Abstract
One biological need for Ni in marine cyanobacteria stems from the utilization of the Ni metalloenzyme urease for the assimilation of urea as a nitrogen source. In many of the same cyanobacteria, including Synechococcus sp. strain WH8102, an additional and obligate nutrient requirement for Ni results from usage of a Ni superoxide dismutase (Ni-SOD), which is encoded by sodN. To better understand the effects of Ni deprivation on WH8102, parallel microarray-based analysis of gene expression and gene knockout experiments were conducted. The global transcriptional response to Ni deprivation depends upon the nitrogen source provided for growth; fewer than 1% of differentially expressed genes for Ni deprivation on ammonium or urea were concordantly expressed. Surprisingly, genes for putative Ni transporters, including one colocalized on the genome with sodN, sodT, were not induced despite an increase in Ni transport. Knockouts of the putative Ni transporter gene sodT appeared to be lethal in WH8102, so the genes for sodT and sodN in WH8102 were interrupted with the gene for Fe-SOD, sodB, and its promoter from Synechococcus sp. strain WH7803. The sodT::sodB exconjugants were unable to grow at low Ni concentrations, confirming that SodT is a Ni transporter. The sodN::sodB exconjugants displayed higher growth rates at low Ni concentrations than did the wild type, presumably due to a relaxed competition between urease and Ni-SOD for Ni. Both sodT::sodB and sodN::sodB lines exhibited an impaired ability to grow at low Fe concentrations. We propose a posttranslational allosteric SodT regulation involving the binding of Ni to a histidine-rich intracellular protein loop.
INTRODUCTION
As the most abundant photoautotrophs on the planet, the cluster 5.1 marine cyanobacteria Synechococcus and Prochlorococcus contribute greatly to the global carbon and nitrogen cycles. In addition to the classical macronutrients nitrogen, phosphorus, and carbon, marine cyanobacteria also require several micronutrients such as Fe and Co in appreciable-enough quantities to affect their distribution and abundance in natural marine systems (32, 51). Recently, Ni additions were also shown to prompt growth in select natural populations of Synechococcus and Prochlorococcus (14), suggesting that Ni also influences the distributions of cyanobacteria in the ocean.
In Synechococcus and Prochlorococcus, the biological requirements for Ni arise from the usage of two known enzymes, though others may be currently unknown. Marine Synechococcus bacteria require a Ni metalloenzyme, urease, to use urea as a nitrogen source (10) and possibly for arginine catabolism (44). Without added Ni, Synechococcus cultures provided with urea as a nitrogen source will cease to grow, with growth being restored by the addition of either Ni, resulting in a functional urease, or NH4+, which does not require Ni for assimilation (13). Some Synechococcus and all Prochlorococcus genomes sequenced or assembled from metagenomic data contain a gene (sodN, synw1626 in WH8102) coding for a Ni-containing superoxide dismutase (Ni-SOD) (15, 39, 50, 64). Growth without Ni for Synechococcus sp. strain WH8102 results in reduced SOD activity, regardless of the nitrogen source used for growth, which is eventually lethal (13, 43). Some Synechococcus strains, like CC9311, have a second SOD containing a Cu/Zn active site, which appears to facilitate growth without Ni, but only when supplied with NH4+ as a nitrogen source. Thus far, the few marine Synechococcus sp. strains lacking a Ni-SOD (e.g., strains WH7803 and WH7805) contain the gene for a Fe-containing SOD (sodB) in the genomic location corresponding to sodN (Fig. 1A). This evolutionary exchange has been hypothesized to result in a loss of an obligate Ni requirement, but with increased Fe requirements as a consequence (15).
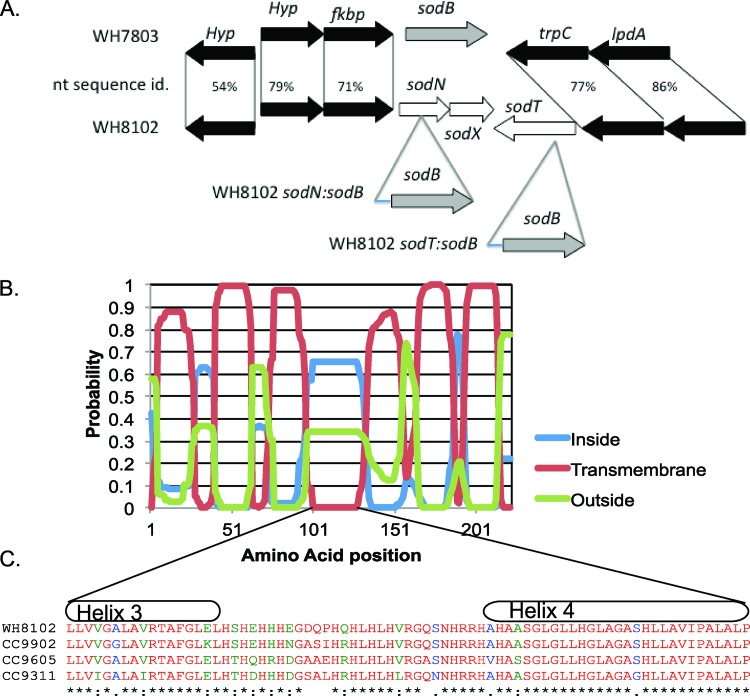
Fig 1.
Genomic locales of sodN and sodT. (A) A comparison of the genomic locations where sodB and sodN are found in WH7803 and WH8102, respectively, as well as the nucleotide sequence identities (nt sequence id.) of flanking genes. Also shown are the sodN::sodB and sodT::sodB lines developed for this study. The short extension at the N terminus of the sodB sequence indicates the 75 bp upstream of this gene in WH7803. Note that Kanr is also inserted into the indicated site along with sodB but is not shown here for clarity. (B) The predicted locations of transmembrane domains in sodT using TMHMM (27). (C) The sequences of the intracellular loop found between transmembrane helices 3 and 4 in WH8102, CC9902, CC9605, and CC9311.
In cyanobacteria, nutrient deprivation often induces a stress response specific to the nutrient but also a general stress response due to the perturbations in the intracellular balance of metabolites (52). In both CC9311 and WH8102, declining intracellular Ni concentrations induce an increase in the activity of a Ni-specific high-affinity (Km = 5 nM) uptake system, maintaining intracellular Ni concentrations at 100 to 300 nM (14). In the simplest sense, this Ni-specific stress response requires a transporter and a regulator. In other bacteria, Ni uptake has been shown to be catalyzed by one of four families of transporters, the Ni/Co permeases with eight transmembrane domains, HupE/UreJ-like permeases, the ABC-type NikABCDE transporters, and the ABC-type NikMNQO transporters (47). In the WH8102 genome, two genes code for proteins in the hupE/ureJ permease family. One gene (synw1628), called sodT in one review (17), colocalizes with sodN in all cyanobacterial genomes that contain it but is absent from all sodB-containing genomes, indicating a clear linkage between sodN and sodT (Fig. 1A). sodT codes for a protein with six predicted transmembrane domains (Fig. 1B), a topology associated with the HupE/UreJ Ni transporter family (17). Between transmembrane helices 3 and 4, an intracellular domain contains a high percentage of the Ni-binding amino acid histidine (Fig. 1C). The other putative Ni/Co permease in the WH8102 genome also has sequence similarity to the hydrogenase-associated Ni transporter hupE (synw2127), and yet the presence of a predicted B12 riboswitch in the upstream promoter region of the gene led to the conclusion that the WH8102 hupE transports Co (47). The WH8102 genome contains a putative transcriptional unit (synw0708-synw0709) coding for an ABC transporter in the nickel/peptide/opine family, where synw0709 codes for the putative periplasmic substrate-binding protein. While a homolog of sodT is found exclusively in sodN-containing Synechococcus and Prochlorococcus genomes, homologs of hupE (synw2127) and synw0709 are also found in the sodB-containing Synechococcus genomes such as those of strains WH7803 and WH7805.
Ni uptake in many bacteria is regulated at the transcriptional level by the protein NikR (12). In Escherichia coli, NikR is a negative repressor of the ABC-type nikABCDE operon expression, thereby controlling Ni uptake activity (12). NikR also controls the expression of a Ni transporter in the Ni/Co permease family in Helicobacter pylori (18), indicating a certain evolutionary ability to exchange regulator and transporter pairs. Another Ni regulatory protein, Nur, is a divergent ferric uptake regulator (Fur) family protein that represses Ni uptake in Streptomyces when intracellular Ni concentrations are sufficient for growth and Ni-SOD function (1). Homologs of NikR and Nur are absent from the genomes of WH8102 and other sodN-containing cyanobacterial genomes, implying a novel form of Ni transport regulation in these organisms.
