Abstract
Protein microarrays provide an efficient method to immunoprofile patients in an effort to rapidly identify disease immunosignatures. The validity of using autoantibodies in diagnosis has been demonstrated in type 1 diabetes, rheumatoid arthritis, and systemic lupus, and is now being strongly considered in cancer. Several types of protein microarrays exist including antibody and antigen arrays. In this chapter, we describe the immunoprofiling application for one type of antigen array called NAPPA (nucleic acids programmable protein array). We provide a guideline for setting up the screening study and designing protein arrays to maximize the likelihood of obtaining quality data.
Keywords: NAPPA protein microarray, Immunoprofile, Immunosignature, Autoantibody, Breast cancer, Diabetes, Autoimmune, Proteomics, Serum screening, Antigen
1. Introduction
Protein microarrays are powerful in their ability to test hundreds to thousands of proteins simultaneously and in parallel in a miniaturized format. Most protein microarrays fall grossly into two categories: antibody and antigen arrays. Antibody arrays, in which numerous antibodies (or other affinity reagents) are printed on a slide, were first developed by Haab et al. (1) and later utilized by Sreekumar et al. (2) to demonstrate feasibility in detecting cancer antigens in a complex cell lysate. Antigen arrays involve the display of proteins on the microarray and can be used in the identification of serological autoantibodies present in patients, but not controls. Their feasibility was first demonstrated by Joos et al. (3) for the Ro and La autoantigens for Sjogren’s syndrome.
Recently, several developments in the use of antigen microarrays in immunoprofiling of patients to identify disease signatures were reported (4–10). One approach used in cancer studies is to identify autoantibodies targeting self-proteins present in cancer patients but not controls (11–21). Antigen microarrays are ideal for this purpose as they provide a set of target antigens to which the autoantibodies can bind. Traditionally, antigen arrays were made by individually purifying proteins and printing them on the microarray, a long, tedious and expensive endeavor. NAPPA (nucleic acids programmable protein array) microarrays offer a platform in which proteins are made from printed cDNA-containing plasmids to produce a just-in-time, fresh protein microarray that is incubated with human serum to detect autoantibody binding (Fig. 1) (22–24). A secondary labeled antihuman antibody is added to visualize the autoantibodies bound to autoantigens.
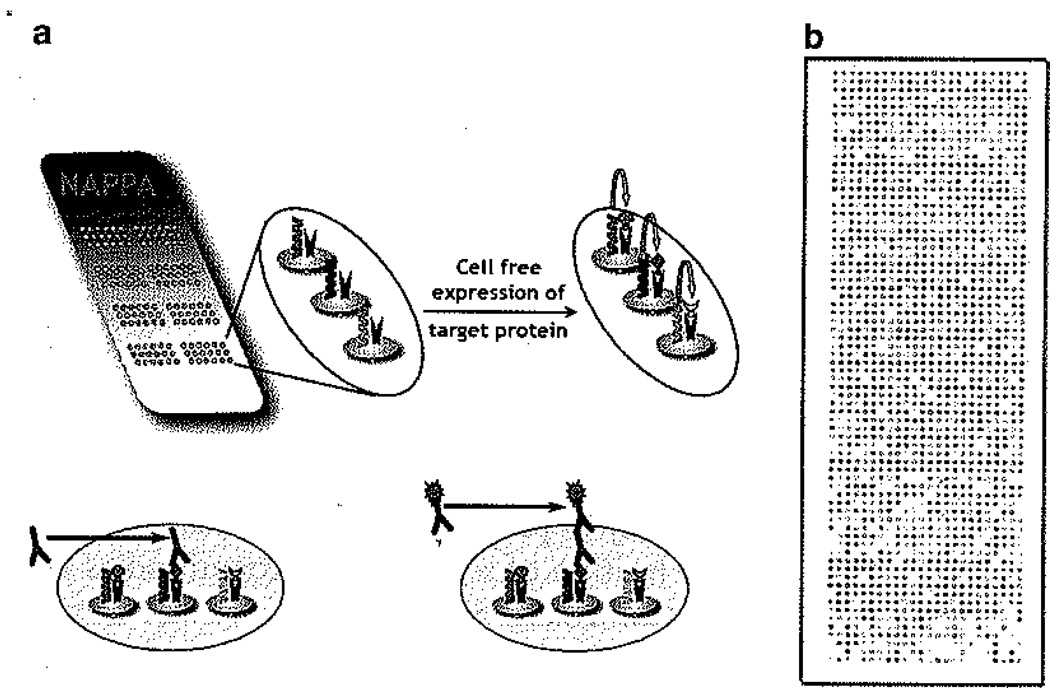
Fig. 1.
