Abstract
Purpose
Obesity is a disease with genetic susceptibility characterized by an increase in storage and irregular distribution of body fat. In obese patients, the decrease in the Adiponectin gene (ADIPOQ) expression has been associated with a systemic low-grade inflammatory state. Our aim was to investigate the relationship between ADIPOQ +45T>G gene simple nucleotide polymorphism (SNP rs2241766) with serum adiponectin (sAdiponectin), distribution of body fat storage, and inflammation markers.
Subjects and methods
In this cross-sectional study, 242 individuals from Western Mexico characterized as Mexican-Mestizo and classified by body mass index (BMI), were included. Anthropometrics, body composition, body fat distribution, and inflammation markers were measured by routine methods. Genotypes were characterized using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technique and sAdiponectin by the ELISA method. A P-value <0.05 was considered the statistically significant threshold.
Results
sAdiponectin is associated with BMI (P < 0.001) and the genotypes (P < 0.001 to 0.0046) GG (8169 ± 1162 ng/mL), TG (5189 ± 501 ng/mL), and TT (3741 ± 323 ng/mL), but the SNP ADIPOQ +45T>G is not associated with BMI. However, the detailed analysis showed association of this SNP with a pattern of fat distribution and correlations (P < 0.05) with inflammation markers and distribution of body fat storage (Pearson’s r = −0.169 to −0.465) were found.
Conclusion
In this study, we have suggested that the ADIPOQ +45G allele could be associated with distribution of body fat storage in obesity. On the other hand, as no association was observed between ADIPOQ +45T>G gene polymorphism and obesity, it cannot be concluded that the ADIPOQ +45G allele is responsible for the increase of adiponectin levels.
Keywords: ADIPOQ gene polymorphism, levels of inflammation markers, body fat distribution, obesity, Mexican-Mestizo population
Introduction
Obesity is a systemic and chronic disease with genetic susceptibility.1,2 It is accompanied by metabolic changes that predispose the presentation of disorders in the health status and is associated in most cases with endocrine, cardiovascular, and orthopedic disorders relating to diverse biological, sociocultural, and psychological factors. In Mexico, the combined prevalence of overweight and obesity in adults is 70.9%.3,4
Obesity is characterized by an increase of the body mass index (BMI) and disproportion of body composition leading to a rise of fat mass.5,6 This increase, represented by an expansion of white adipose tissue (WAT) with an irregular distribution, is the most consistent pathological process in obesity. This WAT increase also promotes a unique microenvironment with a persistent abnormal inflammatory response that leads to a systemic low-grade subacute inflammatory state.7–9
An early phase of this inflammatory response occurs when the adipocytes and infiltrated macrophages (M1) in the WAT increase the release of inflammation markers and decrease adiponectin.10 Adiponectin belongs to the adipokine family and is secreted almost exclusively by adipose tissue. Its function is to maintain stability in glucose homeostasis and insulin sensitivity,11 and is down-regulated in subjects with obesity related comorbidity. Low serum levels of adiponectin (sAdiponectin) were associated with unfavorable serum lipid profiles and high inflammatory markers.12
On the other hand, the susceptibility for development of obesity with an irregular accumulation of WAT in the body depends on the interaction of two factors: (1) exposure to environmental risk factors, and (2) variation in genes (characterized as “obesogenes”) involved in body weight regulation and obesity pathogenesis such as ADIPOQ.1,2 The ADIPOQ gene, linked to the regulation and expansion of WAT, has been investigated as a possible factor associated with obesity and a synonymous (15Gly > Gly) single nucleotide polymorphism (SNP) +45T > G in the exon 2 was identified. The genotype frequencies of this polymorphism show ethnic variation and the +45G polymorphic allele has been associated with insulin resistance and type 2 diabetes (T2D) in several populations;13,14 however, their functional effects are undefined.
In this context, we investigated the distribution of ADIPOQ +45T>G polymorphism in a Mexican-Mestizo population from Western Mexico. This study evaluated the probable association of ADIPOQ genetic variants with blood levels of adiponectin, inflammation markers, and the distribution of body fat storage in obesity.
Subjects and methods
Subjects
In this cross-sectional study, beginning February 2009 and ending January 2011, we included a total of 242 nonrelated adults, aged 24 to 69 years, recruited from a population of Western Mexico. Individuals were classified according to BMI, with World Health Organization (WHO) categories, as (1) overweight: BMI of 25.0 to 29.9 kg/m2, obese: BMI > 30.0 kg/m2, lean: BMI of 18.5 to 24.9 kg/m2; and (2) individuals with excess body weight BMI > 25.0 kg/m2.15
All individuals were Mexican-Mestizos, in accordance with the National Institute of Anthropology classification: they were born in Mexico, with a family last name of Spanish origin, and a family history of Mexican ancestors for at least three generations.16
The group was composed of healthy adult volunteers (ie, without medication and glucose intolerance according to clinical history, and with a stable weight for at least three weeks); individuals with infectious diseases, hypertension, history of cardiovascular disease, malignancy, or renal and metabolic diseases such as T2D, were not included. Before enrollment in this study, informed participants gave their signature in a consent document, in keeping with the Helsinki declaration guidelines17 and the Institutional (University of Guadalajara) Review Committees to ensure appropriate ethical and biosecurity conduct (record No 70/ UG-JAL/2011).
