Abstract
The meningococcal antigen typing system (MATS) sandwich enzyme-linked immunosorbent assay (ELISA) was designed to measure the immunologic cross-reactivity and quantity of antigens in target strains of a pathogen. It was first used to measure the factor H-binding protein (fHbp), neisserial adhesin A (NadA), and neisserial heparin-binding antigen (NHBA) content of serogroup B meningococcal (MenB) isolates relative to a reference strain, or “relative potency” (RP). With the PorA genotype, the RPs were then used to assess strain coverage by 4CMenB, a multicomponent MenB vaccine. In preliminary studies, MATS accurately predicted killing in the serum bactericidal assay using human complement, an accepted correlate of protection for meningococcal vaccines. A study across seven laboratories assessed the reproducibility of RPs for fHbp, NadA, and NHBA and established qualification parameters for new laboratories. RPs were determined in replicate for 17 MenB reference strains at laboratories A to G. The reproducibility of RPs among laboratories and against consensus values across laboratories was evaluated using a mixed-model analysis of variance. Interlaboratory agreement was very good; the Pearson correlation coefficients, coefficients of accuracy, and concordance correlation coefficients exceeded 99%. The summary measures of reproducibility, expressed as between-laboratory coefficients of variation, were 7.85% (fHbp), 16.51% (NadA), and 12.60% (NHBA). The overall within-laboratory measures of variation adjusted for strain and laboratory were 19.8% (fHbp), 28.8% (NHBA), and 38.3% (NadA). The MATS ELISA was successfully transferred to six laboratories, and a further laboratory was successfully qualified.
INTRODUCTION
Serogroup B Neisseria meningitidis (MenB) is a worldwide public health threat that accounts for 90% of infant meningococcal disease cases in many European countries (6, 13, 23, 26). Unlike serogroups A, C, W-135, and Y, for which broad-coverage polysaccharide protein conjugate vaccines are available, MenB has presented unique challenges to vaccine development because its polysaccharide capsule mimics the human neural cell adhesion molecule (10). Therefore, various subcapsular targets have been investigated since the 1980s (1, 5, 10, 15, 25).
To date, three licensed vaccines against invasive serogroup B meningococcal disease based on wild-type outer membrane vesicles (OMVs) have successfully contained clonal outbreaks in Cuba, Norway, New Zealand, parts of Latin America, and Normandy, France (5, 15, 23, 26). However, these OMV vaccines do not provide protection against strains carrying PorA serosubtypes that differ from that in the vaccine (1, 5, 9, 15, 20, 21, 23, 25–27). To provide protection against genetically diverse MenB strains, several investigational vaccines, including formulations with multiple OMVs, recombinant OMVs, multiple PorA and FetA components, or antigens discovered through genome mining, have been developed (1, 5, 9, 10, 15, 20, 21, 23, 25–27). Of the several investigational formulations that have entered the clinic, two vaccines that contain factor H-binding protein (fHbp), a novel protein antigen, have entered late-stage clinical trials (1, 25, 26). Additional investigational MenB vaccines assessed in the clinic include bi- and hexavalent PorA outer membrane vesicle (OMV) vaccines (9, 20, 21, 23, 26, 27). However, only a single vaccine, 4CMenB, has completed phase 3 clinical trials and is under consideration for licensure, while a bivalent fHbp vaccine has entered late-stage clinical trials (1, 25). 4CMenB has four primary antigenic components: OMV from the New Zealand outbreak strain NZ 98/254, fHbp, neisserial adhesin A (NadA), and neisserial heparin-binding antigen (NHBA) (1, 2, 11, 23, 25).
Genotyping based on sequence variation of housekeeping genes (multilocus sequence typing [MLST]) or surface-expressed proteins such as PorA and FetA shows high genetic diversity among meningococcal populations (16, 19). These patterns of variation do not account for additional surface proteins such as the three recombinant antigens included in 4CMenB (2) because of the high rates of recombination affecting the meningococcal chromosome (4, 8). Moreover, MLST and antigen genotyping do not account for expression levels or cross-reactivity to various antigen variants present on pathogenic strains (3, 7, 19). To determine the potential impact of 4CMenB on endemic MenB bacteria in different countries or regions, a new method was needed to account for fHbp, NadA, and NHBA diversity and expression, which varies among strains. The meningococcal antigen typing system (MATS) was developed as a rapid, reliable system that combines conventional genotyping for PorA with a specialized sandwich enzyme-linked immunosorbent assay (ELISA) that determines phenotypic expression and the cross-reactivity, or relative potency (RP), of fHbp, NadA, and NHBA on individual strains (7).
MenB vaccine development requires the use of immunogenic surface proteins, which can vary in sequence, and expression from strain to strain, which can affect their ability to be targeted by bactericidal antibodies (3, 7, 10, 15, 23). This dynamic profile complicates the task of health authorities and recommending agencies, which must assess whether circulating MenB strains are susceptible to killing by a vaccine-induced immune response (5, 7, 13, 15, 23, 26). Conducting serum bactericidal assay using human complement (hSBA), the accepted immunogenicity endpoint in clinical trials of meningococcal vaccines, against large strain panels to assess vaccine effects poses serious logistical and ethical challenges. To mitigate the need to perform hSBA on large panels of isolates, the MATS method established a minimum level of RP, named the positive bactericidal threshold (PBT) that predicts whether a given MenB isolate would be susceptible to killing in hSBA by antibodies induced by 4CMenB (6, 7).
MATS was developed as a reliable and reproducible means to identify individual MenB strains likely to be covered by 4CMenB (7). Obtaining a large body of current data on strain coverage will be required to support national vaccine recommendations. Moreover, decentralizing such typing could accelerate these efforts and provide a surveillance method to detect the potential emergence of vaccine resistance over time. Given that many countries might need this information, Novartis elected to transfer the MATS ELISA to several national meningococcal reference laboratories. To ensure that results would remain consistent and reproducible wherever they were obtained and to determine criteria for validating additional laboratories, we performed the current interlaboratory standardization study using a defined set of test strains. Seven laboratories (A to G) used a standardized procedure for MATS ELISAs and generated RPs for fHbp, NadA, and NHBA that were subject to analysis based on a comprehensive statistical plan.
MATERIALS AND METHODS
Bacterial strains used.