Schematic representation of a NAPPA protein microarray. (a) NAPPA microarrays are made by printing cDNA-harboring plasmids on glass slides that are later transcribed and translated in situ to create a fresh protein microarray (top panel]. When human serum is incubated with the microarray, autoantibodies bind their target antigens and are identified using a labeled secondary antibody (lower panel), (b) An image of an expressed NAPPA microarray. For each experiment, it is highly recommended that the expression efficiency of the microarray is assessed at the onset.
NAPPA microarrays have been successfully used in the detection of autoantibodies to p53 in breast and ovarian cancer, ML-IAP in melanoma patients, BCL2 in prostate cancer, and GAD65 and IA2 in type 1 diabetes patients (25, 26). NAPPA microarrays were shown to express over 94% of their proteins regardless of protein size or type (25). Their testing in the detection of the p53 autoantibody in breast cancer serum provided a CV of 7% for within day testing and 11% day-to-day testing (25). The remainder of this chapter details study design using NAPPA microarrays. Although the production of the microarrays is beyond the scope of this chapter, a detailed protocol was published in 2008 (24).
1.1. Study Design Considerations
In designing serum screening studies, several study design issues should be addressed prior to initiation in order to generate inter-pretable and meaningful results. These include the following.
1.1.1. Serum Samples
In obtaining serum samples for the study, the following need to be considered:
Serum samples from the patient and control groups are best when matched for gender and age to avoid confounding factors arising from differences in these variables.
Samples may also require pairing based on other variables depending on the experiment (e.g., same smoking status when studying lung cancer)
Researchers should ensure that the sera used were collected around the same period of time using the same method. This avoids the confounding effects of different serum collection SOPs that may be used at different institutions, or even different divisions of the same institution (e.g., outpatient clinic vs. hospital phlebotomy staff).
Study coordinators need to obtain IRB approval and informed consent from the patients and controls to be included in the study.
1.1.2. Experimental Setup
Large serum screening studies face several challenges (see Note 1) that may be best handled by dividing the project into three stages:
Pilot screen – A pilot screen of 20–50 patient and control samples against a test microarray is carried out to test signal variation among patients and controls. These data can then be used for a statistical power study that will identify the number of samples required for an expected frequency of an autoantibody in the population, the variability of the signal for autoantibody in the population, and the degree of certainty desired to ensure that a marker will be found if it is there.
Training stage – Patient and control sera are screened against the entire panel of proteins to identify antigens that can discriminate between patients and controls. The number of sera screened depends on the results of the power study conducted during the pilot screen. The sera used during the training stage can overlap those used during the pilot screen. Proteins that pass this step will form the validation set.
Validation stage – The aim here is to validate the antigens identified during the training set using an independent set of sera. The number of sera used here is also dictated by the power calculation from the pilot screen. Key here is that none of the samples used for validation (cases or controls) was used in any of the previous studies. The ideal validation study is performed in a blinded fashion. For antigens confirmed by validation, it is useful to test them by an alternate method. One method would be ELISA, commonly used in clinical laboratories, which also serves the purpose of facilitating adoption of the biomarkers in clinical tests.
1.2. Protein Microarray Considerations
The structure and content of the protein microarrays also need to be considered to ensure good quality data and interpretable results. Items to be contemplated should include the following.
1.2.1. Spot Replicates
These replicates are used to measure two different variations valuable during statistical analysis:
- Zone variation: This type of variation, which can occur with virtually all microarray technologies, results from a microarray printing or processing method that causes one or more region(s) of the array to erroneously display different signal magnitude(s). Examples are illustrated in Fig. 2 where overall spots at the top of the slide are more intense than at the bottom. Such variation can be adjusted for by two different methods:
- Printing of identical features from the same protein sample throughout the microarray as references to monitor zone variations. In the case of NAPPA arrays, this was accomplished with a grid of eight features across the width of the slide by 12 spots along its length. These 48 features typically consist of negative controls discussed below.
- Averages of regional or neighboring features can be used in place of the identical features method described above. In this case, we assume that the majority of proteins do not show reactivity to serological autoantibodies and display similar levels of nonspecific binding. Hence averaging 16 proteins in a 4 × 4 grid, for example, throughout the microarray would identify regions of varying background signals that demonstrate zone variation.
Printing variability: Variation in printing efficiency and spotting chemistry contributes to this type of variability that can be monitored by printing multiple identical features. These features are best placed within a close vicinity to each other to avoid the confounding effects of zone variation.