Methods
According to the inclusion criteria, all subjects had a complete medical history. Distribution of body fat storage measurements included: height, which was measured to the nearest 1 mm by using a stadiometer (Seca GmbH and Co, KG, Hamburg, Germany), weight and body composition (that included total muscle and fat mass and body fat percentage), which were measured by using bio-electrical impedance analysis (TBF-304; Tokyo, Japan) to the nearest 0.1 kg, and BMI according to Lukaski’s technique.18,19 Waist, hip, and arm circumferences were measured using an anthropometric fiberglass tape (GULICK®, length 0–180 cm, accuracy ± 1 mm; North Coast Medical, Inc., Gilroy, CA) and following the procedures recommended by the anthropometric indicators measurement guide.20–22 Four measures of skinfold thickness (ie, biceps, triceps, subscapular, and suprailiac) were obtained on the right side of the body by using a caliper skinfold Harpenden (opening 80 mm and precision of ±0.2 mm, constant pressure of 10 g/mm2; Holtain Ltd, Crosswell, UK) in accordance with the procedures recommended by Ness-Abramof and Apovian,22 Durnin and Rahaman,23 and Lukaski.24 We calculated the sum of the four skinfold thicknesses as an indicator of subcutaneous fatness, body fat ratio as an indicator of adiposity, and waist-hip ratio (WHR) as an indicator of preferential accumulation of fat in the abdomen rather than on extremities.22,25
We confirmed an overnight fast of 12 hours in all subjects, and obtained venous blood samples. After allowing these to clot at room temperature they were centrifuged at 1509 RCF (Rotanta 460R; Andreas Hettich GmbH and Co, KG, Tuttlingen, Germany) for 10 minutes and serum samples were separated and stored at −70°C until analysis. Serum concentrations of sAdiponectin were determined using a commercial kit of enzyme-linked immune-absorbent assay (R&D Systems Inc, Minneapolis, MN), with a limit of detection of 1 ng/mL. The profile of inflammation markers included high-sensitivity C reactive protein (CRP) with a limit of detection of 0.15 mg/L (Random Access Analyzer Clinical Chemistry-Turbidimetry A25 Immunochemistry; Biosystems, Barcelona, Spain); erythrocyte sedimentation rate was measured by the Wintrobe method; and a complete blood count, including platelet count and white blood cell count, were measured with the Cell-Dyn 3700 (Abbott Diagnostics, Abbott Park, IL).
To identify ADIPOQ +45T>G gene polymorphism, genomic DNA was obtained from total blood using a standard protocol for extraction with the modified Miller method26 and was stored at −20°C until use for genotyping. The polymorphic region was amplified by the polymerase chain reaction (PCR) method27 to analyze the SNP +45T>G in ADIPOQ (rs2241766) in the locus 3q27. Primers were: forward 5′-GCAGCTCCTAGAAGTAGACTCTGCTG-3′, reverse 5′-GGAGGTCTGTGATGAAAGAGGCC-3′, and annealing temperature of 50°C. The PCR product was 372 base pairs (bp, Figure 1A); and was performed in a 25 μL total volume mixture (100 ng of DNA, 2 nM of each primer, 0.20 mM of each dNTP, 2.5 mM MgCl2, 0.25 U Taq polymerase, and 1X PCR buffer, Invitrogen®); followed by digestion of PCR products with Bsp HI restriction enzyme (New England Biolabs© Inc, Ipswich, MA). The following digestion fragments were obtained: allele T, 206, and 166 bp; allele G, 372 bp (Figure 1B). Electrophoresis was performed at a constant voltage of 80 volts on 3% agarose gels stained with ethidium bromide 0.01 mg. For quality control, a blank and samples previously confirmed as positive for each genotype were used as controls. To ensure the accuracy of genotype data, we used internal controls and repetitive experiments. Any sample that yielded a weak signal was repeated. In addition, all samples were repeated at random to verify the reproducibility with positive controls in each experiment and we confirmed that the positive controls were genotyped correctly. The genotyping success rate was 100%.
Figure 1.
Identification of ADIPOQ +45T>G polymorphism. (A) PCR amplification fragments. Line 1, molecular weight marker of 50 bp; lines 2–16, samples; line 17, negative control. (B) Bsp HI restriction fragments. Line 1, molecular weight marker of 50 bp; lines 2, 3, 7, 9–11, and 16, TT genotype; lines 4–6 and 12–15, TG genotype; line 8, GG genotype; line 17, negative control.
Abbreviations: PCR, polymerase chain reaction; bp, base pairs.
Statistical analysis
Data were analyzed with the PASW Statistics program (version 18.0; IBM Corporation, Armonk, NY). Results are given as percentages or mean ± standard deviation or standard error of the mean. The distribution of body fat storage measurements, sAdiponectin levels, and profile of inflammation markers were analyzed with the unpaired Student’s t-test, one-way ANOVA, analysis of covariance, the multifactorial ANOVA model corrected by sex among carriers of the three genotypes, and the additive model with the post hoc Bonferroni test.
Data from serum concentrations of sAdiponectin, inflammation markers, and distribution of body fat storage variables were subjected to Pearson correlation tests. After analysis we performed a multiple linear regression analysis with the positive correlation variables. The test for Hardy– Weinberg equilibrium for ADIPOQ loci was performed. Contingency tables with χ2 trend test or Fisher’s exact test, as appropriate, were used for testing the differences of genotype distribution and allele frequencies between all groups. Two models were used for these analyses: (1) the dominant model where the SNP was modeled categorically and separated into three categories, one for each genotype, and (2) the additive model where the SNP was modeled into two categories with two genotypes combined into one category (TG + GG) choosing one genotype (TT) as the reference group for both models. A P-value less than 0.05 was considered statistically significant.
Results
In our study, we found an overweight and obesity prevalence of 40.4% and 23.8%, respectively. Table 1 shows data of anthropometrics measurements in our study group. The main anthropometric characteristics in females were higher percentage of body fat mass and skinfold thicknesses and lower total muscle mass and WHR than males.
Table 1.