MenB strains (see Table S1 in the supplemental material) were obtained under materials transfer agreements from the Norwegian Institute of Public Health (Oslo, Norway), the Health Protection Agency (Manchester, United Kingdom), and the Centers for Disease Control and Prevention (Atlanta, GA). These strains were selected to provide examples of a range of MATS ELISA values and not as representative of any particular country, region, or RP distribution.
MATS ELISA.
To measure RPs for fHbp, NHBA, and NadA, ELISAs were performed as described previously (23). Precoated microtiter plates, antibody reagents, substrate buffer, washing buffer, and a written set of instructions were provided in kit form by Novartis Vaccines & Diagnostics. Briefly, bacteria were cultured overnight on chocolate agar (bioMérieux) and then suspended in Mueller-Hinton broth (Difco) to a defined optical density at 600 nm of 0.4. Empigen BB (Sigma) was added, and then the bacterial suspensions were incubated for 1 h as described below for inactivation by the lysis buffer. Serial dilutions of the bacterial lysate were then transferred to microtiter plates previously coated with rabbit polyclonal antibodies specific for fHbp, NHBA, or NadA. The plates were incubated and washed, and an antigen-specific secondary antibody, also prepared in rabbits and labeled with biotin, was used to detect the bound antigens. The plates were developed with streptavidin-horseradish peroxidase and ortho-phenylene diamine substrate (Sigma), the reactions were stopped with H2SO4, and the plates were read at 492 nm. The results were analyzed with StatLIA (Brendan Technologies, Carlsbad, CA) by calculating the RP of the test strain bacterial lysate compared to that of a reference strain that was treated identically and assayed in each microtiter plate (H44/76 for fHbp, NGH38 for NHBA, and 5/99 for NadA).
In each laboratory, at least 12 assays considered acceptable, based on visual evaluation of the dilution curves, were used to constitute a reference data set used by StatLIA to determine mean-variance regressions and typical variability of the standard curve. On this basis, a weighted five-parameter logistic regression was performed for the optical densities of the test samples and of the reference samples of each assay, and the RP was determined by parallelism analysis of the regressions for the test and the reference strain (14, 18). For all laboratories, acceptance criteria previously defined and based on StatLIA P values (7) were used to determine acceptability of each microtiter plate and individual test strain. The following laboratories participated in the study: the Division of Bacterial Diseases, Centers for Disease Control and Prevention (Atlanta, GA); Novartis Vaccines & Diagnostics (Siena, Italy); the National Reference Centre for Meningococci, IBI Unit, Institut Pasteur (Paris, France), the Reference Laboratory for Meningococci, Institute of Health Carlos III (Madrid, Spain), the National Reference Laboratory for Meningococci, University of Würzburg, Institute for Hygiene and Microbiology (Würzburg, Germany), the Manchester Laboratory of the Health Protection Agency (Manchester, United Kingdom), the Department of Infectious Diseases, Istituto Superiore di Sanità (Rome, Italy), and the Department of Bacteriology and Immunology, Norwegian Institute of Public Health (Oslo, Norway). Laboratories participating to the standardization study were randomly designated A to G. A laboratory that performed the qualification study was designated H.
Study design.
Seven laboratories (A to G) performed multiple MATS ELISAs for fHbp, NHBA, and NadA on 17 shared strains (Table 1; see also Table S1 in the supplemental material).
Table 1.
Laboratories A to G participating in the interlaboratory study and the number of strains assayed at two temperatures for bacterial lysate preparation
| Laboratory | No. of strainsa |
|||||
|---|---|---|---|---|---|---|
| fHbp |
NHBA |
NadA |
||||
| 37°C | 45°C | 37°C | 45°C | 37°C | 45°C | |
| A | 10 (10) | 2 (2) | 17 (17) | 6 (6) | 8 (8) | 3 (3) |
| B | 13 (10) | 3 (2) | 17 (17) | 6 (6) | 8 (8) | 3 (3) |
| C | 17 (10) | 12 (10) | 17 (17) | 17 (17) | 10 (9) | 7 (7) |
| D | 16 (10) | 1 (1) | 17 (17) | 6 (6) | 8 (8) | 2 (2) |
| E | 10 (10) | 2 (2) | 17 (17) | 6 (6) | 8 (8) | 2 (2) |
| F | 17 (10) | 0 (0) | 16 (16) | 0 (0) | 8 (8) | 0 (0) |
| G | 0 (0) | 15 (11) | 0 (0) | 17 (17) | 0 (0) | 13 (11) |
The numbers of strains with assayed values above LLOQ that define the core of reference MATS isolates appear in parentheses. The lysate preparation temperatures are indicated in the column subheadings.
We previously (7) defined provisional lower limits of quantitation (LLOQs) for fHbp, NadA, and NHBA based on data from one laboratory. In the present study, analysis of variance (ANOVA) models using data from laboratories A to G were used to redefine these limits as 0.009, 0.04, and 0.0009 for fHbp, NHBA, and NadA, respectively. RP values less than these were excluded from the present analysis.
During the present study, antigen extraction at 37°C (room temperature) permitted the survival of small numbers of bacteria at some laboratories, which could potentially pose a safety risk for laboratory workers. Retesting revealed that antigen extraction at 45°C for 1 h resulted in complete inactivation of bacterial suspensions for all laboratories. Therefore, the temperature for bacterial lysate preparation was changed to 45°C. Five laboratories assayed strains at both temperatures to determine possible effects on RP values. Laboratory G used 45°C, and laboratory F used 37°C only (Table 1).
Construction of a core of reference MATS strains.
