Abstract
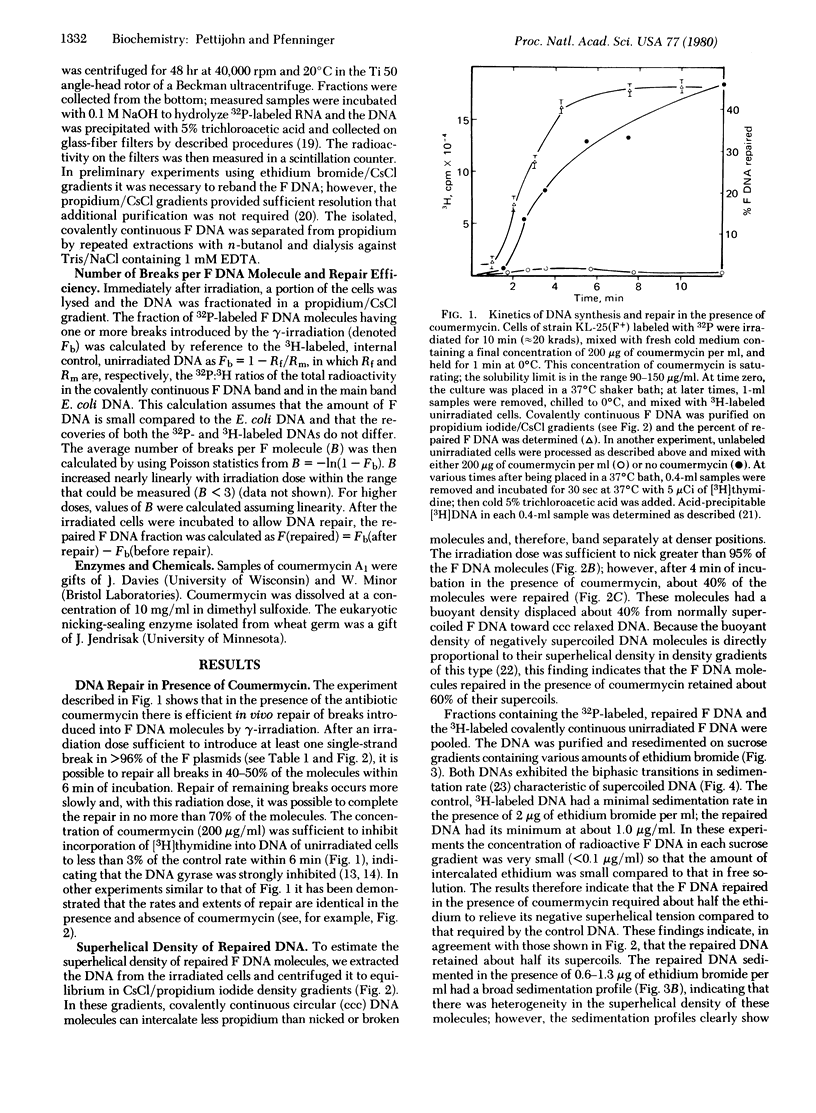
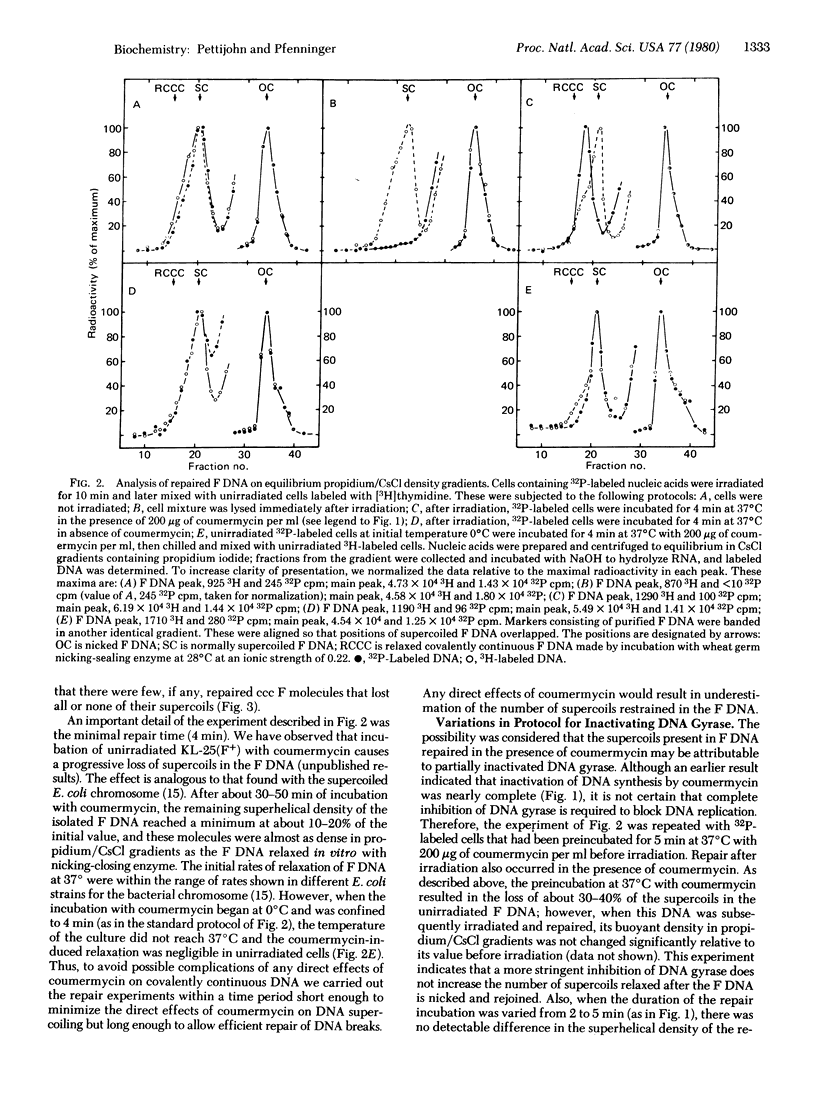
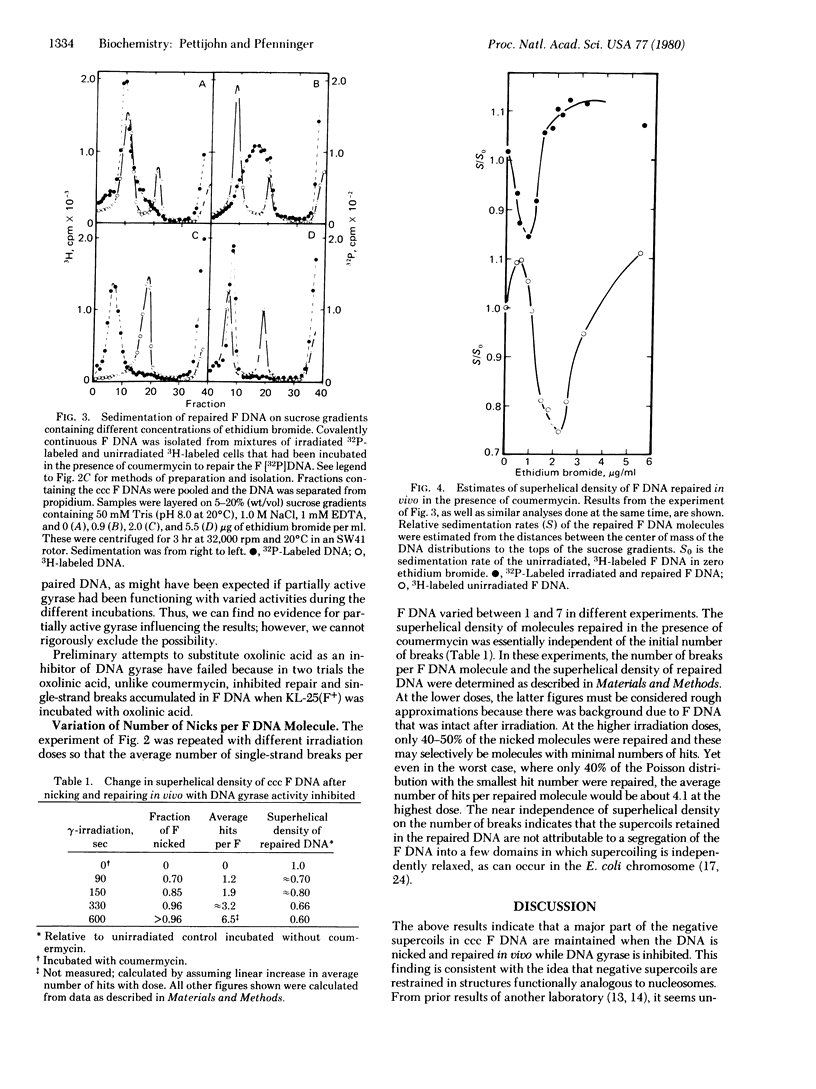
Cells of Escherichia coli containing the plasmid F were gamma-irradiated with various doses to introduce determined numbers of single-strand breaks in the F DNA. The cells were then incubated to permit repair of the breaks while DNA gyrase was inhibited with coumermycin to limit restoration of any relaxed supercoil. Repaired, covalently continuous F DNA was isolated and its superhelical density was measured by two different methods. Both indicated that a major part (50-60%) of the negative superhelical turns were maintained in the repaired molecules, suggesting that the supercoils are restrained in vivo.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bram S., Kouprach S., Baudy P. DNA structure in chromatin and in solution studied by electron microscopy and neutron and X-ray scattering. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):23–29. doi: 10.1101/sqb.1978.042.01.005. [DOI] [PubMed] [Google Scholar]
- Crawford L. V., Waring M. J. Supercoiling of polyoma virus DNA measured by its interaction with ethidium bromide. J Mol Biol. 1967 Apr 14;25(1):23–30. doi: 10.1016/0022-2836(67)90276-8. [DOI] [PubMed] [Google Scholar]
- Drlica K., Snyder M. Superhelical Escherichia coli DNA: relaxation by coumermycin. J Mol Biol. 1978 Apr 5;120(2):145–154. doi: 10.1016/0022-2836(78)90061-x. [DOI] [PubMed] [Google Scholar]
- Eickbush T. H., Moudrianakis E. N. The compaction of DNA helices into either continuous supercoils or folded-fiber rods and toroids. Cell. 1978 Feb;13(2):295–306. doi: 10.1016/0092-8674(78)90198-8. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J. I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Itoh T., Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorno R., Stamato T., Lydersen B., Pettijohn D. Transcription in vitro of DNA in isolated bacterial nucleoids. J Mol Biol. 1975 Aug 5;96(2):217–237. doi: 10.1016/0022-2836(75)90344-7. