Abstract
White matter lesions are a frequent phenomenon in the elderly and contribute to the development of disability. The mechanisms underlying these brain lesions are still not fully understood with age and hypertension being the most well established risk factors. The heritability of white matter lesions is consistently high in different populations. Candidate gene studies strongly support the role of genes involved in the renin–angiotensin system, as well as Notch3 signaling. The recent genome wide association study by the CHARGE consortium identified a novel locus on chromosome 17q25 harboring several genes such as TRIM65 and TRIM47 which pinpoint to possible novel mechanisms leading to white matter lesions.
Keywords: White matter lesions, Leuko-araiosis, Genetics, Polymorphism, Genome wide association study, Risk factor
1. Introduction
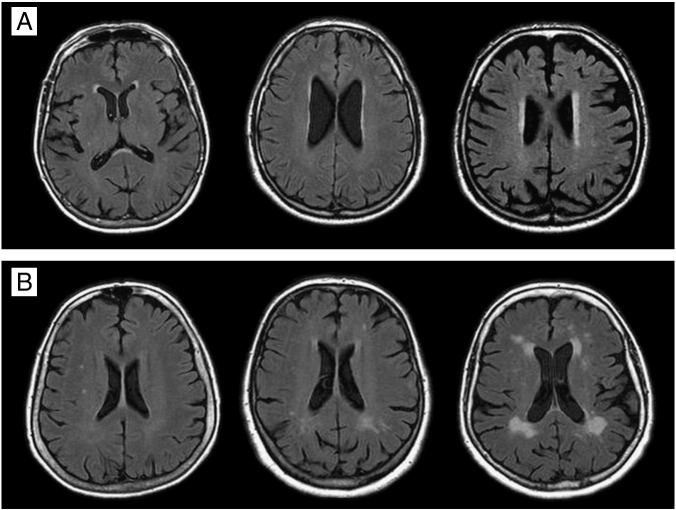
White matter lesions (WML) occur at high frequencies in routine magnetic resonance imaging (MRI) scans of elderly persons. In population-based studies the prevalence of any WML varies between 45% and 95%. Depending on the composition of the investigated cohorts 12 to 33% of subjects have severe changes [1]. According to location patterns WML can be separated in perventricular and deep/subcortical abnormalities (Fig. 1).
Fig. 1.
White matter lesions (WML) in T2-weighted FLAIR MRI scans. Panel A shows periventricular WML: from left to right caps, pencil-thin lining and halo. Panel B shows deep or subcortical WML: from left to right punctate, early-confluent and confluent lesions.
Pathological correlations demonstrated that this nosological distinction reflects etiological differences. Table 1 summarizes the findings of previous histopathological correlations.
Table 1.
The histopathological correlates of different types of white matter lesions.
| Study | Sample | Main findings |
|---|---|---|
| Fazekas 1991 [2] | 2 subjects without and 4 with neurologic disease | Punctate lesions relate to perivascular damage with lipohyalinosis, atrophic neuropil and rarefaction of myelinated fibers, 2 patients had cortical heterotopia. |
| Chimowitz 1992 [3] | 7 subjects with neurologic disease | Periventricular rims relate to ependymal loss and subependymal gliosis. |
| Periventricular caps relate to myelin pallor. | ||
| Punctate DWMH are widened perivascular spaces. | ||
| Fazekas 1993 [4] | 11 subjects partly with neurologic disease | Periventricular rims: Smooth myelin pallor, loose fibers, tortuous venules, no arteriolosclerosis, discontinuity of ependym with mild-moderate gliosis. |
| Irregular periventricular lesions have varying fiber loss, gliosis and cavitation with lipohyalinosis | ||
| Deep white matter lesions: Punctate: no ischemic changes; demyelination, atrophic neuropil around lipohyalionotic arterioles and perivenous damage. | ||
| Early confluent: perivascular rarefaction of myelin, mild to moderate fiber loss, varying gliosis. | ||
| Confluent: irregular areas of incomplete parenchymal destruction with focal transitions to true infarcts. | ||
| Munoz 1993 [5] | 2 Alzheimer cases and 13 controls | Punctate relates to widened perivascular spaces. |
| Extensive white matter lesions are broad areas of loss of myelin, axons, and gliosis. No infarction or vascular wall changes. | ||
| Fernando 2006 [6] | 99 demented subjects and 108 controls | Wall thickening, dilated perivascular spaces in WMH, ependym denudation in PVH. |
As can be seen from this table periventricular hyperintensities including caps around the ventricular horns, periventricular lining and halos are likely to be of non-vascular origin while deep and subcortical WML are often ischemic lesions and there exists a continuum in severity ranging from punctuate to early confluent and confluent changes. Punctate lesions often represent widened perivascular spaces only. By contrast, early confluent and confluent abnormalities correspond to incomplete ischemic tissue destruction including demyelination, axonal degeneration, gliosis and focal transitions to true infarctions. Early confluent and confluent changes progress rapidly over time [7] and these lesion types often lead to clinical consequences including cognitive decline, depression, gait disturbances, and urinary incontinence [8–11]. The LADIS study has shown that early confluent and confluent WML are also important predictors of disability [12].
