Abstract
Direct interspecies electron transfer (DIET) is an alternative to interspecies H2/formate transfer as a mechanism for microbial species to cooperatively exchange electrons during syntrophic metabolism. To understand what specific properties contribute to DIET, studies were conducted with Pelobacter carbinolicus, a close relative of Geobacter metallireducens, which is capable of DIET. P. carbinolicus grew in coculture with Geobacter sulfurreducens with ethanol as the electron donor and fumarate as the electron acceptor, conditions under which G. sulfurreducens formed direct electrical connections with G. metallireducens. In contrast to the cell aggregation associated with DIET, P. carbinolicus and G. sulfurreducens did not aggregate. Attempts to initiate cocultures with a genetically modified strain of G. sulfurreducens incapable of both H2 and formate utilization were unsuccessful, whereas cocultures readily grew with mutant strains capable of formate but not H2 uptake or vice versa. The hydrogenase mutant of G. sulfurreducens compensated, in cocultures, with significantly increased formate dehydrogenase gene expression. In contrast, the transcript abundance of a hydrogenase gene was comparable in cocultures with that for the formate dehydrogenase mutant of G. sulfurreducens or the wild type, suggesting that H2 was the primary electron carrier in the wild-type cocultures. Cocultures were also initiated with strains of G. sulfurreducens that could not produce pili or OmcS, two essential components for DIET. The finding that P. carbinolicus exchanged electrons with G. sulfurreducens via interspecies transfer of H2/formate rather than DIET demonstrates that not all microorganisms that can grow syntrophically are capable of DIET and that closely related microorganisms may use significantly different strategies for interspecies electron exchange.
INTRODUCTION
Since the discovery of the “S organism” (6), microbiologists have tried to understand the mechanisms of electron exchange between microorganisms syntrophically degrading organic compounds under anaerobic conditions. For example Pelobacter carbinolicus, which is a modern-day analog for the S organism, can metabolize ethanol to acetate, H2, and carbon dioxide only when an H2-consuming partner, such as Methanospirillum hungatei, maintains low H2 partial pressures (32). In some syntrophic cultures, formate may be the electron carrier between species (24, 33, 35). Previous studies provided evidence for H2 and formate transfer by evaluating H2- and/or formate-utilizing microorganisms as electron-accepting partners (24, 33, 35) and also by adding exogenous excess H2 or formate to the cocultures to disrupt the syntrophic metabolism, decoupling methanogenesis from utilization of the substrate (1, 2, 40). Thermodynamic calculations have demonstrated that a small window of opportunity exists for the syntrophic partners, where the concentration of H2 or formate provides optimum conditions for both partners (33, 36). Other electron carriers that facilitate electron exchange between syntrophic partners include the humic substance analog anthraquinone-2,6-disulfonate (19, 21) and cysteine (15). Direct interspecies electron transfer (DIET), could be an efficient alternative strategy for microorganisms to cooperate in the anaerobic degradation of organic substrates (20, 27, 37). DIET was discovered in cocultures of Geobacter metallireducens and Geobacter sulfurreducens, which grew with ethanol as the electron donor and fumarate as the electron acceptor (37). G. sulfurreducens cannot metabolize ethanol, whereas G. metallireducens cannot use fumarate as an electron acceptor. Adaptive evolution of the coculture for enhanced ethanol metabolism was associated with the formation of large aggregates of the two species. Although G. sulfurreducens is capable of utilizing either H2 or formate as an electron donor for fumarate reduction when acetate is available as a carbon source (9), cells within the aggregates were not effective in H2 or formate metabolism, and cocultures were readily initiated with a mutant strain of G. sulfurreducens that was unable to use H2 as an electron donor (37). These results suggested that the coculture was functioning via an alternative to interspecies H2 or formate transfer.
In the adapted cocultures, G. sulfurreducens produced large quantities of the multiheme c-type cytochrome OmcS (25, 37), which is localized (18) along the electrically conductive (23, 30) type IV pili of G. sulfurreducens. Increased OmcS expression was attributed to point mutations that accumulated in the gene for the transcriptional regulator PilR (37). Deleting pilR in G. sulfurreducens accelerated aggregate formation and adaption for rapid ethanol metabolism (37). Deletion of genes required for OmcS or pilus expression inhibited ethanol metabolism (37). Furthermore, the aggregates were electrically conductive, likely due to the pili, which have been shown to provide long-range conductivity in G. sulfurreducens biofilms (23, 24). These results suggested that electrons were directly transferred from G. metallireducens to G. sulfurreducens.
There was also substantial evidence for DIET within aggregates from an anaerobic digester converting brewery waste to methane, in which Geobacter was abundant (27). The mixed community aggregates exhibited metal-like conductivity (27) similar to that of Geobacter current-producing biofilms and the pili of G. sulfurreducens (23).