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Upholt W. B., Vinograd J. A buoyant method for the determination of the superhelix density of closed circular DNA. J Mol Biol. 1971 Nov 28;62(1):1–19. doi: 10.1016/0022-2836(71)90127-6. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Visualization of prokaryotic DNA in a regularly condensed chromatin-like fiber. Proc Natl Acad Sci U S A. 1976 Feb;73(2):563–567. doi: 10.1073/pnas.73.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Hayashi M. Template activities of the phi X-174 replicative allomorphic deoxyribonucleic acids. Biochemistry. 1971 Nov;10(23):4212–4218. doi: 10.1021/bi00799a009. [DOI] [PubMed] [Google Scholar]
- Hays J. B., Boehmer S. Antagonists of DNA gyrase inhibit repair and recombination of UV-irradiated phage lambda. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4125–4129. doi: 10.1073/pnas.75.9.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht R. M., Pettijohn D. E. Studies of DNA bound RNA molecules isolated from nucleoids of Escherichia coli. Nucleic Acids Res. 1976 Mar;3(3):767–788. doi: 10.1093/nar/3.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloman W. K., Radding C. M. Recombination promoted by superhelical DNA and the recA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3910–3914. doi: 10.1073/pnas.73.11.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson B., Upholt W. B., Devinny J., Vinograd J. The use of an ethidium analogue in the dye-buoyant density procedure for the isolation of closed circular DNA: the variation of the superhelix density of mitochondrial DNA. Proc Natl Acad Sci U S A. 1969 Mar;62(3):813–820. doi: 10.1073/pnas.62.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. On the degree of unwinding of the DNA helix by ethidium. II. Studies by electron microscopy. Biochim Biophys Acta. 1975 Jul 23;395(4):401–412. [PubMed] [Google Scholar]