The genetic raw model for cerebral small vessel disease is Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy (CADASIL) [13]. CADASIL has been reported in more than 500 families worldwide. The west of Scotland registry estimated the prevalence of NOTCH3 gene mutations, which are responsible for the disease, to be 4.14 per 100 000 subjects in the general population [14]. The MRI abnormalities seen in CADASIL patients are strikingly similar to changes seen in sporadic small vessel disease. MRI changes in CADASIL precede the onset of other symptoms by 10–15 years. The earliest and most frequent abnormalities are WML. Just like in age-related WML CADASIL white matter abnormalities first appear as punctuate changes often in periventricular areas and later become more diffuse, mostly symmetrical. Involvement of the external capsule is less frequent in age-related white matter diseases, but the most distinguishing MRI finding in CADASIL is the involvement of the anterior part of the temporal lobes [15].
In genetic studies WML have been either defined by their localization (periventricular or deep/subcortical) or severity, which can be assessed visually or by semiautomatic volume measurement resulting in a quantitative trait [16–18]. The etiology of WML is still incompletely understood. Conventional risk factors such as age and hypertension explain only a small proportion of the occurrence of WML [19]. Yet, WML load is highly heritable [20–22] and numerous investigators used genetic approaches to further explore genetic factors.
2. Heritability of WML
The heritability of a trait gives the proportion of the total phenotypic variance (Vp) in the population which can be explained by genetic differences among individuals (Vg). Importantly, the heritability index is a quotient (h2 = Vg/Vp) and its estimate is high, when there is a strong phenotypic variability due to genetic factors, as well as when there is low phenotypic variability due to environmental factors in a population. Therefore heritability indices strictly apply to the population they have been estimated for. So far heritability estimates for WML load have been reported in elderly male twins [20], in a large population based sample of middle aged–elderly sibships [21], within non-hispanic white and black hypertensive siblings [22,23], and in siblings discordant for Alzheimer disease [24]. In spite of major differences in the design of these studies, heritability estimates were comparable, within the range of 50–80%. The heritability of WML may depend on gender and age as suggested by the Framingham study [21]. A bivariate heritability analysis, where the genetic correlation between WML load and total and lobar brain volumes has been estimated, indicated that there may be common genetic influences on WML volume and specific brain regions such as frontal lobe volume and parietal lobe volume [25]. The Genoa (Genetic Epidemiology Network of Arteriopathy) study reported significant genetic correlations for WML with mean arterial pressure and with pulse pressure in non-Hispanic white subjects with essential hypertension, indicating shared genetic components between WML and blood pressure traits [23].
