Abstract
Hydrodynamics predicts that swimming bacteria generate a propulsion force when a helical flagellum rotates because rotating helices necessarily translate at a low Reynolds number. It is generally believed that the flagella of motile bacteria are semirigid helices with a fixed pitch determined by hydrodynamic principles. Here, we report the characterization of three mutations in laboratory strains of Escherichia coli that produce different steady-state flagella without losing cell motility. E. coli flagella rotate counterclockwise during forward swimming, and the normal form of the flagella is a left-handed helix. A single amino acid exchange A45G and a double mutation of A48S and S110A change the resting flagella to right-handed helices. The stationary flagella of the triple mutant were often straight or slightly curved at neutral pH. Deprotonation facilitates the helix formation of it. The helical and curved flagella can be transformed to the normal form by torsion upon rotation and thus propel the cell. These mutations arose in the long-term laboratory cultivation. However, flagella are under strong selection pressure as extracellular appendages, and similar transformable flagella would be common in natural environments.
INTRODUCTION
Bacterial flagella mediate cell motility. They are composed of three different parts: a long filament ∼10 μm long and ∼20 nm in diameter is connected, through a curved flexible hook, to the basal body, an intricate set of disks, and rods embedded in the cell wall and membrane that generates the swimming force (5). In 1973, Berg and Anderson proposed that the flagellar filaments must rotate relative to the cell body to propel cells (6). Hydrodynamics also indicates that in the low Reynolds number regime, a helix that rotates due to an external torque necessarily translates (37). The pitch angle of a helix that maximizes translation efficiency has been estimated to be ca. 42° (10), while that of the fluorescence-labeled flagella of the peritrichous Escherichia coli strain AW405 was determined to be 41° on average (43). The flagella of E. coli form a left-handed helical bundle that rotates counterclockwise when viewed from behind during smooth swimming (termed a run) (5, 13, 27). When the rotation is reversed, some flagella transiently change to right-handed helices. The bundle either flies apart (43) or some flagella come out of the bundle (12), and thus cells change to a new direction abruptly (termed a tumble) due to the unbalanced thrust coupling with Brownian motion (5, 33).
The bacteria flagella must be semirigid helices to generate thrust (5). The bending stiffness of the flagella of Salmonella enterica subsp. enterica serovar Typhimurium is estimated to be ca. 3.5 pN μm2 (11). With a counterclockwise rotation motor, a functional flagellum needs to be left-handed (33). The bundle formation also relies on the helix geometry (22). Although a whole variety of flagella shape have been observed on different bacterial species (14), it is generally believed that the normal shape—the helical form observed in runs—is a metastable form. Other polymorphs are transient. They are triggered by external mechanical forces, i.e., in tumble, and by environmental perturbations, such as pH and ionic strength (7, 11). Seven different helical forms of the wild-type Salmonella flagella have been observed (7). The presence of some of them in E. coli was also confirmed (43). A mechanical model based on intrinsic packing of bi-stable subunits predicts 11 permissible canonical waveforms besides the normal helix (7, 8).
The flagellar filament of S. enterica serovar Typhimurium has been extensively studied (30). It is a tubular structure composed of a single protein called flagellin, that is encoded by the fliC gene, which is at 44 min on the E. coli genome (5). Many amino acid exchanges that alter the morphology of stationary filaments have been identified in Salmonella and E. coli (13, 21, 24). These mutants are generally nonmotile, yet provide a useful tool for elucidating flagellum structure (16, 24, 38, 45). The flagella of strain LMG194, an E. coli K-12 derivative, appeared as straight filaments at neutral pH after fluorescence staining, but the cells remain chemotactic (32). On the other hand, it has been observed that at low Reynolds number flexible planar structures can be deformed into reversible helical shapes in an oscillating field to generate propulsion (15); the photosynthetic bacterium Rhodobacter sphaeroides has a polar flagellum that is a flat coil at rest (1) but extends into a helix in swim (33). Here, we report the characterization of three mutations on the fliC gene of E. coli strain LMG194 and it ancestors. They generate flagella with noncanonical equilibrium configurations but are still functional, viz., they can be transformed into the canonical forms by external torque to propel the cell.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
Strain AW405 was a gift from Sandy Parkinson, and strains M65 and KS272 were gifts from Jonathan R. Beckwith. Strain LMG194 was obtained from Invitrogen (Darmstadt, Germany). Strains with the prefix LPE were generated in our lab. Other strains were obtained from the Coli Genetic Stock Center at Yale University. Bacteria were generally cultivated in Luria-Bertani (LB) broth at 37°C unless specified otherwise. The swarm assay was performed by inoculating 2 μl of overnight culture into semisolid agar plates according to a published procedure (19). The plates containing 0.30% agar were incubated at 37°C for 20 h, and the plates containing 0.35% agar were incubated at 37°C for 24 h. Examination of swimming motility was performed directly on cells growing in tryptone broth (2) at 23°C (optical density at 600 nm [OD600] = 0.2) or on cells suspended in chemotaxis medium (19). The pH of the mediums was adjusted from 3.5 to 12.0 with HCl or NaOH. All strains carrying pBD33-derived plasmids were cultivated in medium supplemented with 25 μg/ml of chloramphenicol.
