Abstract
The evolution of multicellularity has occurred in diverse lineages and in multiple ways among eukaryotic species. For plants and fungi, multicellular forms are derived from ancestors that failed to separate following cell division, thus retaining cytoplasmic continuity between the daughter cells. In networked organisms, such as filamentous fungi, cytoplasmic continuity facilitates the long-distance transport of resources without the elaboration of a separate vascular system. Nutrient translocation in fungi is essential for nutrient cycling in ecosystems, mycorrhizal symbioses, virulence, and substrate utilization. It has been proposed that an interconnected mycelial network influences resource translocation, but the theory has not been empirically tested. Here we show, by using mutants that disrupt network formation in Neurospora crassa (Δso mutant, no fusion; ΔPrm-1 mutant, ∼50% fusion), that the translocation of labeled nutrients is adversely affected in homogeneous environments and is even more severely impacted in heterogeneous environments. We also show that the ability to share resources and genetic exchange between colonies (via hyphal fusion) is very limited in mature colonies, in contrast to in young colonies and germlings that readily share nutrients and genetic resources. The differences in genetic/resource sharing between young and mature colonies were associated with variations in colony architecture (hyphal differentiation/diameters, branching patterns, and angles). Thus, the ability to share resources and genetic material between colonies is developmentally regulated and is a function of the age of a colony. This study highlights the necessity of hyphal fusion for efficient nutrient translocation within an N. crassa colony but also shows that established N. crassa colonies do not share resources in a significant manner.
INTRODUCTION
The transition from unicellular to multicellular organisms has occurred on multiple occasions in diverse lineages over considerable evolutionary time (28, 38, 68). While an initial adaptive advantage may have accrued simply from being larger, multicellular organisms subsequently developed increased differentiation and specialization, leading to a more efficient division of labor (8). Multicellularity may have arisen by either the aggregation of individual cells to form a colony or by the failure of daughter cells to separate following division. Comparisons of unicellular animals and their multicellular relatives support the view that multicellularity is associated with expansion of the genetic families involved in cell adhesion, cell-cell signaling, and cell differentiation (63). In contrast, multicellular plants and fungi are derived from ancestors that failed to separate following cell division, providing an opportunity to retain cytoplasmic continuity between daughter cells (75). Thus, plant cells are linked by tissue-specific patterns of plasmodesmata (41, 47), while fungi are either coenocytic or have perforated septa that allow intercompartmental exchange (40).
In ascomycete and basidiomycete fungi, cytoplasmic continuity is increased further through hyphal fusions (anastomoses), leading to an interconnected mycelial network (9, 31, 56–59). Network formation is hypothesized to be an adaptation to foraging, particularly where resources have a heterogeneous distribution in time and space, or to allowing a more rapid capture, exploitation, and defense of new territory. Network architecture is remodeled during growth, branching, and fusion (3, 21, 22), making it highly responsive to variations in resource availability or in the amount of damage incurred (7, 65). In networked organisms, such as filamentous fungi, cytoplasmic continuity facilitates long-distance transport of resources at speeds much faster than those with diffusion alone through cytoplasmic streaming (26, 50, 73) or mass flow (14) without the elaboration of a separate vascular system (32). These data suggest that the architecture of a colony can have an important influence on the physiological state, organelle distribution, and nutrient translocation within the mycelium (64).
While self-fusion within a colony influences network architecture, different colonies can potentially share resources via hyphal fusion events (17, 21, 25, 33, 45). This aspect has been hypothesized to be important for the exploitation of new resources via heterokaryon formation (55) and for the generation of new genetic diversity via parasexual genetics and lateral gene/chromosome transfer (42, 51, 60). The molecular and genetic mechanisms involved in hyphal fusion have been studied extensively in the filamentous ascomycete fungus Neurospora crassa (19, 39, 58). Fusion occurs when N. crassa germlings and/or hyphae grow toward each other through chemotropic interactions and adhere upon physical contact, which is subsequently followed by cell wall breakdown and a membrane merger to create a pore through which the cytoplasm and organelles move (9, 17, 33). Many N. crassa mutants defective in hyphal fusion have been identified (1, 15, 18, 24, 48, 66, 76). Although some fusion mutants show a pleiotropic growth phenotype, two fusion mutants, the Δso and ΔPrm-1 strains, maintain near-wild-type maximal growth rates. The soft (so) locus encodes a filamentous ascomycete-specific protein required for hyphal fusion, dynamic communication between germlings, and septal plugging after injury (16, 18). The Δso mutant also shows a lag in colony establishment (61) due to its inability to undergo germling fusion. In contrast to Δso mutants, which show essentially no germling or hyphal fusion, ΔPrm-1 mutants have an ∼50% reduction in germling and hyphal fusion events compared to the number in the wild type (15). The N. crassa Prm-1 locus encodes a transmembrane protein that has been shown to be important for plasma membrane merging during germling and hyphal fusion. These mutants bring the degree of fusion, and therefore the network architecture, under experimental control, making it feasible to test the importance of hyphal fusion on nutrient transport, genetic mixing, and network function.
Previous work on translocation in fungi, particularly in basidiomycete species, has focused on wild isolates; the requirement of an interconnected network for nutrient translocation and colony interactions has not been tested. We therefore evaluated whether network formation in the N. crassa wild type, versus the Δso and ΔPrm-1 mutants, influenced nutrient transport under increasingly challenging conditions, from initially homogeneous media to heterogeneous systems with strongly asymmetric demands. We investigated further whether fusion that is associated with colony developmental age influences resource sharing between colonies, and we evaluated its potential impact on generating genetic heterogeneity.
MATERIALS AND METHODS
Strains and media.
The soft (FGSC 508), Δso::hph A (FGSC 11293), ΔPrm-1::hph A (A32), and wild-type (FGSC 2489) strains were obtained from the Fungal Genetics Stock Center (43). We used the his-3::H1-dsRed and his-3::Prdi1-rdi-1-sgfp strains (53) for fluorescence imaging of the colonies. The strains were grown on Vogel's minimal medium (VMM) (74) with the required supplements. To obtain inoculum plugs and strips, strains were grown from conidia on plates of VMM (74). The one-tenth sucrose medium used for the stable isotope experiments contained 2 g/liter sucrose and 1.5% agar. The medium used for sucrose resource plugs in radioisotope experiments contained 20 g/liter sucrose and 1.5% agar.
Stable isotope experiments.
Colonies of the Δso, ΔPrm-1, and wild-type strains were grown for approximately 1 week in VMM slants at 25°C in constant light. Conidia were harvested by adding 1 ml of sterile water to a tube and vortexing for 30 s. Twenty microliters of conidial suspension was streaked in a line across the middle of a 24- by 24-cm petri plate filled with VMM. Sections of the colony that were 2 mm wide by 2 mm deep by 22 cm long were taken approximately 2 to 6 mm behind the periphery of the colony and used as inoculum strips. Subsequently, these strips were put on glass slides placed on top of a nonionized Nytran nylon membrane (Whatman) on top of low-sucrose VMM agar prepared as described above. Ten replicates of each strain were prepared. Three hundred microliters of 2-amino[15N]isobutyric acid ([15N]AIB) (Sigma-Aldrich) at a concentration of 1 μg/μl was added evenly to 4 plates of each strain along the inoculum strip. Three hundred microliters of water was added to one plate of each strain as a control. All colonies were then grown until they were 5 cm in linear length. Colonies were grown at 25°C in constant light for the entire experiment. The remaining 5 replicates per strain were used to repeat this experiment with a slight variation: the tracer was applied to the inoculum strip after the colonies had grown 3 cm.