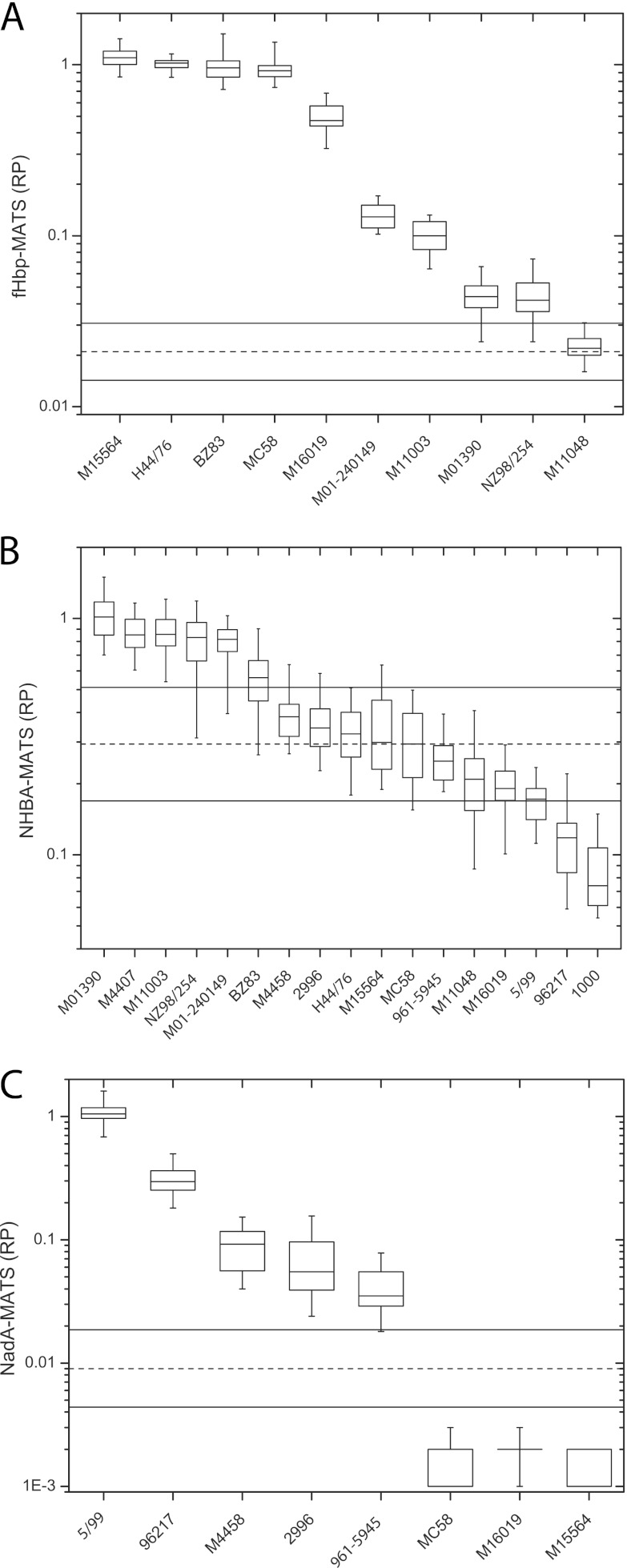
RPs below the LLOQ were excluded from all analyses. For fHbp and NadA, all replicate RPs for several strains were below the LLOQ, leaving core sets of 10 (fHbp), 17 (NHBA), and 8 (NadA) strains (see Fig. 1A, B, and C, respectively).
Fig 1.
Reference MATS strains and robustness of positive bactericidal thresholds (PBTs) for fHbp (A), NHBA (B), and NadA (C). Box plots show the median and interquartile ranges of RPs collected during the present study, combined over laboratories A to G. Vertical lines extend to the most extreme observation that is <1.5 × the interquartile distance (75th to 25th percentiles). Each box represents all replicate assay data for each strain and antigen combined over all laboratories. Dashed and solid horizontal lines indicate PBT and 95% CIs derived according to equation 1.
Statistical analysis.
RPs were log10 transformed prior to analysis. For the 17 test strains, consensus RP values were estimated across the laboratories A to G, using an ANOVA mixed-effects model. Consensus RPs were then used to quantify accuracy, reproducibility, repeatability, precision, and bias within and among the laboratories. Accuracy, defined as the closeness of a laboratory-assayed value to the consensus value, was measured using Lin's coefficient of accuracy (Ca) (25). Precision, a measure of how far a set of observations deviate from a fitted straight line, was quantified using Pearson correlation coefficient (r). Lin's concordance correlation coefficient (rc), a combination of Ca and r, was used to form a single statistic describing accuracy and precision. Repeatability, a measure of intralaboratory variation (for replicate assays), and reproducibility, a measure of interlaboratory variation (an estimate of the overall error) of the assay, were expressed as coefficients of variation (CVs). Bias is a measure of directional error (consistent offset) of the laboratory titer compared to the consensus RP (26).
Linear mixed-effects ANOVA models were used to estimate consensus values for each strain and antigen, as well as assay repeatability and reproducibility. All models were fit independently by antigen and included strain and laboratory as random effects. Since each strain was assayed in replicate, a single predicted RP was estimated using the ANOVA models to represent the replicate values for analysis and comparison of laboratories in the figures and tables. Prediction intervals used to develop acceptance criteria for future laboratories were also derived from these ANOVA models.
The estimated values from the ANOVA models were used to assess and evaluate the ability of laboratories A to G to reproduce RPs and to test the consistency of the RPs with consensus values. Laboratory bias was quantified by comparing observed laboratory values to consensus values. For each individual antigen, the mean bias was expected to be zero.
Development of qualification criteria for new laboratories.
Anticipating the need for additional laboratories to use the MATS ELISA, the present data were used to suggest qualification criteria. A protocol was then set up to ensure that new laboratories produced results within tolerance limits defined by the variability measured in the present study. This approach also would ensure that new laboratories developed adequate proficiency to qualify to use the MATS ELISA. A laboratory that met these specifications would be considered qualified to assay new strains, and the resulting RPs would be considered consistent with those measured by laboratories A to G.
RESULTS
MATS strains span a wide assay range.
Figure 1 displays the distribution of RPs for each strain and antigen. Each box represents all replicate assay data for each strain and antigen combined over all laboratories.
MATS RPs were robust in response to temperature change during lysate preparation.