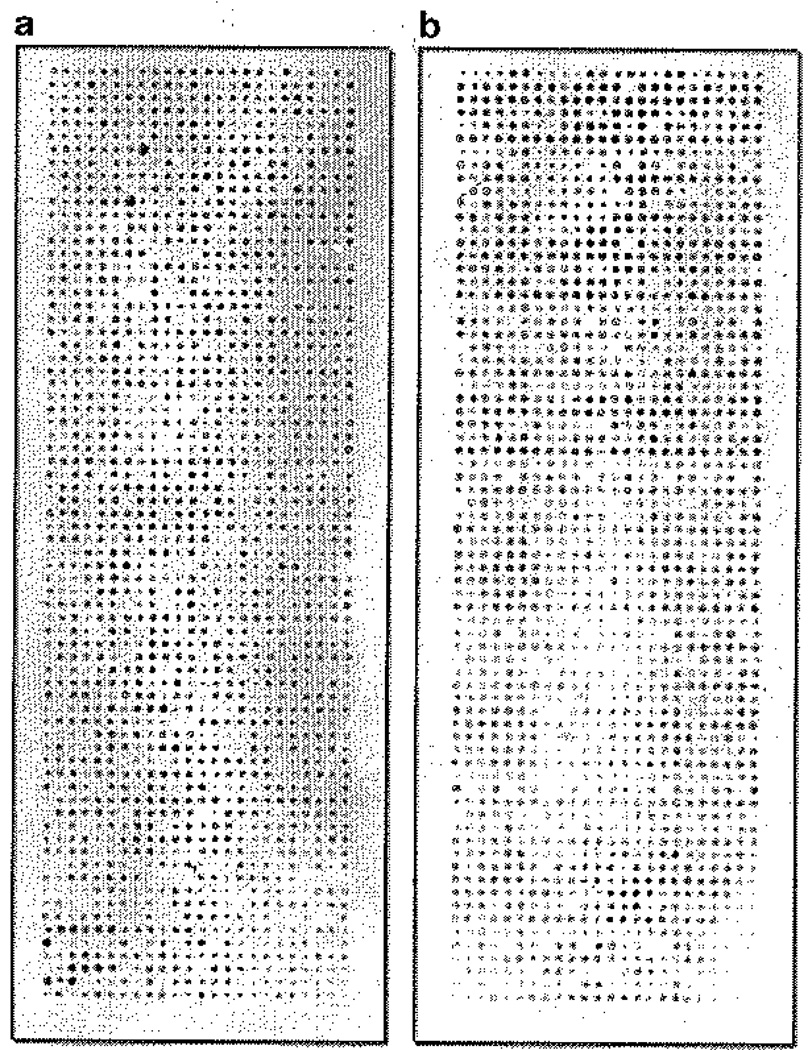
Fig. 2.
Examples of zone variations that can occur on protein microarrays. (a) The zone variation here shows a strong signals at the top of the slide and weaker signals at the bottom, (b) in this case, the top-to-bottom zone variation is additionally confounded with a regional oval-shaped variation observed in the lower third of the slide. Practice of microarray processing technique will alleviate these variations over time.
1.2.2. Controls
Controls are of the utmost importance in monitoring proper microarray processing and technical and biological variability. There are three types of controls that should be included on the arrays:
Processing positive controls. In order to ensure that the arrays are working appropriately, various positive controls should be included on the arrays. To confirm that the antihuman secondary antibodies are working and to provide reference features, human IgG can be included. It is also useful to include a protein that is likely to reveal a response in most individuals, regardless of whether they are patients or controls. Examples of such proteins include the EBNA1 antigen, from the Epstein Barr virus to which approximately 90% of the adult population have antibodies, or childhood vaccines such as tetanus toxoid.
Negative controls. These are used to determine background or noise levels on the microarrays during the data analysis. They should be distributed throughout the microarray and are used to detect and adjust for zone variations.
Disease-specific controls. Whenever possible, it is best to include positive controls for a disease to test the viability of the serum screening conditions. It should be noted, though, that not all diseases have known controls and not all patients will be reactive to such controls, hence their availability and usefulness may be limited.
1.2.3. Technical Reproducibility Test
As with all large screening experiments that are carried out over the course of weeks or months, the degree of technical reproducibility needs to be assessed to ensure that the differences observed between test groups are real. Here are the forms of technical reproducibility that should be considered:
Within Day reproducibility: This tests the microarray-to-microarray variability within one processing run. It is measured by testing each of three or four serum samples on two or three identical microarrays. It is best not to proceed to a full scale screen until the coefficient of variation of such tests is less than 10% for 80% of the features interrogated. Otherwise, the microarray processing protocol needs to be reoptimized.