Sequencing the fliC genes.
A primer pair covering the entire fliC gene (see Table S1 in the supplemental material) was designed according to the genome of E. coli strain MG1655 (accession no. NC_000913). PCR was performed with Platinum Taq DNA polymerase (Invitrogen) and the ingredients provided by the manufacturer. A total of 50 ng of genomic DNA purified from respective E. coli strains was used as templates. Thermo-cycles were as follows: initial denaturation at 94°C for 3 min; 35 cycles of 94°C for 45 s, 55°C for 30 s, and 72°C for 1.5 min; and a final extension at 72°C for 5 min. The PCR products were cloned using a TOPO-TA cloning kit (Invitrogen) and sequenced as previously described (36).
Creating a fliC-null mutant.
The kanamycin-resistant cassette was amplified with the primers Kanf and Kanr (see Table S1 in the supplemental material) and cloned into the pCR2.1 vector to create the plasmid pKK. A 416-bp fragment containing the 5′ end of fliC gene and an upstream region was amplified from the genomic DNA of strain AW405 with the primer pair Cf1 and Cr1. The KpnI and SpeI sites were introduced with primer pair KpnCf1 and SpeCr1 in a second round of PCR. After double digestion, the amplified fragment was separated on a 0.8% agarose gel and purified with a PureLink quick gel extraction and PCR purification combo kit (Invitrogen). The purified fragment was cloned into plasmid pKK digested with the same enzymes to generate plasmid pFK01. A 425-bp fragment containing the 3′ end of the fliC gene and a downstream sequence was amplified with the primers Cf2 and Cr2. The XhoI and XbaI restriction sites were introduced using the primers XhoCf2 and XbaCr2. After double digestion, the PCR product was ligated into linearized pFK01 to create a plasmid pFK02.
The 2,184-bp gene replacement cassette was released from plasmid pFK02 by double digestion with KpnI and XbaI. The ends of the fragments were polished with Pfu polymerase (Fermentas, St. Leon-Rot, Germany) and ligated into the pKOV gene replacement vector (26) linearized with SmaI to generate plasmid pFK03. The plasmid pFK03 was propagated in TOPO 10 chemical competent cells (Invitrogen) and electroporated into strain AW405. Screening of the knockout mutant was performed according to a published procedure (26), and the ΔfliC strain was labeled LPE16.
Cloning and creating fliC alleles.
The entire fliC genes, including the promoter regions from strain LMG194 and strain AW405, were amplified with PCR using the primer pair Flid01 and Flic4r (see Table S1 in the supplemental material). The PCR products were cloned into pCR2.1 vector and sequenced. The fragments were released from the plasmid by double digestion with HindIII and XbaI and ligated into plasmid pBD33 (17) linearized with same enzymes. The produced plasmid was propagated in TOPO10 cells and finally electroporated into LPE16 to create strains LPE28 and LPE30 (Table 1).
Table 1.