To better understand the mechanisms of DIET, it is important to determine if other microorganisms are capable of DIET and what features those microorganisms must have to enable DIET. The potential for P. carbinolicus to participate in DIET was evaluated because both P. carbinolicus and G. metallireducens appear to have evolved from a common ancestor capable of extracellular electron transfer (7), but the two differ significantly in several aspects of their basic physiology and mechanisms for extracellular electron transfer (7, 12, 31). Thus, it was unknown whether the absence of previous evidence for DIET with P. carbinolicus could be attributed to syntrophic growth being evaluated with an electron-accepting partner incapable of DIET or whether P. carbinolicus lacks key physiological features required for DIET. The results indicate that P. carbinolicus is not capable of DIET and must rely on interspecies transfer of H2 or formate for electron exchange with G. sulfurreducens.
MATERIALS AND METHODS
Organisms, media, and growth conditions.
All incubations of pure cultures and cocultures were performed under strict anaerobic culturing techniques as previously described (3). Cultures were incubated in 27-ml pressure tubes or 160-ml serum bottles sealed with butyl rubber stoppers and filled with 10 or 50 ml of medium. The increase in culture turbidity was monitored at 600 nm by placing the culture tubes into a Genesys 5 spectrophotometer (Spectronics Instruments) with a path length of 1.5 cm.
P. carbinolicus (DSM 2380) was regularly transferred under fermentative conditions with 10 mM acetoin as the substrate and 0.02 mM Na2S as the reductant, as previously described (12). G. sulfurreducens PCA (ATCC 51573) and mutants of this microorganism which were tested for the study (ΔhybL, ΔfdnG, double ΔhybL ΔfdnG, ΔomcS, and ΔpilA mutants) were routinely cultured in freshwater medium containing 1 mM cysteine as the reductant, 10 or 15 mM acetate, and 40 mM fumarate as previously described (8). Newly constructed mutants of G. sulfurreducens were tested for growth with H2 (138 KPa) or formate (40 mM and 10 mM) as the electron donor in freshwater medium in the presence of 1 mM acetate as the carbon source.
For cocultures of P. carbinolicus and G. sulfurreducens, 20 mM ethanol and 40 mM fumarate served as substrates for growth in a medium prepared as previously described (12). Cocultures of G. metallireducens and the G. sulfurreducens strain deficient in formate dehydrogenase and hydrogenase activities were initiated using a 2% inoculum of each syntrophic partner added to a freshwater medium prepared as previously described (37) with fumarate and ethanol as substrates.
All cocultures were regularly transferred (2% inocula) under strict anaerobic conditions at least six times prior to monitoring organic acids and ethanol over time. The only exception was a coculture of P. carbinolicus with the G. sulfurreducens double mutant incapable of H2 and formate utilization. This coculture could not grow on ethanol and was therefore analyzed during the initial transfer.
Construction of G. sulfurreducens mutants.
The fdnG gene (GSU0777) was replaced with a kanamycin resistance gene, such that the coding region for amino acid residues from 62Asp to 951Pro was deleted. Double-crossover homologous recombination was carried out by electroporation (8) with the linear DNA fragment consisting of the kanamycin resistance gene flanked by ∼0.7-kbp DNA fragments containing the upstream and the downstream regions of fdnG. These flanking DNA fragments were amplified by PCR with primers fdnG-P1 (TCTCTAGAACGGCTTGGTGACGTAGTC; the XbaI site is underlined) and fdnG-P2 (TCGGATCCTTGGTATGGACGATCAG; the BamHI site is underlined) for the upstream region and fdnG-P3 (TCTAAGCTTCAACGTGCAGGGCAAGC; the HindIII site is underlined) and fdnG-P4 (TCTCTCGAGACCACTTTCACGTAGCGGTC; the XhoI site is underlined) for the downstream region. The kanamycin resistance gene was amplified by PCR with Km-Fwd (GCATGAGAATTCCTGACGGAACAGCGGGAAGTCCAGC; the EcoRI site is underlined) and Km-Rev (GCTATGAAGCTTTCATAGAAGGCGGCGGTGGAATCGAA; the HindIII site is underlined) and using pBBR1MCS-2 (17) as the template. Gene replacement was confirmed by PCR analysis. The ΔfdnG ΔhybL double mutant was constructed in a similar manner by deleting the fdnG gene from a previously characterized ΔhybL uptake hydrogenase mutant (10).
Reverse transcription-quantitative PCR (RT-qPCR).
To quantify the abundance of hydrogenase and formate dehydrogenase transcripts in cocultures of P. carbinolicus with the wild-type strain of G. sulfurreducens, the hydrogenase-deficient strain, and the formate dehydrogenase-deficient strain, four biological replicates of each late mid-exponential-phase coculture, 10 ml each, were treated with 2 ml RNA Later (Ambion), mixed well, and harvested at 4°C by centrifugation at 6,000 × g for 20 min. The tubes were opened, and cocultures were removed for further use for RNA extraction using TRIzol (Invitrogen) with a slight modification of the manufacturer's protocol. Briefly, the cell pellets were mixed homogenously with a 1-ml volume of TRIzol reagent. The mix was transferred to a 2-ml O-ring tube containing 0.5 g of 0.1-mm glass-zirconia beads and homogenized for 20 s on a FastPrep instrument (MoBio Laboratories) at 3 m/s. The tubes were then incubated at room temperature for 5 min before addition of 200 μl chloroform, vortexed for 15 s, and centrifuged at 12,000 × g for 15 min at 4°C. The aqueous layer was then used for the RNA isolation. The RNA thus obtained was purified using the MiniElute PCR purification kit (Qiagen) and further treated with rDNAse I (Ambion) to digest any traces of genomic DNA contamination. A final round of RNA purification was done with a MiniElute PCR purification kit (Qiagen) following the manufacturer's protocol. The quality and the quantity of pure RNA were assessed with the Experion RNA standard sensitivity kit (Bio-Rad). Furthermore, absence of genomic DNA contamination was verified by 16S rRNA gene PCR using 9F and 519R primer sets (34).