Day-to-day reproducibility: This measures the microarray-to-microarray variability between tests, each run on a different day. Since most large scale screening studies are processed over the course of weeks, the daily reproducibility needs to be addressed and the variability minimized. One method to minimize the likelihood of obtaining nonspecific variations between patients and controls is to process the same number of patients and controls daily (such as five patients and five controls every day).
2. Materials
2.1. Activation of cDNA-Based Microarrays
NAPPA microarrays (see Note 2).
HybriWell gaskets (Grace).
TNT® T7 Quick Coupled Transcription/Translation System (Promega).
RNaseOUT (Invitrogen).
DEPC water (Ambion).
EchoTherm™ IN30 Bench Top, Chilling/Heating Programmable Incubator (Torrey Pines Scientific).
SuperBlock (Pierce).
Phosphate buffered saline (1 × PBS): 137 mM NaCl, 2.7 mM KC1, 10 mM Na2HPO4, 1.8 mM KH2PO4. Adjust pH to 7.4 with HC1 if necessary.
5% milk blotto: Dissolve 5 g of nonfat dry milk in 1 l of 1× PBS. Add Tween-20 to final concentration of 0.2% (see Note 3).
2.2. Detection of Protein Display on the Microarrays
Corning® Hybridization Chamber.
Mouse anti-GST antibody (Cell Signaling).
Antimouse HRP-conjugated antibody (Jackson Laboratories).
TSA (tyramide signal amplification) reagent (Perkin Elmer).
Lifter slips, 24 × 65 mm (Erie).
2.3. Serum Antibody Profiling
5% milk blotto: Dissolve 5 g of nonfat dry milk in 1 l of 1× PBS. Add Tween-20 to final concentration of 0.2% (see Note 3).
Corning® Hybridization Chamber (Product).
Mouse antihuman IgG HRP-conjugated antibody (Jackson ImmunoResearch).
TSA reagent (Perkin Elmer).
Lifter slips, 24 × 65 mm (Erie).
ProScan Array Scanner (Perkin Elmer).
3. Methods
Serological autoantibody screening using protein microarrays provides a rapid and efficient method to profile an individual’s humoral immune response to known or unclassified antigens. Loosely based on the broadly utilized ELISA assay, this method of serum screening requires specific optimization to microarrays to avoid artifacts and technical variations that would lead to false data. There are different types of microarrays, and each possess unique advantages and challenges. The remainder of this chapter will focus on a specific type of cDNA-based protein microarrays called NAPPA (Nucleic Acid Programmable Protein Array). NAPPA arrays are built by printing a plasmid containing the cDNA of a protein tagged with GST, along with an anti-GST capture antibody. The arrays are converted to functional protein microarrays through in situ protein production and capture using an in vitro expression system. They are then treated like any other protein microarray with attention to avoiding protein degradation and maintaining stability. Serum samples from patients or controls are diluted in a milk-based buffer and added to the microarrays in an overnight incubation at 4°C. Such long incubations allow low abundance and/or weak affinity autoantibodies adequate time to bind their target proteins on the microarray. The microarrays are then washed, incubated with a secondary antibody, and then visualized for subsequent quantification and analysis.
The study size (i.e., number of serum samples) required to complete a comprehensive and meaningful investigation depends on numerous factors including the frequency with which any one autoantibody is expected within the patient population, the estimated clinical specificity of such an antibody in predicting disease state, the relative affinity of the autoantibodies to their targets, the antigen density on the microarrays, and the technical reproducibility of the microarrays. It is imperative for researchers to work with a biostatistician prior to starting the screen in order to determine the study size and number of replicates that will be needed to obtain statistically significant data.
3.1. Activation of NAPPA Microarrays
Microarrays on which the proteins will be synthesized in situ from a cDNA require activation which will lead to the synthesis and capture of each protein. On NAPPA microarrays, the microarrays are first immersed in Superblock for 30–60 min to block any nonspecific protein binding sites that may exist on the microarray. These would include non-specific binding to the cDNA-containing spots as well as the glass surface on which the microarrays are printed. At the end of this incubation, the slides are rinsed with dH2O and dried using filtered, pressurized air.
Promega’s T7-based coupled transcription-translation rabbit reticulocyte lysate expression system is prepared according to the manufacturer’s instruction with the exception of the addition of both of the provided amino acid mixes so as to obtain a full complement of amino acids. For examples, the amino acid mix lacking methionine and that lacking leucine are combined together. Each microarray will consume 150 µl of the lysate.