Demographics and distribution of body fat mass in study group according to WHO BMI categories
| Measurements | Males | Females | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Lean | Overweight | Obesity | Lean | Overweight | Obesity | |
| n | 28 | 41 | 15 | 60 | 56 | 42 |
| Age (years) | 33.8 ± 8.7 | 37.1 ± 14.3 | 40.4 ± 11.4 | 36 ± 10.9 | 43 ± 13.7 | 42 ± 12.8 |
| Height (cm)* | 172.9 ± 5.5 | 171.9 ± 6.8 | 174.8 ± 6.6 | 161 ± 7.1 | 158.1 ± 6.0 | 159.1 ± 7.9 |
| Weight (kg)* | 66.7 ± 6.9 | 80.5 ± 7.7 | 101.3 ± 11.5 | 58.6 ± 8.4 | 68.4 ± 7.3 | 87.4 ± 12.9 |
| BMI (kg/m2) | 22.3 ± 1.8 | 27.2 ± 1.4 | 33.0 ± 2.7 | 22.3 ± 1.6 | 27.3 ± 1.6 | 34.4 ± 4.0 |
| Body fat mass (%)* | 13.7 ± 4.2 | 25.4 ± 3.6 | 32.0 ± 4.2 | 26.5 ± 5.3 | 36.4 ± 4.4 | 43.1 ± 3.4 |
| Total body fat mass (kg)* | 9.3 ± 4.5 | 20.0 ± 4.4 | 33.4 ± 7.8 | 15.9 ± 5.9 | 25.0 ± 5.7 | 38.0 ± 9.0 |
| Total muscle mass (kg)* | 55.5 ± 6.6 | 59.2 ± 6.0 | 68.3 ± 6.5 | 42.7 ± 2.9 | 43.2 ± 3.8 | 50.2 ± 5.4 |
| Body fat ratio | 2.2 ± 0.72 | 2.0 ± 0.67 | 2.1 ± 0.33 | 2.28 ± 0.50 | 2.57 ± 0.47 | 2.81 ± 0.60 |
| Waist circumference (cm)* | 79.4 ± 6.7 | 95.5 ± 8.6 | 110.5 ± 10.6 | 74.8 ± 8.1 | 90.9 ± 7.2 | 104.9 ± 10.4 |
| Hip circumference (cm) | 93.9 ± 7.3 | 100.5 ± 5.6 | 110.4 ± 8.8 | 97.2 ± 6.8 | 104.2 ± 6.0 | 119.2 ± 10.0 |
| Waist/hip ratio* | 0.84 ± 0.09 | 0.95 ± 0.07 | 1.01 ± 0.09 | 0.77 ± 0.04 | 0.87 ± 0.06 | 0.88 ± 0.09 |
| Arm circumference (cm) | 28.4 ± 2.9 | 32.0 ± 3.3 | 36.9 ± 2.6 | 27.7 ± 3.3 | 31.8 ± 2.2 | 36.1 ± 3.4 |
| Skinfold thickness (mm)* | ||||||
| Biceps | 5.6 ± 3.2 | 9.6 ± 5.2 | 16.4 ± 7.4 | 11.7 ± 5.0 | 17.6 ± 5.4 | 23.3 ± 8.3 |
| Triceps | 11.9 ± 5.7 | 16.5 ± 6.1 | 22.0 ± 7.9 | 20.2 ± 5.1 | 27.3 ± 5.3 | 33.3 ± 8.7 |
| Subscapular | 13.8 ± 6.3 | 25.1 ± 7.3 | 37.3 ± 11.1 | 18.7 ± 7.8 | 30.7 ± 7.2 | 38.7 ± 8.7 |
| Suprailiac | 15.6 ± 8.7 | 26.5 ± 8.4 | 35.0 ± 9.7 | 22.6 ± 6.8 | 30.0 ± 6.2 | 34.8 ± 7.9 |
| Sum of four skinfold thicknesses | 47.1 ± 21.9 | 77.8 ± 21.8 | 110.8 ± 32.2 | 73.3 ± 21.6 | 105.7 ± 20.3 | 130.2 ± 28.6 |
Notes: n = 242: males n = 84, females n = 158. Data are represented as mean ± SD.
Data in bold font were significant with Student’s t-test (P < 0.01) for males versus females, classified by BMI according to WHO categories. Arm circumference and body fat ratio showed no significant differences. Sum of four skinfold thicknesses: biceps, triceps, subscapular, and suprailiac.
Abbreviations: BMI, body mass index; WHO, World Health Organization; SD, standard deviation.
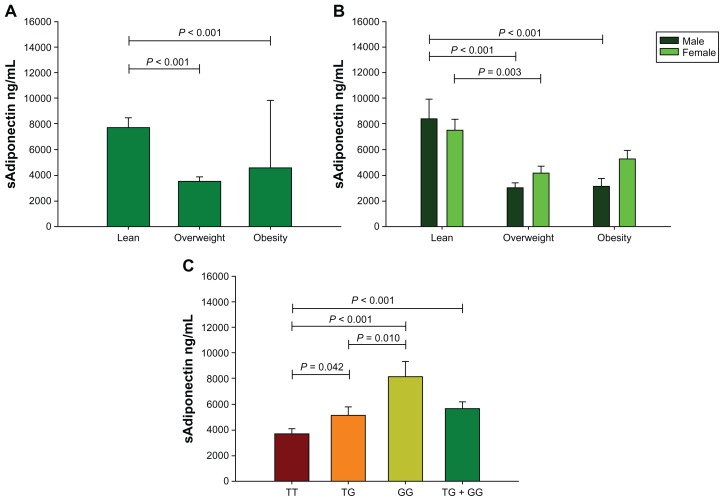
Intergroup comparison according to BMI categories (lean versus overweight versus obesity) displayed higher levels of CRP and erythrocyte sedimentation rate in female than male individuals (Table 2). While sAdiponectin levels were higher in the lean group than the overweight and obesity groups (Figure 2A), this difference was also represented in the males (Figure 2B). A negative correlation was found between sAdiponectin with inflammation markers and distribution of body fat storage. We performed a multiple linear regression analysis and found that 10.6% of the variation of sAdiponectin is explained by fat percentage, age, and sex (P = 0.001). Other correlations of inflammation markers with adiposity are shown in Table 3.
Table 2.
Inflammation markers in study group according to WHO BMI categories
| Inflammation markers | Males | Females | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Lean | Overweight | Obesity | P | Lean | Overweight | Obesity | P | |
| n | 28 | 41 | 15 | – | 60 | 56 | 42 | – |
| BMI (kg/m2) | 22.3 ± 1.8 | 27.2 ± 1.4 | 33.0 ± 2.7 | – | 22.3 ± 1.6 | 27.3 ± 1.6 | 34.4 ± 4.0 | – |
| CRP (mg/L) | 0.65 ± 0.28 | 1.93 ± 0.30 | 3.44 ± 0.62 | 0.007c | 1.04 ± 0.25 | 3.7 ± 0.47 | 4.48 ± 0.47 | <0.001b |
| ESR (mm/h) | 3.8 ± 0.7 | 7.5 ± 1.1 | 10.1 ±1.5 | 0.006b | 15.9 ± 8.99 | 21.7 ± 9.10 | 24.2 ± 9.05 | <0.001b |
| PLT (κ/μL) | 207 ± 47 | 255 ± 46 | 248 ± 87 | 0.004a | 252 ± 64 | 280 ± 72 | 265 ± 63 | 0.080 |
| WBC (κ/μL) | 5.57 ± 1.9 | 6.4 ± 1.2 | 6.8 ± 1.9 | 0.050 | 5.7 ± 1.3 | 6.1 ± 1.2 | 6.4 ± 1.6 | 0.030c |
Notes: n = 242; males n = 84, females n = 158. Data are represented as means ± SD. One-way ANOVA and post hoc Bonferroni tests:
lean versus overweight;
lean versus overweight and obesity;
lean versus obesity.
Abbreviations: CRP, C reactive protein; ESR, erythrocyte sedimentation rate; PLT, platelet count; WBC, white blood cell count; BMI, body mass index.