For whole-transcriptome amplification (WTA), about 300 ng of total RNA was converted into WTA cDNA libraries and amplified by WTA PCR using reagents and protocols supplied with or recommended by Sigma. Briefly, 300 ng of total RNA was mixed with 2.5 μl WTA library synthesis buffer and 2.5 μl WTA library stabilization solution, the total volume was adjusted to 24 μl using nuclease-free water, and the mixture was heated at 70°C for 5 min and immediately cooled. Library synthesis enzyme (1 μl) was added, and WTA cDNA libraries were synthesized using the following thermocycler program: 24°C for 15 min, 42°C for 2 h, and 95°C for 5 min. Aliquots were WTA PCR amplified using JumpStart Taq DNA polymerase (Sigma), WTA amplification master mix, and deoxynucleoside triphosphate (dNTP) mix following the manufacturer's protocol except that the total number of cycles was reduced to 15. The enriched product was then purified using a PCR purification kit (Qiagen) and used as a template in qPCR experiments.
Real-time PCR was carried out using an ABI prism 7900 (Applied Biosystems). Primers designed for G. sulfurreducens (26) were used to target hybA, fdnG, and the housekeeping gene recA: fdnG-F, 5′-ACTTCACCAAGGACGTCACC-3′; fdnG-R, 5′-TCCCTTCGTTGGTGTAGGAG-3′; hybA-F, 5′-CTACGGCGAGAAGGAAGTTG-3′; hybA-R, 5′-CCCCTTGTAGATGGTGTGCT-3′; recA-F, 5′-CACCGGCATAATCTCCAAGT-3′; and recA-R, 5′-ATCTTGCGGATATCGAGACG-3′. Reactions were performed in triplicate for each gene tested in a final volume of 20 μl containing 10 μl of Power Sybr green PCR master mix, 0.6 mM reverse and forward primers were made, and 2 μl of enriched WTA product was added as the template. The real-time PCR was run for 50 cycles using 60°C as the annealing temperature with the absolute quantification option.
Microscopy.
To resolve whether cells grew freely in the medium or whether they were associated in aggregate structures, cells were visualized by phase-contrast microscopy on a Nikon Eclipse E600 microscope.
To resolve the cell abundance and overall distribution of the two microorganisms in the cocultures, cells were fixed [2% paraformaldehyde and 0.5% glutaraldehyde in 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) at pH 7.2] for 1 h at room temperature, and a droplet was placed on a gelatin-coated slide, dried at 46°C for 5 min, and then dehydrated in 70% ethanol for 30 min at 4°C. Dehydrated samples were hybridized as described previously (29) using the probes PCARB1 (5′-Cy3-GCCTATTCGACCACGATA-3′), specific for P. carbinolicus (31), and GEO2 (5′-Cy5-GAAGACAGGAGGCCCGAAA-3′), specific for G. sulfurreducens (37). Samples were visualized on a Leica TCS SP5 confocal fluorescence microscope using consecutive line scanning to detect Cy3 and Cy5 fluorochromes.
Identification of OmcS cytochrome content in cocultures.
OmcS abundance was determined in P. carbinolicus-G. sulfurreducens and G. metallireducens-G. sulfurreducens cocultures versus G. sulfurreducens cells grown on freshwater medium with 40 mM fumarate and 10 mM acetate as substrates (8). Cells were retrieved during the late stages of mid-exponential growth, and the whole-cell lysates obtained (5 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by immunoblotting and probing with an OmcS-specific antiserum as previously described (37).
Analytical techniques.
For determination of substrate depletion and production of metabolic products, samples were withdrawn with hypodermic needles and syringes under strict anaerobic conditions and passed through 0.2-μm Acrodisc filters. A minimum of three biological replicates were analyzed for each coculture type. Volatile fatty acids were monitored by high-performance liquid chromatography (HPLC) as previously described (28). Changes in ethanol concentration over time were monitored by gas chromatography (GC) as previously described (27).
RESULTS AND DISCUSSION
Syntrophic growth on ethanol.