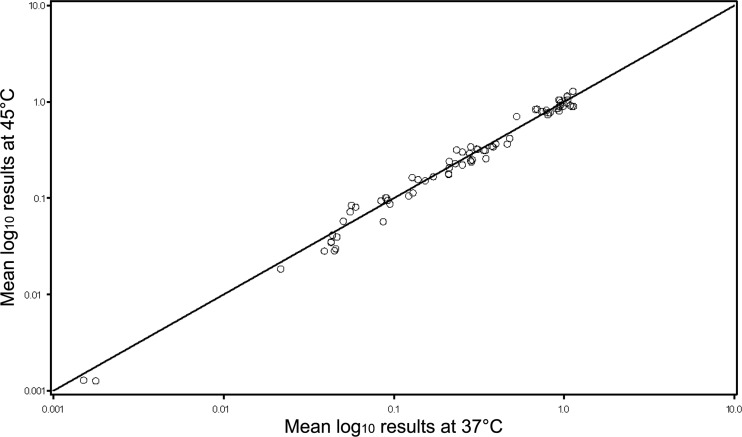
ANOVA models were constructed adjusting for laboratory (A to G), antigen, as well as lysate preparation temperature, from the replicate RPs submitted for analysis. For the five laboratories that assayed the strains at both temperatures, the predicted RPs were derived for each laboratory, strain, antigen, and temperature combination, yielding 75 pairs of measurements that are plotted in Fig. 2. In this scatter plot, the line of identity represents perfect agreement (intercept = 0, slope = 1). Agreement was generally excellent, with data clustered tightly over the line of identity. Lin's coefficient of accuracy (Ca) was 0.999 (95% confidence interval [CI] = 0.997 to 1.00). Precision was high; the Pearson correlation coefficient was 0.993 (95% CI = 0.988 to 0.995), and the combined concordance correlation coefficient (rc) was 0.992 (95% CI = 0.987 to 0.995). Due to this high level of agreement, we combined results at 37 and 45°C for all remaining analyses.
Fig 2.
Comparison RPs at two temperatures for bacterial lysate preparation. For the five laboratories (A to E) that assayed the strains at both temperatures, the predicted RPs were derived for each laboratory, strain, antigen, and temperature combination (n = 75 pairs). The line of identity represents perfect agreement (intercept = 0, slope = 1).
Interlaboratory correlations of MATS RPs were close to identity.
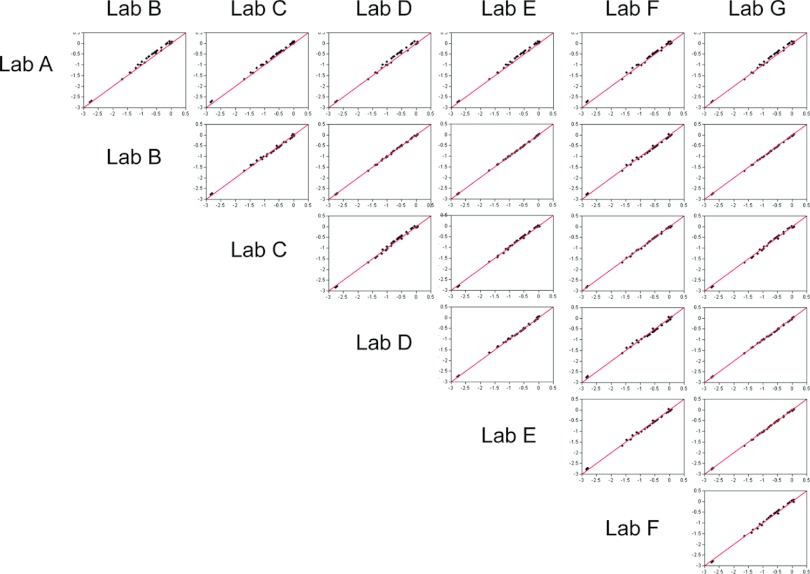
ANOVA models were used to estimate single RPs for each strain by laboratory and antigen. Scatter plots comparing these predicted RPs among laboratories A to G and combined over the three antigens are shown in Fig. 3. Most lab-to-lab comparisons yielded clusters of points centered over the line of identity (which represents perfect agreement), indicating good agreement between laboratories A to G. Table 2 presents a comparison of RPs among laboratories A to G for accuracy (Ca), precision (Pearson r), and concordance correlation coefficient (rc), combined over the three antigens. Accuracy, precision, and concordance were all high and exceeded 0.99. The RPs for laboratory A were slightly higher than those for laboratories B to G, as indicated by the small upward shift in the point clusters in Fig. 3; however, no significant impact on the quality of the interlaboratory correlations, whose 95% lower confidence bounds (LCB) remained ≥0.984, was detected (Table 2).
Fig 3.
Scatter plots of pairwise comparisons of RP values derived from ANOVA models, adjusting for laboratory (A to G), antigen, and lysate preparation temperature, from the replicate RPs submitted for analysis. The data are combined over antigens and plotted on a log10 scale. Predicted RPs were derived from ANOVA random-effects models. The number of strains in common between any two laboratories is listed in Table 2. The solid line indicates perfect agreement (intercept = 0, slope = 1).
Table 2.
RP comparison between laboratories for accuracy, precision, and concordance correlation coefficient combined over antigens for laboratories A to Ga
| Laboratory | Statistic | Laboratory |
|||||
|---|---|---|---|---|---|---|---|
| B | C | D | E | F | G | ||
| A | n | 35 | 35 | 35 | 35 | 34 | 35 |
| Accuracy (Ca) | 0.995 | 0.997 | 0.996 | 0.996 | 0.999 | 0.999 | |
| Precision (r) | 0.997 | 0.999 | 0.995 | 0.998 | 0.997 | 0.997 | |
| CCC (rc) | 0.992 | 0.996 | 0.991 | 0.995 | 0.996 | 0.995 | |
| 95% CI | 0.986–0.996 | 0.993–0.998 | 0.984–0.995 | 0.99–0.997 | 0.992–0.998 | 0.991–0.998 | |
| B | n | 35 | 35 | 35 | 34 | 35 | |
| Accuracy (Ca) | 0.999 | 1 | 1 | 0.998 | 0.999 | ||
| Precision (r) | 0.998 | 1 | 1 | 0.998 | 1 | ||
| CCC (rc) | 0.998 | 1 | 1 | 0.996 | 0.999 | ||
| 95% CI | 0.996–0.999 | 0.999–1 | 1–1 | 0.993–0.998 | 0.998–0.999 | ||
| C | n | 35 | 35 | 34 | 35 | ||
| Accuracy (Ca) | 0.999 | 1 | 0.999 | 0.999 | |||
| Precision (r) | 0.997 | 0.999 | 0.999 | 0.998 | |||
| CCC (rc) | 0.996 | 0.999 | 0.999 | 0.997 | |||
| 95% CI | 0.993–0.998 | 0.997–0.999 | 0.998–0.999 | 0.995–0.999 | |||
| D | n | 35 | 34 | 35 | |||
| Accuracy (Ca) | 1 | 0.998 | 0.999 | ||||
| Precision (r) | 0.999 | 0.998 | 1 | ||||
| CCC (rc) | 0.999 | 0.996 | 0.999 | ||||
| 95% CI | 0.998–1 | 0.993–0.998 | 0.999–1 | ||||
| E | n | 34 | 35 | ||||
| Accuracy (Ca) | 0.999 | 0.999 | |||||
| Precision (r) | 0.998 | 1 | |||||
| CCC (rc) | 0.997 | 0.999 | |||||
| 95% CI | 0.995–0.998 | 0.999–1 | |||||
| F | n | 34 | |||||
| Accuracy (Ca) | 0.999 | ||||||
| Precision (r) | 0.999 | ||||||
| CCC (rc) | 0.998 | ||||||
| 95% CI | 0.996–0.999 | ||||||
Relative potency (RP) comparisons between laboratories for accuracy (Ca), precision (Pearson's r), and concordance correlation coefficient (rc) combined over antigens for laboratories A to G are shown. The predicted RPs were obtained for laboratories A to G by sample within an antigen for each series of replicate values using random-effects ANOVA models. n represents the number of strains in common over the three antigens with a maximum of 35 (10 fHbp + 17 NHBA + 8 NadA).
