pharmaceuticals, Inc. is being tested for kidney disease in type II diabetes patients. Small molecule activators of the Nrf2-ARE pathway have been shown to reduce nigrostriatal degeneration in mouse models of Parkinson’s disease (7, 17). However, identification of such potential compounds through conventional high-throughput screening is slow, extremely expensive and prone to failure in secondary assays. Another confounding factor is that different small molecule activators of the Nrf2-ARE pathway act through different mechanisms (18, 19). Finally, there is great structural diversity in the known Nrf2 activating molecules making it difficult to predict or intelligently design novel small molecule activators of the Nrf2-ARE pathway.
To overcome this obstacle, we used Constellation, an innovative quantum-based computational process for drug discovery developed by Gradient Biomodeling LLC, as recently described (20). Constellation has been shown to identify novel compounds with significant potency against high-profile targets ranging from single macromolecules [renin, glutamate carboxypeptidase II], to complex cellular pathways [Galanin Receptor 2 agonists], to complete organisms [P. falciparum] with high accuracy and efficiency [Sullivan et al.; unpublished data Gradient Biomodeling LLC].
As Constellation does not require any prior, explicit knowledge of the macromolecular targets, the approach allows for simultaneous interrogation of multiple targets within a pathway or multiple pathways within a sub-system (or a system), integrating various exploration of systems behavior, or “phenotypes,” at all levels of structural and functional complexity.
Herein, we report on the experimental in vitro results obtained from this computational in silico prediction approach. A novel chemical class of Nrf2 pathway activators has been identified and validated in primary neuronal cultures. Activation is comparable to that of the existing Nrf2 activators and protects against oxidative stress induced neurotoxicity in primary cortical neuronal culture as well as MPP+-induced dopaminergic neuron death in primary midbrain culture. Thus, the increased throughput of compound screening afforded by this method of quantum drug discovery and design will ultimately result in the de novo synthesis of novel Nrf2 activating molecules with the potential to treat PD as well as other neurodegenerative diseases.
Materials and Methods
Summary of Constellation software
This technique has recently been described (20). Briefly, the metric modeling technology considers the compounds of interest as quantum objects, without explicit dependence on their particular chemical structure. On a theoretical level, these quantum properties serve as powerful descriptors for molecular modeling, compound identification, optimization and de novo design. The computational platform determines essential, rigorous, easily computable molecular attributes related to chemical activity. These attributes are derived from a special representation of quantum fields. Their well-defined mathematical characteristics afford systematic theoretical treatment and property prediction with methods that would otherwise be computationally impossible. Specialized machine-learning algorithms with fuzzy decision-making protocols(21, 22) are applied to identify both active compounds and the corresponding quantum features of chemical and biological interest. Combined with the underlying modeling architecture, the algorithms also provide mechanistic hypothesis for the modeled interactions. Since structurally dissimilar compounds can be similar on a quantum level, our process is particularly good at identifying chemically novel compounds that have significant potency against a known target (protein or biological pathway).
Here, the Constellation computational software developed by Gradient Biomodeling, LLC., previously described in (20), was used to identify novel activators of the Nrf2-ARE. Briefly, a predictive quantum filter was designed using a training set of known small molecule activators of the Nrf2-ARE pathway, including tBHQ, 3H-1,2-dithiole-3-thione and sulforaphane and in vitro data for over 10,000 additional compounds. A series of fuzzy algorithms were developed to classify ideal target properties. The resulting quantum filters were used to identify a set of quantum components for property prediction of prospective molecules (Fig. 1). Although there is no direct theoretical correlation between these scores and compound activity, in practice this is often the case: the activity of a compound greatly increases with the number of interaction constraints it satisfies. Approximately 18 million commercially available structures were screened using sources including Enanmine, ChemBridge, LifeChemicals, ChemDiv, TimTec and the National Cancer Institute. The compounds were rank-ordered by the model and 14 high-ranked hits were procured and tested in vitro.
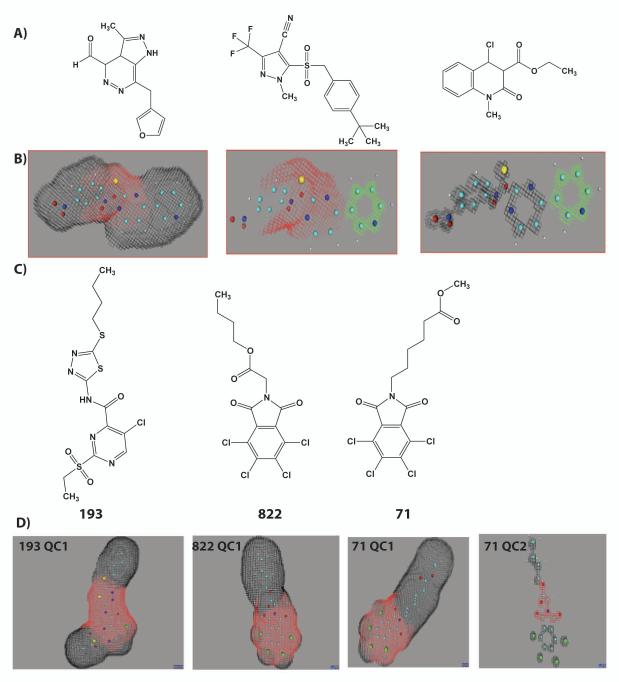
Figure 1.