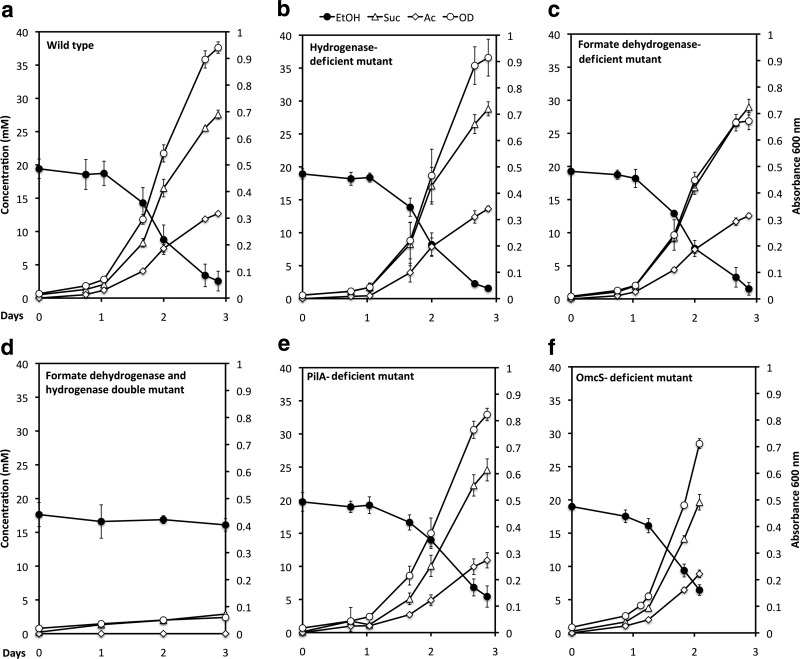
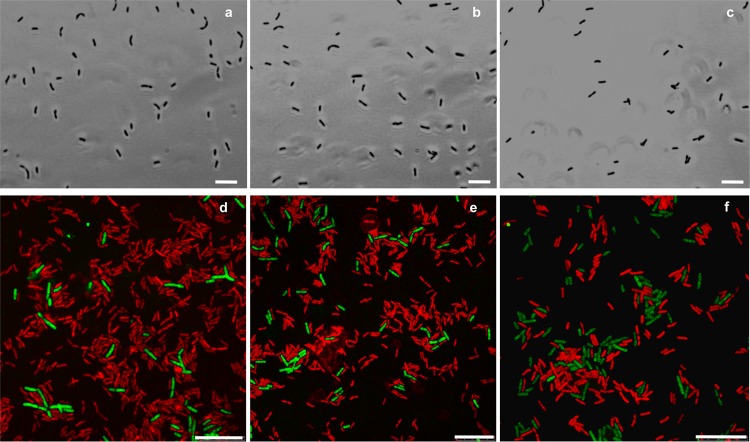
When P. carbinolicus and G. sulfurreducens were simultaneously inoculated into a medium with ethanol as the electron donor and fumarate as the electron acceptor, the coculture grew with the metabolism of ethanol and the reduction of fumarate to succinate (Fig. 1 and 2a). In contrast to the previously described cocultures of G. metallireducens and G. sulfurreducens, which lagged for several weeks before utilizing significant ethanol (37), growth and metabolism of the P. carbinolicus-G. sulfurreducens cocultures typically began within a day (Fig. 1). Furthermore, the P. carbinolicus-G. sulfurreducens cocultures metabolized most of the ethanol provided in 3 days, whereas even after months of adaptation for syntrophic growth, the G. metallireducens-G. sulfurreducens cocultures still required 5 days to metabolize 70% of the added ethanol (37). Although G. metallireducens-G. sulfurreducens cocultures formed large (>1-mm) aggregates (37), the P. carbinolicus-G. sulfurreducens cocultures did not aggregate even after 400 consecutive transfers of the coculture. The cells did not appear to form physical associations, even at the level of individual cells (Fig. 3a). Contact between syntrophic partners is considered to be a requirement for DIET, and although it may also facilitate interspecies H2 or formate transfer (5, 14, 39), long-term coculture studies demonstrated that contact is not necessary for the later (13). Examination of the coculture with fluorescent in situ hybridization (FISH) probes specific for the two species revealed that G. sulfurreducens was more abundant than P. carbinolicus (Fig. 3).
Fig 1.