MATS repeatability supports the within-laboratory discriminatory power for RPs.
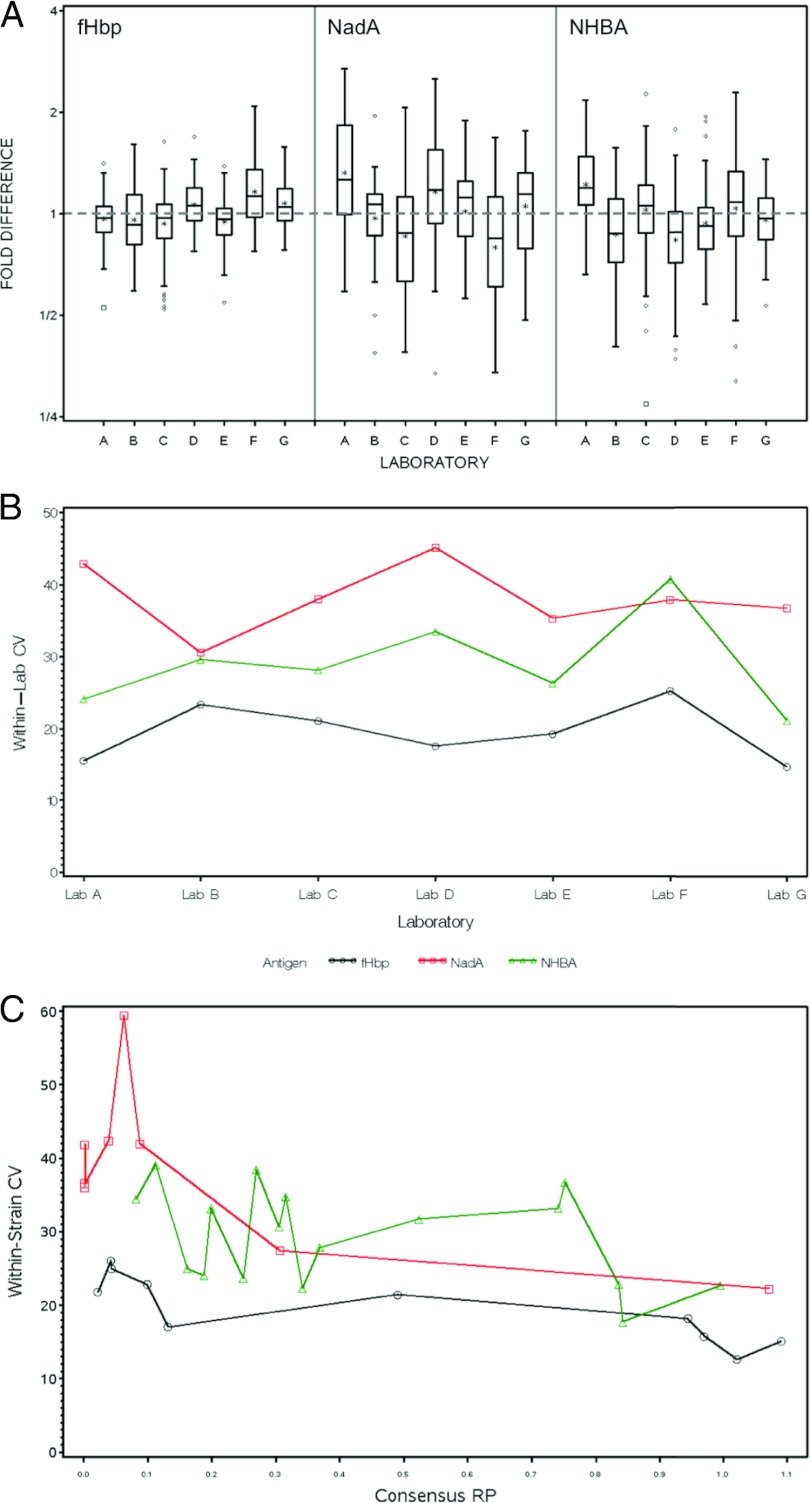
ANOVA models were used to estimate consensus RPs for each antigen and strain. Box plots displaying the distribution of the fold differences between the individual laboratory-reported and consensus RPs by antigen are shown by antigen in Fig. 4A. The total span of the box plot, including the vertical lines above and below the interquartile range, is a direct indicator of within-laboratory variability (repeatability, see Table S2 in the supplemental material for variances and CVs). For example, boxes centered about the gray dashed line in Fig. 4 (which represents the consensus value) with vertical lines extending between 1/2 and 2 indicate an experimental distribution of RPs within a ±2-fold difference from the consensus value. The experimental ranges of the assay from LLOQ to the maximum observed RP were 200-, 50-, and 2,000-fold for fHbp, NHBA, and NadA, respectively The largest within-laboratory variability (or worst repeatability) for NHBA was observed in laboratory F, which had a ±2.3-fold difference between the two ends of the vertical lines of the box. The 50-fold experimental range for NHBA can be subdivided in ∼11 nonoverlapping 4.6-fold ranges from LLOQ to the maximum RP observed, which represent the minimum discriminatory power of a standardized laboratory for NHBA-RPs. For fHbp and NadA, which each had a wider experimental range, a better within-lab discriminatory power can be calculated in the same way. A quantitative measure of within-laboratory CVs by antigen and adjusted for strain is shown in Fig. 4B. For laboratories A to G, fHbp displayed the lowest degree of within-laboratory variance, NHBA showed an intermediate level of variance, and NadA had the highest degree of variance, although laboratory F reported a slightly higher CV for NHBA than for NadA.