Identification of novel Nrf2 activators through quantum modeling. Starting with a training set of compounds shown in vitro to activate the Nrf2 pathway (A), we identified quantum components (QC) that a molecule should possess for activity (red and green shaded areas, B). The virtual screen identified and rank-ordered novel compounds (C) that Gradient’s algorithms predicted to activate the Nrf2 pathway by the presence of one or more quantum components, D.
Neuronally enriched and mixed neuron/astrocyte culture preparation
Cultures were derived from ARE-hPAP reporter (23), Nrf2 wildtype (WT), or Nrf2 knockout (KO)(24) mice as previously described (Kraft et al., 2004). Briefly, cortices from E15 mouse pups were pooled in 10 mL ice-cold Ca2+ and Mg2+ free HBSS (Life Technologies, Carlsbad, CA). Tissue was minced, centrifuged and digested in 0.05% trypsin without EDTA in HBSS for 15 minutes at 37°C. Following trypsinization, cells were rinsed 3 times with HBSS. Cells were then washed with CEMEM (minimum essential media with Earle’s salts; Life Technologies, Carlsbad, CA), 2mM glutamine, 1% penicillin/streptomycin, and 10% each of heat inactivated fetal bovine serum and horse serum (Atlanta Biologicals, Inc., Lawrenceville, GA) and triturated to a single-cell suspension and strained through a 70μM cell strainer (BD Biosciences, San Jose, CA). Cell were counted and assayed for viability using trypan blue and plated at a density of 3×105 cell/cm2 on poly-D-lysine coated plates. Cells were maintained in CEMEM for 45 minutes, followed by media change. After 45 minutes, media for neuronally-enriched cultures (>98% neurons) was changed from CEMEM to NBM (Neurobasal media; Life Technologies, Carlsbad CA) supplemented with B27 with antioxidants and 2mM glutamine. For mixed cultures (~ 40% astrocytes and 60% neurons), media was changed from CEMEM to NBM at 2 days post plating. All cells were left for at least 48 hours in NBM prior to initiating experiments. Cells were incubated at 37°C in a tri-gas incubator with 5% O2, 5% CO2, and 90% N2. Preparations for astrocytes were similar except cells were prepared from the cortices of P1 pups, plated at a density of 3×104 cell/cm2 on collagen coated plates, and maintained in CEMEM throughout with a complete media change every 2-3 days; experiments were performed when the cells were confluent (one week).
Compounds were dissolved in 100% DMSO and administered to cells for 48 hours (final concentration of DMSO was 0.1%). For H2O2 challenge experiments, 48 hours after dosing medium was changed to NBM with B27 minus antioxidants and hydrogen peroxide (H2O2) was administered to the cells for an additional 24 hours.
Dopaminergic Neuronal Culture preparation
Dopaminergic cultures were prepared as in (25) with slight modifications. All chemicals were purchased from Sigma (St. Louis, MO) or Fisher Scientific (Lafayette, CO). Briefly, two litters of P3 ARE-hPAP pups were decapitated, skulls were removed, and the midbrain was dissected out in sterile ice-cold dissociation solution (90mM Na2SO4, 30 mM K2SO4, 6mM MgCl2, 0.25mM CaCl2, 10mM HEPES, 20mM D-glucose, pH 7.4). Midbrains were minced and incubated in papain (Worthington Biochemical Corporation, Lakewood, NJ) activated in 10mL dissociation dilution for 20 minutes at 37°C. Tissue blocks were washed twice with sterile trituration solution (NBM-A (Life Technologies), 1% penicillin/streptomycin, 1% glutaMAX™ (Life technologies), 2% B27, 10% heat inactivated fetal bovine serum, 1mg/mL trypsin inhibitor, 1mg/mL BSA, 10mM HEPES, pH 7.4). Cells were triturated in trituration solution, and gently dropped onto centrifugation solution (NBM-A, 1% penicillin/streptomycin, 1% glutaMAX™, 2% B27 with antioxidants, 10% heat inactivated fetal bovine serum, 10mg/mL trypsin inhibitor, 10mg/mL BSA, 10mM HEPES, pH 7.4) and centrifuged. Cells were resuspended in 500 uL trituration solution, counted and assayed for viability with trypan blue, and plated at 6×104 cells/cm2 in a mixture of 2/3 NBM-A+ (1% penicillin/streptomycin, 1% glutaMAX™, 2% B27 with antioxidants, 10% heat inactivated fetal bovine serum) and 1/3 MEM+ [1% penicillin/streptomycin, 1% glutaMAX™, 10% heat inactivated fetal bovine serum, 0.2mM glucose, 0.5mM sodium pyruvate, 0.05% MITO+ (BD Biosciences, San Jose, CA)]. One day after plating, cultures were treated with 2.5uL/mL of 5-fluoro-2′-deoxyuridine (Sigma) to prevent cell proliferation. Experiments were performed within a week. Cells were treated with 25μM 822 or tBHQ for 48 hours followed by 2.5μM MPP+ (Sigma) for 48 hours, fixed with 4% paraformaldehyde, and stained for tyrosine hydroxylase.