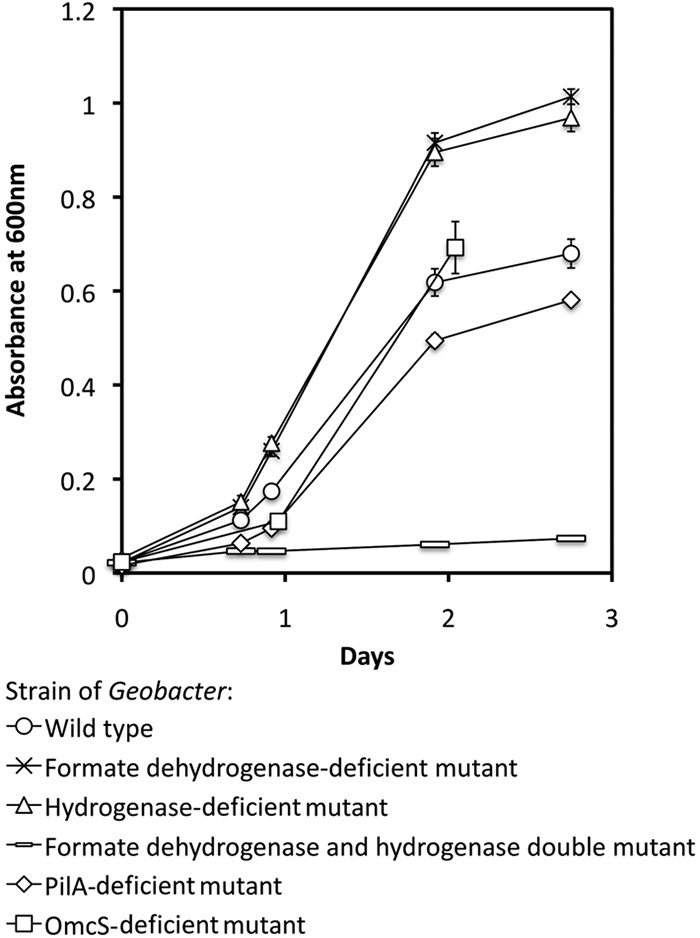
Initial growth of cocultures in ethanol-fumarate medium started with P. carbinolicus and different strains of G. sulfurreducens. The results are the means and standard deviations for triplicate cultures.
Fig 2.
Growth (optical density [OD]), ethanol (EtOH) metabolism, acetate (Ac) accumulation, and succinate (Suc) production from fumarate reduction after more than five consecutive transfers of cocultures of P. carbinolicus with different strains of G. sulfurreducens. Also shown are the data from the initial attempt to start a coculture with a strain of G. sulfurreducens unable to utilize formate or H2. The results are the means and standard deviations for triplicate cultures.
Fig 3.
Phase-contrast (a, b, and c) and epifluorescence (d, e, and f) micrographs of P. carbinolicus cells in coculture with G. sulfurreducens wild-type cells (a and d), the hydrogenase-deficient G. sulfurreducens strain (b and e), or the strain deficient in formate dehydrogenase (c and f). Epifluorescence of in situ hybridized cells with P. carbinolicus is shown as green, and that with G. sulfurreducens is shown as red. Scale bars, 10 μm.
Interspecies electron transfer via H2 or formate.
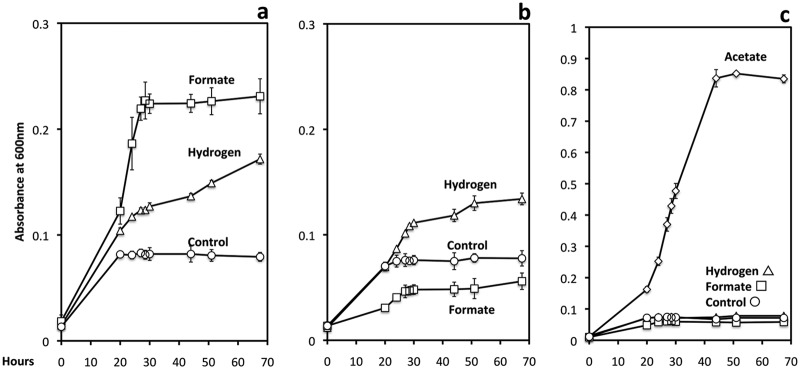
In order to evaluate the possibility of interspecies H2 or formate transfer, cocultures were initiated with one of the following strains of G. sulfurreducens: (i) a strain that could not metabolize H2 because the gene for the large subunit of the uptake hydrogenase (hybL) was deleted (10), (ii) a strain that could not grow on formate because the gene for the catalytic subunit of formate dehydrogenase (fdnG) was deleted (Fig. 4b), or (iii) a strain that could not grow on H2 or formate because both hybL and fdnG were deleted (Fig. 4c). Cocultures initiated with G. sulfurreducens strains that could metabolize only formate (Fig. 2b) or only H2 (Fig. 2c) readily metabolized ethanol with the reduction of fumarate.
Fig 4.
Growth on formate (10 mM) or H2 (138 KPa) in the presence of 1 mM acetate for wild-type G. sulfurreducens (a), a strain deficient in a formate dehydrogenase subunit (b), or a strain deficient in both a formate dehydrogenase subunit and an uptake hydrogenase subunit (c). In controls without added hydrogen or formate, the acetate added as a carbon source could also serve as an electron donor to support growth. Growth of the double mutant growth on 15 mM acetate is also shown (c) to demonstrate that cells were viable yet unable to grow on formate or H2. The results are the means and standard deviations for triplicate cultures.
However, growth and ethanol metabolism did not proceed in cocultures initiated with a strain of G. sulfurreducens that could not metabolize either H2 or formate (Fig. 2d). These results indicate that either H2 or formate can serve as an electron carrier for interspecies electron transfer, and interspecies electron transfer via one of these two electron carriers was the only mechanism by which the coculture could function. In contrast, G. metallireducens formed well-functioning syntrophic cultures with the G. sulfurreducens strain that could not utilize H2 and formate, consistent with the concept of DIET in that coculture system (see Fig. SM1 in the supplemental material).