Fig 4.
(A) Box plots by antigen and laboratory (A to G) for the fold difference between the consensus and observed RPs. Consensus RPs were estimated for each strain and antigen using a random-effects ANOVA model. The box is defined by the 25th and 75th percentiles of the distribution; the horizontal line in the box represents the median (50th percentile) and the asterisks (*) signify the means. Vertical lines extend to the most extreme observation that is <1.5 × the interquartile distance (75th to 25th percentiles). Diamonds and small boxes (♢, □) correspond to moderate and severe outlying assay values, respectively. Each box represents the distribution of all replicate data for all strains within a laboratory and antigen. (B) Plots of within-laboratory CVs by antigen derived using random-effects ANOVA models. (C) Plots of within-strain CVs by antigen using random-effects ANOVA models. For fHbp, NadA, and NHBA, the strains are plotted on the x axis using consensus RPs derived from ANOVA models.
MATS RPs are not affected by a systematic bias.
Within-laboratory bias per antigen was also estimated from the ANOVA models, as illustrated in Fig. 4A by the distance of the mean (*) from the gray dashed line at 1, for each laboratory (A to G) and antigen (FHbp, NadA, and NHBA). The mean bias varied across antigens and laboratories and did not display any systematic patterns or associations between bias and variability. For example, laboratory A had substantial positive bias and high variability for NadA, positive bias and lower variability for NHBA, and virtually no bias and low variability for fHbp. Conversely, laboratory F had negative bias and high variability for NadA, no bias and high variability for NHBA, and positive bias and low variability for fHbp.
Between-laboratory variation had only a minor impact on MATS RPs.
In Fig. 4A, the relative positioning of the boxes about the gray dashed line for a given antigen across laboratories A to G gives a visual indication of the between-laboratory variability. Table S2 in the supplemental material lists the between-laboratory CVs for each antigen. Table S2 also tabulates the within-laboratory CV for each lab and antigen. Finally, Table S2 lists the overall within-laboratory CV for each antigen adjusted for laboratory and strain. The within-strain CV is plotted opposite the predicted RPs in Fig. 4C. This plot provides the opportunity to observe whether there is a relationship between variability and the magnitude of the RP. In general, samples with the lowest RP values exhibited the greatest degree of within-strain variability.
Between-laboratory CVs for each antigen were 7.85% (fHbp), 12.60% (NHBA), and 16.51% (NadA). Overall, the within-laboratory CVs estimated from ANOVA models adjusted for strain and laboratory were 19.8, 28.8, and 38.3% for fHbp, NHBA, and NadA, respectively. The between-laboratory CVs were lower than the within-laboratory CVs for each of the three antigens. This indicates that intrinsic biological variables such as bacterial growth contributed more to the overall variability of the assay than did differences between the laboratories.
Interlaboratory-derived 95% CIs for the PBT provided robust vaccine strain coverage predictions.
As previously described, any strain with an RP above the PBT for any antigen is predicted to be covered by 4CMenB (23). To assess the 95% CIs of strain coverage, a sufficient number of replicate assays for each strain could be performed to estimate the 95% CI values for the RP of each strain to be compared to the PBT; however, this strategy is not feasible with large panels of strains. Alternatively, the 95% CI for the PBT could be used to assess coverage with 95% confidence for single RPs from individual strains. Using data from the present study, we derived an empirical estimate of the 95% CI around the PBT as follows:
| (1) |
Since the PBT is strain independent, in equation 1 we used the overall within-laboratory variances σ2 for fHbp, NadA, and NHBA adjusted for strain and laboratory as estimated by our ANOVA models described above (see Table S2 in the supplemental material). The 95% CIs of the PBT obtained for fHbp (0.014 to 0.031, point estimate = 0.021), NadA (0.004 to 0.019, point estimate = 0.009), and NHBA (0.169 to 0.511, point estimate = 0.294) are shown in Fig. 1.
We propose using the PBT 95% CIs to define empirical limits to predict strain coverage by 4CMenB. Comparing individual RPs from tested strains to the PBT 95% CI bounds will account for boundaries around the estimate of strain coverage, defined in terms of both within- and between-laboratory variances. If the number of strains used to generate the estimate is significantly smaller than the population of strains for which vaccine strain coverage is being estimated, then additional corrections may be needed.
One possible limitation of the PBT 95% CI bounds is that among replicate RPs from a single strain some could be both above and below these bounds, which would indicate that the strain is simultaneously covered and not covered and thereby invalidate the model. To confirm that the PBT 95% CI bounds provide a robust measurement for strain coverage, we examined the empirical distribution of experimental RPs at or above the LLOQ for all laboratories and strains tested, as shown in Fig. 1. No strain had replicate RPs that were both above the 95% upper confidence bound (UCB) and below the 95% LCB, either within or across laboratories, indicating that the empirical PBT 95% CI is robust and capable of accounting for within-laboratory and between-laboratory variation.
Qualification guidelines for new laboratories.
Based on our results, we propose the following criteria be used to qualify a laboratory to perform the MATS ELISA to assess 4CMenB. (i) Use the 12 quality-control strains defined in Table S3 in the supplemental material, spanning the range of RPs for fHbp (8/12 strains), NHBA (12/12 strains), and NadA (6/12 strains). (ii) Collect a minimum of 12 assays (microtiter plates) per antigen that have been deemed reasonable by visual evaluation of the reference dilution curves and then build a reference data set and analyze the assays in StatLIA. (iii) Retest test strains that fail StatLIA acceptance criteria (7) to collect a minimum of five acceptable results above the LLOQ per strain, per antigen for those with positive results. (iv) For fHbp, NadA, and NHBA, ensure that all strains display a geometric mean of RPs within the range reported in Table S3 in the supplemental material, and a CV equal to or lower than the maximum CV reported in Table S3. For failures, identify and solve the source of the problem, and repeat the test. A deviation in one of these criteria by a small amount for a single strain per antigen is acceptable. (v) Finally, for fHbp, NadA, and NHBA, ensure that at least 95% of the RPs are within the 95% prediction intervals reported in Table S3 and also that at most one RP per strain falls outside the 95% prediction intervals. For any failures, identify and solve the source of the problem and then repeat the testing for the failed strains starting with those having the largest deviation from predicted values until both criteria are met.