Viability Assays
Cell viability was assayed using the MTS (3-(4,5-Dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt) assay from Promega (Madison, WI) following the manufacturer’s suggested protocols. For TUNEL staining, cells were fixed for one hour with 4% paraformaldehyde, permeabilized and treated with the Roche (Indianapolis IN) In Situ Cell Death Detection kit according to manufacturer’s instructions. Apoptotic cells are identified by fluorescein-labeling of DNA strand breaks with the TdT enzyme. Cells were counter stained with Hoescht and imaged using a Zeiss microscope.
hPAP activity assays and histochemistry
The hPAP activity assay protocol was based on the protocol described in (23). Briefly, cells were lysed in TMNC lysis buffer (50mM Tris, 5mM MgCl2, 100mM NaCl, 1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)) and freeze-thawed at −20°C. Extracts were incubated with 200mM diethanolamine (DEA) buffer at 65°C to inactivate endogenous alkaline phosphatase activity. hPAP activity was quantified in 200mM DEA with 0.8mM CSPD [disodium 3-(4-methoxyspiro (1,2-dioxetane-3,2′-(5′-chloro)tricycle(3.3.1.1 3,7)decan)-4-yl)phenyl phosphate) (Life technologies), 2× Emerald and 5mM MgCl2]. Luminescence was measured on a Berthold Orion microplate luminometer with one second integration. Baseline signal from hPAP negative control culture samples was subtracted from all value.
To visualize hPAP activity, cells were fixed with 4% paraformaldehyde for 20 minutes and incubated in TMN buffer (50mM Tris, 5mM MgCl2, 100mM NaCl, pH 9.5) at 65°C for 25 minutes to heat inactivate endogenous alkaline phosphatase activity. Cells were stained for hPAP by incubating at 37°C in TMN buffer plus 1mg/mL NBT and 1mg/mL X-phosphate (5-bromo-4-chloro-3-indoyl-phosphate) (BCIP) (EMD Chemicals, Gibbstown, NJ) and counterstained with eosin.
NQO1 histochemistry
To visualize NQO1 activity, cells were prepared as described in (26). Briefly, cells were fixed in 4% paraformaldehyde for 20 minutes, washed with PBS and incubated in reaction buffer (25mM Tris, 0.8% Triton X-100, 2mg/mL BSA) with 100μM NBT (Calbiochem), 1mM NADPH and with or without 100μM of the NQO1 substrate LY83583. The reaction was incubated at 37°C for an hour.
Tyrosine Hydroxylase Histology
For immunohistochemistry, cells were fixed in 4% paraformaldehyde, then incubated for one hour with a blocking solution (PBS with 10% goat serum, 0.4% Triton-X-100, and 0.005mg/mL BSA), then incubated with tyrosine hydroxylase (TH) antibody (Millipore, Billerica, MA) at 1/1000 dilution overnight. Secondary detection was performed with fluorescently labeled antibodies. The Hoescht stain was used to identify nuclear DNA.
Statistics
All values are represented as the mean ± SEM. The n number of values used is presented in the individual figure legends. Significance was determined using an unpaired Student’s t-test (p<0.05) or a one-way ANOVA (p<0.05) followed by posthoc analysis to determine significant paired comparisons (p<0.05) based on experimental design.
Results
Identification of novel molecules
The Constellation computational software developed by Gradient Biomodeling, LLC., previously described in (20), was used to identify novel activators of the Nrf2-ARE. All of the identified hits contained multiple quantum components [Fig. 1, quantum component (QC) 1] that correlated with Nrf2-ARE activation. Some compounds had multiple quantum components: for instance, structure 71 contained an additional component (QC2) not found on 193 and 822 (Fig. 1, C,D). The two oxygens at the end of the tail on 71, but not at the beginning of the tail on 822, create the additional quantum component.
The resulting efficacy of Nrf2-ARE activation was assessed via ARE-hPAP reporter activation (Fig. 2A, structures shown in Fig. 2B). Dose curves were run for all compounds to determine toxicity using the MTS assay (data not shown). The data presented in Fig. 2A represents the fold increase in hPAP activity for the highest non-toxic dose of each compound (ranging from 0.75-50μM). Of the 14 tested compounds, 7 showed significant activation relative to vehicle treated cells at non-toxic doses (822, 071, 193, 797, 186, 543, 881). Of these 7 compounds, 4 had hPAP activity of greater than 2-fold (822, 071, 797, 193). In fact, the strongest activator, 822, had activation that was higher than the positive control used, tert-butyl hydroquinone (tBHQ) at the same dose, although the difference was not statistically significant at this dose. Overall, this is a vast improvement on available screening methods; 50% of the predicted activators were validated in vitro to show Nrf2-ARE activation.