Colonies were cut with clean razor blades into three regions (A, B, and C) that were 1.67 cm wide. Hyphal biomass was scraped off the membrane covering the media for each region and placed in preweighed 5- by 9-mm tin cups (Costech Analytical Inc.) in a 96-well plate. The biomass was dried for 2 days at 60°C, and the dry weight was calculated. The samples were analyzed for C and N with an isotope ratio mass spectrometer at the Colorado Plateau Stable Isotope Laboratory (www.isotope.nau.edu).
Radioisotope experiments.
The ability of wild-type and fusion mutant strains to translocate 2-amino[1-14C]isobutyric acid ([14C]AIB) from the center of a colony to hyphae at the periphery was assessed as the colony grew out from an inoculation plug. Two 8-mm-diameter, 3-mm-thick VMM agar plugs cut from just behind the tip region of a growing colony were placed in opposite corners on translucent scintillation screens (BioMax TRanscreen LE) lining 12-cm by 12-cm-square petri plates. The plugs were approximately 4 cm from the side of the plates, approximately 3 cm from the top or bottom of the plates, and approximately 7.2 cm away from each other on a diagonal. An 8-mm-diameter glucose and agar plug, which we termed a “resource plug,” was placed approximately 4 cm opposite each inoculation plug. Water-soaked pieces of paper towel were placed in the corners of the plates off the screen to maintain humidity. Ten microliters of 0.9 mM [14C]AIB was added to each inoculation plug at the time of placement on the scintillation screen. The colonies were then imaged using a photon-counting camera over 3 days to observe the movement of the [14C]AIB through the colonies as they grew. The specifics on imaging techniques can be found in references 70 and 72. To test whether the strains could transport AIB from the periphery of a colony toward the interior, we used the same experimental design as described above, except that [14C]AIB was added to the uninoculated resource plugs instead of to the inoculation plugs.
To test the transport of [14C]AIB from one colony to another, six 8-mm-diameter inoculation plugs were taken from just behind the tip region of a colony and placed in two rows of three approximately 3 cm apart on a scintillation screen lining a 12-cm by 12-cm-square petri plate. [14C]AIB was added to three of the inoculation plugs in a checkerboard pattern, and photon emission was counted as described in reference 72.
Microscopy and developmental age experiments.
Two different strains with fluorescently labeled proteins were used: histone H1 (H1-dsRed) (20, 54) and rdi-1-gfp (53). The H1-red fluorescent protein (dsRED) localizes to nuclei, and Rho guanosine nucleotide dissociation inhibitor 1 (RDI1)-green fluorescent protein (GFP) localizes to the cytoplasm. Three microliters of approximately 108/ml conidia of each strain was inoculated in a line either 5 mm or 10 mm apart on the center of a rectangle of VMM agar on a 6- by 4-cm glass slide. The colonies were kept in a 25°C constant-light room. The colonies each had two linear growth fronts, one growing toward the other colony, referred to as the inner front, and one growing frontward away from the other colony, referred to as the outer front. We captured fluorescent images (GFP and RFP) of the colonies (i) on the center of the inner front where the two colonies met, (ii) directly next to the inoculation lines on the inner front, and (iii) at the tips of the outer front. Additional transects were imaged on the outer fronts every 5 mm up to every 1 cm. At each point, an image was taken with a GFP filter, and an image was taken with an RFP filter. Three replicates were performed for each experiment. Micrographs were taken with a digital C4742-95 charge-coupled-device camera (Hamamatsu, Japan) using the Openlab software program (Coventry, United Kingdom) and a Zeiss Axioskop II microscope.
Colony architecture images were obtained by covering the hyphae with 10 mM calcofluor. Images were taken after 15 min, allowing the calcofluor to be absorbed into the hyphae and to stain the cell walls. Micrographs were taken using a QIClick camera (QImaging, Surrey, BC, Canada) on a Zeiss AxioImager microscope, and the images were analyzed using iVision Mac 4.5 software. The hyphal diameters and angles were measured for 10 leading hyphae containing ∼30 primary branches and ∼20 secondary branches.
RESULTS
Hyphal fusion is required for efficient nutrient translocation.
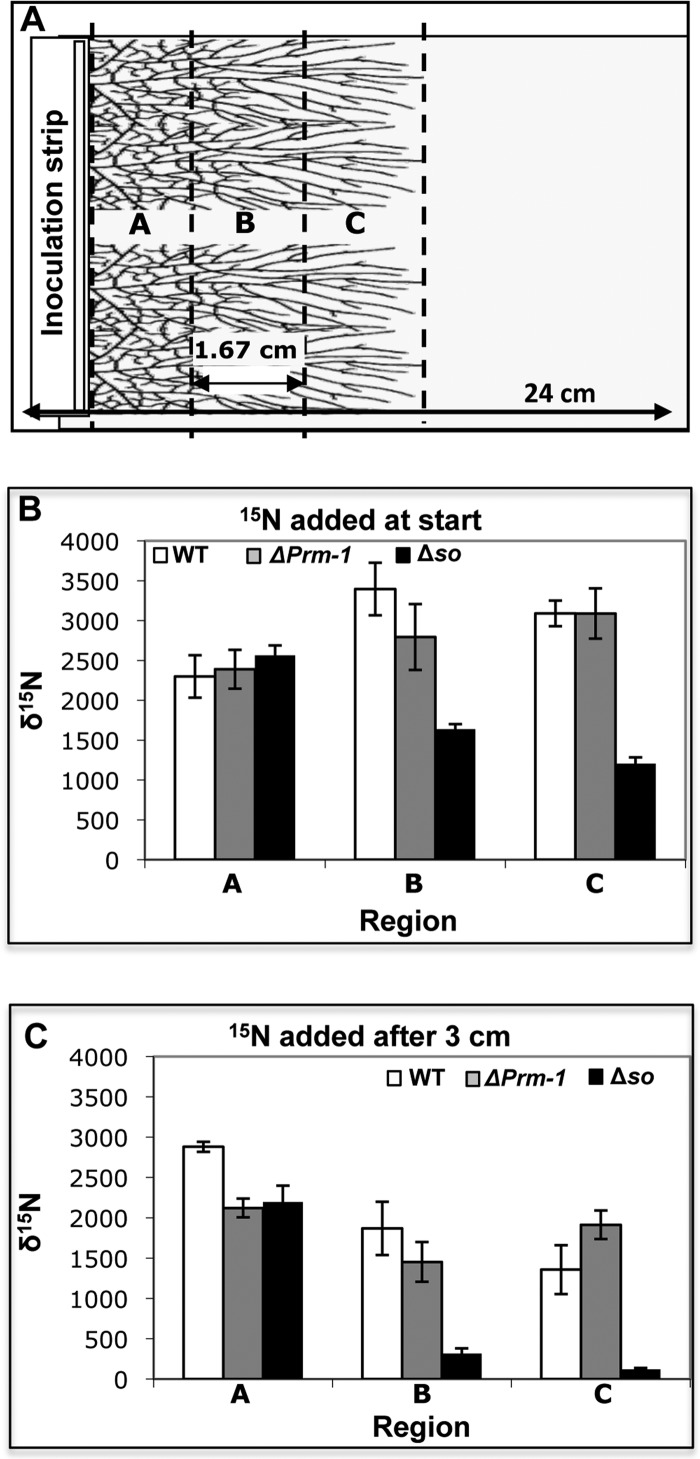
The impact of varying levels of hyphal fusion on nutrient translocation was determined by evaluating the distribution of an enriched stable isotope tracer, [15N]AIB, which was added to an inoculum of a fully interconnected wild-type (WT) colony, a partially connected ΔPrm-1 colony, or an unconnected Δso colony. Following inoculation and [15N]AIB addition, the δ15N distribution across sections of the colony was measured after ∼5 cm of linear growth by harvesting a strip immediately adjacent to the inoculum point (region A), a strip at the midzone (region B), and a strip at the colony margin (region C) (Fig. 1A). The levels of δ15N in the WT, ΔPrm-1, and Δso colonies were similar in region A adjacent to the inoculum point (Fig. 1B). In both the WT and ΔPrm-1 colonies, the δ15N levels significantly increased toward the colony periphery (regions B and C; Fig. 1B). However, Δso colonies had much lower δ15N levels in the midzone and colony margins (Fig. 1B).