Helical parameters of flagella from different E. coli strains
| Allele | Strain(s) examined | Helical form | Handedness | Mean ± SD |
Abundance | |
|---|---|---|---|---|---|---|
| Pitch (μm) | Diam (μm) | |||||
| Wild type | LPE30, AW405 | Normal | Left | 2.33 ± 0.12 | 0.42 ± 0.03 | Almost all |
| M123 | LPE28, LMG194 | Straight | – | ∞ | 0 | Almost all |
| Imperfect helixa | Right | 3.17 ± 0.62 | 0.92 ± 0.33 | Minor | ||
| w-coilb | Right | 2.85 ± 0.13 | 0.84 ± 0.13 | Minor | ||
| M1 | LPE20 | w-coil | Right | 2.81 ± 0.11 | 0.71 ± 0.05 | Major |
| Extended coil | Right | 2.86 ± 0.18 | 0.47 ± 0.10 | Minor | ||
| M2 | LPE21 | L-curly | Left | 1.09 ± 0.07 | 0.43 ± 0.17 | Almost all |
| M3 | LPE22 | Straight | – | ∞ | 0 | Almost all |
| M12 | LPE23 | Super coil | Irregular | Variable | Variable | Major |
| Imperfect helix | Right | 2.41 ± 0.10 | 0.48 ± 0.07 | Minor | ||
| Imperfect helix | Left | 0.91 ± 0.05 | 0.28 ± 0.03 | Minor | ||
| M13 | LPE24 | Straight | – | ∞ | 0 | All |
| M23 | LPE25, M182 | R-normal | Right | 2.47 ± 0.12 | 0.51 ± 0.04 | Major |
| Curly II | Right | 1.06 ± 0.07 | 0.42 ± 0.06 | Minor | ||
Determined on partially helical flagella at pH 7.0.
Determined on perfect helices at pH 8.0.
To create a fliC gene only containing the A45G mutation (M1), a two-step fusion PCR was performed with the cloned sequence from strain AW405 as the template. In the first round, primer Flid01 that contains the target mutation was paired with the primer Mut01r (see Table S1 in the supplemental material); another primer, Mut01f, that contains the same mutation but toward the opposite direction, was paired with the primer Flic5r. The purified fragments were mixed at a molar ratio of 1:1 and used as a template for the second round of PCR in which the primer Flid01 was paired with Flic5r. The fused band purified from gel was cloned and sequenced. The A48S mutation (M2) was introduced with the same procedure using the primers Mut02r and Mut02f that contain the mutation. To introduce the S110A mutation (M3), the primers Mut03r and Mut03f that contain the expected mutation were paired with the primers E3flid and Mut03r, respectively. Double mutations were obtained by introducing a second mutation onto the single-mutation sequences. These fliC alleles were cloned into pBD33 vector (17) and expressed in LPE16 (Table 1).
Flagellum examination.
Single colonies were inoculated into 4 ml of LB medium and grown at 37°C overnight. A 10-μl portion of overnight culture was mixed with 10 ml of fresh medium, followed by growth at 23°C to the early exponential phase (OD600 = 0.4). Bacteria were precipitated for 1 min at the following centrifugation forces: wild type, 10,000 × g; M123 mutants, 6,000 × g; and other strains, 8,000 × g. The pellet was resuspended in 5 ml of flagellum staining buffer (43) and washed three times with the buffer. Staining with Alex Fluor 594 and microscopic observation were performed as described previously (32). The helical parameters were determined on detached flagella. Images were analyzed with ImageJ (http://rsb.info.nih.gov/ij/). Values of 15 to 50 independent measurements were averaged.
To preserve the flagellum configuration on swimming cells, the bacterial pellet was resuspended in 1× phosphate-buffered saline. After standing on the bench for 5 min, the cell suspension was fixed with 4% glutaraldehyde (Sigma), followed by incubation at room temperature for 1 h. The cells were stored at 4°C after washing away the fixative. Staining and examination were as described for living bacteria. For electron microscopy, aliquots of fixed and unfixed bacteria were adsorbed to carbon coated 400-mesh copper grids. The samples were washed with distilled water and negatively stained with 2% uranyl acetate for 1 min. Specimens were examined with an EM 902 transmission electron microscope (Zeiss, Oberkochen, Germany).
Mapping the mutations on flagellin.
The atomic coordinates for the L-type flagellin (accession no. 3A5X) (28) and the R-type flagellin (accession no. 1UCU) (45) from S. enterica serovar Typhimurium were retrieved from the RCSB Protein Data bank (http://www.rcsb.org/pdb/home/home.do). The coordinates for 0-start, 5-start, 6-start, 11-start, and 16-start subunits of the L-type and R-type straight filaments were kindly provided by Keiichi Namba. Residue localization and interaction analysis were performed using the software Vega ZZ (31).