3. Dissecting the genetic architecture of WML
The consistently high heritability index reported for WML indicates that genetic variants play an important role in the development of these lesions. Different genetic variations in the human genome, such as single nucleotide polymorphisms (SNPs), copy number variations (CNVs), and epigenetic modifications may account for the genetic component. The majority of the studies tested common SNPs (minor allele frequency, MAF ≥ 5%) in the nuclear genome, one study investigated common SNP in the mitochondrial genome [26] and one study systematically tested one candidate gene, NOTCH3 for both common and rare SNPs [27]. There are two alternative hypotheses existing to explain the genetics of common diseases. The “common disease common variant” hypothesis states that common genetic variants in the population with relatively small effect sizes, typically odds ratios (OR) < 2, explain most of the heritability. The “common disease rare variant” hypothesis supposes that common diseases can be explained by rare variants (MAF ≤ 1–5%) segregating in families and enhancing the risk of their carriers substantially (OR > 2). Linkage studies investigate the co-segregation of markers and phenotype in family-based settings and have highest power to test the “common disease rare variant” hypothesis. Association studies performed in unrelated subjects have high power to identify common alleles with small effect sizes. In spite of the success in detecting many new loci by GWA studies, these common variants often explain only a small proportion of the variation and are not useful in clinical settings. Although linkage studies had detected significant loci for WML, genetic variants responsible for the linkage signals have not yet been identified. With the evolving ‘next-generation’ DNA sequencing techniques, sequencing these regions in a large number of individuals became feasible. Rare variants seem to be quite common in the general population [28], and it is now possible to combine new DNA sequencing methods with linkage studies to identify rare causal variants with a large effect size.
4. Linkage studies
So far significant logarithm of the odds (LOD) scores for WML has been found at 4 centiMorgan (cM) on chromosome 4 [29,30] in healthy elderly white subjects, and at chromosome 1q24 in hypertensive sibships [23]. Suggestive LOD scores by these studies have been reported at 95 cM on chromosome 17 (LOD = 1.78) [29], at 118 cM on chromosome 11, and at 13 cM on chromosome 21 in white, and at 36 cM on chromosome 22 as well as at 58 cM on chromosome 21 in black subjects [23]. Bivariate linkage studies identified significant (chromosome 1q24) and suggestive (chromosome 1q42, 10q22-q26, 15q26) shared loci for WML volumes and BP measurements, such as pulse pressure and systolic blood pressure, indicating the presence of genes with pleiotropic effects at these regions [23,31].
5. Candidate gene association studies
For more than a decade, the only feasible approach to identify genetic variants for WML was the candidate gene approach. According to a recent review 46 candidate gene studies investigating variants in 19 genes have been performed [32]. Here we focus on those genetic variants which are strongly supported by replication studies or functional data.
5.1. Genes involved in blood pressure regulation
The renin–angiotensin system (RAS) is an excellent candidate for WML, as it is a major regulator of systemic blood pressure, and cerebral blood flow [33]. Plasma angiotensinogen (AGT) synthesized by the liver is processed to angiotensin II (Ang II) by the serial action of renin and angiotensin-converting enzyme (ACE). Ang II exerts its effect through the angiotensin receptors (AGTR). The association of ACE I/D polymorphism with WML had been investigated in nine studies including 2396 individuals. Upon meta-analysis the DD genotype remained a significant predictor of WML (OR = 1.95; 95% CI 1.09:3.48) [32]. The most frequently investigated SNP in the AGT gene is M235T. Meta-analyses of 6 candidate gene studies including 2702 individuals showed no effect of this SNP on WML [32]. In contrast, linkage studies repeatedly implicated the AGT locus (1q42) [23,31] and the recent GWA study had replicated the association between the 235T allele and WML [34]. Most probably the 235T is not by itself a causal variant, but rather is in linkage disequilibrium (LD) with functional variants in its neighborhood. One such variant might be a haplotype (-6:a, -20:c, -153:g, -218:g) at the AGT promoter, which significantly enhanced the risk for WML. The effect was independent of hypertension [35]. We later showed that the haplotype enhances the basal transcriptional activity of the AGT promoter in astrocytes, which are the major source of AGT in the brain but not in hepatocytes [36], suggesting that its association with WML is mediated by altered activity of the cerebral RAS. Recently, a longitudinal study reported that progression rate of WML in elderly men is influenced by polymorphisms in the AGTR1 and AGTR2 genes. Homozygotes for the 1166A allele at AGTR1 showed less change over time than 1166C carriers [37]. There is also some indication for interaction on WML between the 573T allele at the AGTR1 gene and the 235T allele at the AGT gene [38].