Figure 2.
Structures and Nrf2-ARE activity in preliminary screen. A) Average fold hPAP activity of compounds. All compounds were dosed for 48 hours, n=3, mean ± SEM. The hPAP fold increase is listed as determined by the highest non-toxic dose for each compound: black bars, 50μM; dark grey bars, 25 μM; light grey bars, 12.5μM; checkered bars, 6.25μM; dotted bars, 3μM; lined bars, 1.5 μM; white bars, 0.75μM. B) Structures of compounds, organized according to highest hPAP activation, followed by non-activating compounds in order of lowest toxicity. *Significantly different than the corresponding vehicle-treated value (p<0.05).
Secondary Screen on 822 family molecules
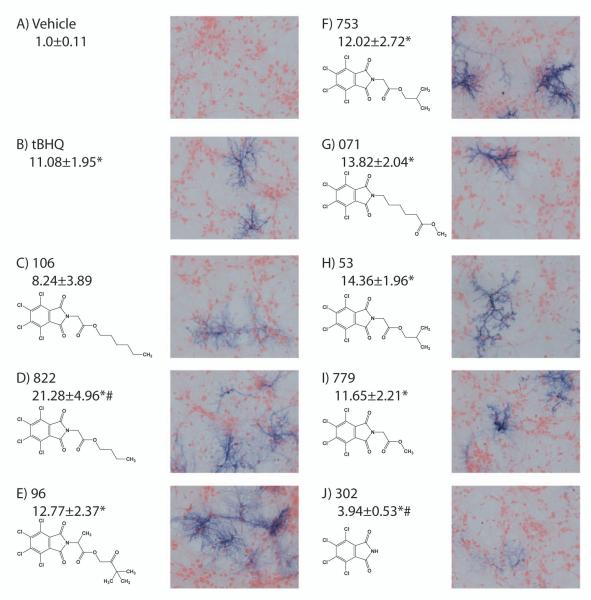
Interestingly, two of the best activators of the Nrf2-ARE pathway in this quantum screen were also structurally similar (Fig. 1C, 2B, 822 and 071). Since this structure has never previously been reported to activate the Nrf2-ARE pathway, a family of similar structures was obtained and screened for Nrf2-ARE activation using cultures derived from the ARE-hPAP reporter mice. An initial dose response analysis revealed that all compounds were non-toxic at 25μM (MTS assay; data not shown). This dose was used for further studies comparing all compounds. The structures, hPAP activity (fold change), and hPAP histochemistry for this set of compounds are shown in Fig. 3. Compounds are arranged in order of their partition coefficient or LogP, a measure of hydrophobicity, with the lowest, 302 having a LogP of 3.2 and the highest, 106, having a LogP of 5.7. All of the compounds except the base structure (302) show comparable or greater increases in hPAP activity relative to tBHQ. At this dose, 822 exhibited significantly higher activation than tBHQ.
Figure 3.
hPAP histochemistry and fold activation in mixed cultures treated with 25μM doses of compounds. A) vehicle-treated cells, B) tBHQ-treated cells C-J) Structures of novel compounds, representative hPAP histochemistry and average fold hPAP activation. Compounds arranged according to LogP. Average fold change (mean ± SEM; n=4-10) shown underneath compound name. *Significantly different the than corresponding vehicle-treated value (p<0.05). #Significantly different than the corresponding tBHQ-treated value (p<0.05).
Activation is Nrf2-dependent
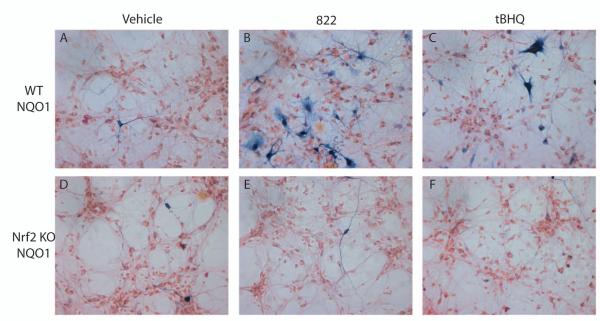
To determine whether the activation of Nrf2-ARE proteins was Nrf2-dependent, changes in the Nrf2-dependent gene NQO1 were evaluated by histochemical staining. 822 was used as a representative compound because it exhibited the greatest activation. Nrf2 WT cultures were compared to Nrf2 KO cultures treated with 822, tBHQ or vehicle. NQO1 histochemical staining was dramatically increased by both 822 and tBHQ in Nrf2 WT cultures (Fig. 4, A-C). In contrast, NQO1 staining of Nrf2 KO cultures was not changed by treatment with either 822 or tBHQ relative to the vehicle treated control (Fig. 4, D-F). Pretreatment with the antioxidant n-acetyl cysteine resulted in approximately a 50% reduction in hPAP activity in 822 treated mixed cultures, but there was no change in hPAP activity in n-acetyl cysteine pretreated tBHQ treated cortical cultures (data not shown).