A hybriwell gasket is applied to the slide with the outline sealed by the adhesive material, thus forming a thin chamber above the slides. This chamber is filled with rabbit reticulocyte lysate and incubated at 30°C for 90 min in a programmable incubator. Each spot will synthesize its target protein that gets captured through its tag by an anti-tag antibody present in the spot. To ensure proper immobilization of these proteins, the slides are incubated at 15°C for at least 30 min following the expression protocol. It is absolutely essential for this step to be completed in order to capture the greatest amount of protein on the spot and to get the highest density possible.
At the end of the expression and capture incubations, the hybriwell is gently removed without disrupting the proteins on the slide (see Note 4) and immersed into a 5% milk blotto solution for three 5 min washes, followed by an hour’s incubation at room temperature.
3.2. Detection of Protein Display on the Microarrays
For each daily experiment, at least one array should be processed to measure the amount of protein displayed (i.e. expressed and captured) per spot. This will be used for future analysis (see Note 5). Each slide is incubated in 2 ml of anti-GST antibody diluted 300-fold in 5% blotto. The slides are placed in a Corning hybridization chamber and an end-over-end rotator and allowed to incubate for 1 h at room temperature (see Note 6).
Slides are removed from the Corning hybridization chamber and washed three 5 min washes with 5% milk blotto, followed by another incubation with the HRP-conjugated secondary antimouse antibody diluted 500-fold in 5% milk blotto. The secondary is incubated for 1 h with end-over-end mixing in the Corning hybridization chamber.
Slides are removed from the secondary antibody and washed three 5 min washes in 1× PBS. They are rinsed with distilled water, and 500 µl of a 50-fold dilution of Cy3 TSA is added to each slide. A lifter slip is applied to spread the TSA, and the slides are incubated for 10 min in the dark.
The lifter slip is removed, and the slide is rinsed with dH2O, dried using filtered pressurized air, placed in a slide box, and stored in a dark dry place until ready to scan.
3.3. Serum Antibody Profiling
Antibody profiling is carried out at the end of step 4 of Subheading 3.1, when the microarrays have been expressed and blocked. The serum (or plasma) is centrifuged at 14,000 rpm for 10 min at 4°C in a microcentrifuge to separate out any leftover lipids and cellular debris (see Note 7).
Serum is diluted 200–900-fold into 2 ml 5% milk/0.3× PBS-T buffer (see Note 8) and applied to the microarray in a Corning hybridization chamber. The slides are placed in an end-over-end rotator and incubated overnight at 4°C to allow low abundance and/or weak affinity antibodies to bind their target proteins.
The next morning, the slides are taken out of the Corning hybridization chamber and washed 3 times with 5% milk/blotto for 5 min each. 2 ml of HRP-conjugated antihuman IgG antibody, diluted 500-fold in 5% milk/blotto, is applied to the slides in a Corning hybridization chamber. The slides are incubated for 1 h at room temperature with end-over-end rotation.
The slides are removed from the slide chamber and washed 3 times in 1× PBS, each wash of 5 min. The slides are rinsed with dH2O and 500 ml of 50-fold diluted TSA reagent is applied to each slide. A lifter slip is placed on top to spread the TSA across the entire slide and incubated for 10 min at room temperature in the dark.
The lifter slip is removed and the slides are rinsed with dH2O, dried with filtered pressurized air, and stored in a slide box in a dark dry place until ready to scan.
Slides are scanned using the red laser for Cy3 at an intensity that will not cause signal saturation (see Note 9). Quantify the spots using Microvigene and analyze by advanced statistical methods such as the binomial proportion, Wilcoxon, or Fisher tests. An example of strongly reactive antigens p53, EBNA, and GAD65 are shown in Fig. 3.