Figure 2.
Soluble levels of sAdiponectin. Study group (A); sex-specific study group (B). Group comparisons between genotypes of ADIPOQ +45T>G polymorphism (C), which included: (1) the dominant model modeled into three categories, with TT genotype chosen as the reference group; and (2) the additive model modeled into two categories with TT genotype as the reference group and the other two genotypes combined into one category (TG + GG).
Note: Data presented as mean ± SEM; n = 242.
Abbreviations: sAdiponectin, serum adiponectin; SEM, standard error of the mean.
Table 3.
sAdiponectin correlations with inflammation markers and distribution of body fat mass in study group
| Measurements | sAdiponectin | CRP | ESR | PLT | WBC |
|---|---|---|---|---|---|
|
|
|||||
| Pearson correlation coefficient (r) (P) | |||||
| CRP (mg/L) | −0.229 (0.006) | – | – | – | – |
| ESR (mm/h) | −0.267 (0.003) | 0.366 (0.000) | – | – | – |
| PLT (κ/μL) | −0.287 (0.032) | 0.196 (0.012) | 0.246 (0.000) | – | – |
| WBC (κ/μL) | −0.134 (0.102) | 0.174 (0.026) | 0.009 (0.891) | 0.151 (0.020) | – |
| Weight (kg) | −0.196 (0.016) | 0.236 (0.002) | 0.243 (0.025) | 0.251 (0.025) | 0.273 (0.000) |
| BMI (kg/m2) | −0.390 (0.002) | 0.379 (0.000) | 0.281 (0.000) | 0.230 (0.000) | 0.251 (0.000) |
| Body fat mass (%) | −0.444 (0.001) | 0.372 (0.000) | 0.536 (0.000) | 0.298 (0.000) | 0.150 (0.021) |
| Total body fat mass (kg) | −0.430 (0.002) | 0.220 (0.029) | 0.178 (0.018) | 0.170 (0.024) | 0.212 (0.003) |
| Body fat ratio | 0.085 (0.550) | 0.226 (0.037) | 0.053 (0.366) | 0.151 (0.047) | −0.053 (0.453) |
| Waist circumference (cm) | −0.384 (0.005) | 0.344 (0.000) | 0.132 (0.014) | 0.151 (0.005) | 0.286 (0.000) |
| Hip circumference (cm) | −0.316 (0.023) | 0.355 (0.000) | 0.300 (0.000) | 0.169 (0.001) | 0.173 (0.014) |
| Waist/hip ratio | −0.169 (0.041) | 0.158 (0.017) | 0.316 (0.000) | 0.052 (0.309) | 0.234 (0.000) |
| Arm circumference (cm) | −0.401 (0.003) | 0.451 (0.000) | 0.224 (0.000) | 0.080 (0.190) | 0.233 (0.002) |
| Skinfold thickness (mm) Biceps | −0.401 (0.003) | 0.525 (0.000) | 0.439 (0.000) | 0.164 (0.007) | 0.163 (0.021) |
| Triceps | −0.402 (0.003) | 0.489 (0.000) | 0.486 (0.000) | 0.275 (0.004) | 0.134 (0.058) |
| Subscapular | −0.194 (0.025) | 0.564 (0.000) | 0.389 (0.000) | 0.177 (0.004) | 0.277 (0.000) |
| Suprailiac | −0.191 (0.028) | 0.442 (0.000) | 0.313 (0.000) | 0.131 (0.031) | 0.267 (0.000) |
| Sum of four skinfold thicknesses | −0.465 (0.001) | 0.214 (0.037) | 0.116 (0.124) | 0.098 (0.196) | 0.240 (0.001) |
Notes: n = 242. Pearson’s correlation coefficient: data in bold font were significant at P < 0.05; sum of four skinfold thicknesses: biceps, triceps, subscapular, and suprailiac. After multiple linear regression analysis it was found that 10.6% of the variation of sAdiponectin is explained by fat percentage, age, and sex (P = 0.001).
Abbreviations: sAdiponectin, serum adiponectin; CRP, C reactive protein; ESR, erythrocyte sedimentation rate; PLT, platelet count; WBC, white blood cell count; BMI, body mass index.
In a different analysis of ADIPOQ +45T>G SNP we found that the independent segregation of the alleles tested in this Mexican-Mestizo group kept with the Hardy–Weinberg equilibrium (P = 0.377). The association of the genotypes and alleles with overweight and obesity was analyzed: no significant association was found between lean, overweight, obesity, or BMI > 25.0 kg/m2 individuals (Table 4).
Table 4.
Distribution of ADIPOQ +45T>G gene polymorphism in Mexican-Mestizo population
| Study group | Genotype n (%) |
Hardy–Weinberg equilibrium | Allele n (%) |
||||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| TT | TG | GG | TG + GG | P | T | G | |
| Lean n = 88 |
54 (61.4) | 28 (31.8) | 6 (6.8) | 34 (38.6) | 0.368 | 136 (77.3) | 40 (22.7) |
| Overweight n = 97 |
64 (66.0) | 28 (28.9) | 5 (5.1) | 33 (34.0) | 0.515 | 156 (80.4) | 38 (19.6) |
| Obese n = 57 |
37 (64.9) | 17 (29.8) | 3 (5.3) | 20 (35.9) | 0.677 | 91 (79.8) | 23 (20.2) |
| BMI > 25 kg/m2 individuals n = 154 |
101 (65.6) | 45 (29.2) | 8 (5.2) | 53 (34.4) | – | 247 (80.2) | 61 (19.8) |
Notes: n = 242 total group. Genotypes and alleles intergroup comparison in ADIPOQ +45T>G polymorphism yielded a nonsignificant difference (χ2 trend test or Fisher’s exact test).
Abbreviation: BMI, body mass index.
We detected different sAdiponectin levels in genotype carriers as follows: GG (8169 ± 1162 ng/mL), TG (5189 ± 501 ng/mL), and TT (3741 ± 323 ng/mL) (Figure 2C). In TG genotype carriers we detected lower measures of hip circumference and biceps skinfold thickness than TT genotype carriers. However, in the additive model the TG + GG genotype carriers showed lower measures of total body fat mass, hip circumference, biceps and triceps skinfold thicknesses, and sum of four skinfold thicknesses than the TT genotype carriers (Table 5). Additionally, the females carrying the G allele (additive model) with an excess of body weight (BMI > 25.0 kg/m2) had low measures of total body fat mass, hip circumference, arm circumference, and skin fold thickness and a high WHR as compared to TT genotype carriers with similar BMI. In lean female individuals carrying the G allele we observed a lower BMI and body fat ratio than TT genotype carriers (Table 6).