Little is known about general stress responses to Ni deprivation, though an examination of the hypothetical biochemical consequences of Ni deprivation provides several hypotheses. A priori, Ni starvation should induce increased oxidative stress due to the reduced amounts of active Ni-SOD. Ni deprivation could also reduce the amount of active urease in the cell, affecting anabolic (10) and catabolic (44) N cycling and, as a result, the balance between carbon and nitrogen assimilation. The nitrogen source provided for growth should modulate the relative importance of catabolic and anabolic urease roles; growth on NH4+ should reduce the anabolic role of urease. To elucidate the molecular dynamics behind enhanced Ni uptake and the general stress responses to Ni deprivation, the global changes in gene expression prompted by Ni deprivation were studied using full-genome microarrays. In parallel, gene knockouts were used to determine the identity of the Ni transporter in WH8102.
MATERIALS AND METHODS
Strains and culturing techniques.
E. coli strains MC1061(pRL528, pRK24) and DH5α (Table 1) were grown in Luria-Bertani medium. When appropriate, ampicillin (100 μg/ml), kanamycin (50 μg/ml), and chloramphenicol (30 μg/ml) were used for the selection and maintenance of plasmids in E. coli. Synechococcus sp. strains WH8102, CC9605, CC9902, and CC9311 (Table 1) were grown in SOW medium made using the trace metal clean methods described in the work of Dupont et al. (13). Unless indicated, all cultures were grown at a constant light level of 50 μmol photons m−2 s−1 and a constant temperature of 19°C (CC9311) or 24°C (WH8102, CC9605, and CC9902). In the experiments presented here, variations were made in the nickel concentrations and the nitrogen source added for growth. NiCl2 was added in final concentrations of 500 pM, 1 nM, 5 nM, or 50 nM to achieve Ni2+ concentrations of 0.5, 1, 5, and 50 pM, respectively, as determined using MINEQL (63). Another set of experiments varied Fe concentrations from 740 to 7.4 μM total Fe. Chelex-100-purified 1 M NH4Cl, 1 M NaNO3, or 1 M urea was added to a final concentration of 2 mM N for growth on the different N sources.
Table 1.
Organisms and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference(s) or source |
|---|---|---|
| Bacterial strains | ||
| Synechococcus | ||
| WH8102 | Clade III, used for microarray studies | 39 |
| WH8102 sodN::sodB1 | WH8102 conjugated with pMsodN::sodB | This work |
| WH8102 sodT::sodB4 | WH8102 conjugated with pMsodT::sodB | This work |
| WH8102 sodT::sodB5 | WH8102 conjugated with pMsodT::sodB | This work |
| CC9311 | Clade I, coastal isolate, chromatic adapting | 39a |
| CC9605 | Clade II, abundant serotype in stratified California current | 59a |
| CC9902 | Clade IV, coastal isolate | 57 |
| E. coli | ||
| MC1061 | Host for pRK24, pRL528; donor in pMUT conjugations | 17a |
| DH5α | Recipient in transformations | |
| Plasmids | ||
| pMUT100 | Kanr Tetr; suicide vector | 6, 7 |
| pRK24 | Tcr Ampr; conjugal plasmid, RK2 derivative | 6, 7 |
| pRL528 | Cmr; helper plasmid, carries mob | 6, 7 |
| pCR2.1-TOPO | Kanr Ampr; PCR product cloning vector | Invitrogen |
| pMSYNWsodT | pMUT100 containing synw1628 fragment of nta 174584 to 174839 | This work |
| pMSYNCsodT | As above: sync0753 fragment of nt 703532 to 703832 | This work |
| pM9902sodT | As above: SynCC9902_0871 fragment of nt 842654 to 842947 | This work |
| pM9605sodT | As above: SynCC9605_1528 fragment of nt 14748101 to 1478370 | This work |
| pMSYNWsodN | As above: synw1626 fragment of nt 173466 to 173758 | This work |
| pMsodN::sodB | pMSYNWsodN containing WH7803 sodB and promoter (nt 1594036 to 1594704) | This work |
| pMsodT::sodB | pMSYNWsodT containing WH7803 sodB and promoter (nt 1594036 to 1594704) | This work |
nt, nucleotide.
The cultures for the growth rate and microarray experiments were grown in “semicontinuous” fashion, with constant Ni-limited exponential growth being achieved and maintained through transfer. With macronutrients added in great excess and bioavailable inorganic trace metal concentrations being buffered by EDTA, stable exponential growth rates and intracellular metal concentrations can be achieved, particularly if cultures are maintained at modest cell density (42, 55). When grown in this fashion, the cultures are essentially chemostats, where the limiting nutrient is the free metal concentration (55). Culture phycoerythrin fluorescence (Turner 10-AU; Turner Instruments, CA) was monitored as a proxy for growth, and rates were determined by slope of the linear ln (phycoerythrin fluorescence)-versus-time relationship. Cultures were transferred during exponential growth to media with the same [Ni2+] until growth rates stabilized, at which time they were allowed to grow for 4 to 5 more generations prior to harvesting.
One experiment examined the transcriptional and Ni uptake response of Ni-starved Synechococcus WH8102 to the addition of Ni. Here, a 2-liter culture of WH8102 was grown on NH4+ without any added Ni. Following the cessation of growth, increasing additions of NiCl2 were made over the next 96 h. Two-hundred-fifty-milliliter samples were removed using trace metal clean techniques at each time point in a HEPA-filtered laminar flow bench.
RNA isolation.
Cultures were harvested by centrifugation (10,000 × g for 10 min) in acid-washed and autoclaved 250-ml polyethylene bottles. Cells were lysed in TRIzol (Invitrogen, Carlsbad, CA) by incubation at 60°C for 1 h, followed by RNA isolation according to the manufacturer's instructions. DNA was removed, and RNA was further purified using a Qiagen RNeasy kit with DNase digestion according to the manufacturers' instructions.
Microarray hybridizations and analysis.
The WH8102 whole-genome microarray and the methods for cDNA synthesis, labeling, hybridization, and scanning have been presented previously (54, 58). Here, RNAs from eight urea-grown cultures and six ammonium-grown cultures were used. Fourteen different hybridizations were carried out, of which seven used different RNA pools and seven were replicates of these seven experiments, either dye swapping experiments or direct replicates. Statistical analyses were carried out using the Significance Analysis of Microarrays (SAM) software package (60), with the hybridizations treated as independent experiments. SAM orders the genes by using a modified t statistic called relative difference based on the ratio of change in gene expression to standard deviation in the data for that gene. It declares a gene to be up- or downregulated if the difference (D value) between the observed relative difference and expected relative difference is above (D value > 0) or below (D value < 0) the global cutoff point, respectively (60). This procedure allows estimation of the median of false discovery rates, and a gene was considered down- or upregulated by Ni deprivation if it displayed a negative or positive D value beyond the selected 1% median false discovery rate. An additional fold change cutoff of |0.5| was used.
Reverse transcriptase quantitative PCR (RT-Q-PCR).
Gene-specific PCR primers (Table 2) were designed to amplify a 100- to 150-bp internal region of each gene of interest using Primer3. DNA-free RNA (100 ng) was converted to cDNA by reverse transcriptase PCR (SuperScript II reverse transcriptase; Invitrogen, Carlsbad, CA) with random hexamer primers (50 ng per reaction; IDT, Inc.) according to the manufacturer's instructions. Quantitative PCR was carried out with each gene-specific primer pair at a concentration of 400 nM each with 2 μl of the cDNA synthesis reactions using the Brilliant SYBR green Q-PCR Master Mix kit and the Mx3000p real-time PCR thermocycler and fluorescence detection system (Stratagene, Santa Clara, CA). PCRs were carried out in 96-well plates in 25-μl volumes following the activation of the SureStart Taq DNA polymerase included in the kit at 95°C for 10 min. The cycling conditions were as follows: 95°C for 30 s, 60°C for 60 s, and 72°C for 60 s for 40 cycles, followed by a single cycle of 95°C for 30 s and 55°C for 30 s and a ramp to 95°C at 0.2°C s−1 to determine the melting temperature of the amplified cDNA and to verify the absence of secondary products. Also analyzed for each primer pair was a standard curve of 0.001 to 1 ng genomic DNA from Synechococcus. All reactions were run in duplicate. Threshold fluorescences of unknown samples were converted to cDNA concentrations using the standard curves for each primer pair. Agarose gels (1.2%) and Q-PCR disassociation curves were examined to verify single amplicons for each primer pair. At the time of this work, a suitable housekeeping gene was not known for WH8102; therefore, changes were quantified as the ratio of expression between experimental (Ni-deprived) and control (Ni-sufficient) samples.
Table 2.