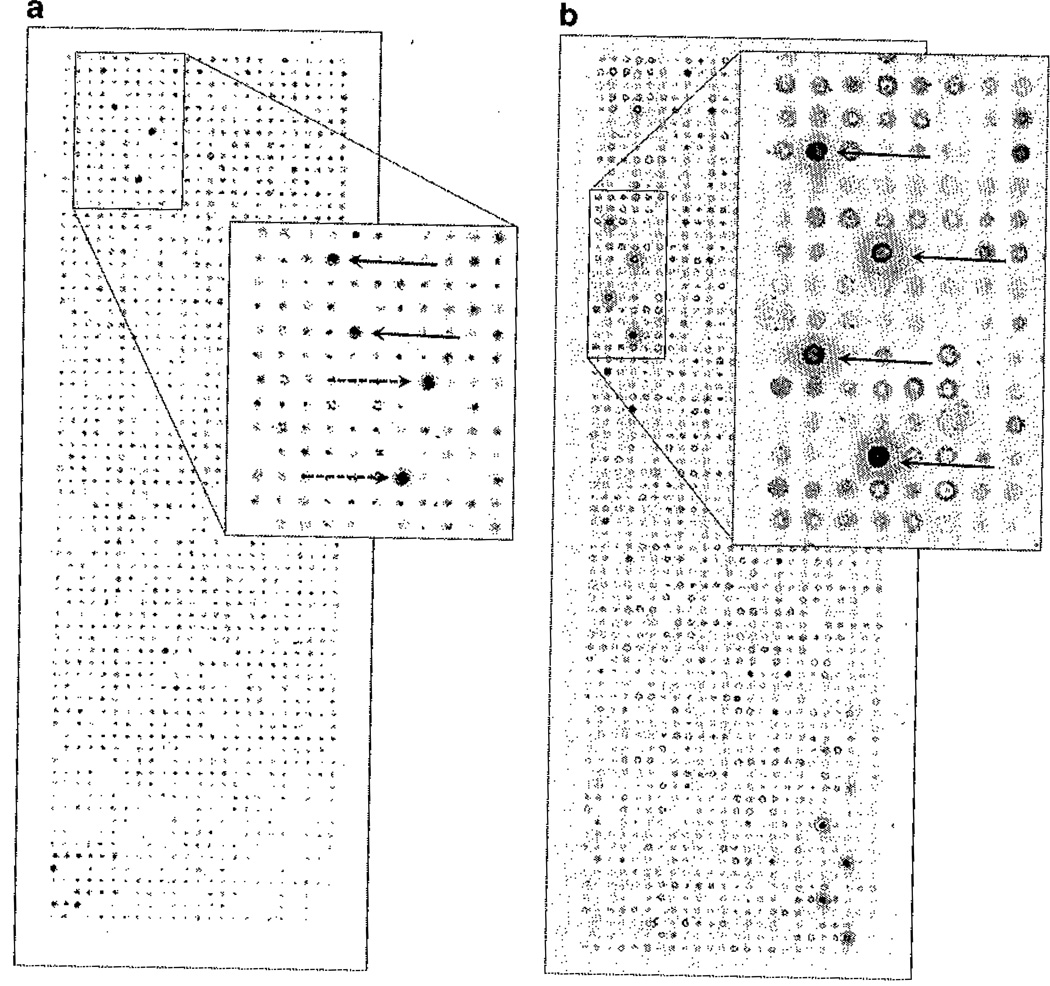
Fig. 3.
Examples of strong antigen signals for EBNA (solid arrow) and p53 (dashed arrow) in a breast cancer patient (a) and GAD65 (solid arrows) in a type 1 diabetes patient (b).
Acknowledgments
The authors would like to acknowledge support for this work by the NIH “Biomarker Detection Using NAPPA Tumor Antigen Arrays” U01 CA117374 and from the Juvenile Diabetes Research Foundation, “The use of protein microarrays to study autoimmunity and diabetes” 17-2007-1045.
Footnotes
Screening many thousands of proteins against hundreds of patient samples may be difficult to manage physically, analytically, and economically. Challenges include the risk of overfitting the data, high false discovery rates, and dismissal of potentially legitimate biomarkers. To avoid such problems and decrease the financial costs associated with large serum screening studies, a subset of 50–100 sera can be screened against the entire panel of protein antigens with the goal of removing any proteins that do not show a signal higher than background or do not show a difference between patients and controls. In this strategy, the hope is to reduce the number of antigens about tenfold in order to have a more focused set of proteins to begin the training studies.
NAPPA microarrays are custom designed to contain up to 2,532 features/spots of which up to 2,304 spots are protein-encoding features. They are DNA arrays that are converted into proteins arrays on the day of use, thus displaying fresh protein for each experiment.
The milk needs to be stirred for over an hour to ensure that it is completely dissolved. Any particulate matter that is present in the milk can adhere to the glass slide leading to either masking of real protein spots or false signals.
Researchers need to be careful not to place any pressure on the middle of the slide when removing the hybriwell so as not to smear proteins away from their designated spots.
Measurements of protein display are used to determine the success of the expression and capture on the array. Typically, over 90% of the features display their target protein.
When the Corning hybridization chamber is closed, the antibody/milk must cover at least half the slide to avoid drying in the middle of the slide.
In our experience, the source of antibody, whether plasma or serum, did not impact the efficiency of antibody binding to the antigens presented on the arrays.