Fig 1.
Experimental design and translocation of δ15N in N. crassa colonies. (A) Diagram of plates used for stable isotope translocation experiments. The plates were 24 by 24 cm and filled with low-sucrose agar covered with a nylon membrane, with glass slides placed at one edge of the plate. An inoculum strip of minimal medium agar containing a strip of hyphae was placed on the edge of the glass slides. [15N]AIB was added to the inoculum strip either at the start of the experiment or after the colonies had grown 3 cm. The letters indicate the 1.67-cm regions from which the colony biomass was harvested. (B) The amounts of δ15N in regions A, B, and C of the WT, ΔPrm-1, and Δso colonies were determined when a [15N]AIB tracer was added to the inoculation strip at the time of colony inoculation. (C) The amounts of δ15N in regions A, B, and C of the wild-type, ΔPrm-1, and Δso colonies were determined when a [15N]AIB tracer was added to the inoculation strip after 3 cm of linear growth. Bars indicate the standard errors.
To determine if [15N]AIB could be transported within an already established colony, a [15N]AIB tracer was added to the inoculation strip of the WT, ΔPrm-1, and Δso colonies after 3 cm of linear growth. As described above, strips of the colony were harvested after 5 cm of linear growth. The ΔPrm-1 and Δso colonies showed similar δ15N levels in the region immediately adjacent to the inoculum point, while WT colonies had significantly higher levels (Fig. 1C). However, a very significant decrease in 15N levels in the Δso colonies toward the colony margin was observed compared to that in the WT and ΔPrm-1 colonies. These data indicate that the WT and the ΔPrm-1 mutant are capable of the translocation of nutrients during growth or in an already-established network, while the unconnected Δso mutant colonies are incapable of significant translocation.
Network formation affects carbon/nitrogen ratios in colonies.
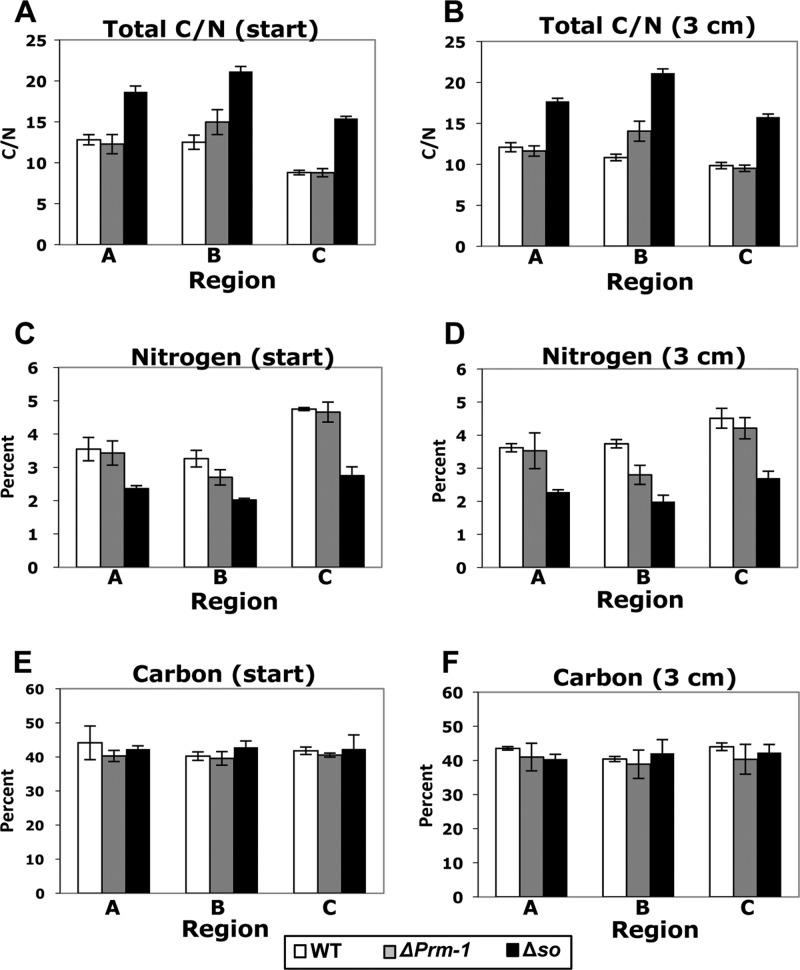
An important trait of filamentous fungi is their ability to rapidly distribute carbon and nitrogen from sources to sinks (6, 21, 32), a trait that is presumably mediated by the interconnected mycelial network. We therefore tested the hypothesis that the total C and N content in Δso colonies, which show a severe defect in resource translocation, would be significantly different from that in the wild-type colonies. As predicted, the Δso mutant had a higher C/N ratio than both the ΔPrm-1 and WT colonies (Fig. 2A and B). Carbon, nitrogen, or the synergy of C and N could drive these differences in C/N ratios. We therefore analyzed the total C and N content separately (see Materials and Methods). A statistically significant trend in total C amounts was not observed between the strains or treatments (Fig. 2E and F). However, differences in the total N percentages were observed. For all three strains, the tip region had higher total N levels. For the Δso colonies, all treatments showed a significantly lower total N level (Fig. 2C and D). Because we did not use an enriched 13C tracer, we were also able to analyze the natural abundance of δ13C. There was a small but significantly more negative δ13C abundance in all the Δso regions than that in the corresponding region of the ΔPrm-1 or wild-type colonies (see Fig. S1 in the supplemental material). These observations indicate that Δso colonies have an altered carbon metabolism or utilization that leads to the fractionation of C, which is significantly different than that in the ΔPrm-1 and WT colonies.
Fig 2.