5.2. Genes involved in vascular dysfunction
Chronic hypoperfusion and impaired cerebral auto-regulation are considered to be important contributors to the development of WML. Recently, Markus reviewed studies investigating endothelial activation and dysfunction in cerebral small vessel disease including also candidate genes such as ET1, MHTFR, NOS1 [39]. Due to small sample size of the studies and lack of replications, there is little statistical evidence presently that these genes harbor causal variants for WML. Also none of these genes had been indicated by the recent GWA study [34].
CADASIL is a monogenic form of systemic small vessel disease primarily affecting the brain [15,40–45]. It is caused by mutations in the NOTCH3 gene [42], which plays a key role in the functional and structural integrity of small arteries [46–48]. We recently sequenced the promoter, coding and 3′-untranslated region of the NOTCH3 gene in a community-dwelling elderly cohort and showed that the NOTCH3 gene is highly variable with both common and rare SNPs spreading across the gene. Among the 9 common SNPs 4 were significantly associated with WML. The association was confined to hypertensives. The most significant SNP, rs10404382 was replicated in the Cohorts for Heart and Ageing Research in Genomic Epidemiology Consortium (CHARGE) on an independent sample of 4773 stroke-free hypertensive elderly individuals of European descent. We also found support that rare SNPs might be relevant in WML. From the 33 rare SNPs that were identified, 9 non-synonymous SNPs were only detected in subjects with severe WML and 6 of these SNPs (H170R, P496L, V1183M, L1518M, D1823N and V1952M) were predicted to be functional by protein modeling tools. The results on NOTCH3 resemble other studies on complex traits, such as dyslipidemia where genes carrying common variants associated with modest effect in the population also carry rare variants with large effects segregating in families [49].
6. Genome wide association studies
GWA studies have become a well established and successful method to identify genes for common diseases. In order to enhance the power, results from different cohorts are combined in meta-analyses. Although there have been concerns regarding the effect of heterogeneity in meta-analyses of GWA studies, it has been shown that the approach is fairly robust, with little differences between the respective studies regarding different measurements of the trait, different environmental exposures or different genotyping chips, etc. [50].
The recent GWA study on WML performed by the CHARGE consortium [34], included 9361 elderly individuals of European descent from 7 community-based cohort studies: the Aging Gene-Environment Susceptibility-Reykjavik Study, the Atherosclerosis Risk in Communities Study, the Austrian Stroke Prevention Study, the Cardiovascular Health Study, the Framingham Heart Study, and 2 cohorts from the Rotterdam Study. Genome wide significant association was identified for 6 SNPs mapping to a locus on chromosome 17q25. The findings of the discovery meta-analyses had been confirmed in an independent sample of 1607 AGES-Reykjavik participants and in 1417 elderly white participants from the 3C-Dijon Study in the original report [34] and later in 1677 persons of the Rotterdam Study III [51]. The region on chromosome 17 is ~ 100 kb long and harbors several genes with diverse functions such as the 2 tripartite motif-containing genes (TRIM65 and TRIM47) the WW domain binding protein 2 gene (WBP2), the mitochondrial ribosomal protein L38 gene (MRPL38), the Fas-binding factor 1 gene (FBF1), the acyl-coenzyme A oxidase 1 gene (ACOX1) and the C-Elegans homolog (UNC13D) gene. The GWA study identified 3 additional SNPs which reached highly suggestive p-values of < 5 ∗ 10− 5 and were located within genes coding for the polyamine-modulated factor 1 gene (PMF1), the collagen type XXV alpha gene (COL25A1), and the methylenetetrahydrofolate dehydro-genase 1 gene (MTHFD1).
7. Summary
The last decade of research on the genetics of WML showed that this phenotype is highly heritable over different populations and subgroups. Candidate gene studies provided strong evidence for at least two pathways involved in WML, namely for the renin angiotensin system and for the Notch3 signaling pathway. The recent GWA study was successful in identifying common variants at chromosome 17 pinpointing to possible novel mechanisms leading to WML. It also brought together the largest population based cohorts on the field to work together in a concerted effort.
