Abstract
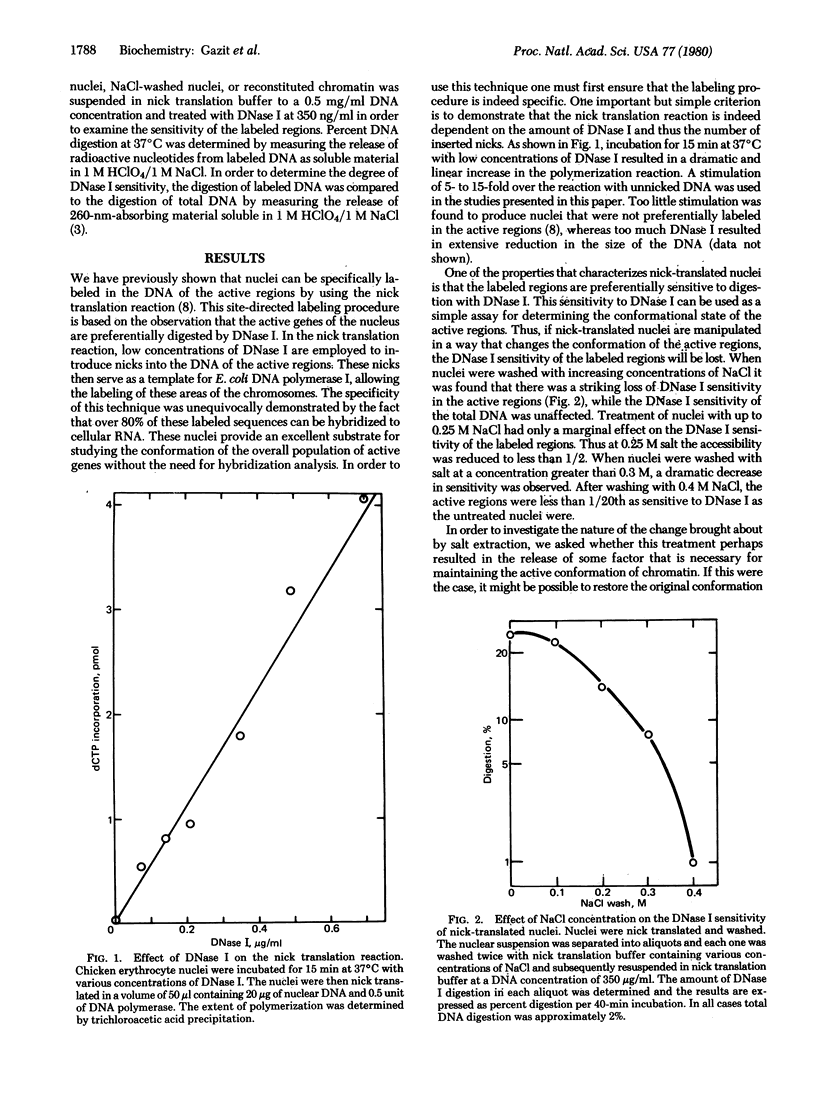
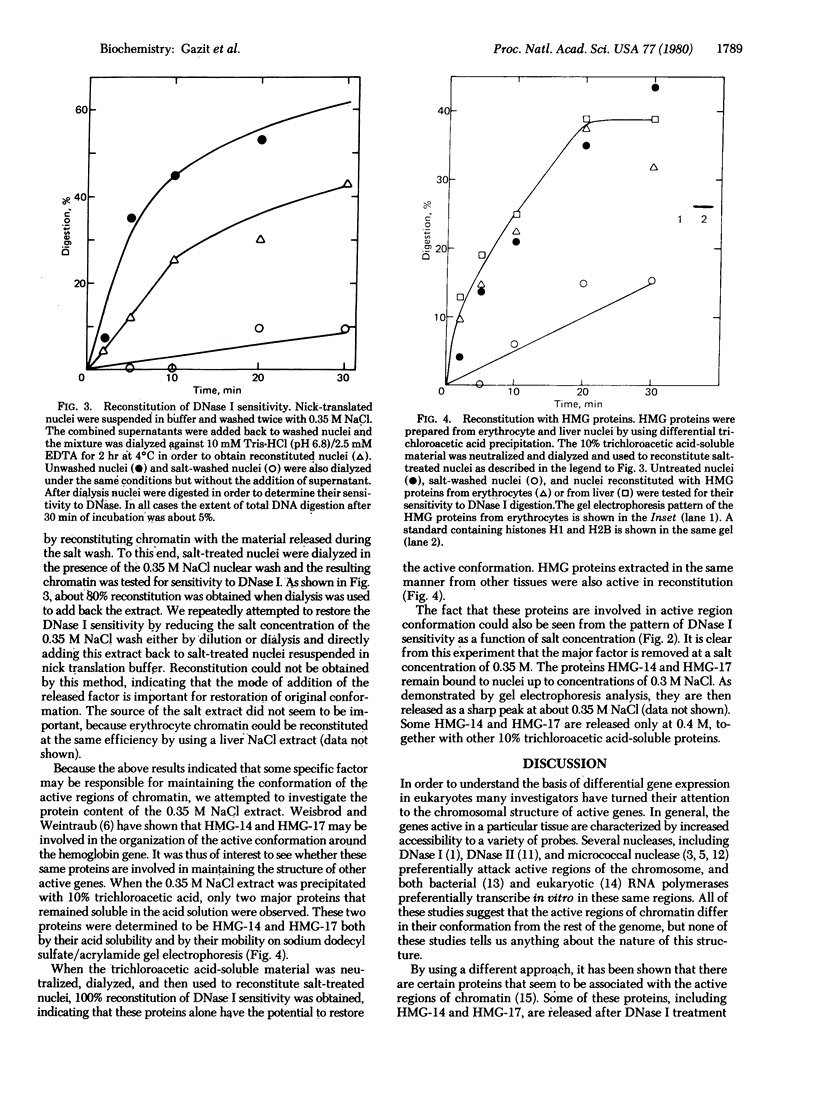
Chicken erythrocyte nuclei have been labeled in the active regions of the chromosome by using the nick translation reaction. In this procedure, accessible areas of the genome are preferentially nicked by the action of pancreatic DNase I and subsequently labeled by using DNA polymerase I from Escherichia coli. These nuclei were employed as a substrate for studying the factors responsible for maintaining the special chromatin conformation of the overall population of active genes. Treatment of nuclei with 0.35 M NaCl resulted in the loss of DNase I sensitivity in the active genes, but this sensitivity could be restored when nuclei were reconstituted with the NaCl eluate. Further purification of the released factors revealed that the HMG (high-mobility group) proteins HMG-14 and HMG-17 are involved in maintaining the conformation of the active regions. These factors are not tissue specific and seem to be involved in the chromosomal structure of most of the active genes.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R., Cedar H., Felsenfeld G. Synthesis of globin ribonucleic acid from duck-reticulocyte chromatin in vitro. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2029–2032. doi: 10.1073/pnas.70.7.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axel R., Cedar H., Felsenfield G. The structure of the globin genes in chromatin. Biochemistry. 1975 Jun 3;14(11):2489–2495. doi: 10.1021/bi00682a031. [DOI] [PubMed] [Google Scholar]
- Bellard M., Gannon F., Chambon P. Nucleosome structure III: the structure and transcriptional activity of the chromatin containing the ovalbumin and globin genes in chick oviduct nuclei. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):779–791. doi: 10.1101/sqb.1978.042.01.078. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Zolan M., Axel R. Genes transcribed at diverse rates have a similar conformation in chromatin. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4867–4871. doi: 10.1073/pnas.74.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M., Garrard W. T., Bagi G., Wilson R. F., Bonner J. Partial purification of the template-active fraction of chromatin: a preliminary report. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2193–2197. doi: 10.1073/pnas.71.6.