RESULTS
Three mutations accumulated during laboratory cultivation.
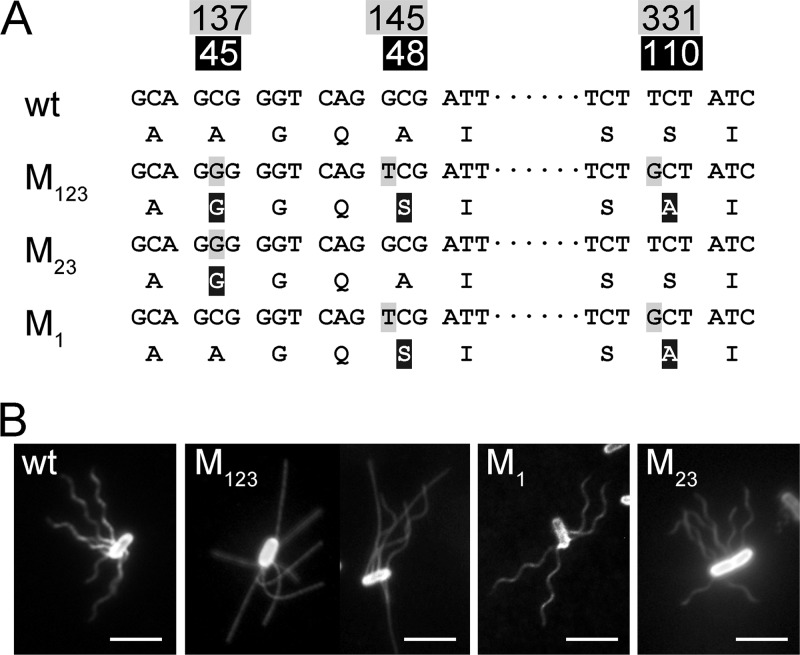
When the entire fliC genes of strain LMG 194 and strain AW405 were sequenced, three different nucleotides were detected (Fig. 1A). They encode flagellins that differ at position 45, where the alanine in strain AW405 was changed to a glycine in strain LMG194 (hitherto named M1); at position 48, where alanine was changed to serine (M2); and at position 110, where serine was changed to alanine (M3). We also selectively sequenced 14 ancestral strains of LMG194 according to the pedigree published by Bachmann (3) and others (9, 17, 42). The sequence of strain AW405 was identical to that of strain K-12 and many of its derivatives (see Fig. S1 in the supplemental material). This allele is therefore regarded as the wild type. Two mutations, M2 and M3, most likely arose simultaneously (referred to here as M23 for convenience), when strain M65 and Hfr3000 X74 were crossed to create strain M182 (J. R. Beckwith, personal communication). They were maintained in the widely used strain MC1000. The M1 mutation was introduced when strain LMG194 was isolated from strain KS272 (17). The triple mutation in strain LMG194 is referred to as M123, and the same nomenclature applies to other mutations in the present study.
Fig 1.
Motile E. coli strains with different fliC genes. (A) DNA and protein sequences of the wild-type (wt) and mutant alleles. The numbers at the top show the positions of the mutated nucleotides. The corresponding amino acids in flagellin are shown in the second line. Genotypes are named as discussed in the text. (B) Immobilized cells of the wild type (strain AW405), M123 mutant (strain LMG194; on the left is a cell at pH 7.0 and on the right a cell at pH 11.0), M1 mutant (strain LPE20), and M23 mutant (strain M182). Scale bar, 5 μm.
Morphologies of mutated flagella.
To reveal the influence of each mutation on flagellar morphology, artificial fliC alleles containing different combinations of mutations were constructed (Table 1). The handedness of detached fluorescent flagella was determined by moving the sample stage of the microscope up and down as described in reference 41, and the helical parameters were measured at the middle focal plane (see Fig. S2 in the supplemental material). The flagella of strain AW405 are left-handed helices with the same parameters as previously determined (12). Most flagella of strain LMG194 growing at neural pH were near-straight filaments as previously reported (32).