The presently identified genetic risk factors explain only a small proportion of the heritability of WML and further larger studies are needed to identify more common SNPs and to test their interactions with genetic and environmental factors. Finding associations with low frequency and rare variants will be certainly facilitated by the comprehensive catalog of these variants by the 1000 Genomes Project (http://www.1000genomes.org/page.php) and by the availability of next generation sequencing methods allowing deep sequencing of regions of interest, such as those identified by linkage or GWA studies or sequencing the whole genome. There is also need for studies investigating other genetic variants such as structural variants and epigenetic marks in WML.
Dissecting the genetic architecture of WML will contribute to a better understanding of the etiology of conditions characterized by WML and may facilitate the identification of new biomarkers and therapeutic targets with implications to prevention, diagnosis and treatment of stroke and dementia.
Acknowledgment
This work was supported by the Austrian Science Fond (FWF) grant number P20545-P05 to HS and P13180 to RS.
References
- 1.van Dijk E.J., Prins N.D., Vermeer S.E., Koudstaal P.J., Breteler M.M. Frequency of white matter lesions and silent lacunar infarcts. J Neural Transm Suppl. 2002;62:25–39. doi: 10.1007/978-3-7091-6139-5_2. [DOI] [PubMed] [Google Scholar]
- 2.Fazekas F., Kleinert R., Offenbacher H., Payer H., Schmidt R., Kleinert G. Themorphologic correlate of incidental punctate MR white matter hyperintensities. Am J Neuroradiol. 1991;12(5):915–921. [PMC free article] [PubMed] [Google Scholar]
- 3.Chimowitz M.I., Estes M.L., Furlan A.J., Awad I.A. Further observations on the pathology of subcortical lesions identified on magnetic resonance imaging. Arch Neurol. 1992;49(7):747–752. doi: 10.1001/archneur.1992.00530310095018. [DOI] [PubMed] [Google Scholar]
- 4.Fazekas F., Kleinert R., Offenbacher H., Schmidt R., Kleinert G., Payer F. Pathologic correlates of incidental MRI white matter signal hyperintensities. Neurology. 1993;43(9):1683–1689. doi: 10.1212/wnl.43.9.1683. [DOI] [PubMed] [Google Scholar]
- 5.Munoz D.G., Hastak S.M., Harper B., Lee D., Hachinski V.C. Pathologic correlates of increased signals of the centrum ovale on magnetic resonance imaging. Arch Neurol. 1993;50(5):492–497. doi: 10.1001/archneur.1993.00540050044013. [DOI] [PubMed] [Google Scholar]
- 6.Fernando M.S., Simpson J.E., Matthews F., Brayne C., Lewis C.E., Barber R. White matter lesions in an unselected cohort of the elderly: molecular pathology suggests origin from chronic hypoperfusion injury. Stroke. 2006;37(6):1391–1398. doi: 10.1161/01.STR.0000221308.94473.14. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt R., Enzinger C., Ropele S., Schmidt H., Fazekas F. Progression of cerebral white matter lesions: 6-year results of the Austrian Stroke Prevention Study. Lancet. 2003;361(9374):2046–2048. doi: 10.1016/s0140-6736(03)13616-1. [DOI] [PubMed] [Google Scholar]
- 8.Baezner H., Blahak C., Poggesi A., Pantoni L., Inzitari D., Chabriat H. Association of gait and balance disorders with age-related white matter changes: the LADIS study. Neurology. 2008;70(12):935–942. doi: 10.1212/01.wnl.0000305959.46197.e6. [DOI] [PubMed] [Google Scholar]
- 9.Vermeer S.E., Prins N.D., den Heijer T., Hofman A., Koudstaal P.J., Breteler M.M. Silent brain infarcts and the risk of dementia and cognitive decline. N Engl J Med. 2003;348(13):1215–1222. doi: 10.1056/NEJMoa022066. [DOI] [PubMed] [Google Scholar]