These proposed laboratory qualification criteria were tested using laboratory H. Figure S1 in the supplemental material shows the distribution of geometric means, CVs, and assay values relative to the range of geometric means, of CVs and to the 95% prediction intervals listed in Table S3 in the supplemental material. The RPs produced in laboratory H were consistent with those produced in the standardized laboratories and qualify laboratory H to perform the MATS ELISA and therefore to use the MATS method to assess strain coverage by 4CMenB. As additional laboratories provide data, refinements may be made to the proposed qualification criteria.
DISCUSSION
Statistical methodologies used in this investigation have been applied previously in similar studies. In characterizing a new human pneumococcal standard reference serum, Goldblatt et al. (12) performed a study bridging the new reference sera (007sp) with the one in current use (89SF). Five laboratories performed the WHO reference pneumococcal ELISAs on a panel of 12 WHO calibration sera for 13 pneumococcal serotypes. Kapasi et al. (17) enrolled four laboratories to perform a comparative study of four different sources of pertussis toxin (PT) in an IgG anti-PT ELISA. Rose et al. (22) examined the level of agreement among six laboratories measuring antibody-mediated killing of Streptococcus pneumoniae (pneumococcus) by phagocytes using each laboratory's own optimized opsonophagocytic assay (OPA) on a panel of 16 WHO calibration reference sera for 13 pneumococcal serotypes. In previous studies (12, 22), concordance correlation results were very good since the laboratory-to-laboratory rc values exceeded 0.96 and 0.97, respectively, among all laboratories. In the study by Kapasi et al. (17), due to the lack of assay standardization and the inherent variability of OPA, concordance was somewhat reduced: laboratory-to-laboratory rc values ranged from 0.67 to 0.99 for all six laboratories with five of the six laboratories exhibiting an rc of >0.80.
The laboratory-to-laboratory rc values measured during the present interlaboratory standardization of the MATS ELISA exceeded 0.99 for all labs, indicating an excellent agreement within and across all participating laboratories for all three antigens, which was equal to or higher than that observed in the similar studies mentioned above.
Of note, no predefined gold-standard RP values were used for the MenB strains examined in this study, and the ANOVA mixed model provided a mechanism for estimating consensus values that served as assigned values for each strain for the duration of the study. The choice to use a consensus value rather than establishing a definitive RP in the laboratory where MATS was initially developed reflects the desire to develop a real-world assay whose standardization and qualification criteria are co-owned by the laboratories that perform it.
Mixed-model analysis of variance allowed us to partition the total variance to measure reproducibility and repeatability. The within-laboratory variation was 2 to 2.5 times higher than the between-laboratory variation for each antigen, indicating a very good level of assay standardization across laboratories but also a significant level of intrinsic variability in the assay. Within-strain CVs measured for each antigen show that variability is slightly increased for low RPs and that CVs varied significantly from strain to strain, but also that, for each antigen, the less variable strain had a CV higher than the between-laboratory CV. Taken together, these results suggest that, even though strain-specific characteristics and the biochemistry of the assay at low concentrations may have an impact on MATS variability, a major source of assay variation is intrinsic to the experimental procedure and may be associated with the quantification of the bacterial suspension that is processed in the assay due to the biological variability of bacterial growth.
One interesting and unanticipated finding observed here was that a small number of bacteria survived Empigen lysis at room temperature in some collaborating laboratories. Exposure to live meningococci is an unacceptable health risk to laboratory workers; therefore, the lysate preparation temperature was adjusted to ensure the safety of MATS operators. This effect is probably strain dependent and indicates that safety precautions need to be evaluated for larger panels of meningococcal strains. Subsequent analysis showed that changing the lysate preparation temperature to 45°C had no significant effect on RPs and supported combining data from all of the assays in this study.
One important goal of the present study was to provide additional national laboratories with an opportunity to obtain the MATS ELISA without additional interlaboratory studies. Therefore, a set of qualification criteria were established to permit a laboratory to test its proficiency in MATS relative to the laboratories (A to G) standardized here. A volunteer national laboratory (laboratory H) followed the qualification procedure and obtained MATS ELISA results that were highly consistent with those in the seven standardized laboratories. As additional laboratories continue to provide assay data, the qualification criteria presented here may be refined.
Strain coverage is a critical component in estimating the potential clinical effects of vaccines against MenB, a pathogen characterized by dynamic mutability and strain epidemiology, as well as a propensity among its more virulent encapsulated strains to cause prolonged epidemic disease. Due to this diversity, the amount of serum required for testing using the existing correlate of protection, the hSBA, is prohibitively high, particularly in trials of infants. In a previous study (7), a minimum MATS RP, the PBT, was established to indicate that a given MenB strain was susceptible to killing in the SBA by antibodies induced by 4CMenB. In the present study, a heuristic method based on interlaboratory variation was developed to derive 95% CIs for strain coverage estimates. The results presented here demonstrate that the MATS RPs produced in each standardized or qualified laboratory can be compared to the PBTs defined in one of the participating laboratories to obtain estimates of vaccine strain coverage that are consistent among them, within the 95% intervals defined.
The PBTs used to define vaccine strain coverage were derived by comparing MATS to pooled hSBA titers on a panel of 57 MenB strains. The use of a broader strain panel and comparison to hSBA data from individual subjects may further support the use of MATS to assess MenB strain coverage independent of clinical sera. This might allow clinical trials to predict vaccine efficacy using a limited number of strains. In addition, the extremely good agreement observed here among different laboratories suggests that the technology adopted in the MATS ELISA could be successfully used to perform similar testing with different antigens or pathogens, providing a general platform for bacterial antigen phenotyping.
A possible area for future investigation with MATS is as a means of postimplementation surveillance of the genetic profiles of fHbp, NadA, and NHBA on circulating meningococcal strains, allowing reference laboratories to monitor the antigenic profiles of pathogenic isolates in real time (24). In addition, MATS might be used to assess the actual nature of potential vaccine failures, which has added importance for outer membrane protein vaccines against MenB, given that not all circulating strains will necessarily be covered. Postimplementation surveillance data based on a standardized assay could allow an indirect comparison of immunization policies across countries and regions, providing a valuable basis for rapid adaptation of public health policies based on worldwide quantitative data.