Medium pH plays an important role in determining the shape of the M123 flagella. At pH 7.0, ca. 5% of the cells had partially or entirely helical flagella (Fig. 1B). When the pH was decreased to 6.0, helical flagella were rarely observable. At pH 3.5, almost all flagella were straight filaments. When the pH was increased to 8.0, the fraction of cells with at least one significantly curved filament increased to ∼11%, and ∼7% of the cells have circular helices (Table 1). At pH 11.0, ca. 80% of the cells had helical flagella, and 50% of the flagella were perfect helices (Fig. 1B). At pH 12.0, all flagella were circular helices (see Fig. S2 in the supplemental material), and most of them detached from the cell body because of severe cell lysis. This right-handed helical form is about three times larger than the semicoil form of the same handedness previously observed (7) and is thus named the “w-coil” for wide coil (18). This pH-dependent polymorphism was observed on flagella from both strain LMG194 (genome borne) and LPE28 (plasmid borne). The number of flagella on cells of strain LPE28 is slightly higher than that of strain LMG194; however, its cell body, like strain AW405, is slightly larger than strain LMG194 (35).
The stationary M1 flagella are also w-coil (Fig. 1B). In the M1 mutant, right-handed extended coils with larger pitch and smaller amplitude were sometimes observed (Table 1). The M23 flagella are another type of right-handed helix with pitch and diameter values almost equal to those of the normal form (Fig. 1B). This form is therefore named R-normal. A stabilized right-handed curly II helix, which was previously observed on tumbling bacteria (22), was sometimes found in M23 mutant cells (Table 1). It is worth noting that on living cells of these mutants, the canonical helical forms involved in run and tumble, including normal, curly II, curly I, and semicoil (43) were all observable.
The M2 flagella are left-handed helices with very small pitch (see Fig. S2 and S3 in the supplemental material). Their helical parameters are close to those of the canonical curly II form (7, 43), but the handedness is opposite (Table 1). This kind of helix has been observed in some polar flagellated bacteria and was named L-curly (14). The M12 flagella are often stable mosaic filaments appearing as highly twisted supercoils, i.e., some parts were right-handed long helices and other parts were left-handed short helices (see Fig. S3 in the supplemental material). Compared to the two single mutations, the pitches and amplitudes were, however, smaller (Table 1). The M3 and the M13 flagella are completely straight (see Fig. S3 in the supplemental material), but the latter were slightly more flexible.
Motility and flagellar polymorphic transition.
Among the fliC mutants, only cells of the M1, M23, and M123 mutations showed relatively normal free-swimming behavior (Table 2). In liquid culture, only a subpopulation of M123 cells actively swam. The motility of different mutants is quantitatively represented by the size of their chemotaxis rings on semisolid agar plates (Table 2). It is worth mentioning that M123 and M12 mutants only spread in 0.30% agar and not in 0.35% agar.
Table 2.
Motility of the flagellar mutants in liquid and semisolid agar compared to cells carrying the wild-type allele
| Allele | Swimming behavior in liquid medium | Relative chemotaxis ring size on: |
|
|---|---|---|---|
| 0.35% agar | 0.30% agar | ||
| Wild type | Alternative run and tumble | 1 | 1 |
| M1 | Cell bodies wobble more | 0.5 | 0.5 |
| M2 | Only tumble, never swim in a straight line | 0 | 0 |
| M3 | Nonmotile | 0 | 0 |
| M12 | Tumble, cells may swim in a line of <2 μm | 0 | 0.1 |