Total C/N ratios found in different regions of WT and mutant N. crassa colonies. (A and B) Total carbon/nitrogen ratios obtained from regions A, B, and C (Fig. 1A) from the WT, ΔPrm-1, and Δso colonies when [15N]AIB tracer was added to the inoculation strip at the time of inoculation (start) (A) or after 3 cm of linear growth (B); (C and D) total percentages of nitrogen found in regions A, B, and C in the WT, ΔPrm-1, and Δso colonies when [15N]AIB tracer was added to the inoculation strip at the time of inoculation (start) (C) or after 3 cm of linear growth (D); (E and F) total percentages of carbon found in regions A, B, and C in the WT, ΔPrm-1, and Δso colonies when [15N]AIB tracer was added at inoculation (E) or after 3 cm of linear growth (F). Error bars indicate the standard errors. The average natural abundance of 15N for the unenriched control colonies was ∼4.5‰.
Dynamic measurements of N distribution during exploration of a heterogeneous environment.
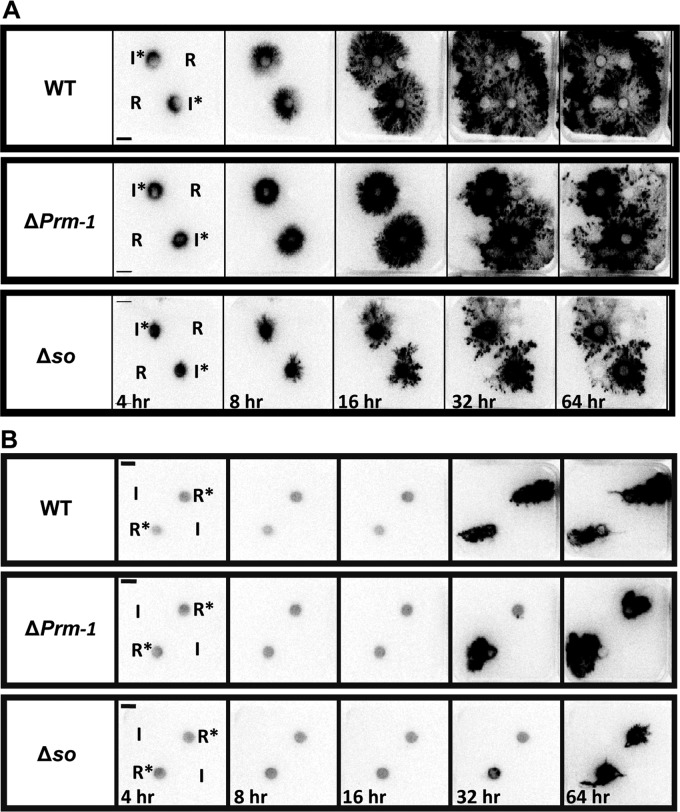
The requirement for harvesting samples destructively (Fig. 1 and 2) restricts the temporal and spatial resolution that can be achieved with 15N isotope studies and makes it challenging to observe dynamic behavior in response to perturbation. We therefore imaged the rates and patterns of resource distribution in living WT, ΔPrm-1, and Δso colonies grown in a heterogeneous environment (media plugs placed on a scintillation screen) using photon-counting scintillation imaging (PCSI) of 14C-labeled AIB. Distinctly different patterns of [14C]AIB distribution were observed in all three strains. In the WT colonies, the majority of the [14C]AIB was distributed toward the periphery of the colony (Fig. 3A, top), consistent with the [14N]AIB tracer data from mass spectrometry analyses of the colony sections. Furthermore, the distribution of [14C]AIB in the WT was not affected by the presence of an additional resource, with no evidence for preferential resource allocation to this sector of the colony. In the ΔPrm-1 mutant colonies, a significant fraction of [14C]AIB remained in the interior of the colonies (Fig. 3A, middle), while in the Δso colonies, the majority of the tracer was concentrated around the original inoculum plugs (Fig. 3A, bottom). These observations indicate that even an ∼50% decrease in network formation, as observed in the ΔPrm-1 colonies, significantly affected the ability of a colony to translocate resources in a more polarized heterogeneous environment.
Fig 3.
Images of [14C]AIB in colonies of WT and mutant N. crassa strains over time. (A) Images of photon emissions were captured at 4, 8, 16, 32, and 64 h from the WT, ΔPrm-1, and Δso colonies where [14C]AIB was added to the inoculation plugs (I*) prior to growth of the colony. Unlabeled sucrose resource plugs (R) were placed ∼4 cm from the inoculation plugs. (B) Photon emissions captured at 4, 8, 16, 32, and 64 h from the WT, ΔPrm-1, and Δso colonies where [14C]AIB had been added to a sucrose resource plug (R*) prior to growth, but the inoculum plug (I) remained unlabeled. Resource plugs were placed ∼4 cm from the inoculation plugs.
There is no detectable retrograde transport of resources in Neurospora.
A WT N. crassa strain was very efficient at translocating AIB from the inoculum to the periphery of the colony (Fig. 3A). We therefore asked whether colonies of WT or the ΔPrm-1 and Δso fusion mutants were affected in the retrograde transport of [14C]AIB from the tips toward the interior of the colony. In this case, the [14C]AIB was added to resource plugs so that the hyphae at the periphery of a colony originating from the inoculation plug would “discover” the resource. However, minimal reverse translocation of the [14C]AIB from the resource plug to the interior of the colony was observed in all three of the strains (Fig. 3B). We infer that retrograde translocation of resources from the hyphal tips to the colony interior does not occur at a significant level in N. crassa under the conditions tested here.
Mature networked colonies of N. crassa do not share resources.
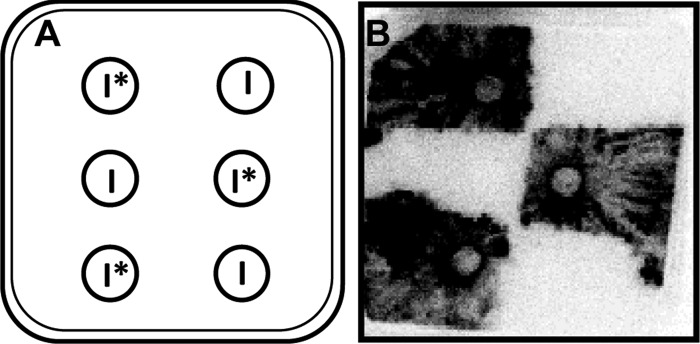
To assess whether fusion might impact resource sharing between adjacent genetically identical colonies, [14C]AIB was added to alternating plugs of the WT colonies in two rows such that every other plug was labeled (Fig. 4A). After 3 days, fungal growth was confluent, and the individual colonies could no longer be distinguished from each other. However, when examined for [14C]AIB distribution, only the colonies that emerged from the [14C]AIB plugs were labeled, resulting in a checkerboard pattern of labeled and unlabeled colonies (Fig. 4B). The labeled colonies did not grow substantially into the unlabeled colonies or translocate [14C]AIB an appreciable distance into neighboring colonies. These data indicate that no or only limited resource sharing occurs between mature N. crassa colonies.
Fig 4.