Table 5.
Comparison of ADIPOQ +45T>G gene polymorphism with distribution and storage of body fat
| Measurements | ADIPOQ +45T>G genotypes | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| TT | TG | GG | P | TG + GG | Pa | |
| Total body fat mass (kg) | 24.2 ± 11.5 | 20.6 ± 10.1 | 22.0 ± 7.9 | NS | 22.6 ± 9.6 | <0.001 |
| Hip circumference (cm) | 105.5 ± 11.1 | 100.5 ± 10.3 | 102.7 ± 8.5 | 0.006b | 100.9 ± 10.0 | <0.001 |
| Skinfold thickness (mm) | ||||||
| Biceps | 15.3 ± 8.5 | 12.9 ± 7.2 | 11.9 ± 6.5 | 0.047b | 12.8 ± 7.1 | <0.001 |
| Triceps | 23.5 ± 9.6 | 21.2 ± 8.8 | 21.2 ± 7.4 | NS | 21.3 ± 8.6 | <0.001 |
| Sum of four skinfold thicknesses | 94.6 ± 36.2 | 85.5 ± 32.1 | 84.4 ± 30.0 | NS | 85.3 ± 31.8 | <0.001 |
Notes: n = 242. Data are represented as means ± SD. Genotype group comparisons were performed by multifactorial ANOVA corrected model with Bonferroni post hoc test. Group comparisons: (1) the dominant model was modeled into three categories, with TT genotype chosen as the reference group; and (2) the additive model was modeled into two categories with TT genotype as the reference group and the other two genotypes combined into one category (TG + GG).
TT versus TG + GG;
TT versus TG. Data in bold font were significant at P < 0.05.
Abbreviations: SD, standard deviation; ANOVA, analysis of variance.
Table 6.
Comparisons of the distribution of body fat mass between females ADIPOQ +45T>G genotype carriers
| Measurements | Female group | ADIPOQ +45T>G genotypes | ||
|---|---|---|---|---|
|
|
||||
| TT | TG + GG | P | ||
| BMI (kg/m2) | Lean | 22.7 ± 1.4 | 21.7 ± 1.7 | 0.026 |
| BMI > 25 kg/m2 individuals | 30.5 ± 4.6 | 29.9 ± 4.3 | 0.574 | |
| Body fat ratio | Lean | 2.4 ± 0.52 | 2.1 ± 0.43 | 0.037 |
| BMI > 25 kg/m2 individuals | 2.6 ± 0.53 | 2.6 ± 0.55 | 0.772 | |
| Total body fat mass (kg) | Lean | 16.8 ± 6.1 | 14.7 ± 5.6 | 0.231 |
| BMI > 25 kg/m2 individuals | 32.0 ± 10.0 | 27.3 ± 8.2 | 0.024 | |
| Hip circumference (cm) | Lean | 98.5 ± 7.1 | 95.5 ± 6.2 | 0.158 |
| BMI > 25 kg/m2 individuals | 112.3 ± 10.9 | 106.6 ± 9.6 | 0.018 | |
| Waist/hip ratio | Lean | 0.77 ± 0.053 | 0.76 ± 0.037 | 0.587 |
| BMI > 25 kg/m2 individuals | 0.86 ± 0.065 | 0.90 ± 0.089 | 0.006 | |
| Arm circumference (cm) | Lean | 27.8 ± 3.3 | 27.6 ± 3.4 | 0.812 |
| BMI > 25 kg/m2 individuals | 34.1 ± 3.1 | 32.4 ± 3.8 | 0.024 | |
| Skinfold thickness (mm) | ||||
| Biceps | Lean | 12.3 ± 5.7 | 10.8 ± 3.6 | 0.310 |
| BMI > 25 kg/m2 individuals | 21.4 ± 7.1 | 17.3 ± 6.9 | 0.013 | |
| Triceps | Lean | 20.4 ± 5.7 | 19.8 ± 4.2 | 0.665 |
| BMI > 25 kg/m2 individuals | 31.4 ± 6.4 | 26.9 ± 8.3 | 0.007 | |
| Suprailiac | Lean | 22.0 ± 8.1 | 23.5 ± 4.7 | 0.441 |
| BMI > 25 kg/m2 individuals | 33.7 ± 6.9 | 28.83 ± 7.1 | 0.003 | |
| Sum of four skinfold thicknesses | Lean | 74.3 ± 26.8 | 71.98 ± 11.9 | 0.719 |
| BMI > 25 kg/m2 individuals | 121.7 ± 24.4 | 105.06 ± 27.8 | 0.005 | |
Notes: n = 158. Data are represented as means ± SD. TT versus TG + GG, estimated by analysis of covariance using genotype as a factor and age as covariate. Additive model was modeled into two categories with one genotype (TT) chosen as the reference group, and the other two genotypes combined into one category (TG + GG). Sum of four skinfold thickness: biceps, triceps, and suprailiac. Data in bold font were significant at P < 0.05.
Abbreviations: BMI, body mass index; SD, standard deviation.
Finally, we independently analyzed the female obesity group (BMI > 30 kg/m2), finding the following differences: (1) body fat mass (P = 0.009) in carriers of TT genotype (43.8% ± 3.09%) was lower as compared to GG genotype carriers (37.5% ± 1.65%), and (2) WHR (P = 0.005) in carriers of TT genotype (0.8625 ± 0.07) was lower as compared to carriers of TG genotype (0.9711 ± 0.11).
Discussion
We investigated the distribution of ADIPOQ +45T>G polymorphism in a Mexican-Mestizo population from Western Mexico. This study evaluated the probable association of ADIPOQ genetic polymorphism with blood levels of adiponectin, inflammation markers, and the distribution of body fat storage in obesity. We found a high prevalence of overweight (40.4%) and obesity (23.8%) in our study group, classified by the BMI according to WHO categories. These findings are in accordance with those reported by the National Survey of Health and Nutrition 2006 for Western Mexico. Obesity is considered a public health problem in Mexico, worsened in the early 1990s. The National Institute of Public Health reported a 47% increase in the prevalence of obesity in adults (older than 20 years) from 1994 through 2006. This increased tendency is not exclusive to Mexico, mainly because obesity is considered a pandemic disease.5,28
The difference between overweight and obese individuals for the anthropometric measurements and body composition, with the exception of the BMI and arm circumference, suggests the following: (1) physiologic factors are involved in obesity development, and (2) there does not exist an equal distribution of body fat storage during the transition from overweight to obesity.