| M13 | Nonmotile | 0 | 0 |
| M23 | Almost indistinguishable from the wild type | 0.9 | 0.9 |
| M123 | Cell bodies wobbled more and with longer runs | 0 | 0.4 |
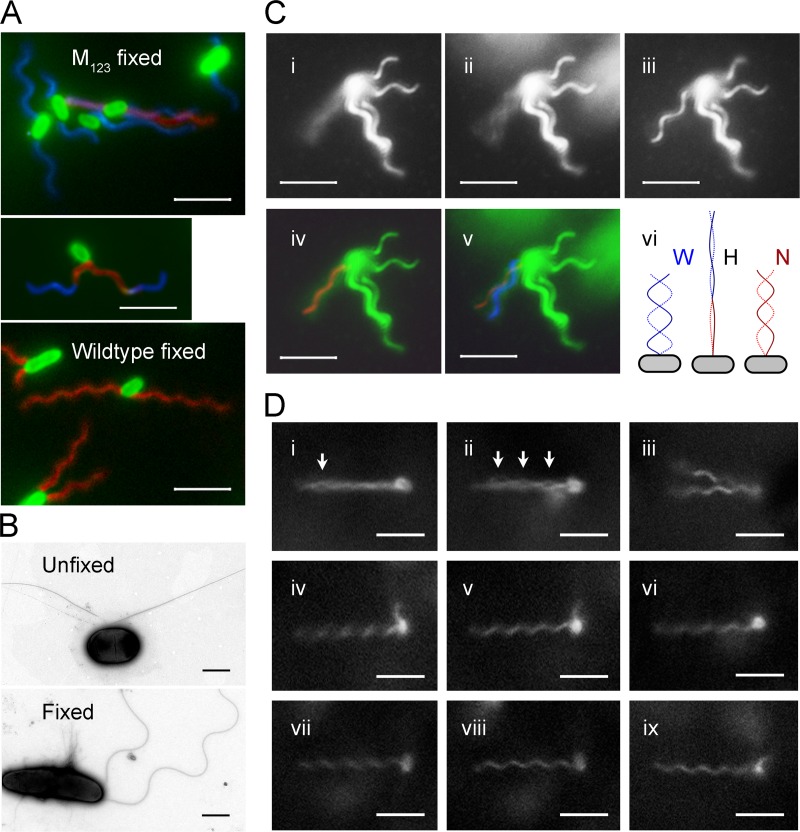
None of these mutants produced flagella that are the left-handed normal form at rest. Their flagella must be converted to the normal form when swim forward. To test this assumption, glutaraldehyde was applied to swimming M123 cells to preserve the helical shape (13). To our surprise, most flagella were transformed into w-coil helices after fixation (Fig. 2A). However, left-handed normal helices (often with slight distortion), flagella undergoing polymorphic transition, and even partially cross-linked bundles were observed. The flagella of fixed and unfixed bacteria were also examined with transmission electron microscopy (TEM) after negative staining (Fig. 2B). The results of fluorescence staining were confirmed.
Fig 2.
Polymorphic transition of mutated flagella. The scale bar indicates 5 μm for fluorescence images and 1 μm for TEM images. (A) Fluorescence-labeled M123 cells (top) and wild-type cells (bottom) after fixation. Flagella are false-colored: red for normal form, blue for right-handed forms, and magenta for near-straight filament and transition segments. The middle panel shows a M123 cell with flagella undergoing polymorphic transition. (B) TEM images of negative-stained unfixed (top) and fixed (bottom) M123 cells. (C) Polymorphic transition of a flagellum on a tethered M123 cell at pH 8.0. Snapshots of a flagellum that was rotating fast (i), slowing down (ii), and stopped (iii) are on the top. The contrast was enhanced to show the flagellar image. (iv) Fitting a normal helix into the image cylinder of the rotating flagellum. (v) The w-coil helix formed as the rotation slowed down. (vi) Diagram showing the transformation of a w-coil helix during counterclockwise rotation. At the beginning (left), the flagellum is w-coil (W); when the threshold speed reached (middle), the flagella became hyperextended (H). Finally, the normal helix (N) is formed (right). (D) Snapshots of a M23 cell. (i) The reversion of a filament produced an eye in the bundle (arrow) when the swimming cell slowed down. (ii) More eyes formed before stop. (iii) Both flagella reverted to R-normal when stopped. (iv) One flagellum began to rotate. (v) Normal flagellum. (vi) Rotation reversed. (vii) Rotating clockwise. (viii) R-normal flagellum. (ix) Flagellum undergoing a polymorphic transition.
The pH of the medium has a significant impact on the motility of M123 cells. When the pH was increased from 7.0 to 8.0 the number of motile cells increased severalfold, whereas at pH 6.0 only a few swimming cells were observed. Although E. coli cells can be cultivated in media with pH values of 6.0 to 8.0, the effect took place within 5 min after resuspension of cells, regardless of their initial growth condition. Right-handed flagella are transformed into the normal form more easily than slightly curved filaments. Deprotonation certainly enhances helix formation of the M123 flagella and, subsequently, the cell motility. If only cells with w-coil flagella were considered, there is no difference between the subpopulation of M123 mutants and the M1 mutants (Fig. 2C). When a stationary w-coil flagellum began counterclockwise rotation, the flagellum first resumed a hyperextended form (diagram). It retracted when the left-handed normal helix propagated from the basal part. These instable normal helices then form a bundle to propel the cells. As rotation slowed down to the threshold value, the flagella quickly reverted to w-coil helices (Fig. 2C).