- Lydersen B. K., Pettijohn D. E. Interactions stabilizing DNA tertiary structure in the Escherichia coli chromosome investigated with ionizing radiation. Chromosoma. 1977 Jul 8;62(3):199–215. doi: 10.1007/BF00286044. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Gellert M., Nash H. A. Involement of supertwisted DNA in integrative recombination of bacteriophage lambda. J Mol Biol. 1978 May 25;121(3):375–392. doi: 10.1016/0022-2836(78)90370-4. [DOI] [PubMed] [Google Scholar]
- Rouvière-Yaniv J., Gros F. Characterization of a novel, low-molecular-weight DNA-binding protein from Escherichia coli. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3428–3432. doi: 10.1073/pnas.72.9.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvière-Yaniv J. Localization of the HU protein on the Escherichia coli nucleoid. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):439–447. doi: 10.1101/sqb.1978.042.01.047. [DOI] [PubMed] [Google Scholar]
- Rouvière-Yaniv J., Yaniv M., Germond J. E. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979 Jun;17(2):265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Kubo M., Imamoto F. Promoter-specific inhibition of transcription by antibiotics which act on DNA gyrase. Nature. 1978 Oct 5;275(5679):420–423. doi: 10.1038/275420a0. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Nedospasov S. A., Georgiev G. P. On the structure of eukaryotic, prokaryotic, and viral chromatin. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):457–473. doi: 10.1101/sqb.1978.042.01.049. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Degree of superhelicity of covalently closed cyclic DNA's from Escherichia coli. J Mol Biol. 1969 Jul 28;43(2):263–272. doi: 10.1016/0022-2836(69)90266-6. [DOI] [PubMed] [Google Scholar]
- Womble D. D., Taylor D. P., Rownd R. H. Method for obtaining more-accurate covalently closed circular plasmid-to-chromosome ratios from bacterial lysates by dye-buoyant density centrifugation. J Bacteriol. 1977 Apr;130(1):148–153. doi: 10.1128/jb.130.1.148-153.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]
- Worcel A., Burgi E. Properties of a membrane-attached form of the folded chromosome of Escherichia coli. J Mol Biol. 1974 Jan 5;82(1):91–105. doi: 10.1016/0022-2836(74)90576-2. [DOI] [PubMed] [Google Scholar]