[14C]AIB translocation between N. crassa colonies. (A) Six colonies were grown on one 12- by 12-cm petri plate, with three inoculum plugs labeled with [14C]AIB (I*) and three inoculum plugs that were left unlabeled (I) in a checkerboard pattern. (B) Photon emissions were captured from WT colonies from an experimental design shown in panel A after growth for 64 h.
Genetic mixing in WT colonies is affected by colony age.
As fusion does not appear to assist long-distance transport between colonies (Fig. 4), we tested whether it had an impact on genetic mixing. We assessed the movement of fluorescently labeled proteins targeted to the nucleus and cytoplasm of WT N. crassa colonies that were allowed to come into physical contact with each other. Conidia from a wild-type strain bearing a nuclear fluorescence marker (H1-dsRED) (20) and conidia from a wild-type strain bearing a cytoplasmic GFP marker (RDI1-GFP) (53) were inoculated in opposing parallel lines. To assess whether genetic mixing occurred, fluorescent images were taken at transects throughout both colonies. Mixed red (H1-dsRED; rhodamine filter) and green (RDI1-GFP; GFP filter) hyphae are a consequence of hyphal fusion events between the H1-dsRed and rdi-1-gfp colonies.
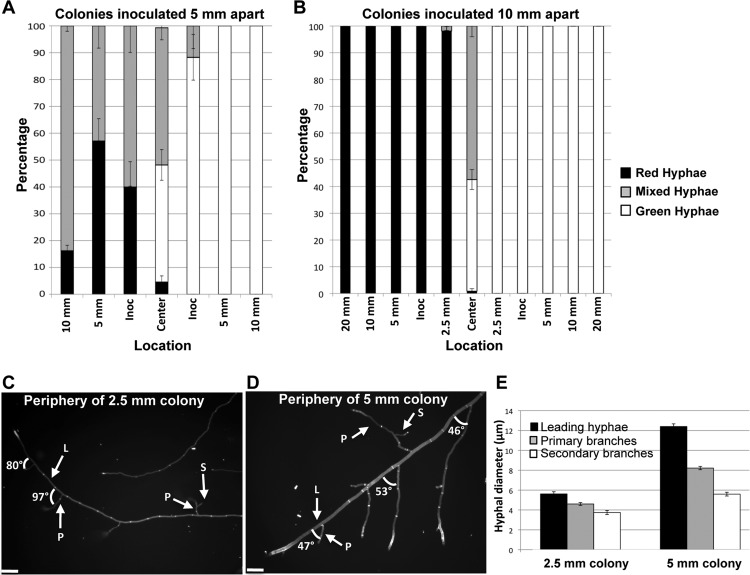
Conidial lines inoculated 5 mm apart germinated and grew ∼2.5 mm before they encountered each other. The contact area between the rdi-1-gfp and H1-dsRed colonies showed ∼50% mixed hyphae (Fig. 5A). In addition, the entire H1-dsRed colony showed significant red and green fluorescence, with the outer periphery of the colony opposite of the contact zone containing 85% mixed hyphae. In the rdi-1-gfp colony, red nuclear fluorescence extended to the inoculation point (∼2.5 mm) but was not observed in the remainder of the rdi-1-gfp colony. The differences in mixing ratios between the two colonies could be due to an inability to detect small numbers of H1-dsRed nuclei, histone H1 turnover, or simply that the H1-dsRed nuclei did not migrate as far as the cytoplasmically localized RDI1. As observed with the [14C]AIB labeling (Fig. 4B), there was little growth of the hyphae of the rdi-1-gfp strain (“green-only” hyphae) into the H1-dsRed colony or growth of H1-dsRed (“red-only”) hyphae into the rdi-1-gfp colony.
Fig 5.
Cytoplasmic mixing between N. crassa colonies of different ages. (A) Graph showing average percentage of hyphae containing only green fluorescence (RDI1-GFP), average percentage of hyphae containing only red fluorescence (H1-dsRED), and average percentage of hyphae with both green and red fluorescence (mixed) across H1-dsRED and rdi-1-gfp colonies inoculated 5 mm apart. Error bars indicate the standard errors. (B) Graph showing average percentage of hyphae containing only green fluorescence (RDI1-GFP), average percentage of hyphae containing only red fluorescence (H1-dsRED), and average percentage of hyphae with both green and red fluorescence (mixed) across H1-dsRED and rdi-1-gfp colonies inoculated 1 cm apart. Bars indicate the standard errors. (C) Micrograph of the periphery of a 2.5-mm colony stained with 10 mM calcofluor; angle values between leading hyphae and primary branches are indicated. Abbreviations: L, leading hyphae; P, primary branch; S, secondary branch. (D) Micrograph of the periphery of a 5-mm colony stained with 10 mM calcofluor; angle values between leading hyphae and primary branches are indicated. Scale bars indicate 100 μm. (E) A graph showing average hyphal diameters at different locations within the colonies. Bars indicate the standard errors.
When the H1-dsRed and rdi-1-gfp conidial lines were plated 10 mm apart, they germinated and grew ∼5 mm from the inoculation point before encountering each other. Unlike with the colonies that were inoculated 5 mm apart, mixing was limited to the interaction zone only (Fig. 5B). No nuclear dsRed fluorescence was detected in the rdi-1-gfp colony past the contact zone, and a very low percentage of hyphae showed cytoplasmic GFP fluorescence 2.5 mm from the contact zone in the H1-dsRed colony. These data suggest that, after 5 mm of linear growth, N. crassa colonies have reached a developmental age at which extensive cytoplasmic and nuclear exchange is restricted.
Hyphal architecture changes with colony age and is associated with the capacity to share resources.
Germlings in N. crassa and young, undifferentiated hyphae show developmental and morphological differences from hyphae in a mature colony (2, 44, 62, 67). For example, while germ tubes have uniform hyphal diameters (∼3.5 μm) (62), in a mature colony at least 4 different hyphal types occur (5), which are characterized by differences in hyphal diameter, branching angles, extension rates, and compartment lengths (67). To test the hypothesis that a developmental time point during colony establishment is associated with resource sharing, conidia/hyphae were stained with 10 mM calcofluor, and images were captured from the conidial inoculation point to the 2.5-mm growth front in colonies inoculated 5 mm apart and from the inoculation point to 5 mm of growth in colonies inoculated 10 mm apart (Fig. 5C and D; see also Fig. S2 in the supplemental material). There were striking differences in hyphal architecture at the colony periphery of a 2.5-mm colony compared to that of the hyphae at the periphery of a 5-mm colony. All hyphae at the periphery of a 2.5-mm colony had similar hyphal diameters and displayed branching angles of ∼90° (right angles) (Fig. 5C and E). In contrast, hyphae at the periphery of a 5-mm colony differentiated into leading (or trunk) hyphae and primary and secondary branch hyphae that had significantly different hyphal diameters (Fig. 5E). In particular, the branch angles of the primary and secondary hyphae were no longer at 90° but showed branch angles of ∼50° (Fig. 5D; see also Fig. S3 in the supplemental material). These observations suggest that while germlings and undifferentiated colonies of N. crassa readily share resources between colonies via hyphal fusion, their differentiation into mature hyphal forms restricts resource sharing to the interaction zones of mature colonies.