At near-neural pH, a large amount of M123 flagella have an innate curvature (imperfect helix), often only slightly curved. When such a filament rotated, the tip first swirls and bending in the middle follows. As the bending amplitude became large enough, the normal helix began to form at the proximal end. A straight or nearly straight filament could also join a rotating bundle passively. It often trailed along the axis of the bundle for a while and suddenly collapsed into the bundle. The R-normal flagella of the M23 mutant also require an external torque to deform into the normal helix, but with a lower speed threshold than the w-coil flagella (Fig. 2D). The polymorphic transition on them could be directly observed on slowly rotating flagella.
On swimming cells of the M1 and M123 mutants, some flagella instantaneously went back to the w-coil or near-straight forms at slow speed. When a w-coil helix came out of the bundle, a “pseudo-tumble” was resulted that make the cell bodies wobble; when a straight or curved flagellum came out of the bundle, it stabilized the swimming direction, which led to a long run. In the case of the M23 mutant, often only part of a flagellum was converted to the R-normal form at slow rotation. The reverted part of the filament and the normal helical bundle were completely out of phase. They form “eyes” which often start at the distal end of the bundle. The partial splitting of the bundle does not influence swim significantly (Fig. 2D). Upon fast clockwise rotation, these mutant flagella were transformed into the semicoil and curly forms that can trigger a real tumble (12). The tumbles of the mutants differ from those of the wild type in that the transitions between canonical polymorphs were interrupted by spontaneous conversions to noncanonical forms.
Interaction between amino acid residues.
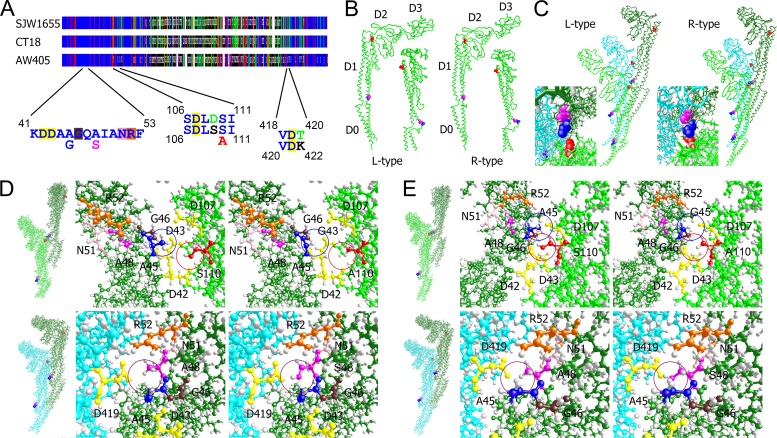
All three mutations locate in the highly conserved N-terminal on flagellin (Fig. 3A). The wild-type E. coli flagellin shares 53% identity with the Salmonella proteins. The structures of the latter have been successfully determined (44). Although the Salmonella mutants also generate straight filaments like the M3 mutant, the left-handed (L) type is caused by a mutation G426A in strain SJW1660 (28), and the right-handed (R) type is caused by a mutation A449V in strain SJW1655 (45).
Fig 3.
Interactions between mutated residues and surrounding amino acids. (A) Graphic representation of the aligned sequences from S. enterica serovar Typhimurium strain SJW1655, E. coli strain AW405, and the reference Salmonella strain CT18. Dark blue, conserved residues; red, similar residues; green, residues shared by two sequences. The peptides containing the mutations and the interacting residues are enlarged. Mutations are shown under the conserved ones. Both the Salmonella sequence (above) and the E. coli sequence (below) are shown when different. (B) Side views of the flagellins. Mutated residues are shown in the space-filling mode: A45, blue; A48, magenta; and S110, red. Insets are views from the concave-surface side. (C) Interactions between the 0-start, 11-start, and 16-start subunits. Insets are views from above. (D) Side-chain interactions in the L-type configuration at the 16-start (above) and the 11-start interfaces (below). The left panels are whole-molecule views; the middle panels are enlarged images showing the side chains of wild-type residues; the right panels showing mutated residues. (E) Side-chain interactions in the R-type configuration. The presence or absence of mutated methyl and hydroxyl groups is highlighted by circles.