- 10.Grool A.M., van der Graaf Y., Mali W.P., Geerlings M.I. Location of cerebrovascular and degenerative changes, depressive symptoms and cognitive functioning in later life: the SMART-Medea study. J Neurol Neurosurg Psychiatry. 2011;82(10):1093–1100. doi: 10.1136/jnnp.2010.232413. [DOI] [PubMed] [Google Scholar]
- 11.Vermeer S.E., Hollander M., van Dijk E.J., Hofman A., Koudstaal P.J., Breteler M.M. Silent brain infarcts and white matter lesions increase stroke risk in the general population: the Rotterdam Scan Study. Stroke. 2003;34(5):1126–1129. doi: 10.1161/01.STR.0000068408.82115.D2. [DOI] [PubMed] [Google Scholar]
- 12.Inzitari D., Pracucci G., Poggesi A., Carlucci G., Barkhof F., Chabriat H. Changes in white matter as determinant of global functional decline in older independent outpatients: three year follow-up of LADIS (leukoaraiosis and disability) study cohort. BMJ. 2009;339:b2477. doi: 10.1136/bmj.b2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tournier-Lasserve E., Joutel A., Melki J., Weissenbach J., Lathrop G.M., Chabriat H. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy maps to chromosome 19q12. Nat Genet. 1993;3(3):256–259. doi: 10.1038/ng0393-256. [DOI] [PubMed] [Google Scholar]
- 14.Dong Y., Hassan A., Zhang Z., Huber D., Dalageorgou C., Markus H.S. Yield of screening for CADASIL mutations in lacunar stroke and leuko-araiosis. Stroke. 2003;34(1):203–205. doi: 10.1161/01.str.0000048162.16852.88. [DOI] [PubMed] [Google Scholar]
- 15.Chabriat H., Joutel A., Dichgans M., Tournier-Lasserve E., Bousser M.G. Cadasil. Lancet Neurol. 2009;8(7):643–653. doi: 10.1016/S1474-4422(09)70127-9. [DOI] [PubMed] [Google Scholar]
- 16.Scheltens P., Barkhof F., Leys D., Pruvo J.P., Nauta J.J., Vermersch P. A semiquantative rating scale for the assessment of signal hyperintensities on magnetic resonance imaging. J Neurol Sci. 1993;114(1):7–12. doi: 10.1016/0022-510x(93)90041-v. [DOI] [PubMed] [Google Scholar]
- 17.Fazekas F., Chawluk J.B., Alavi A., Hurtig H.I., Zimmerman R.A. MR signal abnormalities at 1.5 T in Alzheimer's dementia and normal aging. AJR Am J Roentgenol. 1987;149(2):351–356. doi: 10.2214/ajr.149.2.351. [DOI] [PubMed] [Google Scholar]
- 18.Manolio T.A., Kronmal R.A., Burke G.L., Poirier V., O'Leary D.H., Gardin J.M. Magnetic resonance abnormalities and cardiovascular disease in older adults. The Cardiovascular Health Study. Stroke. 1994;25(2):318–327. doi: 10.1161/01.str.25.2.318. [DOI] [PubMed] [Google Scholar]
- 19.O'Brien J.T., Erkinjuntti T., Reisberg B., Roman G., Sawada T., Pantoni L. Vascular cognitive impairment. Lancet Neurol. 2003;2(2):89–98. doi: 10.1016/s1474-4422(03)00305-3. [DOI] [PubMed] [Google Scholar]
- 20.Carmelli D., DeCarli C., Swan G.E., Jack L.M., Reed T., Wolf P.A. Evidence for genetic variance in white matter hyperintensity volume in normal elderly male twins. Stroke. 1998;29(6):1177–1181. doi: 10.1161/01.str.29.6.1177. [DOI] [PubMed] [Google Scholar]
- 21.Atwood L.D., Wolf P.A., Heard-Costa N.L., Massaro J.M., Beiser A., D'Agostino R.B. Genetic variation in white matter hyperintensity volume in the Framingham Study. Stroke. 2004;35(7):1609–1613. doi: 10.1161/01.STR.0000129643.77045.10. [DOI] [PubMed] [Google Scholar]
- 22.Turner S.T., Jack C.R., Fornage M., Mosley T.H., Boerwinkle E., de Andrade M. Heritability of leukoaraiosis in hypertensive sibships. Hypertension. 2004;43(2):483–487. doi: 10.1161/01.HYP.0000112303.26158.92. [DOI] [PubMed] [Google Scholar]