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M., Partington G. A. Distribution of messenger RNA-coding sequences in fractionated chromatin. Cell. 1977 Dec;12(4):953–962. doi: 10.1016/0092-8674(77)90160-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levitt A., Axel R., Cedar H. Nick translation of active genes in intact nuclei. Dev Biol. 1979 Apr;69(2):496–505. doi: 10.1016/0012-1606(79)90307-5. [DOI] [PubMed] [Google Scholar]
- Levy-Wilson B., Dixon G. H. Limited action of micrococcal nuclease on trout testis nuclei generates two mononucleosome subsets enriched in transcribed DNA sequences. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1682–1686. doi: 10.1073/pnas.76.4.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy B., Dixon G. H. Renaturation kinetics of cDNA complementary to cytoplamic polyadenylated RNA from rainbow trout testis. Accessibility of transcribed genes to pancreatic DNase. Nucleic Acids Res. 1977 Apr;4(4):883–898. doi: 10.1093/nar/4.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall A. J., Burgoyne L. A. Interpretation of the properties of chromatin extracts from mammalian nuclei. Nucleic Acids Res. 1976 Apr;3(4):1101–1110. doi: 10.1093/nar/3.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield J. E., Serunian L. A., Silver L. M., Elgin S. C. A protein released by DNAase I digestion of drosophila nuclei is preferentially associated with puffs. Cell. 1978 Jul;14(3):539–544. doi: 10.1016/0092-8674(78)90240-4. [DOI] [PubMed] [Google Scholar]
- Panet A., Cedar H. Selective degradation of integrated murine leukemia proviral DNA by deoxyribonucleases. Cell. 1977 Aug;11(4):933–940. doi: 10.1016/0092-8674(77)90304-x. [DOI] [PubMed] [Google Scholar]
- Steggles A. W., Wilson G. N., Kantor J. A., Picciano D. J., Falvey A. K., Anderson W. F. Cell-free transcription of mammalian chromatin: transcription of globin messenger RNA sequences from bone-marrow chromatin with mammalian RNA polymerase. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1219–1223. doi: 10.1073/pnas.71.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Weintraub H. Isolation of a subclass of nuclear proteins responsible for conferring a DNase I-sensitive structure on globin chromatin. Proc Natl Acad Sci U S A. 1979 Feb;76(2):630–634. doi: 10.1073/pnas.76.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]