Figure 4.
Mixed cultures treated with 25μM tBHQ or 822. A-C, histochemistry for NQO1 in WT cultures. D-F, representative images of NQO1 histochemical staining in Nrf2 KO cultures. A,D, vehicle treated cells; B, E, 822 treated cells; C,F, tBHQ treated cells.
Astrocytes and neurons respond independently to 822-dependent Nrf2-ARE activation
Since all initial screening was done in mixed neuron/astrocyte co-cultures and tBHQ has been found to selectively activate Nrf2 in astrocytes (23, 27), we prepared enriched cultures (> 98% of cells) of astrocytes and neurons to determine if 822 could also activate Nrf2 in neurons as well as astrocytes. Neuron or astrocyte cultures were incubated for 48 hours with 822. Neuronal-enriched cultures treated with 822 exhibit an increased histochemical staining for both hPAP (Fig. 5A,B) and NQO1 (Fig. 5C,D) relative to vehicle treated cells. hPAP activity also showed a significant increase in neurons following 822 with no observed toxicity (Fig. 5E,F). Similarly, astrocytes had a significant increase in hPAP activity with no associated toxicity (Fig. 5G,H).
Figure 5.
Neuron and astrocyte specific activation of Nrf2-ARE after 822 treatment. Panel A and B, hPAP histochemical staining of vehicle (A) or 20μM 822 (B) in neuronally enriched culture. NQO1 histochemical staining of vehicle (C) or 20μM 822 (D) in neuronally enriched culture. hPAP activity was quantified in neuron cultures at 20μM (E) and astrocytes at 50μM (G), with significant activation of hPAP activity and no corresponding toxicity (F, H). Values are the mean ± SEM (n=3). *Significantly different than the corresponding vehicle-treated value (p<0.05).
Nrf2-ARE dependent protection from H2O2-induced neurotoxicity
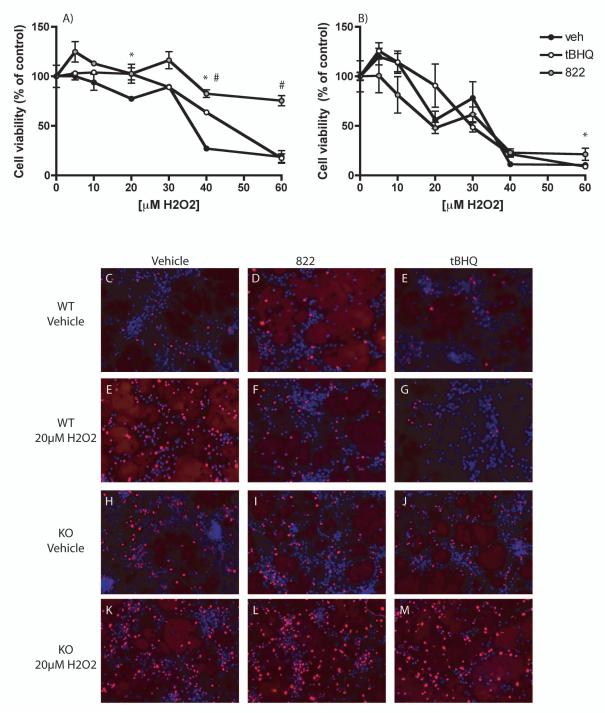
To determine whether the activation of the Nrf2-ARE pathway by 822 treatment was sufficient to coordinate a protective response against oxidative stress-induced neurotoxicity, mixed neuron/astrocyte cultures were treated with 822 or tBHQ for 48 hours followed by challenge with H2O2 for 24 hours. Significant protection was observed in cells treated with 822 or tBHQ relative to vehicle in WT cells (Fig. 6A). No protection was observed in Nrf2 KO cells treated with 822 or tBHQ relative to vehicle (Fig. 6B). In addition, TUNEL staining was evaluated after treatment with tBHQ or 822 for 48 hours followed by treatment with 20μM H2O2 for 24 hours. There was a modest increase in TUNEL staining in KO cells relative to WT cells in the vehicle-treated group (Fig. 6 C-E vs H-J). In WT cells treated with H2O2 , there was a dramatic reduction in TUNEL staining associated with 822 or tBHQ treatment (Fig. 6, E-G), while no reduction in TUNEL staining was observed in the H2O2-treated KO cells (Fig. 6, K-M).
Figure 6.