- 23.Turner S.T., Fornage M., Jack C.R., Jr., Mosley T.H., Knopman D.S., Kardia S.L. Genomic susceptibility loci for brain atrophy, ventricular volume, and leukoaraiosis in hypertensive sibships. Arch Neurol. 2009;66(7):847–857. doi: 10.1001/archneurol.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuenco K.T., Green R.C., Zhang J., Lunetta K., Erlich P.M., Cupples L.A. Magnetic resonance imaging traits in siblings discordant for Alzheimer disease. J Neuroimaging. 2008;18(3):268–275. doi: 10.1111/j.1552-6569.2007.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeStefano A.L., Seshadri S., Beiser A., Atwood L.D., Massaro J.M., Au R. Bivariate heritability of total and regional brain volumes: the Framingham Study. Alzheimer Dis Assoc Disord. 2009;23(3):218–223. doi: 10.1097/WAD.0b013e31819cadd8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson C.D., Biffi A., Rahman R., Ross O.A., Jagiella J.M., Kissela B. Common mitochondrial sequence variants in ischemic stroke. Ann Neurol. 2011;69(3):471–480. doi: 10.1002/ana.22108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt H., Zeginigg M., Wiltgen M., Freudenberger P., Petrovic K., Cavalieri M. Genetic variants of the NOTCH3 gene in the elderly and magnetic resonance imaging correlates of age-related cerebral small vessel disease. Brain. 2011;134(Pt 11):3384–3397. doi: 10.1093/brain/awr252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey-Wilson J.E., Wilson A.F. Linkage analysis in the next-generation sequencing era. Hum Hered. 2011;72(4):228–236. doi: 10.1159/000334381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeStefano A.L., Atwood L.D., Massaro J.M., Heard-Costa N., Beiser A., Au R. Genome-wide scan for white matter hyperintensity: the Framingham Heart Study. Stroke. 2006;37(1):77–81. doi: 10.1161/01.STR.0000196987.68770.b3. [DOI] [PubMed] [Google Scholar]
- 30.Seshadri S., DeStefano A.L., Au R., Massaro J.M., Beiser A.S., Kelly-Hayes M. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham Study. BMC Med Genet. 2007;8(Suppl. 1):S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kochunov P., Glahn D., Lancaster J., Winkler A., Kent J.W., Jr., Olvera R.L. Whole brain and regional hyperintense white matter volume and blood pressure: overlap of genetic loci produced by bivariate, whole-genome linkage analyses. Stroke. 2010;41(10):2137–2142. doi: 10.1161/STROKEAHA.110.590943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paternoster L., Chen W., Sudlow C.L. Genetic determinants of white matter hyperintensities on brain scans: a systematic assessment of 19 candidate gene polymorphisms in 46 studies in 19,000 subjects. Stroke. 2009;40(6):2020–2026. doi: 10.1161/STROKEAHA.108.542050. [DOI] [PubMed] [Google Scholar]
- 33.Wright J.W., Harding J.W. Regulatory role of brain angiotensins in the control of physiological and behavioral responses. Brain Res Brain Res Rev. 1992;17(3):227–262. doi: 10.1016/0165-0173(92)90018-h. [DOI] [PubMed] [Google Scholar]
- 34.Fornage M., Debette S., Bis J.C., Schmidt H., Ikram M.A., Dufouil C. Genome-wide association studies of cerebral white matter lesion burden: the CHARGE consortium. Ann Neurol. 2011;69(6):928–939. doi: 10.1002/ana.22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt H., Fazekas F., Kostner G.M., van Duijn C.M., Schmidt R. Angiotensinogen gene promoter haplotype and microangiopathy-related cerebral damage: results of the Austrian Stroke Prevention Study. Stroke. 2001;32(2):405–412. doi: 10.1161/01.str.32.2.405. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt H., Aulchenko Y.S., Schweighofer N., Schmidt R., Frank S., Kostner G.M. Angiotensinogen promoter B-haplotype associated with cerebral small vessel disease enhances basal transcriptional activity. Stroke. 2004;35(11):2592–2597. doi: 10.1161/01.STR.0000144646.96121.d2. [DOI] [PubMed] [Google Scholar]