Primers used in this study
| Name | Sequence | Purpose |
|---|---|---|
| Qsynw1628for | TCTGGTCCATGTTGAGACGA | Q-PCR of synw1628 |
| Qsynw1628rev | TGACTGTGCAGCTCAAGACC | Q-PCR of synw1628 |
| Qsynw2127for | GACCAATCTTCTGGCTTCTCC | Q-PCR of synw2127 |
| Qsynw2127rev | GGGCTGGAGATCAATAGCAA | Q-PCR of synw2127 |
| Qsynw1709for | TGAAGATTGCAGACCGACAG | Q-PCR of synw0709 |
| Qsynw1709rev | AGTTGCGAAAGGCTGAAAAG | Q-PCR of synw0709 |
| Incsynw0635for | GTTGCTGCACCCTCCA | Inactivation of synw0635 |
| Incsynw0635rev | AGCAGCCCACAAGGATCA | Inactivation of synw0635 |
| Incsynw1628for | CAGCATCCTCACCGGTTT | Inactivation of WH8102 sodT |
| Incsynw1628rev | GGCACCCACCACCAGTAG | Inactivation of WH8102 sodT |
| Incsynw1626for | GTGTCGCGGCTGAAGCC | Inactivation of WH8102 sodN |
| Incsynw1626rev | CAGTTCCTCAGCTTTGGC | Inactivation of WH8102 sodN |
| Incsync0753for | AGCCATGAAGTCAGGAATGG | Inactivation of CC9311 sodT |
| Incsync0753rev | CTGGTCTTGGATTGCTCCAT | Inactivation of CC9311 sodT |
| IncSyncc9902_1528for | CCAGAAGATGACTCGCACCT | Inactivation of CC9902 sodT |
| IncSyncc9902_1528rev | CGATTGGCTTGAAAGACCTC | Inactivation of CC9902 sodT |
| IncSyncc9605_0871for | GCAGAGCATCGTCATCTTCA | Inactivation of CC9605 sodT |
| IncSyncc9605_0871rev | GATCGAGAGAGCACCTGTCC | Inactivation of CC9605 sodT |
| pMUTgibsonFor | CTGTTGCCCGCTTGGCGGGGCTGCTAGCGCTATATGCGTTGATG | Linearization of pMUT vectors with sodB sticky ends |
| pMUTgibsonRev | CCAACCTGGCTGCCGCCTGACCGCAAGAGGCCCGGCAGTA | Linearization of pMUT vectors with sodB sticky ends |
| Synwh7803_1742gibsonFor | CGCATATAGCGCTAGCAGCCCCGCCAAGCGGGCAACAG | Adding pMUT sticky ends to WH7803 sodB+ promoter sequence |
| Synwh7803_1742gibsonRev | TACTGCCGGGCCTCTTGCGGTCAGGCGGCAGCCAGGTTGG | Adding pMUT sticky ends to WH7803 sodB+ promoter sequence |
| VerSynw1626For | ATGCTGCGCTCGGCCCTCACCG | PCR verification of sodN::sodB |
| VerSynw1626Rev | GCCCTTGGATTGCCAGAACATGCC | PCR verification of sodN::sodB |
| Versynw1628 for | GGATCAGGGCTCGTTGCTGGTGA | PCR verification of sodT::sodB |
| Versynw1628 rev | GTGCAGCTCAAGACCGAAG | PCR verification of sodT::sodB |
Genetic manipulations.
Inactivations were performed and validated according to the methods presented in reference 7. Briefly, for pMSYNWsodT, pMSYNCsodT, pM9902sodT, pM9605sodT, and pMSYNWsodN, a gene-specific internal fragment for each target (Table 2) was cloned into the EcoRI site (bp 0) of the suicide vector pMUT100 (7). To generate the pMsodN::sodB and pMsodT::sodB vectors, the sodB gene and 70 bp of the upstream noncoding region were amplified from WH7803 genomic DNA (primers, Synwh7803_1742gibsonFor and Synwh7803_1742gibsonRev, bp 1594036 to 1594704) while the pMSYNWsodT and pMSYNWsodN vectors were linearized at bp 174 using inverse PCR (primers, pMUTgibsonFor and pMUTgibsonRev). All primers contained 20-bp overhangs to desired cloning sites (Table 2). After isothermal cloning (22), the resulting plasmids were verified by restriction screening and sequencing. Each of the pMUT constructions was transformed into MC1061(pRL528, pRK24) and introduced into Synechococcus by conjugation with subsequent selection of exconjugants on pour-plated SN medium (6, 62) supplemented with 25 μg/ml filter-sterilized kanamycin sulfate, unless noted otherwise. Colonies were inoculated into liquid SN supplemented with 25 μg/ml kanamycin and maintained in exponential growth until axenic, as determined by spotting on LB agar plates. The complete segregation of mutant chromosomes was verified using PCR as described in reference 34 with the verification primers from Table 2. In addition, the presence of the sodB genes was also examined by PCR. To facilitate the isolation of Ni-uptake-impaired exconjugants, SN-agar medium was amended with 5 nM NiCl2 (10× the amount present in normal SN agar). In the experiments with Cu/Zn-SOD-containing CC9311, CC9902, and CC9605, SN-agar medium was amended with 5 nM CuCl2 and 0.5 mM NH4+ to mimic the conditions where these strains can grow on NH4+ without Ni. Cell lines were also screened using native protein gels with SOD activity screening (4), and the different SODs were identified by protein size.
Ni uptake rates.
Cells were harvested by centrifugation (8,000 × g for 10 min) in acid-washed polycarbonate bottles, rinsed twice with SOW containing no added EDTA or trace metals, and resuspended in 10 ml SOW with no added EDTA or trace metals in 70-ml acid-washed polycarbonate bottles. To measure high-affinity Ni uptake rates, 63Ni was added to a final concentration of 5 nM. Following a 30-min incubation under standard growth conditions, experiments were terminated by filtration onto 0.2-μm-pore-size filters (Supor; polyethersulfone). The filters were rinsed with 5 ml of sulfoxime (a Ni chelator, 1 mM, in SOW; Avocado Biochemicals, Lancashire, United Kingdom) and 5 ml of SOW to remove surface-bound 63Ni prior to being added to 15 ml of scintillation fluid (Ecolyte; MP Biomedicals, Solon, OH). To determine the total added 63Ni, small aliquots (150 μl) of unfiltered medium were added to 15 ml of scintillation fluid. Radioactivity was assayed using standard scintillation counting with quench correction. Disintegrations per minute were converted to 63Ni concentrations using a dilution curve of a 63Ni of known Ni concentration, as done in reference 13. The single time point uptake rate measurements are possible for this Ni concentration based on more detailed time course experiments published in reference 13, where Ni uptake was constant through 60 min.
Microarray data accession numbers.
Microarray data have been deposited under GEO accession numbers GSM982627 to GSM982639.
RESULTS
Overall physiological and transcriptional effects of Ni deprivation.
Ni deprivation was examined in cultures of WH8102 growing on NH4+ (here referred to as NH4+-Ni) and urea (urea-Ni). To induce Ni deprivation, cultures were maintained in exponential growth at constant [Ni2+] of 5 and 50 pM until growth rates were stable (at least 20 doublings), at which point they were harvested and mRNA was isolated for use in microarray experiments. At the lower [Ni2+], growth rates were suppressed 15 to 20% and Ni uptake rates (with 5 nM Ni) increased 400 to 500% (Table 3). Using a maximum false-positive rate of 1% and a fold change of >|0.5| as selection criteria for the transcriptomic analysis (see Materials and Methods), 50 to 115 open reading frames were determined to be significantly up- or downregulated in response to Ni deprivation in both experiments (see Table S1 in the supplemental material).
Table 3.
Growth rates and nickel uptake rates for cultures used in microarray experimentsa
| Strain and nitrogen source | 5 pM [Ni2+] |
50 pM [Ni2+] |
||
|---|---|---|---|---|
| μ (day−1) | Ni uptake (zmol cell−1 min−1) | μ (day−1) | Ni uptake (zmol cell−1 min−1) | |
| WH8102 and NH4 | 0.43 ± 0.03 | 3.5 ± 0.05 | 0.49 ± 0.03 | 0.7 ± 0.05 |
| WH8102 and urea | 0.375 ± 0.25 | 4.0 ± 0.05 | 0.52 ± 0.3 | 1 ± 0.05 |
Ranges are for duplicate cultures. Uptake rates were determined at a Ni concentration of 5 nM for cells that had been precultured at the indicated Ni concentrations.
The nitrogen source for growth drastically affected the transcriptional response of WH8102 to Ni deprivation (see Fig. S1 in the supplemental material), as only 18 of the 290 differentially expressed genes were shared between treatments. Of this subset, 11 genes were downregulated in both experiments, three were upregulated in NH4+-Ni and downregulated in urea-Ni, three were downregulated in NH4+-Ni and upregulated in urea-Ni, and only synw1592 was upregulated in both experiments. Many (114/290) of the differentially expressed genes colocalize on the genome (see Table S1), though 38 of the genes converge tail to tail from opposite strands and thus are not operons.
ATP and protein synthesis.