Protection in WT vs KO cultures after tBHQ or 822 administration. Figure A) WT cultures treated with vehicle, 50 μM tBHQ or 50 μM 822 for 48 hours, followed by H2O2 challenge for 24 hours and viability assay as determined by MTS. B) Nrf2 KO cultures treated as in A. Figure C-M are representative TUNEL staining images of vehicle (C,E,H,K) 25 μM 822 (D,F,I,L), or 25 μM tBHQ (E,G,J,M) treated mixed cultures. C-E and H-I are treated with vehicle; E-G and K-M are treated with 20 μM H2O2. WT cultures are panels C-G; KO cultures are panels H-M. Values are the mean ± SEM (n=3). *Significantly different than corresponding vehicle-treated value for tBHQ (p<0.05). #Significantly different than the corresponding vehicle-treated value for 822 (p<0.05).
Protection of dopaminergic neurons from MPP+ toxicity in primary midbrain cultures
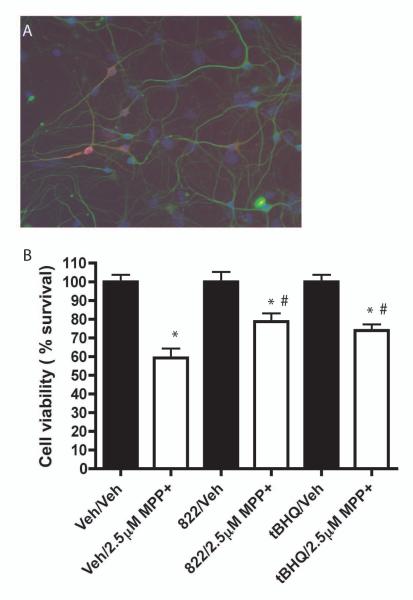
Primary midbrain cultures were stained for tyrosine hydroxylase, a marker for dopaminergic neurons, to verify the presence of dopaminergic neurons (Fig. 7A). Cultures were treated with vehicle, 822 or tBHQ for 48 hours, followed by a 48 hour administration with MPP+. MPP+ is a mitochondrial complex I inhibitor that is selectively taken up by the dopamine transporter, which is highly expressed on dopaminergic neurons, and thus useful as an in vitro model for dopaminergic neuronal death that is the cause in PD. Following treatment, the cultures were fixed and stained for tyrosine hydroxylase and surviving tyrosine-hydroxylase positive cells were counted. There was a significant loss to dopaminergic neurons associated with MPP+ treatment. There was also a significant reduction in dopaminergic neuronal loss in cultures treated with either 822 or tBHQ (Fig. 7B).
Figure 7.
Protection in dopaminergic cell culture treated with MPP+. A) Representative staining of dopaminergic cultures for beta III tubulin (green), tyrosine hydroxylase (red) and hoescht (blue). Cells were treated for 48 hours with vehicle, 25 μM 822 or 25 μM tBHQ , followed by 48 hour treatment with vehicle or 2.5μM MPP+. Cells were stained with antibodies against tyrosine hydroxylase and counted to determine percent survival. Values are the mean ± SEM (n=3). *Significantly different than corresponding vehicle-treated value (p<0.05). #Significantly different than the corresponding Veh/MPP+-treated value (p<0.05).
Discussion
The Nrf2-ARE pathway is a viable therapeutic target for protection against oxidative stress in neurodegenerative disease. Many Nrf2-ARE activating small molecules have been identified (as recently reviewed in (28)) and most have a highly reactive chemical moiety that can be grouped broadly into families of isothiocyanates, Michael acceptors, organosulfur compounds, electrophilic compounds or heavy metal containing compounds (28). However, the complexity of the protein interactions in the Nrf2-ARE pathway makes the identification of novel activators difficult. For instance, it has been shown that the activators of Nrf2 function through different mechanisms: tBHQ treatment is associated with the formation of a Keap1 dimer and increased Keap1 ubiquitination(19); whereas sulforaphane appears to modulate the Nrf2-ARE pathway primarily through blocking the interaction between Keap1 and the Cul3, the ubiquitin ligase adaptor protein that is associated with Nrf2 ubiquitization (13). Other compounds have been shown to activate the Nrf2-ARE pathway in a phostphatidyl-inositol 3 kinase PI3K dependent mechanism (23, 27, 29, 30).
The multiple interactions possible in the Nrf2-ARE pathway makes intelligent design of novel activators of the Nrf2-ARE pathway especially difficult. A previous article detailed de novo development of Nrf2-ARE activators and found a modest 2-3 fold activation in cell lines (31), which was independently confirmed in our lab using primary cortical cultures (unpublished data). The difficulty of identifying novel Nrf2-ARE activators is further evidenced by the results of several recent chemical screens. In each, a single hit with activity comparable to that of tBHQ was identified out either 117 (32) or 9400 (33) screened compounds. In contrast, an initial in vitro screen in this work identified a compound with greater activation than that of tBHQ out of only 14 initial molecules, greatly improving the screening efficacy. Also, half of the initial screened compounds showed significant activation in vitro in this work. Thus, the in silico screening method described here was able to identify far more potent hits from existing chemical libraries that should translate to the eventual synthesis of novel potent Nrf2 activating compound based on the final quantum model.