- 37.Taylor W.D., Steffens D.C., Ashley-Koch A., Payne M.E., MacFall J.R., Potocky C.F. Angiotensin receptor gene polymorphisms and 2-year change in hyperintense lesion volume in men. Mol Psychiatry. 2010;15(8):816–822. doi: 10.1038/mp.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Rijn M.J., Bos M.J., Isaacs A., Yazdanpanah M., Arias-Vásquez A., Stricker B.H. Polymorphisms of the renin–angiotensin system are associated with blood pressure, atherosclerosis and cerebral white matter pathology. J Neurol Neurosurg Psychiatry. 2007;78(10):1083–1087. doi: 10.1136/jnnp.2006.109819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markus H.S. Genes, endothelial function and cerebral small vessel disease in man. Exp Physiol. 2008;93(1):121–127. doi: 10.1113/expphysiol.2007.038752. [DOI] [PubMed] [Google Scholar]
- 40.Adib-Samii P., Brice G., Martin R.J., Markus H.S. Clinical spectrum of CADASIL and the effect of cardiovascular risk factors on phenotype: study in 200 consecutively recruited individuals. Stroke. 2010;41(4):630–634. doi: 10.1161/STROKEAHA.109.568402. [DOI] [PubMed] [Google Scholar]
- 41.Chabriat H., Vahedi K., Iba-Zizen M.T., Joutel A., Nibbio A., Nagy T.G. Clinical spectrum of CADASIL: a study of 7 families. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Lancet. 1995;346(8980):934–939. doi: 10.1016/s0140-6736(95)91557-5. [DOI] [PubMed] [Google Scholar]
- 42.Joutel A., Corpechot C., Ducros A., Vahedi K., Chabriat H., Mouton P. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383(6602):707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 43.Dichgans M., Mayer M., Uttner I., Brüning R., Müller-Höcker J., Rungger G. The phenotypic spectrum of CADASIL: clinical findings in 102 cases. Ann Neurol. 1998;44(5):731–739. doi: 10.1002/ana.410440506. [DOI] [PubMed] [Google Scholar]
- 44.Desmond D.W., Moroney J.T., Lynch T., Chan S., Chin S.S., Mohr J.P. The natural history of CADASIL: a pooled analysis of previously published cases. Stroke. 1999;30(6):1230–1233. doi: 10.1161/01.str.30.6.1230. [DOI] [PubMed] [Google Scholar]
- 45.Reyes S., Viswanathan A., Godin O., Dufouil C., Benisty S., Hernandez K. Apathy: a major symptom in CADASIL. Neurology. 2009;72(10):905–910. doi: 10.1212/01.wnl.0000344166.03470.f8. [DOI] [PubMed] [Google Scholar]
- 46.Artavanis-Tsakonas S., Rand M.D., Lake R.J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284(5415):770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 47.Domenga V., Fardoux P., Lacombe P., Monet M., Maciazek J., Krebs L.T. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004;18(22):2730–2735. doi: 10.1101/gad.308904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang T., Baron M., Trump D. An overview of Notch3 function in vascular smooth muscle cells. Prog Biophys Mol Biol. 2008;96(1–3):499–509. doi: 10.1016/j.pbiomolbio.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 49.Wang J., Cao H., Ban M.R., Kennedy B.A., Zhu S., Anand S. Resequencing genomic DNA of patients with severe hypertriglyceridemia (MIM 144650) Arterioscler Thromb Vasc Biol. 2007;27(11):2450–2455. doi: 10.1161/ATVBAHA.107.150680. [DOI] [PubMed] [Google Scholar]
- 50.Lin D.Y., Zeng D. Meta-analysis of genome-wide association studies: no efficiency gain in using individual participant data. Genet Epidemiol. 2010;34(1):60–66. doi: 10.1002/gepi.20435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhaaren B.F., de Boer R., Vernooij M.W., Rivadeneira F., Uitterlinden A.G., Hofman A. Replication study of chr17q25 with cerebral white matter lesion volume. Stroke. 2011;42(11):3297–3299. doi: 10.1161/STROKEAHA.111.623090. [DOI] [PubMed] [Google Scholar]