In Mexico, the National Department of Health established a cut-off value of the waist circumference measurement (80.0 cm for women and 90.0 cm for men) as a clinical criterion to estimate the risk of comorbidities in obesity; in this sense, all individuals with obesity in our study group had abdominal obesity, a state in which IL-6 production increases. This finding is important because the increase of IL-6 is a key inductor of the hepatic synthesis of CRP.10,29 Our CRP data coincides with previous reports where it was established that obesity is characterized by a systemic low-grade subacute inflammatory state,7 also considered the clinical interphase between obesity and the appearance of multiple illnesses.
The present study shows that when sAdiponectin levels have decreased the BMI and body fat storage increase. Our data are consistent with previous findings: low sAdiponectin levels have been observed in overweight, obesity, and comorbidities.12,30
Low levels of sAdiponectin reflected the adiposity in our groups of study; this finding is underlined because we identified a negative correlation between sAdiponectin with body fat storage and inflammation markers. These findings therefore suggest that the relationship between low sAdiponectin concentrations and obesity may be in part mediated by the accumulation of adipose tissue. The possible explanation is that sAdiponectin is an exclusive product of the adipose tissue.
On the other hand, controversial results have been reported for ADIPOQ gene polymorphism in different populations. ADIPOQ +45T>G polymorphism has been associated with obesity, insulin resistance, metabolic syndrome, T2D, and variation in BMI and sAdiponectin in Chinese,31,32 Korean,33,34 Japanese,27,35 Canadian,25 Spanish,36 Iranian,37 Finnish,38 and Greek39,40 populations, while in Taiwanese,41 French,42 Swedish Caucasian,43 Czech,44 Malaysian,45 Pima Indian,14 and Italian46,47 populations, there has been no association.45
Significant associations between obesity and polymorphisms in the gene coding for ADIPOQ have been reported in other studies (Table 7). The genetic contribution to susceptibility of obesity is well accepted, although, in our study we did not find an association of the ADIPOQ +45T>G gene polymorphism with obesity, but we found in the additive model that the TT genotype carriers had a higher total body fat mass and biceps and triceps skinfold thickness than the TG + GG genotype carriers which also had higher sAdiponectin levels. The distribution of body fat storage can play different roles in adiponectin production and/or function during the maintenance of the inflammatory process in obesity. As a result of this association of the polymorphic allele with distribution of body fat storage, we suggest that genetic variability in the ADIPOQ gene may modulate the levels of sAdiponectin. However, a potential limitation of our study is the possibility that other variants in other ADIPOQ loci than in those we studied may play a role in obesity. Nevertheless, we chose +45T>G polymorphism that has been implicated functionally and associated with obesity comorbidities in previous studies. A limitation regarding the association analysis of our study is that our sample of only 242 individuals may reduce the ability to detect associations.
Table 7.
Comparison of the ADIPOQ +45T>G gene polymorphism distribution in Mexican-Mestizo population with other studies
| Study | Population | Sample size | Genotype distributions | PHWE | Pχ2 | Main findings | ||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| TT | TG | GG | ||||||
| This study | Mexican- Mestizos | 242 | 155 | 73 | 14 | 0.377 | – | +45G allele may increase the levels of sAdiponectin. |
| Katsuda et al26 | Obese Japanese | 64 | 34 | 25 | 5 | 0.236a | 0.278 | +45G allele could be a risk factor of metabolic syndrome and the development of atherosclerosis. |
| Jang et al33 | Koreans | 333 | 156 | 146 | 31 | 0.703 | <0.000 | +45G allele did not associate with IR and CVD risk than SNP in nonobese, nondiabetic Korean men. |
| Gonzalez- Sanchez et al36 | Spanish | 862 | 554 | 282 | 26 | 0.179 | 0.112 | SNP + 45T>G, further increasing the risk of impaired glucose tolerance and of low circulating sAdiponectin concentrations. |
| Melistas et al39 | Greek white women | 343 | 249 | 87 | 7 | 0.850 | 0.016 | ADIPOQ variants at positions +45 and +276 in the development of IR in healthy Greek women. |
| Filippi et al47 | Italians | 250 | 189 | 52 | 9 | 0.036a | 0.020 | SNP + 45T>G did not influence circulating levels of sAdiponectin. |
| Li et al30 | Chinese Uygur | 94 | 75 | 16 | 3 | 0.105a | 0.024 | The +45G allele carriers who have reduced plasma concentrations of sAdiponectin may have associated IR. |
| Park et al32 | Koreans | 986 | 533 | 372 | 81 | 0.240 | 0.018 | Suggest the contribution of the ADIPOQ gene toward susceptibility to obesity in healthy Koreans. |
| Vendramini et al34 | Japanese Brazilians | 200 | 100 | 85 | 15 | 0.597a | 0.012 | We identified in the ADIPOQ gene a risk haplotype for type 2 diabetes in the Japanese-Brazilian population. |
| Vasseur et al42 | French Caucasians | 680 | 500 | 156 | 24 | 0.001a | 0.017 | Our results suggest that an at-risk haplotype of common variants located in the promoter and rare mutations in exon 3 contribute to the variation of the adipocyte-secreted sAdiponectin hormone level, and may be part of the genetic determinants for T2D in the French-Caucasian population. |
| Lau and Muniandy45 | Malaysians | 809 | 501 | 264 | 44 | 0.274 | 0.768 | SNP + 45T>G, did not influence circulating levels of sAdiponectin. |
Notes: All studies are cross-sectional except Vendramini et al, which was a case-control study; χ2 was performed to compare frequencies found with those reported in other studies.
Calculated data not shown in the original papers.
Abbreviations: PHWE, P values of Hardy–Weinberg equilibrium; Pχ2, P values of Chi-square test.
Conclusion
As the end point of this study, we have suggested that the ADIPOQ +45G allele could be associated with the distribution of body fat storage in obesity. On the other hand, as no association was observed between the ADIPOQ +45T>G gene polymorphism and obesity it cannot be concluded if the ADIPOQ +45G allele is responsible for the increase of adiponectin levels.