DISCUSSION
This study used multiple techniques, including radioisotope tracers, stable isotope tracers, and fluorescently labeled proteins, to visualize and quantify nutrient flows and cytoplasmic and nuclear movement within and between N. crassa colonies. Using the Δso and ΔPrm-1 fusion mutants, we determined that hyphal fusion influences nutrient distribution within N. crassa colonies. The soft mutant, which lacks hyphal fusion (18), is severely decreased in its ability to transport resources. These results imply that an interconnected network is important for nutrient translocation within a fungal colony. The soft mutant is also different than the ΔPrm-1 mutant and the WT in terms of carbon and nitrogen composition and ratios, but it is unclear whether this is a result of the lack of hyphal fusion and a reduced ability to translocate nutrients.
A hallmark of true filamentous fungi is the ability to form interconnected networks (9). Mathematical models of interconnected networks show that the exchange of energy resources provides an adaptive advantage to these organisms (49). Fungal interconnected networks form a contiguous lumen, which may facilitate information flow through biophysical principles, such as growth-induced mass flows and hydraulic coupling (30, 31), or through cytoplasmic streaming, leading to significant advection (26, 50, 73). However, different fungal species show marked differences in septation (29) that can restrict these flows, particularly those of nuclei. In N. crassa, the septal pores are open, and the cytoplasm and all organelles, including nuclei, show dramatic flow rates through interconnected networks (33). Other species, such as many filamentous basidiomycetes, have complex structures at the septa that allow cytoplasmic and presumably some organellar transfer but which restrict nuclear mixing (46). Additionally, some basidiomycete species can form multihyphal aggregates, termed cords or rhizomorphs, that show some tissue differentiation, with larger-vessel hyphae acting as conduits for long-distance transport (14, 32).
Nutrient translocation and the rates of movement in fungal species are of particular interest in many fields, but research linking network morphology to measured nutrient movement is scarce (23). Most analyses use theoretical models to predict transport efficiency and network resilience from an empirically determined network architecture system (3, 30). In theory, efficient and robust translocation networks can be achieved by differential reinforcement of the main transport pathways coupled with the introduction of cross-linking pathways, analogous to fusion events, which add robustness to the system. During grazing by Collembola on cord-forming colonies of the basidiomycete Phanerochaete velutina, the network responded to attack by increasing the degree of cross-linking, dynamically adjusting the resilience of the system at the expense of further exploratory growth (65). In this study, we removed or reduced the fusion component of an N. crassa colony through genetic manipulation by using mutants that differed in fusion frequency. Our results show that hyphal fusion is necessary for effective nutrient translocation, and even an ∼50% reduction in fusion frequency in the ΔPrm-1 mutant resulted in decreased translocation rates (Fig. 4).
Using the concepts advanced by Grime (27), saprotrophic basidiomycetes, such as P. velutina, are persistent stress-tolerant (S-selected) or combative (C-selected) species, while N. crassa is a ruderal (R-selected) ascomycete species (52) adapted for the rapid colonization of fire-damaged trees (35). Our results show that N. crassa colonies can easily translocate a tracer when labeled from an inoculation point, as well as through an established colony. However, retrograde transport was not observed. When encountering a carbon-rich resource plug under nutrient-poor conditions, the N. crassa colonies did not transport resources to the interior of the colony but instead sent these resources outward toward foraging tips (Fig. 3). These observations are in contrast to those of mycorrhizal fungi, where bidirectional transport is required from the tips to the roots and the roots to the tips (11). In persistent basidiomycete species, such as P. velutina, rapid pulsatile fluxes operate both acropetally and basipetally between the inoculum and added resources (69, 71, 72). In addition, unlike in N. crassa, rapid shifts in nutrient distribution occurred in P. velutina in response to new resource additions (70) and even to long-distance translocation between different individuals (21). These differences in colony transport may be primarily due to the formation of cords in some basidiomycete species, such as P. velutina or Armillaria mellea, that facilitate retrograde movement (70). Indeed, some degree of hyphal differentiation and tissue organization may be essential to allow more complex patterns of nutrient distribution, such as retrograde translocation and strategic reallocation of nutrients from sources to sinks. Differences in colony transport potentially have a further effect on C and N composition and metabolism in colonies, as is seen in the significantly decreased N and altered C isotope fractionations in N. crassa colonies lacking fusion.
Germlings readily share cytoplasm and nuclei when they fuse (17, 62). However, our results show that genetically identical ∼5-mm colonies that physically interact do not significantly share cytoplasm and nutrients. There are a number of developmental differences between germlings and mature N. crassa colonies. For example, the transcriptional profile of germlings is much different from that of older colonies (36, 37), and some hyphal fusion mutants that do not show germling fusion will resume fusion after colony development (66). N. crassa germlings lack a defined Spitzenkörper and a nuclear exclusion zone in the hyphal apex, but once these germlings grow to 150 μm, they have acquired both (2). Similarly, young undifferentiated N. crassa mycelia have uniform hyphal diameters (∼3 to 4 μm), uniform hyphal extension rates and compartment lengths (67), and near-90° branch angles (44). By contrast, hyphae in mature colonies show differentiation, with “leader or trunk” hyphae (∼10 μm in diameter) versus primary (∼7 μm in diameter) and secondary (∼5 μm in diameter) branches, with branching angles of ∼60° (44, 67). These observations indicate that germlings and young colonies are physiologically and developmentally distinct from mature colonies, which dramatically affects their capacity for resource sharing. Ecologically, it may be important for germlings and young colonies to pool their resources via fusion to outcompete others for the utilization of a new resource (61). However, once a colony is established, cooperation may be less important, and other aspects, such as resource plundering and the transfer of mycoviruses (4, 10, 12, 13), both of which occur via hyphal fusion between colonies, may play an important role in the restriction of interaction between colonies. Interestingly, it was recently shown that a non-self-recognition system in filamentous fungi, termed heterokaryon or vegetative incompatibility, which restricts heterokaryon formation between mature colonies, is suppressed in germlings in the filamentous ascomycete Colletotrichum lindemuthianum (34). Thus, our data indicate that hyphal architecture, which is regulated developmentally, plays an important role in the capacity of individual colonies to share resources. Further research will be necessary to determine the signals and processes affected by hyphal fusion within and between fungal colonies and its capacity for genetic exchange and resource sharing, especially between fungi with differing life histories.
Supplementary Material
ACKNOWLEDGMENTS
A.S., J.P.-G., and N.L.G. were supported by grants MCB-0817615 and MCB-1121311 from the National Science Foundation (USA).
We acknowledge technical assistance from Greg Iwahashi and Marcus Roper, and we thank Abigail Leeder and Marcus Roper for their thoughtful discussions and suggestions.