Serum samples from different individuals display different response magnitudes. We have observed that approximately 10–15% of the sera have a high nonspecific background regardless of their patient or control designation. Moreover, for such large scale studies, samples were typically collected on different days by different persons introducing variation in serum reactivity solely due to the collection method. To adjust for both of these issues, we titer each serum prior to screening it by testing it on mini-arrays of 36 protein spots to identify an optimal dilution factor that provides an acceptable background that will not overwhelm the true signals. This is combined with the use of lower PBS concentrations to promote the binding of low affinity autoantibodies. We have found that in the majority of cases, a 600-fold dilution into 5% milk/0.3× PBS-T worked best.
It is important to avoid saturation of the signal in order to obtain a valid quantifiable result. Most scanners provide a method to monitor signal intensity, including a warning for signal saturation.
References
- 1.Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2001;2:1–13. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sreekumar A, Nyati MK, Varambally S, Barrette TR, Ghosh D, Lawrence TS, Chinnaiyan AM. Profiling of cancer cells using protein microarrays: discovery of novel radiation-regulated proteins. Cancer Res. 2001;61:7585–7593. [PubMed] [Google Scholar]
- 3.Joos TO, Schrenk M, Hopfl P, Kroger K, Chowdhury U, Stoll D, Schorner D, Durr M, Herick K, Rupp S, Sohn K, Hammerle H. A microarray enzyme-linked immunosorbent assay for autoimmune diagnostics. Electrophoresis. 2000;21:2641–2650. doi: 10.1002/1522-2683(20000701)21:13<2641::AID-ELPS2641>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 4.Tan HT, Low J, Lim SG, Chung MC. Serum autoantibodies as biomarkers for early cancer detection. Febs J. 2009;276(23):6880–6904. doi: 10.1111/j.1742-4658.2009.07396.x. [DOI] [PubMed] [Google Scholar]
- 5.Yu X, Schneiderhan-Marra N, Hsu HY, Bachmann J, Joos TO. Protein microarrays: effective tools for the study of inflammatory diseases. Methods Mol Biol. 2009;577:199–214. doi: 10.1007/978-1-60761-232-2_15. [DOI] [PubMed] [Google Scholar]
- 6.Song Q, Liu G, Hu S, Zhang Y, Tao Y, Han Y, Zeng H, Huang W, Li F, Chen P, Zhu J, Hu C, Zhang S, Li Y, Zhu H, Wu L. Novel autoimmune hepatitis-specific autoantigens identified using protein microarray technology. J Proteome Res. 2009;9(l):30–39. doi: 10.1021/pr900131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lorenz P, Kreutzer M, Zerweck J, Schutkowski M, Thiesen HJ. Probing the epitope signatures of IgG antibodies in human serum from patients with autoimmune disease. Methods Mol Biol. 2009;524:247–258. doi: 10.1007/978-1-59745-450-6_18. [DOI] [PubMed] [Google Scholar]
- 8.Quintana FJ, Farez MF, Viglietta V, Iglesias AH, Merbl Y, Izquierdo G, Lucas M, Basso AS, Khoury SJ, Lucchinetti CF, Cohen IR, Weiner HL. Antigen microarrays identify unique serum autoantibody signatures in clinical and pathologic subtypes of multiple sclerosis. Proc Natl Acad Sci U S A. 2008;105:18889–18894. doi: 10.1073/pnas.0806310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auger I, Balandraud N, Rak J, Lambert N, Martin M, Roudier J. New autoantigens in rheumatoid arthritis (RA): screening 8268 protein arrays with sera from patients with RA. Ann Rheum Dis. 2009;68:591–594. doi: 10.1136/ard.2008.096917. [DOI] [PubMed] [Google Scholar]
- 10.Roche S, Dauvilliers Y, Tiers L, Couderc C, Piva MT, Provansal M, Gabelle A, Lehmann S. Autoantibody profiling on high-density protein microarrays for biomarker discovery in the cerebrospinal fluid. J Immunol Methods. 2008;338:75–78. doi: 10.1016/j.jim.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Qiu J, Hanash S. Autoantibody profiling for cancer detection. Clin Lab Med. 2009;29:31–46. doi: 10.1016/j.cll.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Kijanka G, Murphy D. Protein arrays as tools for serum autoantibody marker discovery in cancer. J Proteomics. 2009;72:936–944. doi: 10.1016/j.jprot.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Liu W, Wang P, Li Z, Xu W, Dai L, Wang K, Zhang J. Evaluation of tumour-associated antigen (TAA) miniarray in immunodiagnosis of colon cancer. Scand J Immunol. 2009;69:57–63. doi: 10.1111/j.1365-3083.2008.02195.x. [DOI] [PubMed] [Google Scholar]