Consistent with the decline in growth rates, genes coding for ATP synthase (synw0488 to synw0495) were downregulated in both experiments (see Table S1 in the supplemental material), but not in a coordinated fashion. Three subunits were downregulated in NH4+-Ni while four other subunits were downregulated in urea-Ni. All of these genes occur in the same genomic region, and a closer examination reveals that the entire region exhibits negative fold changes in both experiments, though individual genes do not make the statistical cutoff. Two genes coding for ribosomal proteins (50S L7/L12 and L10) were also downregulated in both experiments, with a further one and five repressed in NH4+-Ni and urea-Ni, respectively. One ribosomal protein (50S protein L28) was upregulated in urea-Ni. No tRNA synthases were downregulated in the NH4+-Ni experiment, while only one was downregulated in the urea-Ni experiment. Genes coding for two subunits of Clp protease (synw0938 and synw1503) were upregulated by Ni deprivation for cells growing on NH4+.
Transcription and regulatory elements.
Sigma factors are global transcriptional regulators. One alternative type II σ factor (synw1509) was downregulated in NH4+-Ni while another (synw1621) was downregulated in urea-Ni. The putative principal sigma factor (synw1783) was upregulated in urea-Ni. Another system of cellular regulation in cyanobacteria involves histidine kinases and response regulators, of which WH8102 has only 5 and 9, respectively. While no histidine kinases were differentially expressed in these experiments, 5 of the response regulators were differentially expressed, but again not in a shared fashion. One response regulator (synw2289) was upregulated in NH4+-Ni, while synw0126 was downregulated. Two response regulators (synw2236 and synw2246) were downregulated by urea-Ni, while another was induced (synw0808). The only gene upregulated in both experiments is a putative transcription factor (synw1592) containing an AraC domain (PFAM 00165). The helix-turn-helix AraC domain is located at the C terminus of the putative protein, while the N terminus is uncharacterized.
Expression of the ntc regulon and carbon assimilation-related genes.
In cyanobacteria, the cyclic AMP (cAMP) receptor protein NtcA controls the expression of genes involved in nitrogen assimilation (19). Recently, the subset of genes regulated in WH8102 by ntcA was examined using a combination of bioinformatic and microarray-based approaches (WH8102 ntcA regulon [54]). Putative ntcA promoter sequences are located upstream of 18 genes differentially regulated in one of the experiments, with 12 in NH4+-Ni and 7 in urea-Ni. Only one gene was shared: a putative allophycocyanin beta-18 subunit was downregulated in both experiments. Most (11/12) of the differentially expressed ntcA-associated genes in NH4+-Ni were downregulated, with only a putative ferredoxin being induced. In the urea-Ni experiment, two genes involved in molybdenum cofactor biosynthesis were upregulated, while several genes involved in nitrogen assimilation were downregulated. These include a putative ammonium transporter (synw0253), a putative cyanate transporter (synw2487), and glutamine synthase (synw1073). This downregulation of canonical elements of nitrogen assimilation is contrary to the expectation that nickel deprivation would induce a nitrogen starvation-like response for growth on urea.
The expression of genes predicted to be part of the carbon fixation pathway in WH8102 by an in silico analysis (19) suggests that Ni deprivation affects carbon assimilation in a nitrogen-dependent manner. In the NH4+-Ni experiment, downregulated carbon fixation genes included fabF (synw0142), phosphoribulose kinase (prk, synw0785), cbbE (synw1115), hemA (synw1117), and glyC (synw1118). In contrast, only fabF and hemA were downregulated in urea-Ni. Together, this suggests that carbon fixation may be repressed in the NH4+-Ni experiment relative to that in the urea-Ni experiment.
Photosynthesis, phycobiliproteins, and porphyrins.
Ni deprivation resulted in a downregulation of photosynthetic reaction centers and associated proteins in both experiments but in a more extreme fashion for growth on urea than for growth on ammonium. No photosynthesis genes were upregulated. In all, 13 genes for photosynthetic reaction centers were downregulated in urea-Ni, while only three were downregulated in NH4+-Ni. In both cases, these were split relatively evenly between photosystems I and II. In both experiments, five genes related to the light-harvesting phycobilisomes were downregulated, with one (synw1074, allophycocyanin beta-18 subunit) shared in the two experiments.
Ni deprivation on urea prompted Synechococcus WH8102 to downregulate four of the eight genes coding for proteins involved in the synthesis of the precursor of chlorophyll and heme, protoporphyrinogen IX, from glutamate, specifically hemA, hemC, hemE, and hemL. hemA and hemE were also downregulated in the NH4+-Ni experiment. Mg-chelatase (chlH), which catalyzes the first unique step in the synthesis of chlorophyll, was downregulated in both experiments. Ferrochelatase (hemH) was downregulated for Ni deprivation on urea only. In both experiments, nearly every gene in the pathway exhibited negative fold changes, though only the genes noted here were statistically significant for the cutoffs used. The downregulation of this pathway can reduce the amount of chlorophyll a (Chl a) and therefore light absorption at the photosynthetic reaction centers. Alternatively, reduced heme synthesis may indicate a reduced amount of electron transport chaperones in the photosynthetic reaction pathway.
Electron flow to terminal oxidases.
Several of the WH8102 terminal oxidases were regulated by Ni availability. In the NH4+-Ni experiment, subunits for a putative cytochrome c oxidase were upregulated in response to Ni deprivation (synw1861 to synw1863; see Table S1 in the supplemental material). The Synechococcus sp. strain PCC7002 has a pair of functionally characterized cytochrome c oxidases; one (ctaI) was shown to act as a sink for excess electrons and oxygen in the thylakoids (38). The second, ctaII, appears to regulate the cellular response to oxidative stress from one of the cellular membranes (37). A phylogenetic analysis reveals that ctaDIEIFI from PCC7002 corresponds to synw1861 to synw1863 while PCC7002 ctaDIIEIIFII matches synw1528 to synw1530. While on the strand opposite that of ctaD1E1F1, the colocalized genes synw1859-synw1860 were also upregulated. Also upregulated by Ni deprivation for growth on NH4+ was a gene coding for a putative plastoquinone terminal oxidase (PTOX), based upon the sequence similarity to the well-characterized PTOX found in plant chloroplasts (35).
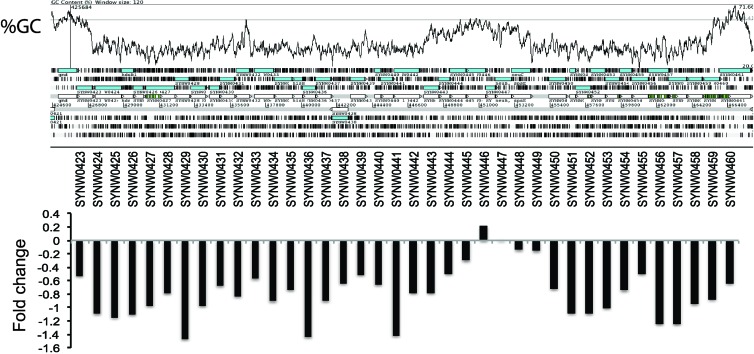
Repression of “island” genes by Ni deprivation.
In the NH4+-Ni experiment, nearly every gene of a 38-kbp low-percentage guanine-cytosine stretch of the genome was downregulated (Fig. 2). Relative to the rest of the genome, this region recruits poorly from the Global Ocean Sampling data set (49), indicating that it is not highly conserved in natural populations of Synechococcus with otherwise high nucleotide identity (>90%). Regions with similar characteristics have been identified in Synechococcus WH8102 as being similar to pathogenicity islands (39), and similar regions have been found in Prochlorococcus (9). In Prochlorococcus, these regions have been found to be transcriptionally hyperresponsive to phosphate limitation (33). The observation of Ni influencing the expression of a genomic island is novel, though Fe acquisition systems are often found in the genomic islands of pathogenic bacteria (11). Here, the majority of the island contents are involved in the synthesis of a cell surface polysaccharide or of unknown function, suggesting that the concerted downregulation may be due to an ancillary effect of Ni starvation for growth on NH4+, like the downregulation of carbon fixation.
Fig 2.
Genomic island downregulated by Ni deprivation. Shown are the genome organization and percent guanine-cytosine of a 38-kbp region of the WH8102 genome. Also shown is the fold change of each gene within the island associated with Ni deprivation when WH8102 is grown on NH4+.
Identification of genes involved in Ni uptake in WH8102.