Acknowledgments
This work was supported by grant number PS-2009-552 to Rosa Elena Navarro of the State Council of Science and Technology (COECyTJal-University of Guadalajara). We thank Michele Brennan, MSc in Clinical Chemistry, for the review of the English language editing.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Blakemore AI, Froguel P. Is obesity our genetic legacy? J Clin Endocrinol Metab. 2008;93(11 Suppl 1):S51–S56. doi: 10.1210/jc.2008-1676. [DOI] [PubMed] [Google Scholar]
- 2.Das UN. Obesity: genes, brain, gut, and environment. Nutrition. 2010;26(5):459–473. doi: 10.1016/j.nut.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 3.Shamah-Levy TV-HS, Rivera-Dommarco JA. Públic INdS, editor. National Health and Nutrition Survey, 2006. Cuernavaca, Mexico: National Institute of Public Health; 2007. Nutrition Results. Spanish. [Google Scholar]
- 4.Olaiz-Fernández G, Rivera-Dommarco J, Shamah-Levy T, et al. National Health and Nutrition Survey, 2006. Cuernavaca, México: National Institute of Public Health; 2006. Spanish. [Google Scholar]
- 5.Dixon JB. The effect of obesity on health outcomes. Mol Cell Endocrinol. 2010;316(2):104–108. doi: 10.1016/j.mce.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Mutch DM, Clément K. Genetics of human obesity. Best Pract Res Clin Endocrinol Metab. 2006;20(4):647–664. doi: 10.1016/j.beem.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Mathis D, Shoelson SE. Immunometabolism: an emerging frontier. Nat Rev Immunol. 2011;11(2):81. doi: 10.1038/nri2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galic S, Oakhill JS, Steinberg GR. Adipose tissue as an endocrine organ. Mol Cell Endocrinol. 2010;316(2):129–139. doi: 10.1016/j.mce.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 9.Poulos SP, Hausman DB, Hausman GJ. The development and endocrine functions of adipose tissue. Mol Cell Endocrinol. 2010;323(1):20–34. doi: 10.1016/j.mce.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Ouchi N, Parker JL, Lugus JJ, Walsh K. Adipokines in inflammation and metabolic disease. Nat Rev Immunol. 2011;11(2):85–97. doi: 10.1038/nri2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerre-Millo M. Adiponectin: an update. Diabetes Metab. 2008;34(1):12–18. doi: 10.1016/j.diabet.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Chang LC, Huang KC, Wu YW, et al. The clinical implications of blood adiponectin in cardiometabolic disorders. J Formos Med Assoc. 2009;108(5):353–366. doi: 10.1016/S0929-6646(09)60079-6. [DOI] [PubMed] [Google Scholar]
- 13.Woo JG, Dolan LM, Deka R, et al. Interactions between noncontiguous haplotypes in the adiponectin gene ACDC are associated with plasma adiponectin. Diabetes. 2006;55(2):523–529. doi: 10.2337/diabetes.55.02.06.db05-0446. [DOI] [PubMed] [Google Scholar]
- 14.Vozarova de Courten B, Hanson RL, Funahashi T, et al. Common polymorphisms in the adiponectin gene acdc are not associated with diabetes in Pima Indians. Diabetes. 2005;54(1):284–289. doi: 10.2337/diabetes.54.1.284. [DOI] [PubMed] [Google Scholar]
- 15.WHO Technical Report Series. Geneva: World Health Organization; 2004. The International Classification of adult underweight, overweight and obesity according to BMI. [Google Scholar]
- 16.Gorodezky C, Alaez C, Vázquez-García MN, et al. The genetic structure of Mexican Mestizos of different locations: tracking back their origins through MHC genes, blood group systems, and microsatellites. Hum Immunol. 2001;62(9):979–991. doi: 10.1016/s0198-8859(01)00296-8. [DOI] [PubMed] [Google Scholar]
- 17.World Medical Association. Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. [Accessed January 8, 2009]. (revised October 2008). Available from: http://www.wma.net/en/30publications/10policies/b3/
- 18.Lukaski HC. Body mass index, bioelectrical impedance, and body composition. Nutrition. 2001;17(1):55–56. doi: 10.1016/s0899-9007(00)00499-8. [DOI] [PubMed] [Google Scholar]
- 19.Lukaski HC. Regional bioelectrical impedance analysis: applications in health and medicine. Acta Diabetol. 2003;40(Suppl 1):S196–S199. doi: 10.1007/s00592-003-0064-4. [DOI] [PubMed] [Google Scholar]
- 20.van der Ploeg GE, Gunn SM, Withers RT, Modra AC. Use of anthropometric variables to predict relative body fat determined by a four-compartment body composition model. Eur J Clin Nutr. 2003;57(8):1009–1016. doi: 10.1038/sj.ejcn.1601636. [DOI] [PubMed] [Google Scholar]
- 21.Durnin JV, Womersley J. Body fat assessed from total body density and its estimation from skinfold thickness: measurements on 481 men and women aged from 16 to 72 years. Br J Nutr. 1974;32(1):77–97. doi: 10.1079/bjn19740060. [DOI] [PubMed] [Google Scholar]
- 22.Ness-Abramof R, Apovian CM. Waist circumference measurement in clinical practice. Nutr Clin Pract. 2008;23(4):397–404. doi: 10.1177/0884533608321700. [DOI] [PubMed] [Google Scholar]
- 23.Durnin JV, Rahaman MM. The assessment of the amount of fat in the human body from measurements of skinfold thickness. Br J Nutr. 1967;21(3):681–689. doi: 10.1079/bjn19670070. [DOI] [PubMed] [Google Scholar]
- 24.Lukaski HC. Methods for the assessment of human body composition: traditional and new. Am J Clin Nutr. 1987;46(4):537–556. doi: 10.1093/ajcn/46.4.537. [DOI] [PubMed] [Google Scholar]
- 25.Loos RJ, Ruchat S, Rankinen T, Tremblay A, Pérusse L, Bouchard C. Adiponectin and adiponectin receptor gene variants in relation to resting metabolic rate, respiratory quotient, and adiposity-related phenotypes in the Quebec Family Study. Am J Clin Nutr. 2007;85(1):26–34. doi: 10.1093/ajcn/85.1.26. [DOI] [PubMed] [Google Scholar]
- 26.Miller DN, Bryant JE, Madsen EL, Ghiorse WC. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microbiol. 1999;65(11):4715–4724. doi: 10.1128/aem.65.11.4715-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katsuda Y, Asano A, Murase Y, et al. Association of genetic variation of the adiponectin gene with body fat distribution and carotid atherosclerosis in Japanese obese subjects. J Atheroscler Thromb. 2007;14(1):19–26. doi: 10.5551/jat.14.19. [DOI] [PubMed] [Google Scholar]