Footnotes
Published ahead of print 7 September 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Aldabbous MS, et al. 2010. The ham-5, rcm-1 and rco-1 genes regulate hyphal fusion in Neurospora crassa. Microbiology 156:2621–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Araujo-Palomares CL, Castro-Longoria E, Riquelme M. 2007. Ontogeny of the Spitzenkörper in germlings of Neurospora crassa. Fungal Genet. Biol. 44:492–503 [DOI] [PubMed] [Google Scholar]
- 3. Bebber DP, Hynes J, Darrah PR, Boddy L, Fricker MD. 2007. Biological solutions to transport network design. Proc. Biol. Sci. 274:2307–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Biella S, Smith ML, Aist JR, Cortesi P, Milgroom MG. 2002. Programmed cell death correlates with virus transmission in a filamentous fungus. Proc. Biol. Sci. 269:2269–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bistis GN, Perkins DD, Read ND. 2003. Different cell types in Neurospora crassa. Fungal Genet. Newsl. 50:17–19 [Google Scholar]
- 6. Boddy L, Watkinson SC. 1995. Wood decomposition, higher fungi, and their role in nutrient redistribution. Can. J. Bot. 73:S1377–S1383 [Google Scholar]
- 7. Boddy L, Wood J, Redman E, Hynes J, Fricker MD. 2010. Fungal network responses to grazing. Fungal Genet. Biol. 47:522–530 [DOI] [PubMed] [Google Scholar]
- 8. Bonner JT. 1998. The origins of multicellularity. Integr. Biol. 1:27–36 [Google Scholar]
- 9. Buller AHR. 1933. Researches on fungi, vol 5 Longman, London, England [Google Scholar]
- 10. Caten CE. 1972. Vegetative incompatibility and cytoplasmic infection in fungi. J. Gen. Microbiol. 72:221–229 [DOI] [PubMed] [Google Scholar]
- 11. Cruz C, et al. 2007. Enzymatic evidence for the key role of arginine in nitrogen translocation by arbuscular mycorrhizal fungi. Plant Physiol. 144:782–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Debets AJM, Griffiths AJF. 1998. Polymorphism of het-genes prevents resource plundering in Neurospora crassa. Mycol. Res. 102:1343–1349 [Google Scholar]
- 13. Debets F, Yang X, Griffiths AJ. 1994. Vegetative incompatibility in Neurospora: its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr. Genet. 26:113–119 [DOI] [PubMed] [Google Scholar]
- 14. Eamus D, Thompson W, Cairney JWG, Jennings DH. 1985. Internal structure and hydraulic conductivity of basidiomycete translocating organs. J. Exp. Bot. 36:1110–1116 [Google Scholar]
- 15. Fleissner A, Diamond S, Glass NL. 2009. The Saccharomyces cerevisiae PRM1 homolog in Neurospora crassa is involved in vegetative and sexual cell fusion events but also has postfertilization functions. Genetics 181:497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fleissner A, Glass NL. 2007. SO, a protein involved in hyphal fusion in Neurospora crassa, localizes to septal plugs. Eukaryot. Cell 6:84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fleissner A, Leeder AC, Roca MG, Read ND, Glass NL. 2009. Oscillatory recruitment of signaling proteins to cell tips promotes coordinated behavior during cell fusion. Proc. Natl. Acad. Sci. U. S. A. 106:19387–19392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fleissner A, et al. 2005. The so locus is required for vegetative cell fusion and postfertilization events in Neurospora crassa. Eukaryot. Cell 4:920–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fleissner A, Simonin AR, Glass NL. 2008. Cell fusion in the filamentous fungus, Neurospora crassa. Methods Mol. Biol. 475:21–38 [DOI] [PubMed] [Google Scholar]
- 20. Freitag M, Hickey PC, Raju NB, Selker EU, Read ND. 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41:897–910 [DOI] [PubMed] [Google Scholar]
- 21. Fricker M, Boddy L, Nakagaki T, Bebber DP. 2009. Adaptive biological networks, p 51–70 In Gross T, Sayama H. (ed), Adaptive networks: theory, models and applications. Springer, Heidelberg, Germany [Google Scholar]
- 22. Fricker MD, Boddy L, Bebber D. 2007. Network organization of filamentous fungi, p 309–330 In Howard RJ, Gow NAR. (ed), Biology of the fungal cell. Springer-Verlag, Berlin, Germany [Google Scholar]
- 23. Fricker MD, et al. 2008. Imaging complex nutrient dynamics in mycelial networks. J. Microsc. 231:317–331 [DOI] [PubMed] [Google Scholar]
- 24. Fu C, et al. 2011. Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryot. Cell 10:1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Giovannetti M, Azzolini D, Citernesi AS. 1999. Anastomosis formation and nuclear and protoplasmic exchange in arbuscular mycorrhizal fungi. Appl. Environ. Microbiol. 65:5571–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goldstein RE, Tuval I, van de Meent JW. 2008. Microfluidics of cytoplasmic streaming and its implications for intracellular transport. Proc. Natl. Acad. Sci. U. S. A. 105:3663–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grime JP. 1977. Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. Am. Nat. 111:1169–1194 [Google Scholar]
- 28. Grosberg RK, Strathmann RR. 2007. The evolution of multicellularity: a minor major transition? Annu. Rev. Ecol. Evol. Syst. 38:621–654 [Google Scholar]
- 29. Gull K. 1978. Form and function of septa in filamentous fungi, p 78–93 In Smith JE, Berrys DR. (ed), Developmental mycology. John Wiley and Sons, New York, NY [Google Scholar]
- 30. Heaton LL, Lopez E, Maini PK, Fricker MD, Jones NS. 2010. Growth-induced mass flows in fungal networks. Proc. Biol. Sci. 277:3265–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heaton LL, et al. 2012. Analysis of fungal networks. Fungal Biol. Rev. 26:12–29 [Google Scholar]
- 32. Heaton LLM, López E, Maini PK, Fricker MD, Jones NS. 2012. Advection, diffusion and delivery over a network. Phys. Rev. E 86:021905 doi:10.1103/PhysRevE.86.021905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hickey PC, Jacobson D, Read ND, Louise Glass NL. 2002. Live-cell imaging of vegetative hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 37:109–119 [DOI] [PubMed] [Google Scholar]
- 34. Ishikawa FH, et al. Heterokaryon incompatibility is suppressed following conidial anastomosis tube fusion in a fungal plant pathogen. PLoS One 7:e31175 doi:10.1371/journal.pone.0031175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jacobson DJ, et al. 2004. Neurospora in temperate forests of western North America. Mycologia 96:66–74 [PubMed] [Google Scholar]
- 36. Kasuga T, Glass NL. 2008. Dissecting colony development of Neurospora crassa using mRNA profiling and comparative genomics approaches. Eukaryot. Cell 7:1549–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kasuga T, et al. 2005. Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination. Nucleic Acids Res. 33:6469–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Knoll AH. 2011. The multiple origins of complex multicellularity. Annu. Rev. Earth Planet. Sci. 39:217–239 [Google Scholar]