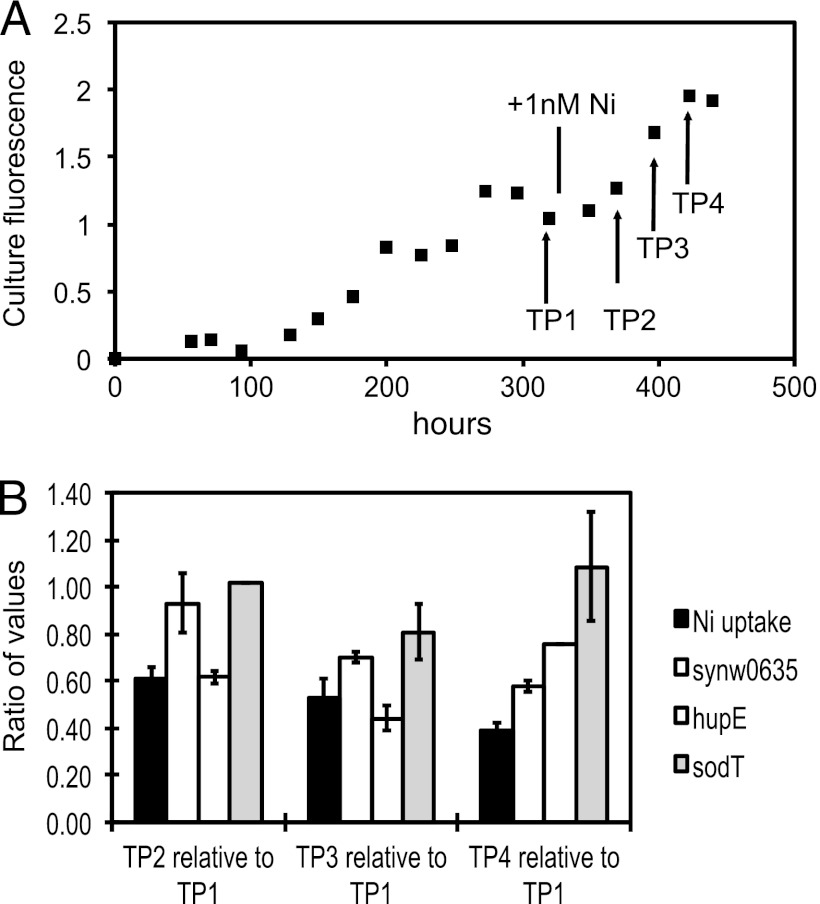
Ni uptake rates (at Ni concentrations of 5 nM) are greatly enhanced by the decreases in extracellular Ni concentrations (Table 3); therefore, it came as some surprise that none of the three hypothetical Ni transporters, hupE, sodT, and synw0709, were upregulated in response to Ni deprivation in any treatment. synw0709 was downregulated in the NH4+-Ni experiment, providing evidence that this gene is unlikely to be involved in high-affinity Ni uptake. Two further experiments were designed to manipulate the biological Ni uptake rates while examining the expression of sodT and hupE using RT-Q-PCR. One putative sodium ion-bile symporter (synw0635) was also examined because it exhibited positive fold changes in both microarray experiments, though not in a statistically significant fashion. In the first experiment, cultures of WH8102 were grown on NH4+ at constant [Ni2+] of 0.5, 5, and 50 pM. While Ni uptake rates with 5 nM Ni increased nearly 5-fold, sodT and hupE expression levels were relatively constant (Tables 4 and 5). synw0635 was upregulated, though not substantially. In a second experiment, WH8102 was grown on NH4+ without any Ni added to the medium, resulting in a cessation of growth due to Ni limitation (Fig. 3A). At this point, the culture received a Ni addition of 5 nM, stimulating a return to exponential growth (Fig. 3A) and a decline in Ni uptake rates (Fig. 3B). The transcription of synw0635, sodT, and hupE did not follow the changes in Ni uptake rates.
Table 4.
Ni uptakea
| [Ni2+] (pM) | pNi | Ni uptake rate with 5 nM Ni (zmol cell−1 min−1) |
|---|---|---|
| 0.5 | 12.3 | 4.75 |
| 5 | 11.3 | 1.90 |
| 50 | 10.3 | 1.00 |
The ratios or fold changes of Ni uptake rates at 5 nM total Ni and the expression of the indicated genes were determined for semicontinuous cultures grown on NH4+ at the indicated [Ni2+] where pNi = −log10[Ni2+]. The values are shown as the ratios of the measured rates and gene transcript abundance for the indicated cultures. Ranges for triplicate Q-PCR measurements on duplicate cultures are shown.
Table 5.
Ni transporter expression at constant [Ni2+]a
| Comparison | Fold change |
||
|---|---|---|---|
| SYNW1628 | SYNW0635 | hupE | |
| pNi 12.3 vs pNi 10.3 | 0.80 ± 0.1 | 1.33 ± 0.05 | 0.62 ± 0.1 |
| pNi 11.3 vs pNi 10.3 | 0.86 ± 0.08 | 1.10 ± 0.03 | 0.63 ± 0.07 |
The ratios or fold changes of Ni uptake rates at 5 nM total Ni and the expression of the indicated genes were determined for semicontinuous cultures grown on NH4+ at the indicated [Ni2+] where pNi = −log10[Ni2+]. The values are shown as the ratios of the measured rates and gene transcript abundance for the indicated cultures. Ranges for triplicate Q-PCR measurements on duplicate cultures are shown.
Fig 3.
Time course of Ni limitation and putative Ni transporter expression. (A) The top panel shows the culture fluorescence of a culture of WH8102 grown on NH4+ without added Ni until limitation (TP1). At this point, Ni was added, resulting in a return to exponential growth. Culture fluorescence is in arbitrary units. (B) Ni uptake rates and the expression of the indicated genes were determined at the indicated time points. Ratios of each time point with the initial Ni-limited values are shown. Ranges are for duplicate cultures.
Inactivation of genes in Synechococcus WH8102 can be achieved via conjugation with E. coli (7), providing a valuable tool for functional identification. synw0635 was targeted despite the modest correlation with Ni uptake rates. While constitutively expressed, sodT also was chosen for inactivation due to the lack of functional information, the possibility of posttranslational regulation, and the genomic colocalization with sodN in all genomes. Exconjugants with an interrupted synw0635 were obtained on the first attempt. Like the wild-type Synechococcus or the motility-impaired swmA knockout (6), this strain exhibited a normal tolerance to Ni deprivation and inducible Ni uptake (data not shown). This provides strong evidence either that synw0635 is not a nickel transporter or that a functional alternative is present.
Several attempts to insertionally inactivate sodT in WH8102 were unsuccessful. Conjugations were performed in parallel with constructions specific for swmA that were successful, providing experimental positive controls for conjugation conditions. Given the obligate Ni requirement for growth, the inactivation of a Ni transporter could be fatal to this strain. Exconjugants of sodT were also not recovered with supplementation of selective plates with 10× normal Ni concentrations, which is close to the toxicity threshold. Alternatively, sodT may be required for sodN activity through a non-transport-based mechanism. Synechococcus sp. strains CC9311, CC9605, and CC9902 can grow on NH4+ without Ni, presumably due to the usage of a Cu/Zn-containing SOD, though not on nitrate or urea (13). Therefore, pMUT plasmids were constructed for conjugation-based inactivation of sodT in Synechococcus CC9311, CC9902, and CC9605 (Table 1), yet exconjugants were not attained. While results were exclusively negative, the consistent lack of success suggests that inactivation of sodT in these strains is lethal, potentially due to an interaction of SodT and SodN. Attempts to interrupt sodN also were unsuccessful, suggesting that this gene is similarly necessary.
Hypothesizing that mature SodT is required for Ni-SOD activity, conjugation vectors were designed to interrupt either sodN or sodT in WH8102 with a construct containing both Kanr and the Fe-SOD-encoding sodB and its attendant upstream region from WH7803 (8) (Fig. 1). Exconjugants were isolated, and the genomic interruption was validated via PCR (see Fig. S2 in the supplemental material) for both sodN::sodB and sodT::sodB. On native SOD activity gels, both knockout strains exhibited SOD activity associated by protein size with Fe-SOD, while only the wild-type strain exhibited Ni-SOD activity (data not shown).