- 14.Gure AO, Altorki NK, Stockert E, Scanlan MJ, Old LJ, Chen YT. Human lung cancer antigens recognized by autologous antibodies: definition of a novel cDNA derived from the tumor suppressor gene locus on chromosome 3p21.3. Cancer Res. 1998;58:1034–1041. [PubMed] [Google Scholar]
- 15.Gure AO, Stockert E, Scanlan MJ, Keresztes RS, Jager D, Altorki NK, Old LJ, Chen YT. Serological identification of embryonic neural proteins as highly immunogenic tumor antigens in small cell lung cancer. Proc Natl Acad Sci U S A. 2000;97:4198–4203. doi: 10.1073/pnas.97.8.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scanlan MJ, Chen YT, Williamson B, Gure AO, Stockert E, Gordan JD, Tureci O, Sahin U, Pfreundschuh M, Old LJ. Characterization of human colon cancer antigens recognized by autologous antibodies. Int J Cancer. 1998;76:652–658. doi: 10.1002/(sici)1097-0215(19980529)76:5<652::aid-ijc7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 17.Jager D, Stockert E, Gure AO, Scanlan MJ, Karbach J, Jager E, Knuth A, Old LJ, Chen YT. Identification of a tissue-specific putative transcription factor in breast tissue by serological screening of a breast cancer library. Cancer Res. 2001;61:2055–2061. [PubMed] [Google Scholar]
- 18.Forti S, Scanlan MJ, Invernizzi A, Castiglioni F, Pupa S, Agresti R, Fontanelli R, Morelli D, Old LJ, Pupa SM, Menard S. Identification of breast cancer-restricted antigens by antibody screening of SKBR3 cDNA library using a preselected patient’s serum. Breast Cancer Res Treat. 2002;73:245–256. doi: 10.1023/a:1015854415746. [DOI] [PubMed] [Google Scholar]
- 19.Jager D, Unkelbach M, Frei C, Bert F, Scanlan MJ, Jager E, Old LJ, Chen YT, Knuth A. Identification of tumor-restricted antigens NY-BR-1, SCP-1, and a new cancer/testis-like antigen NW-BR-3 by serological screening of a testicular library with breast cancer serum. Cancer Immun. 2002;2:5. [PubMed] [Google Scholar]
- 20.Kilic A, Schuchert MJ, Luketich JD, Landreneau RJ, Lokshin AE, Bigbee WL, El-Hefhawy T. Use of novel autoantibody and cancer-related protein arrays for the detection of esophageal adenocarcinoma in serum. J Thorac Cardiovasc Surg. 2008;136:199–204. doi: 10.1016/j.jtcvs.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 21.Taylor BS, Pal M, Yu J, Laxman B, Kalyana-Sundaram S, Zhao R, Menon A, Wei JT, Nesvizhskii AI, Ghosh D, Omenn GS, Lubman DM, Chinnaiyan AM, Sreekumar A. Humoral response profiling reveals pathways to prostate cancer progression. Mol Cell Proteomics. 2008;7:600–611. doi: 10.1074/mcp.M700263-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Ramachandran N, Hainsworth E, Bhullar B, Eisenstein S, Rosen B, Lau AY, Walter JC, LaBaer J. Self-assembling protein microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- 23.Ramachandran N, Hainsworth E, Demirkan G, LaBaer J. On-chip protein synthesis for making microarrays. Methods Mol Biol. 2006;328:1–14. doi: 10.1385/1-59745-026-X:1. [DOI] [PubMed] [Google Scholar]
- 24.Ramachandran N, Raphael JV, Hainsworth E, Demirkan G, Fuentes MG, Rolfs A, Hu Y, LaBaer J. Next-generation high-density self-assembling functional protein arrays. Nat Methods. 2008;5:535–538. doi: 10.1038/nmeth.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramachandran N, Anderson KS, Raphael JV, Hainsworth E, Sibani S, Montor WR, Pacek M, Wong J, Eljanne M, Sanda MG, Hu Y, Logvinenko T, LaBaer J. Tracking humoral responses using self assembling protein microarrays. Proteomics Clin Appl. 2008;2:1518–1527. doi: 10.1002/prca.200800034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson KS, Ramachandran N, Wong J, Raphael JV, Hainsworth E, Demirkan G, Cramer D, Aronzon D, Hodi FS, Harris L, Logvinenko T, LaBaer J. Application of protein microarrays for multiplexed detection of antibodies to tumor antigens in breast cancer. J Proteome Res. 2008;7:1490–1499. doi: 10.1021/pr700804c. [DOI] [PMC free article] [PubMed] [Google Scholar]