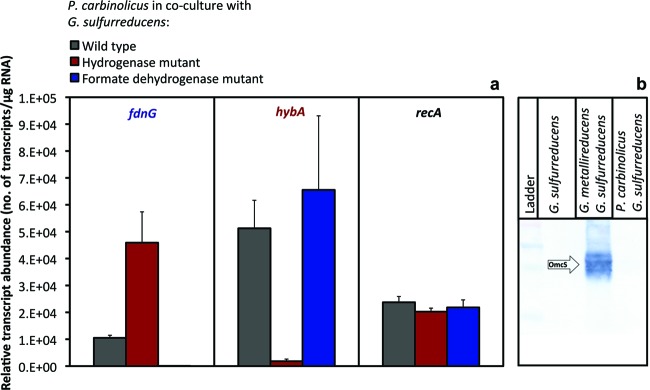
In order to evaluate the potential contributions of H2 and formate as electron carriers between P. carbinolicus and G. sulfurreducens, the transcript abundance of an uptake hydrogenase subunit (hybA product) and the large subunit of formate dehydrogenase (fdnG product) were monitored (Fig. 5a). When H2 uptake was not possible, G. sulfurreducens adapted with increased expression of fdnG (P = 0.009). In contrast, when formate metabolism was inhibited, the transcript abundance of hybA was not significantly different (P = 0.5) from that in the wild type (Fig. 5a). These results, and the fact that hybA transcripts were much more abundant than fdnG transcripts in the wild type, suggest that although the cocultures could function via either interspecies H2 or formate transfer, H2 was the primary electron carrier between species in cocultures with wild-type G. sulfurreducens.
Fig 5.
Molecular analysis of cocultures. (a) Relative transcript abundances of the formate dehydrogenase gene (fdnG), the hydrogenase gene (hybA), and the housekeeping gene recA in P. carbinolicus-G. sulfurreducens cocultures as determined by RT-qPCR. Results are the means and standard deviations for triplicate cultures. (b) Western blot analysis of OmcS in equivalent cell protein of G. sulfurreducens grown with fumarate as the electron acceptor or ethanol-fumarate cocultures of P. carbinolicus-G. sulfurreducens or G. metallireducens-G. sulfurreducens cocultures.
In contrast to the case for G. metallireducens-G. sulfurreducens cocultures (see Fig. SM1 in the supplemental material), acetate accumulated over time in P. carbinolicus-G. sulfurreducens cocultures (Fig. 1). The likely explanation for this difference is that the expression of citrate synthase in G. sulfurreducens is inhibited in the presence of H2, preventing acetate metabolism (4, 38). Thus, the availability of H2 in P. carbinolicus-G. sulfurreducens cocultures would be expected to limit acetate metabolism of G. sulfurreducens, whereas no such inhibition of acetate metabolism is expected in G. metallireducens-G. sulfurreducens cocultures because of the lack of H2 production during DIET.
Pili and OmcS are not required during H2/formate electron transfer.
Deleting the gene for PilA or OmcS in G. sulfurreducens did not prevent P. carbinolicus from forming effective cocultures (Fig. 2e and 2f, respectively). This contrasts with the previous finding (37) that G. metallireducens-G. sulfurreducens cocultures could not be established if either pilA or omcS was deleted from G. sulfurreducens (37). As previously reported (37), G. sulfurreducens expressed OmcS at high levels in G. metallireducens-G. sulfurreducens cocultures, but OmcS was not detected in P. carbinolicus-G. sulfurreducens cocultures (Fig. 5b). These results suggest that the model for DIET between G. metallireducens and G. sulfurreducens, in which OmcS and pili are important components of the electrical connection between the two species (20, 37), does not apply to the P. carbinolicus-G. sulfurreducens coculture.
Implications.
These findings demonstrate that not all microorganisms that can grow syntrophically via interspecies electron exchange are capable of DIET and that even closely related microorganisms may differ in their modes of syntrophic growth. The finding that P. carbinolicus was not able to directly transfer electrons to another species capable of DIET is consistent with previous findings which suggest that P. carbinolicus is poorly suited for direct electron transfer to insoluble extracellular electron acceptors, such as electrodes (31) and Fe(III) oxide (12). The ability to growth syntrophically via interspecies hydrogen/formate transfer but not DIET may be common in laboratory cocultures. For example a syntrophic coculture of Desulfovibrio vulgaris and Methanococcus maripaludis did not form aggregates even after 300 generations (13), suggesting a lack of DIET in that system as well.
Although there is evidence for DIET in microbial aggregates from methanogenic wastewater digesters (27) the prevalence of DIET in natural environments and the factors that might favor DIET over interspecies H2 and formate transfer are unknown. It may be that G. metallireducens interacts with G. sulfurreducens via DIET because it is well suited for extracellular electron transfer (22) but has limited ability to produce H2 (11).