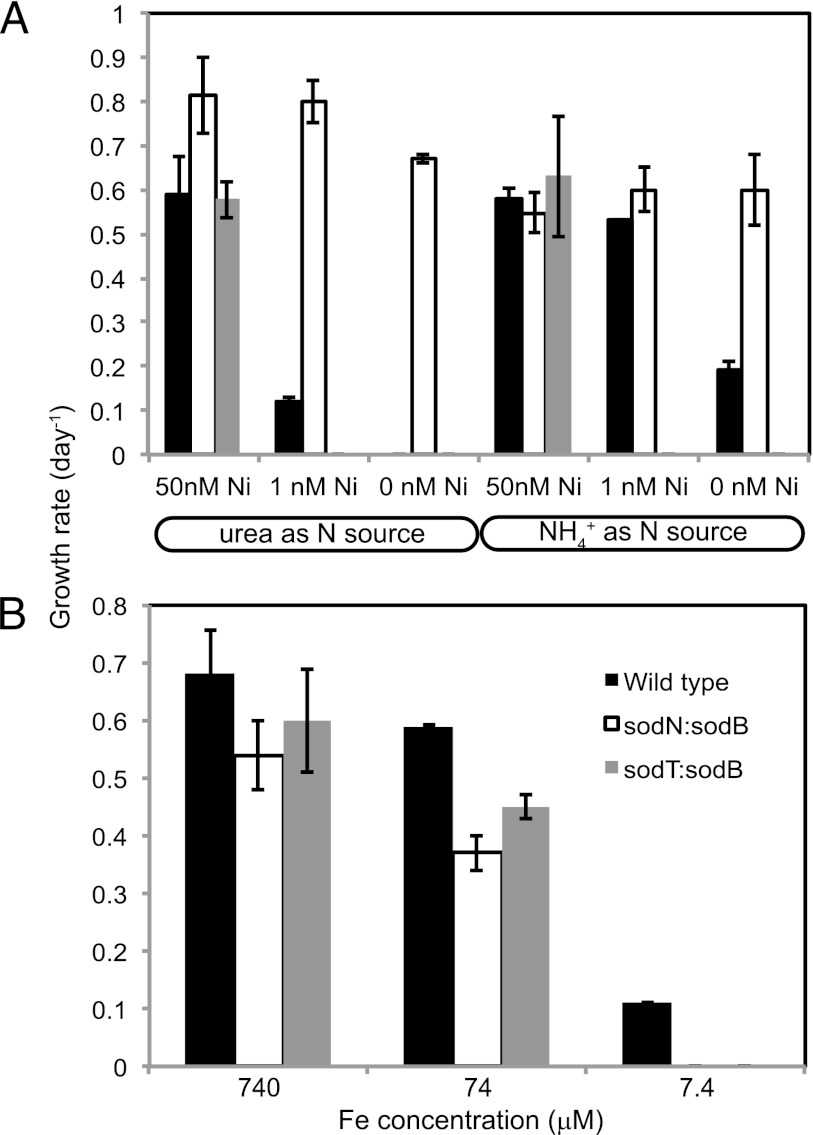
When grown at 50 nM, 1 nM, and 0 nM total added Ni on either urea or NH4+, each strain exhibited unique growth characteristics (Fig. 4A). The wild-type WH8102 exhibited the previously described characteristics: optimal growth at high Ni concentrations regardless of N source, with declining growth rates at 1 pM where urea levels were lower than NH4+ levels, and no growth at 0 pM Ni2+. The sodT::sodB line was unable to grow at 1 pM or 0 pM Ni2+ regardless of nitrogen source. The sodN::sodB cell line displayed heightened ability to grow at low Ni concentrations relative to that of the wild type. Near-maximal growth rates were maintained at 1 pM Ni2+. With zero added Ni for growth on urea, over 20 generations were required to induce Ni-urea colimitation compared to 5 in the wild type and 1 for the sodT::sodB line. Ni limitation on NH4+ was not achieved for the sodN::sodB line. For Chelex-treated artificial seawater, the background Ni concentrations were determined to be 10 to 20 pM total using competing ligand exchange cathodic stripping voltammetry (10 to 20 fM Ni2+ [14]); thus, the sodN::sodB line has vanishingly low or no Ni requirements for growth on NH4+.
Fig 4.
Growth rates of WH8102 wild type and sod knockouts. (A) Growth rates at the indicated nitrogen source and nickel concentration. (B) Growth rates on nitrate at the indicated Fe concentration. Error bars are for triplicate cultures.
Given the relaxation of Ni requirements, we examined the converse effects of increased Fe requirements. Here, each line was grown on nitrate at 740 μM, 74 μM, or 7.4 μM total Fe (Fig. 4B). In the wild-type WH8102, 74 μM total Fe results in a 10% reduction in growth rates while 7.4 μM total Fe results in a 90% reduction relative to Fe-replete conditions. In both sodN::sodB and sodT::sodB strains, 74 μM total Fe suppressed growth by 25%, and neither strain was able to grow at 7.4 μM total Fe (multiple inoculations). This is consistent with the hypothesis that the usage of sodB instead of sodT increases Fe requirements, but more detailed experiments directly examining Fe and Ni quotas and uptake rates are necessary for complete validation.
Ni uptake rates in response to low Ni concentrations could not be directly assessed in the sodT::sodB mutant as the biomass required could not be attained. However, the lack of growth of the sodT::sodB lines at low Ni concentrations is consistent with the role of sodT as a Ni transporter. As with the wild type, sodN::sodB lines exhibited enhanced Ni uptake when grown at 5 pM Ni2+ (see Fig. S3 in the supplemental material). Therefore, the sodN::sodB line provides ancillary evidence that sodT is a Ni transporter; by removing an intracellular sink for Ni while retaining transport function, an increased resistance to Ni starvation could be achieved. While Ni uptake rates were induced in sodN::sodB bacteria, they were not induced to the extent observed in wild-type WH8102. As discussed below, this is consistent with a competition between urease and Ni-SOD for intracellular Ni; with removal of Ni-SOD, lower uptake rates are required to provide Ni for just urease.
DISCUSSION
Influence of Ni deprivation on gene expression in WH8102.
The full-genome microarray analyses provided limited insight into the Ni-specific stress response to Ni deprivation. First, Ni uptake and the transcription of the verified Ni transporter sodT are uncoupled. Second, only one gene, synw1592, was upregulated in both experiments. This AraC domain-containing putative transcription factor may be involved in sensing Ni concentrations within the cell. However, as there were few genes regulated in a concordant fashion between the two experiments, either the regulon of synw1592 is modulated by other transcription factors or synw1592 senses another metabolite. Further supporting the latter hypothesis, homologs of synw1592 are found in nearly every other cyanobacterial genome, including WH7803, which lacks both urease and Ni-SOD, though Ni-requiring proteins not identified at this time may be present. As synw1592 is found in strains that apparently do not require Ni homeostasis, it seems unlikely that it is a Ni-sensing regulatory protein, though functional verification is necessary.
The general stress response to Ni deprivation is heavily influenced by nitrogen source (see Fig. S1 in the supplemental material). The only consistency between the two experiments was a downregulation of ATP synthase, porphyrin synthesis, and photosynthesis-related genes, which is consistent with the lower growth rates. The few sigma factors and response regulators found in the WH8102 genome were either divergently expressed or at least not shared. Carbon assimilation and nitrogen assimilation pathways were affected differently. An entire 38-kbp genomic “island” was downregulated in response to Ni deprivation on NH4+ only. Potentially, this lack of overlap is due to a role of urease in central metabolism and a competition for Ni between urease and Ni-SOD. Ni deprivation for growth on NH4+ may result in a deficiency of catabolically produced NH4+ from the cyanobacterial urea cycle, while Ni deprivation on urea reduces both catabolic and anabolic NH4+ production. By removing the competitive sink for Ni in Ni-SOD, the sodN::sodB cell line will be invaluable for elucidating the possible catabolic role of urease and the cyanobacterial urea cycle. Specifically, Ni deprivation of the sodN::sodB line for growth on ammonium or nitrate would limit just urease without the ancillary oxidative stress associated with the Ni-SOD using wild-type WH8102.
One consistent response was the upregulation of terminal oxidases CtaI and PTOX. The upregulation of ctaI may be important in detoxifying superoxide anions in the absence of Ni-SOD or reducing the generation of superoxide by acting as a sink for oxygen. While the exact functional role in cyanobacteria remains unknown, in plastids PTOX couples the oxidation of plastoquinol with the reduction of oxygen to water (2). PTOX may act as a “safety valve” preventing the overreduction of photosystem I electron acceptors in plastids (48). In cyanobacteria, PTOX may stabilize the redox state of the plastoquinone pool, which is important in the regulation of NADPH dehydrogenase (NADI)-catalyzed cyclic electron flow around photosystem I in cyanobacteria (31). The physiological trademarks of a water-water cycle have been observed in marine cyanobacteria and evoked as a mechanism to scavenge oxygen radicals (3, 24, 25, 36), yet the machinery involved is not known. Either the thylakoid-localized PTOX or CtaI may catalyze a water-water cyclic electron flow similar to the Mehler reaction, generating ATP while compensating for the reduced Ni-SOD activity.
A theoretical model of Ni uptake and homeostasis involving sodT.
As transcription of sodT and high-affinity Ni uptake are uncoupled (Fig. 3 and Tables 4 and 5), the activity of the protein must be controlled at a posttranscriptional or posttranslational level. The posttranscriptional regulation of metal transporter genes has not been observed in bacteria, though positive regulation of genes like sodB and ferritin by Fur is achieved indirectly through Fur-dependent repression of an antisense regulatory small RNA that acts posttranscriptionally to reduce translation of sodB and ferritin (28). WH8102 does not contain a nur homolog, and the fur orthologs found in the genome were not differentially expressed.