- 28.Rokholm B, Baker JL, Sørensen TI. The levelling off of the obesity epidemic since the year 1999 – a review of evidence and perspectives. Obes Rev. 2010;11(12):835–846. doi: 10.1111/j.1467-789X.2010.00810.x. [DOI] [PubMed] [Google Scholar]
- 29.Hodge AM, Maple-Brown L, Cunningham J, et al. Abdominal obesity and other risk factors largely explain the high CRP in indigenous Australians relative to the general population, but not gender differences: a cross-sectional study. BMC Public Health. 2010;10:700. doi: 10.1186/1471-2458-10-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staiger H, Tschritter O, Machann J, et al. Relationship of serum adiponectin and leptin concentrations with body fat distribution in humans. Obes Res. 2003;11(3):368–372. doi: 10.1038/oby.2003.48. [DOI] [PubMed] [Google Scholar]
- 31.Li LL, Kang XL, Ran XJ, et al. Associations between 45T/G polymorphism of the adiponectin gene and plasma adiponectin levels with type 2 diabetes. Clin Exp Pharmacol Physiol. 2007;34(12):1287–1290. doi: 10.1111/j.1440-1681.2007.04713.x. [DOI] [PubMed] [Google Scholar]
- 32.Tso AW, Sham PC, Wat NM, et al. Polymorphisms of the gene encoding adiponectin and glycaemic outcome of Chinese subjects with impaired glucose tolerance: a 5-year follow-up study. Diabetologia. 2006;49(8):1806–1815. doi: 10.1007/s00125-006-0324-2. [DOI] [PubMed] [Google Scholar]
- 33.Park JW, Park J, Jee SH. ADIPOQ gene variants associated with susceptibility to obesity and low serum adiponectin levels in healthy Koreans. Epidemiol Health. 2011;33:e2011003. doi: 10.4178/epih/e2011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jang Y, Lee JH, Kim OY, et al. The SNP276G > T polymorphism in the adiponectin (ACDC) gene is more strongly associated with insulin resistance and cardiovascular disease risk than SNP45T > G in nonobese/nondiabetic Korean men independent of abdominal adiposity and circulating plasma adiponectin. Metabolism. 2006;55(1):59–66. doi: 10.1016/j.metabol.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Vendramini MF, Pereira AC, Ferreira SR, Kasamatsu TS, Moisés RS Japanese Brazilian Diabetes Study Group. Association of genetic variants in the adiponectin encoding gene (ADIPOQ) with type 2 diabetes in Japanese Brazilians. J Diabetes Complications. 2010;24(2):115–120. doi: 10.1016/j.jdiacomp.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 36.González-Sánchez JL, Martínez-Calatrava MJ, Martinez-Larrad MT, et al. Interaction of the −308G/A promoter polymorphism of the tumor necrosis factor-alpha gene with single-nucleotide polymorphism 45 of the adiponectin gene: effect on serum adiponectin concentrations in a Spanish population. Clin Chem. 2006;52(1):97–103. doi: 10.1373/clinchem.2005.049452. [DOI] [PubMed] [Google Scholar]
- 37.Mohammadzadeh G, Zarghami N. Associations between single-nucleotide polymorphisms of the adiponectin gene, serum adiponectin levels and increased risk of type 2 diabetes mellitus in Iranian obese individuals. Scand J Clin Lab Invest. 2009;69(7):764–771. doi: 10.3109/00365510903137237. [DOI] [PubMed] [Google Scholar]
- 38.Siitonen N, Pulkkinen L, Lindström J, et al. Association of ADIPOQ gene variants with body weight, type 2 diabetes and serum adiponectin concentrations: the Finnish Diabetes Prevention Study. BMC Med Genet. 2011;12:5. doi: 10.1186/1471-2350-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melistas L, Mantzoros CS, Kontogianni M, Antonopoulou S, Ordovas JM, Yiannakouris N. Association of the +45T > G and +276G > T polymorphisms in the adiponectin gene with insulin resistance in nondiabetic Greek women. Eur J Endocrinol. 2009;161(6):845–852. doi: 10.1530/EJE-09-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xita N, Georgiou I, Chatzikyriakidou A, et al. Effect of adiponectin gene polymorphisms on circulating adiponectin and insulin resistance indexes in women with polycystic ovary syndrome. Clin Chem. 2005;51(2):416–423. doi: 10.1373/clinchem.2004.043109. [DOI] [PubMed] [Google Scholar]
- 41.Leu HB, Chung CM, Lin SJ, Jong YS, Pan WH, Chen JW. Adiponectin gene polymorphism is selectively associated with the concomitant presence of metabolic syndrome and essential hypertension. PloS one. 2011;6(5):e19999. doi: 10.1371/journal.pone.0019999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasseur F, Helbecque N, Dina C, et al. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet. 2002;11(21):2607–2614. doi: 10.1093/hmg/11.21.2607. [DOI] [PubMed] [Google Scholar]
- 43.Gu HF, Abulaiti A, Ostenson CG, et al. Single nucleotide polymorphisms in the proximal promoter region of the adiponectin (APM1) gene are associated with type 2 diabetes in Swedish caucasians. Diabetes. 2004;53(Suppl 1):S31–S35. doi: 10.2337/diabetes.53.2007.s31. [DOI] [PubMed] [Google Scholar]
- 44.Bienertova-Vasku J, Bienert P, Hlinomaz O, Vasku A. Common polymorphism +45T/G in adiponectin gene as potential modulator of in-stent restenosis development. Int J Cardiol. 2010;145(2):351. doi: 10.1016/j.ijcard.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 45.Lau CH, Muniandy S. Adiponectin and resistin gene polymorphisms in association with their respective adipokine levels. Ann Hum Genet. 2011;75(3):370–382. doi: 10.1111/j.1469-1809.2010.00635.x. [DOI] [PubMed] [Google Scholar]
- 46.Menzaghi C, Ercolino T, Salvemini L, et al. Multigenic control of serum adiponectin levels: evidence for a role of the APM1 gene and a locus on 14q13. Physiological Genomics. 2004;19(2):170–174. doi: 10.1152/physiolgenomics.00122.2004. [DOI] [PubMed] [Google Scholar]
- 47.Filippi E, Sentinelli F, Trischitta V, et al. Association of the human adiponectin gene and insulin resistance. Eur J Hum Genet. 2004;12(3):199–205. doi: 10.1038/sj.ejhg.5201120. [DOI] [PubMed] [Google Scholar]