Metabolizing substrates with the release of electrons as H2 or formate requires less coordination with syntrophic partners than DIET and may account for the ability of the P. carbinolicus-G. sulfurreducens cocultures to initiate syntrophic growth much faster and to metabolize ethanol more rapidly than G. metallireducens-G. sulfurreducens cocultures. Another consideration is that consortia cooperating via DIET must bear the additional energetic investment of producing the proteins necessary to establish the electrical connections required for DIET. However, the high abundance of Geobacter species in electrically conductive aggregates from methanogenic digesters (27) and the finding that addition of conductive or semiconductive supplementary materials enhance DIET with increased rates of methanogenesis in sediments (16) and methanogenic digester aggregates (19) suggest that DIET can be more favorable than interspecies H2/formate transfer in important methane-producing environments. Genome-scale metabolic modeling might offer an approach for calculating the cost/benefit relationships of the different strategies for interspecies electron transfer under diverse environmental conditions, as evidenced by the ability of this approach to effectively predict the outcome of microbial competition in different subsurface environments (41).
The physiological differences between microorganisms that are effective in DIET and those that rely on interspecies H2/formate transfer are important considerations when attempting to enrich and isolate syntrophic microorganisms capable of DIET. Common procedures for the isolation of syntrophic microorganisms, such as the use of fermentable substrates (33) or coculturing with an H2-consuming partner (24), may fail to recover organisms that specialize in DIET. Thus, new approaches for the isolation and study of syntrophic interactions are required to better assess the diversity and environmental relevance of microorganisms capable of DIET.
Supplementary Material
ACKNOWLEDGMENTS
We thank Trevor Woodard and Jaclyn Izbicki for facilitating HPLC and GC analysis, Stefan Hansen for developing the chromatography data extractor software for HPLC data screening, Muktak Aklujkar for helpful comments on the manuscript, and Dan Carlo Flores and Molie Murnane for lab assistance.
This research was supported by the Office of Science (BER), U.S. Department of Energy, award no. DE-SC0004485.
Footnotes
Published ahead of print 24 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ahring BK, Westermann P. 1988. Product inhibition of butyrate metabolism by acetate and hydrogen in a thermophilic coculture. Appl. Environ. Microbiol. 54:2393–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahring BK, Westermann P. 1987. Thermophilic anaerobic degradation of butyrate by a butyrate-utilizing bacterium in coculture and triculture with methanogenic bacteria. Appl. Environ. Microbiol. 53:429–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bond DR, et al. 2005. Characterization of citrate synthase from Geobacter sulfurreducens and evidence for a family of citrate synthases similar to those of eukaryotes throughout the Geobacteraceae. Appl. Environ. Microbiol. 71:3858–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boone DR, Johnson RL, Liu Y. 1989. Diffusion of the interspecies electron carriers H2 and formate in methanogenic ecosystems and its implications in the measurement of Km for H2 or formate uptake. Appl. Environ. Microbiol. 55:1735–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bryant MP, Wolin EA, Wolin MJ, Wolfe RS. 1967. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch. Mikrobiol. 59:20–31 [DOI] [PubMed] [Google Scholar]
- 7. Butler JE, Young ND, Lovley DR. 2009. Evolution from a respiratory ancestor to fill syntrophic and fermentative niches: comparative fenomics of six Geobacteraceae species. BMC Genomics 10:103 doi:10.1186/1471-2164-10-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coppi MV, Leang C, Sandler SJ, Lovley DR. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67:3180–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coppi MV, et al. 2007. Involvement of Geobacter sulfurreducens SfrAB in acetate metabolism rather than intracellular, respiration-linked Fe(III) citrate reduction. Microbiology 153:3572–3585 [DOI] [PubMed] [Google Scholar]
- 10. Coppi MV, O'Neil RA, Lovley DR. 2004. Identification of an uptake hydrogenase required for hydrogen-dependent reduction of Fe(III) and other electron acceptors by Geobacter sulfurreducens. J. Bacteriol. 186:3022–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cord-Ruwisch R, Lovley DR, Schink B. 1998. Growth of Geobacter sulfurreducens with acetate in syntrophic cooperation with hydrogen-oxidizing anaerobic partners. Appl. Environ. Microbiol. 64:2232–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haveman SA, et al. 2008. Genome-wide gene expression patterns and growth requirements suggest that Pelobacter carbinolicus reduces Fe(III) indirectly via sulfide production. Appl. Environ. Microbiol. 74:4277–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hillesland KL, Stahl DA. 2010. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc. Natl. Acad. Sci. U. S. A. 107:2124–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ishii S, Kosaka T, Hori K, Hotta Y, Watanabe K. 2005. Coaggregation facilitates interspecies hydrogen transfer between Pelotomaculum thermopropionicum and Methanothermobacter thermautotrophicus. Appl. Environ. Microbiol. 71:7838–7845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaden J, SG A, Schink B. 2002. Cysteine-mediated electron transfer in syntrophic acetate oxidation by cocultures of Geobacter sulfurreducens and Wolinella succinogenes. Arch. Microbiol. 178:53–58 [DOI] [PubMed] [Google Scholar]