Alternatively, Ni uptake in WH8102 may be regulated posttranslationally. The protein coded for by sodT contains an intracellular loop laden with histidine residues (Fig. 1), which have imidazole side chains with high affinity for Ni. Constitutively expressed, SodT would presumably be present in the cytoplasmic membrane, and with sufficient intracellular Ni concentrations, the His-rich loop would be complexed with Ni. With declining intracellular Ni concentrations, Ni would disassociate from the His-rich loop as it is bound by Ni-SOD and urease. Potentially, the structural changes associated with Ni disassociation would activate the transport activities of the enzyme. The reduction in Ni uptake rates for the sodN::sodB lines relative to those for the wild type supports this model; by removing Ni-SOD, only urease is removing Ni from SodT, thus resulting in lower Ni uptake rates. The reduction of competition between Ni metalloenzymes has been shown to influence the induction of Ni uptake in Helicobacter pylori (5). Similar allosteric regulation of transporter activity has been observed for transporters of methionine (23) and molybdate (21), though not for Ni. In this scenario, the genomic locale displayed in Fig. 1 contains all of the elements required for marine cyanobacteria to use a Ni-containing SOD: (i) sodN, coding for apo-Ni-SOD; (ii) ppIase and sodX, protein chaperones required for posttranslational modifications of the apo-Ni-SOD following Ni binding (16, 26); and (iii) sodT, which we hypothesize both senses and transports Ni. Further, this would represent a form of streamlining, where one protein, SodT, performs roles performed by several proteins in other organisms.
The transcription of specific stress-responsive genes has been suggested as a way to assay the limitation state of communities of marine microbes. For example, within the cyanobacteria, genes that are responsive to nitrogen, phosphate, Fe, or Cu stress have been identified (29, 46, 53, 58, 59). The proposed posttranslational regulatory mechanism would stymie such approaches to studying Ni stress and possibly other stresses in natural cyanobacterial populations.
Implications for the biogeography of Synechococcus and Prochlorococcus.
Another implication of this model is the lack of inducible high-affinity Ni uptake in all of the sodB-containing strains of marine Synechococcus, including WH7803, WH7805, RSS9917, RSS9916, and WH5701. While these strains do not need Ni for SOD activity, all but WH7803 have the genes for a Ni-containing urease. As a low-affinity Ni uptake system may be present, these strains may be able to grow on urea if provided with high-enough Ni concentrations. However, one would expect that strains like WH8102 with high-affinity Ni uptake and lower Fe requirements would have a competitive advantage in oligotrophic conditions where urea features prominently in community metabolism (61). Essentially, the exchange of sodN-sodX-sodT for sodB may have created a major physiological divide in the marine cyanobacteria.
The usage of Ni-SOD versus Fe-SOD mostly lies along phylogenetic lines of the cluster 5 cyanobacteria. Specifically, all Prochlorococcus and most clade 5.1 Synechococcus bacteria, including clades I to IV, contain sodN. Synechococcus bacteria in clusters 5.2 and 5.3 (e.g., WH5701 and RCC307) and cluster 5.1B clades V, VI, VIII, and IX contain sodB instead (15). It is generally well accepted that Prochlorococcus dominates in the open ocean or stratified nutrient-poor waters, along with Synechococcus clades II and III (65). Clades I and IV are often found in mesotrophic waters with intermediate nutrient concentrations (30, 57). In contrast, the sodB-containing strains of Synechococcus are numerically dominant in upwelling systems (20, 41), where mixing delivers nitrate, phosphate, and Fe to the euphotic zone. Given the shift in Fe and Ni requirements, it is tempting to suggest that the choice of sodB constrains some Synechococcus strains to either low abundance or specific locations where Fe is enhanced by mixing or dust delivery. From the perspective of geological time, the modern ocean is Fe poor, with concentrations likely falling several orders of magnitude with the shift to large-scale oxic conditions around 500 million years ago (40, 45). Freshwater Synechococcus strains such as PCC7942 that are basal to cluster 5 (56) utilize Fe-SOD, suggesting that this is the ancestral state. Potentially, the replacement of a Fe-SOD with a Ni-SOD associated with cluster 5.1 cyanobacteria facilitated the proliferation of these Synechococcus lineages in the modern ocean.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Center for Bioinorganic Chemistry (to C.L.D.) and the U.S. Department of Energy's Genomes to Life program (www.doegenomestolife.org) under project Carbon Sequestration in Synechococcus Sp.: From Molecular Machines to Hierarchical Modeling (DOE DE-FG03-O1ER63148 to B.P., B.B., and I.T.P.). C.L.D. was also supported by a NASA Astrobiology Institute Directors Discretionary fund during some of the work.
We thank Katherine Barbeau from the Scripps Institution of Oceanography, UCSD, for access to laboratory facilities.
Footnotes
Published ahead of print 17 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ahn B-E, et al. 2006. Nur, a nickel-responsive regulator of the Fur family regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 59: 1848–1858 [DOI] [PubMed] [Google Scholar]
- 2. Aluru MR, Rodermel SR. 2004. Control of chloroplast redox by the IMMUTANS terminal oxidase. Physiol. Plant. 120: 4–11 [DOI] [PubMed] [Google Scholar]
- 3. Bailey S, et al. 2008. Alternative photosynthetic electron flow to oxygen in marine Synechococcus. Biochim. Biophys. Acta 1777: 269–276 [DOI] [PubMed] [Google Scholar]
- 4. Beauchamp C, Fridovich I. 1971. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44: 276–287 [DOI] [PubMed] [Google Scholar]
- 5. Benanti EL, Chivers PT. 2009. An intact urease assembly pathway is required to compete with NikR for nickel ions in Helicobacter pylori. J. Bacteriol. 191: 2405–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brahamsha B. 1996. An abundant cell-surface polypeptide is required for swimming by the non-flagellated marine cyanobacterium Synechococcus. Proc. Natl. Acad. Sci. U. S. A. 93: 6504–6509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brahamsha B. 1996. A genetic manipulation system for oceanic cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 62: 1747–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chadd HE, Newman J, Mann NH, Carr NG. 1996. Identification of iron superoxide dismutase and a copper/zinc superoxide dismutase enzyme activity with the marine cyanobacterium Synechococcus sp. WH7803. FEMS Microbiol. Lett. 138: 161–165 [DOI] [PubMed] [Google Scholar]
- 9. Coleman ML, et al. 2006. Genomic islands and the ecology and evolution of Prochlorococcus. Science 311: 1768–1770 [DOI] [PubMed] [Google Scholar]
- 10. Collier JL, Brahamsha B, Palenik B. 1999. The marine cyanobacterium Synechococcus sp WH 7805 requires urease (urea amidohydrolase, EC 3.5.1.5) to utilize urea as a nitrogen source: molecular-genetic and biochemical analysis of the enzyme. Microbiology 145: 447–459 [DOI] [PubMed] [Google Scholar]
- 11. Dobrindt U, Hochhut B, Hentschel U, Hacker J. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2: 414–424 [DOI] [PubMed] [Google Scholar]
- 12. Dosanjh NS, Michel SLJ. 2006. Microbial nickel metalloregulation: NikRs for nickel ions. Curr. Opin. Chem. Biol. 10: 123–130 [DOI] [PubMed] [Google Scholar]
- 13. Dupont CL, Barbeau K, Palenik B. 2008. Ni uptake and limitation in marine Synechococcus strains. Appl. Environ. Microbiol. 74: 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dupont CL, Buck KN, Palenik B, Barbeau K. 2010. Nickel utilization in phytoplankton assemblages from contrasting oceanic regimes. Deep Sea Res. I 57: 553–566 [Google Scholar]
- 15. Dupont CL, Neupane KP, Shearer J, Palenik B. 2008. Diversity, function, and evolution of genes coding for putative Ni-containing superoxide dismutases. Environ. Microbiol. 10: 1831–1843 [DOI] [PubMed] [Google Scholar]
- 16. Eitinger T. 2004. In vivo production of active nickel superoxide dismutase from Prochlorococcus marinus MIT9313 is dependent on its cognate peptidase. J. Bacteriol. 186: 7821–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eitinger T, Suhr J, Moore J, Smith JAC. 2005. Secondary transporters for nickel and cobalt ions: themes and variations. Biometals 18: 399–405 [DOI] [PubMed] [Google Scholar]
- 17a. Elhai J, Wolk CP. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167: 747–754 [DOI] [PubMed] [Google Scholar]
- 18. Ernst FD, et al. 2005. The nickel-responsive regulator NikR controls activation and repression of gene transcription in Helicobacter pylori. Infect. Immun. 73: 7252–7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flores E, Herrero A. 2005. Nitrogen assimilation and nitrogen control in cyanobacteria. Biochem. Soc. Trans. 33: 164–167 [DOI] [PubMed] [Google Scholar]