In summary, the results reported indicate that MATS is a standardized, reproducible antigen typing system that robustly predicts 4CMenB strain coverage in different geographical regions. These results suggest that MATS may have utility in epidemiologic surveillance of meningococci and could be adapted for assessing other pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Active Bacterial Core Surveillance Team and the Emerging Infection Programs Network, E. Richard Moxon, Diana Martin, and Geoff Hogg for providing N. meningitidis strains used in the present study. We thank Stefania Bambini, Sara Comandi, and Maurizio Comanducci for molecular typing and Fabio Rigat for useful discussions.
Footnotes
Published ahead of print 8 August 2012
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1. Bai X, Findlow J, Borrow R. 2001. Recombinant protein meningococcal serogroup B vaccine combined with outer membrane vesicles. Expert Opin. Biol. Ther. 11:969–985 [DOI] [PubMed] [Google Scholar]
- 2. Bambini S, et al. 2009. Distribution and genetic variability of three vaccine components in a panel of strains representative of the diversity of serogroup B meningococcus. Vaccine 27:2794–2803 [DOI] [PubMed] [Google Scholar]
- 3. Brehony C, Jolley KA, Maiden MC. 2007. Multilocus sequence typing for global surveillance of meningococcal disease. FEMS Microbiol. Rev. 31:15–26 [DOI] [PubMed] [Google Scholar]
- 4. Budroni S, et al. 2011. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 108:4494–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caron F, et al. 2011. From tailor-made to ready-to-wear meningococcal B vaccines: longitudinal study of a clonal meningococcal B outbreak. Lancet Infect. Dis. 11:455–463 [DOI] [PubMed] [Google Scholar]
- 6. CDC 2009. Surveillance of invasive bacterial diseases in Europe 2008–2009 Centers for Disease Control and Prevention, Atlanta, GA: http://www.ecdc.europa.eu/en/publications/Publications/1107_SUR_IBD_2008-09.pdf/ [Google Scholar]
- 7. Donnelly J, et al. 2010. Qualitative and quantitative assessment of meningococcal antigens to evaluate the potential strain coverage of protein-based vaccines. Proc. Natl. Acad. Sci. U. S. A. 107:19490–19495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Feil EJ, et al. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. U. S. A. 98:182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Findlow J, et al. 2005. Effect of sequence variation in meningococcal PorA outer membrane protein on the effectiveness of a hexavalent PorA outer membrane vesicle vaccine in toddlers and school children. Vaccine 23:2623–2627 [DOI] [PubMed] [Google Scholar]
- 10. Finne J, Bitter-Suermann D, Goridis C, Finne U. 1987. An IgG monoclonal antibody to group B meningococci cross-reacts with developmentally regulated polysialic acid units of glycoproteins in neural and extraneural tissues. J. Immunol. 138:4402–4407 [PubMed] [Google Scholar]
- 11. Giuliani MM, et al. 2006. A universal vaccine for serogroup B meningococcus. Proc. Natl. Acad. Sci. U. S. A. 103:10834–10839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goldblatt D, et al. 2011. Establishment of a new human pneumococcal standard reference serum, 007sp. Clin. Vaccine Immunol. 18:1728–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harrison LH, Trotter CL, Ramsay ME. 2009. Global epidemiology of meningococcal disease. Vaccine 27(Suppl 2):B51–B63 [DOI] [PubMed] [Google Scholar]
- 14. Heyden Y, Smeyers-Verbeke J. 2007. Set-up and evaluation of interlaboratory studies. J. Chromatogr. A 1158:158–167 [DOI] [PubMed] [Google Scholar]
- 15. Holst J, et al. 2009. Properties and clinical performance of vaccines containing outer membrane vesicles from Neisseria meningitidis. Vaccine 27(Suppl 2):B3–B12 [DOI] [PubMed] [Google Scholar]
- 16. Jolley KA, Brehony C, Maiden MC. 2007. Molecular typing of meningococci: recommendations for target choice and nomenclature. FEMS Microbiol. Rev. 31:89–96 [DOI] [PubMed] [Google Scholar]
- 17. Kapasi A, et al. 2012. Comparative study of different sources of pertussis toxin (PT) as coating antigens in IgG anti-PT enzyme-linked immunosorbent assays. Clin. Vaccine Immunol. 19:64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin LIK. 1989. A concordance correlation coefficient to evaluate reproducibility. Biometrics 45:255–268 [PubMed] [Google Scholar]
- 19. Maiden MC, et al. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin DR, Ruijne N, McCallum L, O'Hallahan J, Oster P. 2006. The VR2 epitope on the PorA P1.7-2,4 protein is the major target for the immune response elicited by the strain-specific group B meningococcal vaccine MeNZB. Clin. Vaccine Immunol. 13:486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin SL, et al. 2000. Effect of sequence variation in meningococcal PorA outer membrane protein on the effectiveness of a hexavalent PorA outer membrane vesicle vaccine. Vaccine 18:2476–2481 [DOI] [PubMed] [Google Scholar]
- 22. Rose CE, et al. 2011. Multilaboratory comparison of Streptococcus pneumoniae opsonophagocytic killing assays and their level of agreement for the determination of functional antibody activity in human reference sera. Clin. Vaccine Immunol. 18:135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sadarangani M, Pollard AJ. 2010. Serogroup B meningococcal vaccines: an unfinished story. Lancet Infect. Dis. 10:112–124 [DOI] [PubMed] [Google Scholar]
- 24. Snape MD, et al. 2012. The challenge of post-implementation surveillance for novel meningococcal vaccines, Vaccine 30(Suppl 2):B67–B72 [DOI] [PubMed] [Google Scholar]
- 25. Su EL, Snape MD. 2011. A combination recombinant protein and outer membrane vesicle vaccine against serogroup B meningococcal disease. Expert Rev. Vaccines 10:575–588 [DOI] [PubMed] [Google Scholar]
- 26. Tan LK, Carlone GM, Borrow R. 2010. Advances in the development of vaccines against Neisseria meningitidis. N. Engl. J. Med. 362:1511–1520 [DOI] [PubMed] [Google Scholar]
- 27. Tappero JW, et al. 1999. Immunogenicity of 2 serogroup B outer-membrane protein meningococcal vaccines: a randomized controlled trial in Chile. JAMA 281:1520–1527 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.