- 39. Leeder AC, Palma-Guerrero J, Glass NL. 2011. The social network: deciphering fungal language. Nat. Rev. Microbiol. 9:440–451 [DOI] [PubMed] [Google Scholar]
- 40. Lew RR. 2011. How does a hypha grow? The biophysics of pressurized growth in fungi. Nat. Rev. Microbiol. 9:509–518 [DOI] [PubMed] [Google Scholar]
- 41. Lucas WJ, Ham B-K, Kim J-Y. 2009. Plasmodesmata—bridging the gap between neighboring plant cells. Trends Cell Biol. 19:495–503 [DOI] [PubMed] [Google Scholar]
- 42. Ma LJ, et al. 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464:367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McCluskey K. 2003. The Fungal Genetics Stock Center: from molds to molecules. Adv. Appl. Microbiol. 52:245–262 [DOI] [PubMed] [Google Scholar]
- 44. McLean KM, Prosser JI. 1987. Development of vegetative mycelium during colony growth of Neurospora crassa. Trans. Br. Mycol. Soc. 88:489–495 [Google Scholar]
- 45. Mikkelsen BL, Rosendahl S, Jakobsen I. 2008. Underground resource allocation between individual networks of mycorrhizal fungi. New Phytol. 180:890–898 [DOI] [PubMed] [Google Scholar]
- 46. Muller WH, et al. 1998. Structural differences between two types of basidiomycete septal pore caps. Microbiology 144:1721–1730 [DOI] [PubMed] [Google Scholar]
- 47. Niklas KJ, Kutschera U. 2010. The evolution of the land plant life cycle. New Phytol. 185:27–41 [DOI] [PubMed] [Google Scholar]
- 48. Pandey A, Roca MG, Read ND, Glass NL. 2004. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot. Cell 3:348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pfeffer PE, Douds DD, Jr, Bucking H, Schwartz DP, Shachar-Hill Y. 2004. The fungus does not transfer carbon to or between roots in arbuscular mycorrhizal symbiosis. New Phytol. 163:617–627 [DOI] [PubMed] [Google Scholar]
- 50. Pickard WF. 2003. The role of cytoplasmic streaming in symplastic transport. Plant Cell Environ. 26:1–15 [Google Scholar]
- 51. Pontecorvo G. 1956. The parasexual cycle in fungi. Annu. Rev. Microbiol. 10:393–400 [DOI] [PubMed] [Google Scholar]
- 52. Pugh GJF, Boddy L. 1988. A view of disturbance and life strategies in fungi. Proc. R. Soc. Edinb. Biol. 94:3–11 [Google Scholar]
- 53. Rasmussen CG, Glass NL. 2007. Localization of RHO-4 indicates differential regulation of conidial versus vegetative septation in the filamentous fungus Neurospora crassa. Eukaryot. Cell 6:1097–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rasmussen CG, Morgenstein RM, Peck S, Glass NL. 2008. Lack of the GTPase RHO-4 in Neurospora crassa causes a reduction in numbers and aberrant stabilization of microtubules at hyphal tips. Fungal Genet. Biol. 45:1027–1039 [DOI] [PubMed] [Google Scholar]
- 55. Rayner ADM. 1996. Interconnectedness and individualism in fungal mycelia. Cambridge University Press, Cambridge, England [Google Scholar]
- 56. Rayner ADM, Griffith GS, Ainsworth AM. 1994. Mycelial interconnectedness, p 21–40 In Gow NAR, Gadd GM. (ed), The growing fungus. Chapman and Hall, London, England [Google Scholar]
- 57. Rayner ADM, Watkins ZR, Beeching JR. 1999. Self-integration—an emerging concept from the fungal mycelium, p 1–24 In Gow NAR, Robson GD, Gadd GM. (ed), The fungal colony. Cambridge University Press, Cambridge, England [Google Scholar]
- 58. Read ND, Fleißner A, Roca MG, Glass NL. 2010. Hyphal fusion, p 260–273 In Borkovich K, Ebbole DJ. (ed), Cellular and molecular biology of filamentous fungi. ASM Press, Washington, DC [Google Scholar]
- 59. Read ND, Lichius A, Shoji JY, Goryachev AB. 2009. Self-signalling and self-fusion in filamentous fungi. Curr. Opin. Microbiol. 12:608–615 [DOI] [PubMed] [Google Scholar]
- 60. Rep M, Kistler HC. 2010. The genomic organization of plant pathogenicity in Fusarium species. Curr. Opin. Plant Biol. 13:420–426 [DOI] [PubMed] [Google Scholar]
- 61. Richard R, Glass NL, Pringle A. 2012. Cooperation among germinating spores facilitates the growth of the fungus, Neurospora crassa. Biol. Lett. 8:419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Roca M, Arlt J, Jeffree C, Read N. 2005. Cell biology of conidial anastomosis tubes in Neurospora crassa. Eukaryot. Cell 4:911–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rokas A. 2008. The molecular origins of multicellular transitions. Curr. Opin. Genet. Dev. 18:472–478 [DOI] [PubMed] [Google Scholar]
- 64. Roper M, Ellison C, Taylor JW, Glass NL. 2011. Nuclear and genome dynamics in multinucleate ascomycete fungi. Curr. Biol. 21:R786–R793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rotheray TD, Jones TH, Fricker MD, Boddy L. 2008. Grazing alters network architecture during interspecific mycelial interactions. Fungal Ecol. 1:124–132 [Google Scholar]
- 66. Simonin AR, Rasmussen CG, Yang M, Glass NL. 2010. Genes encoding a striatin-like protein (ham-3) and a forkhead associated protein (ham-4) are required for hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 47:855–868 [DOI] [PubMed] [Google Scholar]
- 67. Steele GC, Trinci APJ. 1975. Morphology and growth kinetics of hyphae of differentiated and undifferentiated mycelia of Neurospora crassa. J. Gen. Microbiol. 91:362–368 [DOI] [PubMed] [Google Scholar]
- 68. Szathmary E, Smith JM. 1995. The major evolutionary transitions. Nature 374:227–232 [DOI] [PubMed] [Google Scholar]
- 69. Tlalka M, Bebber DP, Darrah PR, Watkinson SC, Fricker MD. 2007. Emergence of self-organised oscillatory domains in fungal mycelia. Fungal Genet. Biol. 44:1085–1095 [DOI] [PubMed] [Google Scholar]
- 70. Tlalka M, Bebber DP, Darrah PR, Watkinson SC, Fricker MD. 2008. Quantifying dynamic resource allocation illuminates foraging strategy in Phanerochaete velutina. Fungal Genet. Biol. 45:1111–1121 [DOI] [PubMed] [Google Scholar]
- 71. Tlalka M, Hensman D, Darrah PR, Watkinson SC, Fricker MD. 2003. Noncircadian oscillations in amino acid transport have complementary profiles in assimilatory and foraging hyphae of Phanerochaete velutina. New Phytol. 158:325–335 [Google Scholar]
- 72. Tlalka M, Watkinson SC, Darrah PR, Fricker MD. 2002. Continuous imaging of amino-acid translocation in intact mycelia of Phanerochaete velutina reveals rapid, pulsatile fluxes. New Phytol. 153:173–184 [Google Scholar]
- 73. Verchot-Lubicz J, Goldstein RE. 2010. Cytoplasmic streaming enables the distribution of molecules and vesicles in large plant cells. Protoplasma 240:99–107 [DOI] [PubMed] [Google Scholar]
- 74. Vogel HJ. 1956. A convenient growth medium for Neurospora. Microbiol. Genet. Bull. 13:42–46 [Google Scholar]
- 75. Waggoner B. 2001. Eukaryotes and multicells: origin. eLS. John Wiley & Sons Ltd, Chichester, United Kingdom: doi:10.1038/npg.els.0001640 [Google Scholar]
- 76. Xiang Q, Rasmussen C, Glass NL. 2002. The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics 160:169–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.