Similarly, work modifying known activators has limited success, with most modified compounds showing no improvement in activity relative to the parent compound, and only a few modifications showing some improvement in activation over the parent structure (34, 35). In contrast, 7 of the 8 related family members of the initial hit identified here activated the Nrf2-ARE pathway similarly to that of tBHQ.
Here, we showed that the compounds were activating the Nrf2-ARE pathway through comparison to Nrf2 knockout cells. However, we have not yet identified the mechanism of compound interaction with Nrf2 and/or Keap1. We hypothesize that the mechanism of action is similar to that of a previously published structure, as the chemical features present in the 822 family are similar to those observed in a screen published previously (32). Specifically, the authors identified that a halide group on a hexane ring was critical for ARE activation. In addition, the presence of esters and ketones rather than amides on the tail promotes Nrf2-ARE activation. Further analysis of the ARE-activating compound, AI-1, suggests that the halide group in AI-1 interacts with C151 in the Cul3 interacting region on Keap1. This suggests a proposed sulforaphane-like mechanism of Nrf2-ARE activation via Cul3-Keap1 disruption. The family of compounds identified here have both halide groups and ester/ketone groups; these features are also evident on many of he structures identified in the first screen. This suggests that a similar mechanism may be promoting Nrf2-ARE activation in both AI-1 and these compounds; however, additional work will be required for characterization of the mechanism of Nrf2-ARE activation by the new chemicals identified here.
The data generated will significantly refine the quantum model and subsequent work will add quantum properties associated with the ability to cross the blood-brain-barrier and reduce first pass metabolism. Once optimized, novel Nrf2 activating compounds will be synthesized and tested for neuroprotective properties in mouse models of neurodegenerative diseases. The increased ability to generate potent activators of Nrf2 will increase the possibility of finding a new therapeutics with the potential to slow or halt the progression of PD and other neurodegenerative diseases.
Acknowledgements
Thanks to Jennifer Kutzke for generation of the animals used in these experiments. TPW was supported by award number T32GM008349. This work was additionally supported by the Michael J. Fox foundation, R01ES08089 and R01ES10042 from the National Institute of Environmental Health Sciences.
References
- 1.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–22. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 2.Rushmore TH, Pickett CB. Transcriptional regulation of the rat glutathione S-transferase Ya subunit gene. Characterization of a xenobiotic-responsive element controlling inducible expression by phenolic antioxidants. J Biol Chem. 1990;265:14648–53. [PubMed] [Google Scholar]
- 3.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266:11632–9. [PubMed] [Google Scholar]
- 4.Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem. 2003;278:12029–38. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 5.Shih AY, Johnson DA, Wong G, Kraft AD, Jiang L, Erb H, et al. Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress. J Neurosci. 2003;23:3394–406. doi: 10.1523/JNEUROSCI.23-08-03394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen PC, Vargas MR, Pani AK, Smeyne RJ, Johnson DA, Kan YW, et al. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson’s disease: Critical role for the astrocyte. Proc Natl Acad Sci U S A. 2009;106:2933–8. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jazwa A, Rojo AI, Innamorato NG, Hesse M, Fernandez-Ruiz J, Cuadrado A. Pharmacological targeting of the transcription factor Nrf2 at the basal ganglia provides disease modifying therapy for experimental parkinsonism. Antioxid Redox Signal. 2011;14:2347–60. doi: 10.1089/ars.2010.3731. [DOI] [PubMed] [Google Scholar]
- 8.Vargas MR, Johnson DA, Sirkis DW, Messing A, Johnson JA. Nrf2 activation in astrocytes protects against neurodegeneration in mouse models of familial amyotrophic lateral sclerosis. J Neurosci. 2008;28:13574–81. doi: 10.1523/JNEUROSCI.4099-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alam ZI, Daniel SE, Lees AJ, Marsden DC, Jenner P, Halliwell B. A generalised increase in protein carbonyls in the brain in Parkinson’s but not incidental Lewy body disease. J Neurochem. 1997;69:1326–9. doi: 10.1046/j.1471-4159.1997.69031326.x. [DOI] [PubMed] [Google Scholar]
- 10.Alam ZI, Jenner A, Daniel SE, Lees AJ, Cairns N, Marsden CD, et al. Oxidative DNA damage in the parkinsonian brain: an apparent selective increase in 8-hydroxyguanine levels in substantia nigra. J Neurochem. 1997;69:1196–203. doi: 10.1046/j.1471-4159.1997.69031196.x. [DOI] [PubMed] [Google Scholar]
- 11.Dexter DT, Sian J, Rose S, Hindmarsh JG, Mann VM, Cooper JM, et al. Indices of oxidative stress and mitochondrial function in individuals with incidental Lewy body disease. Ann Neurol. 1994;35:38–44. doi: 10.1002/ana.410350107. [DOI] [PubMed] [Google Scholar]