- 20. Fuller NJ, Tarran GA, Yallop M, Orcutt KM, Scalan DJ. 2006. Molecular analyses of picocyanobacterial community structure along an Arabian Sea transect reveals distinct spatial separation of lineages. Limnol. Oceanogr. 51: 2515–2526 [Google Scholar]
- 21. Gerber S, Comellas-Bigler M, Goetz BA, Locher KP. 2008. Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter. Science 321: 246–250 [DOI] [PubMed] [Google Scholar]
- 22. Gibson DG, et al. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- 23. Kadaba NS, Kaiser JT, Johnson E, Lee A, Rees DC. 2008. The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation. Science 321: 250–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kana TM. 1993. Rapid oxygen cycling in Trichodesmium thiebautii. Limnol. Oceanogr. 38: 18–24 [Google Scholar]
- 25. Kana TM. 1992. Relationship between photosynthetic oxygen cycling and carbon assimilation in Synechococcus WH7803. J. Phycol. 28: 304–308 [Google Scholar]
- 26. Kim E-J, Chung H-J, Suh B, Hah YC, Roe J-H. 1998. Transcriptional and post-translational regulation by nickel of sodN gene encoding nickel-containing superoxide dismutase from Streptomyces coelicolor Muller. Mol. Microbiol. 27: 187–195 [DOI] [PubMed] [Google Scholar]
- 27. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Prediction transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- 28. Lee J-W, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20: 485–499 [DOI] [PubMed] [Google Scholar]
- 29. Lindell D, et al. 2005. Expression of the nitrogen stress response gene ntcA reveals nitrogen-sufficient Synechococcus populations in the oligotrophic northern Red Sea. Limnol. Oceanogr. 50: 1932–1944 [Google Scholar]
- 30. Lucas AJ, et al. 2011. The green ribbon: multiscale physical control of phytoplankton productivity and community structure over a narrow continental shelf. Limnol. Oceanogr. 56: 611–626 [Google Scholar]
- 31. Ma W, Deng Y, Mi H. 2008. Redox of plastoquinone pool regulates the expression and activity of NADPH dehydrogenase supercomplex in Synechocystis sp. strain 6803. Curr. Microbiol. 56: 189–193 [DOI] [PubMed] [Google Scholar]
- 32. Mann EL, Chisholm SW. 2000. Iron limits the cell division rate of Prochlorococcus in the eastern equatorial Pacific. Limnol. Oceanogr. 45: 1067–1076 [Google Scholar]
- 33. Martiny AC, Coleman ML, Chisholm SW. 2006. Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc. Natl. Acad. Sci. U. S. A. 103: 12552–12557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCarren J, Brahamsha B. 2005. Transposon mutagenesis in a marine Synechococcus strain: isolation of swimming motility mutants. J. Bacteriol. 187: 4457–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McDonald AE, Vanlerberghe GC. 2006. Origins, evolutionary history, and taxonomic distribution of alternative oxidase and plastoquinol terminal oxidase. Comp. Biochem. Physiol. D 1: 357–364 [DOI] [PubMed] [Google Scholar]
- 36. Milligan AJ, Berman-Frank I, Gerchman Y, Dismukes GC, Falkowski PG. 2007. Light-dependent oxygen consumption in nitrogen-fixing cyanobacteria plays a key role in nitrogenase protection. J. Phycol. 43: 845–852 [Google Scholar]
- 37. Nomura CT, Persson S, Shen G, Inoue-Sakamoto K, Bryant DA. 2006. Characterization of two cytochrome oxidase operons in the marine cyanobacterium Synechococcus sp. PCC 7002: inactivation of ctaDI affects the PSI:PSII ratio. Photosynth. Res. 87: 215–228 [DOI] [PubMed] [Google Scholar]
- 38. Nomura CT, Sakamoto T, Bryant DA. 2006. Roles for heme-copper oxidases in extreme high-light and oxidative stress response in the cyanobacterium Synechococcus sp. PCC7002. Arch. Microbiol. 185: 471–479 [DOI] [PubMed] [Google Scholar]
- 39. Palenik B, et al. 2003. The genome of a motile marine Synechococcus. Nature 424: 1037–1042 [DOI] [PubMed] [Google Scholar]
- 39a. Palenik B, et al. 2006. The genome of Synechococcus CC9311: insights into adaptation to a coastal environment. Proc. Natl. Acad. Sci. U. S. A. 103: 13555–13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Planavsky NJ, et al. 2011. Widespread iron-rich conditions in the mid-Proterozoic ocean. Nature 477: 448–451 [DOI] [PubMed] [Google Scholar]
- 41. Post AF, et al. 2011. Long term seasonal dynamics of Synechococcus population structure in the Gulf of Aqaba, Northern Red Sea. Front. Microbiol. 2: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Price NM, et al. 1988. Preparation and chemistry of the artificial algal culture medium Aquil. Biol. Oceanogr. 6: 443–461 [Google Scholar]
- 43. Qiu B, Price NM. 2009. Different physiological responses of four marine Synechococcus strains (cyanophyceae) to nickel starvation under iron-replete and iron-deplete conditions. J. Phycol. 45: 1062–1071 [DOI] [PubMed] [Google Scholar]
- 44. Quintero MJ, Muro-Pastor AM, Herrero A, Flores E. 2000. Arginine catabolism in the cyanobacterium Synechocystis sp. strain PCC6803 involves the urea cycle and arginase pathway. J. Bacteriol. 182: 1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reinhard CT, Raiswell R, Scott C, Anbar AD, Lyons TW. 2009. A late Archean sulfidic sea stimulated by early oxidative weathering of the continents. Science 326: 713–716 [DOI] [PubMed] [Google Scholar]
- 46. Rivers AR, Wisniewski-Jakuba R, Webb EA. 2009. Iron stress genes in marine Synechococcus and the development of a flow cytometric iron stress assay. Environ. Microbiol. 11: 382–396 [DOI] [PubMed] [Google Scholar]
- 47. Rodionov DA, Hebbeln P, Gelfand MS, Eitinger T. 2006. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 188: 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rumeau D, Peltier G, Cournac L. 2007. Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 30: 1041–1051 [DOI] [PubMed] [Google Scholar]
- 49. Rusch DB, et al. 2007. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 5: 398–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rusch DB, Martiny A, Dupont CL, Halpern AL, Venter JC. 2010. Characterization of Prochlorococcus clades from iron depleted oceanic regimes. Proc. Natl. Acad. Sci. U. S. A. 107: 16184–16189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Saito MA, Rocap G, Moffett JW. 2005. Production of cobalt binding ligands in a Synechococcus feature at the Costa Rica upwelling dome. Limnol. Oceanogr. 50: 279–290 [Google Scholar]
- 52. Schwarz R, Forchhammer K. 2005. Acclimation of unicellular cyanobacteria to macronutrient deficiency: emergence of a complex network of cellular responses. Microbiology 151: 2503–2514 [DOI] [PubMed] [Google Scholar]
- 53. Stuart RK, Dupont CL, Johnson DA, Paulsen IT, Palenik B. 2009. Coastal strains of marine Synechococcus species exhibit increased tolerance to copper shock and a distinctive transcriptional response. Appl. Environ. Microbiol. 75: 5047–5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Su Z, et al. 2006. Computational inference and experimental validation of the nitrogen assimilation regulatory network in cyanobacterium Synechococcus WH8102. Nucleic Acids Res. 34: 1050–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sunda WG, Price NM, Morel FMM. 2005. Trace metal ion buffers and their use in culture studies, p 35–64 In Anderson RA. (ed), Algal culturing techniques. Elsevier, Oxford, United Kingdom [Google Scholar]
- 56. Swingley WD, Blankenship RE, Raymond J. 2008. Integrating Markov clustering and molecular phylogenetics to reconstruct the cyanobacterial species tree from conserved protein families. Mol. Biol. Evol. 25: 643–654 [DOI] [PubMed] [Google Scholar]
- 57. Tai V, Palenik B. 2009. Temporal variation of Synechococcus clades at a coastal Pacific Ocean monitoring site. ISME J. 3: 903–915 [DOI] [PubMed] [Google Scholar]
- 58. Tetu SG, et al. 2009. Microarray analysis of phosphate regulation in the marine cyanobacterium Synechococcus sp. WH8102. ISME J. 3: 835–849 [DOI] [PubMed] [Google Scholar]
- 59. Thompson AW, Huang K, Saito MA, Chisholm SW. 2011. Transcriptome response of high- and low-light adapted Prochlorococcus strains to changing iron availability. ISME J. 5: 1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59a. Toledo G, Palenik B. 2003. A Synechococcus serotype is found preferentially in surface marine waters. Limnol. Oceanogr. 48: 1744–1755 [Google Scholar]
- 60. Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wafar MVM, Lecorre P, Lhelguen S. 1995. F-ratios calculated with and without urea uptake in nitrogen uptake by phytoplankton. Deep Sea Res. I 42: 1669–1674 [Google Scholar]
- 62. Waterbury JB, Watson SW, Valois FW, Franks DG. 1986. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. Can. Bull. Fish. Aquat. Sci. 214: 71–120 [Google Scholar]
- 63. Westall JC, Zachary JL, Morel FMM. 1976. MINEQL: a computer program for the calculation of chemical equilibrium composition of aqueous systems. Technical note 18; R. M. Parsons Laboratory, Massachusetts Institute of Technology, Cambridge, MA [Google Scholar]
- 64. Youn H-D, Kim E-J, Roe J-H, Hah EC, Kang S-O. 1996. A novel nickel-containing superoxide dismutase from Streptomyces sp. J. Biochem. 318: 889–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zwirglmaier K, et al. 2008. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 10: 147–161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.