- 16. Kato S, Hashimoto K, Watanabe K. 2012. Methanogenesis facilitated by electric syntrophy via (semi)conductive iron-oxide minerals. Environ. Microbiol. 14:1646–1654 [DOI] [PubMed] [Google Scholar]
- 17. Kovach ME, Phillips RW, Elzer PH, Roop RM, 2nd, Peterson KM. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800–802 [PubMed] [Google Scholar]
- 18. Leang C, Qian X, Mester T, Lovley DR. 2010. Alignment of the c-type cytochrome OmcS along pili of Geobacter sulfurreducens. Appl. Environ. Microbiol. 76:4080–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu F, et al. Promoting direct interspecies electron transfer with activated carbon. Energy Environ. Sci., in press [Google Scholar]
- 20. Lovley DR. 2011. Live wires: direct extracellular electron exchange for bioenergy and the bioremediation of energy-related contamination. Energy Environ. Sci. 4:4896–4906 [Google Scholar]
- 21. Lovley DR, Fraga JL, Coates JD, Blunt-Harris EL. 1999. Humics as an electron donor for anaerobic respiration. Environ. Microbiol. 1:89–98 [DOI] [PubMed] [Google Scholar]
- 22. Lovley DR, et al. 2011. Geobacter: the microbe electric's physiology, ecology, and practical applications. Adv. Microb. Physiol. 59:1–100 [DOI] [PubMed] [Google Scholar]
- 23. Malvankar NS, et al. 2011. Tunable metallic-like conductivity in microbial nanowire networks. Nat. Nanotechnol. 6:573–579 [DOI] [PubMed] [Google Scholar]
- 24. McInerney MJ, et al. 2008. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann. N. Y. Acad. Sci. 1125:58–72 [DOI] [PubMed] [Google Scholar]
- 25. Mehta T, Coppi MV, Childers SE, Lovley DR. 2005. Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 71:8634–8641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Methe BA, et al. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302:1967–1969 [DOI] [PubMed] [Google Scholar]
- 27. Morita M, et al. 2011. Potential for direct interspecies electron transfer in methanogenic wastewater digester aggregates. mBio 2(4):e00159–11 doi: 10.1128/mBio.00159-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nevin KP, et al. 2008. Power output and columbic efficiencies from biofilms of Geobacter sulfurreducens comparable to mixed community microbial fuel cells. Environ. Microbiol. 10:2505–2514 [DOI] [PubMed] [Google Scholar]
- 29. Pernthaler J, Glockner FO, Schonhuber W, Amann R. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207–226 [Google Scholar]
- 30. Reguera G, et al. 2005. Extracellular electron transfer via microbial nanowires. Nature 435:1098–1101 [DOI] [PubMed] [Google Scholar]
- 31. Richter H, Lanthier M, Nevin KP, Lovley DR. 2007. Lack of electricity production by Pelobacter carbinolicus indicates that the capacity for Fe(III) oxide reduction does not necessarily confer electron transfer ability to fuel cell anodes. Appl. Environ. Microbiol. 73:5347–5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schink B. 1984. Fermentation of 2,3-butanediol by Pelobacter carbinolicus sp. nov. and Pelobacter propionicus sp. nov., and evidence for propionate formation from C2 compounds. Arch. Microbiol. 137:33–41 [Google Scholar]
- 33. Schink B, Stams AJM. 2006. Syntrophism among prokaryotes, p 309–335 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes, 3rd ed, vol 2 Springer, Berlin, Germany [Google Scholar]
- 34. Shrestha M, Shrestha PM, Frenzel P, Conrad R. 2010. Effect of nitrogen fertilization on methane oxidation, abundance, community structure, and gene expression of methanotrophs in the rice rhizosphere. ISME J. 4:1545–1556 [DOI] [PubMed] [Google Scholar]
- 35. Sieber JR, McInerney MJ, Gunsalus RP. 2012. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu. Rev. Microbiol. 66:429–452 [DOI] [PubMed] [Google Scholar]
- 36. Stams AJ. 1994. Metabolic interactions between anaerobic bacteria in methanogenic environments. Antonie Van Leeuwenhoek 66:271–294 [DOI] [PubMed] [Google Scholar]
- 37. Summers ZM, et al. 2010. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330:1413–1415 [DOI] [PubMed] [Google Scholar]
- 38. Ueki T, Lovley DR. 2010. Genome-wide gene regulation of biosynthesis and energy generation by a novel transcriptional repressor in Geobacter species. Nucleic Acids Res. 38:810–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu W, Jain MK, Zeikus JG. 1996. Formation of fatty acid-degrading, anaerobic granules by defined species. Appl. Environ. Microbiol. 62:2037–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu WM, Jain MK, Hickey RF, Zeikus JG. 1996. Perturbation of syntrophic isobutyrate and butyrate degradation with formate and hydrogen. Biotechnol. Bioeng. 52:404–411 [DOI] [PubMed] [Google Scholar]
- 41. Zhuang K, et al. 2011. Genome-scale dynamic modeling of the competition between Rhodoferax and Geobacter in anoxic subsurface environments. ISME J. 5:305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.