- 12.Kang MI, Kobayashi A, Wakabayashi N, Kim SG, Yamamoto M. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc Natl Acad Sci U S A. 2004;101:2046–51. doi: 10.1073/pnas.0308347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–53. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, et al. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–82. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci U S A. 2004;101:2040–5. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–51. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burton NC, Kensler TW, Guilarte TR. In vivo modulation of the Parkinsonian phenotype by Nrf2. Neurotoxicology. 2006;27:1094–100. doi: 10.1016/j.neuro.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 18.Zhang DD, Lo SC, Sun Z, Habib GM, Lieberman MW, Hannink M. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem. 2005;280:30091–9. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- 19.Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J Biol Chem. 2005;280:31768–75. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan DJ, Jr., Kaludov N, Martinov MN. Discovery of potent, novel, non-toxic anti-malarial compounds via quantum modelling, virtual screening and in vitro experimental validation. Malar J. 2011;10:274. doi: 10.1186/1475-2875-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan YF, Shaw MJ. Induction of Fuzzy Decision Trees. Fuzzy Sets and Systems. 1995;69:125–39. [Google Scholar]
- 22.Janikow CZ. Fuzzy decision trees: Issues and methods. Ieee Transactions on Systems Man and Cybernetics Part B-Cybernetics. 1998;28:1–14. doi: 10.1109/3477.658573. [DOI] [PubMed] [Google Scholar]
- 23.Johnson DA, Andrews GK, Xu W, Johnson JA. Activation of the antioxidant response element in primary cortical neuronal cultures derived from transgenic reporter mice. J Neurochem. 2002;81:1233–41. doi: 10.1046/j.1471-4159.2002.00913.x. [DOI] [PubMed] [Google Scholar]
- 24.Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A. 1996;93:13943–8. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fasano C, Thibault D, Trudeau LE. Culture of postnatal mesencephalic dopamine neurons on an astrocyte monolayer. Curr Protoc Neurosci. 2008 doi: 10.1002/0471142301.ns0321s44. Chapter 3: Unit 3 21. [DOI] [PubMed] [Google Scholar]
- 26.Murphy TH, So AP, Vincent SR. Histochemical detection of quinone reductase activity in situ using LY 83583 reduction and oxidation. J Neurochem. 1998;70:2156–64. doi: 10.1046/j.1471-4159.1998.70052156.x. [DOI] [PubMed] [Google Scholar]
- 27.Kraft AD, Johnson DA, Johnson JA. Nuclear factor E2-related factor 2-dependent antioxidant response element activation by tert-butylhydroquinone and sulforaphane occurring preferentially in astrocytes conditions neurons against oxidative insult. J Neurosci. 2004;24:1101–12. doi: 10.1523/JNEUROSCI.3817-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hur W, Gray NS. Small molecule modulators of antioxidant response pathway. Curr Opin Chem Biol. 2011;15:162–73. doi: 10.1016/j.cbpa.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 29.Chen HH, Chen YT, Huang YW, Tsai HJ, Kuo CC. 4-Ketopinoresinol, a novel naturally occurring ARE activator, induces the Nrf2/HO-1 axis and protects against oxidative stress-induced cell injury via activation of PI3K/AKT signaling. Free Radic Biol Med. 2012;52:1054–66. doi: 10.1016/j.freeradbiomed.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 30.Lee JM, Hanson JM, Chu WA, Johnson JA. Phosphatidylinositol 3-kinase, not extracellular signal-regulated kinase, regulates activation of the antioxidant-responsive element in IMR-32 human neuroblastoma cells. J Biol Chem. 2001;276:20011–6. doi: 10.1074/jbc.M100734200. [DOI] [PubMed] [Google Scholar]
- 31.Wu JH, Miao W, Hu LG, Batist G. Identification and characterization of novel Nrf2 inducers designed to target the intervening region of Keap1. Chem Biol Drug Des. 2010;75:475–80. doi: 10.1111/j.1747-0285.2010.00955.x. [DOI] [PubMed] [Google Scholar]
- 32.Hur W, Sun Z, Jiang T, Mason DE, Peters EC, Zhang DD, et al. A small-molecule inducer of the antioxidant response element. Chem Biol. 2010;17:537–47. doi: 10.1016/j.chembiol.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 33.Zhu M, Baek H, Liu R, Song A, Lam K, Lau D. LAS0811: from combinatorial chemistry to activation of antioxidant response element. J Biomed Biotechnol. 2009;2009:420194. doi: 10.1155/2009/420194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandey MK, Kumar S, Thimmulappa RK, Parmar VS, Biswal S, Watterson AC. Design, synthesis and evaluation of novel PEGylated curcumin analogs as potent Nrf2 activators in human bronchial epithelial cells. Eur J Pharm Sci. 2011;43:16–24. doi: 10.1016/j.ejps.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Wondrak GT, Cabello CM, Villeneuve NF, Zhang S, Ley S, Li Y, et al. Cinnamoyl-based Nrf2-activators targeting human skin cell photo-oxidative stress. Free Radic Biol Med. 2008;45:385–95. doi: 10.1016/j.freeradbiomed.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]