Flagellins have four globular domains termed D0, D1, D2, and D3 (45). Domain D1 fills most of the outer-tube of the filament and plays a crucial role in helix formation (38). All three mutated residues are in domain D1 (Fig. 3B), but none of them in the 11 residues involved in permanent interactions between adjacent subunits (23). M1 and M2 are at the end of an α-helix that forms the edge of the spoke region between domains D1 and D0 (45). M3 is in another short α-helix on the edge of the convex surface. These three residues clustered at the tripartite interface of 0-start, 11-start, and 16-start subunits but are not involved in the 5-start and 6-start interaction (Fig. 3C). In the R-type straight filaments, they were closer to each other than in the L type.
The mutations perturbed local protein packaging, while the global side chain interactions along the flagella remains largely unchanged (44). At the 16-start interface, the side chain of Ser110 is surrounded by three aspartic acids and Ala45. The side chain of Ala45 is surrounded by Ser110, two aspartic acids, and a glycine (Fig. 3D). The M1 mutant lost a methyl group at position 45, which potentially reduced the spatial constraint in the more compacted R-type helix. However, in the M3 mutation, a hydroxyl group was missing, which might also influence hydrogen bonding or ionic interactions. Although the M2 and M3 mutations attenuate the effects of each other to some extent, they interact with different subunits. Ala48 is surrounded by Ala45, an arginine and an asparagine from the same subunits and an aspartic acid (Asp419) from the 11-start subunit. The M2 mutation potentially introduced a de novo hydrogen bond in the R-type filaments or ionic repulsion in the L-type filaments (Fig. 3E).
DISCUSSION
One of the major mechanisms in divergent evolution is the accumulation of point mutations. Here we characterized three mutations on the fliC gene in E. coli that arose in laboratory cultivation. Amino acid exchange in flagellin is known to influence flagellar morphology and mobility. An alanine-to-valine mutation in Bacillus subtilis 168 causes a straight flagellum phenotype and the loss of motility (29). E. coli strain W3623 (ha-177) that produces straight flagella does not swarm in 0.35% semisolid agar (24). In the present study, E. coli strains that contain the M3 mutation, unless compensated by the M2 mutation, also produce straight flagella and lost motility in liquid and semisolid agar.
The M23 double mutant retained 90% of the wild-type motility. However, its resting flagella are right-handed R-normal. Some curative second mutations have been found to restore the motility of Salmonella mutant strain SJW1660 (18), but none of the 19 amino acid exchanges in Salmonella coincides with our mutations. The M1 mutation alone produced another right-handed form, the w-coil that has been observed in a Salmonella strain with the G426A R431S double mutation (18). After introduction of the M1 mutation into the M23 background, straight flagella reappear, and the motility further decreased. However, this effect is reversible to some extent by alkaline pH that increases the relative amount of helical and curved flagella in M123 mutants. Real straight filament cannot deform into the normal form through rotation. The motility of M123 cells increase as the medium pH is increased as far as nondetrimental.
It is known that the straight flagella of Salmonella mutant SJ814 can be converted to the curly form at pH lower than 4.0 (20). The straight M123 flagella could not be transformed at such an acidic pH but instead were converted to w-coil at alkaline pH and by glutaraldehyde fixation. In the present study, polymorphic transitions were also observable on slowly rotating M23 flagella. Similar polymorphism has been demonstrated previously in low-pH, low-salt, and low-temperature conditions (11) and fluorescence-labeled swimming bacteria (12, 43). Interactions between hydrophilic amino acid pairs and between hydrophobic pairs determine the configuration of the flagella (23). These three mutations clustered at a tripartite interface where a hydrophilic microenvironment composed of four aspartic acids, an arginine, and an asparagine is formed. It makes intuitive sense that pH significantly influences the polymorphism of the M123 flagella. When the surrounding side chains were deprotonated, ionic interaction became strong and some potential steric hindrance disappeared.
Flagella assemble through the axial intersubunit interaction between the concave and convex surfaces of D1 domain (38). None of the studied mutations involves in the permanent interaction between subunits, and thus the rigidity of the flagella preserved and swim was not severely handicapped. The M1 and M2 mutations are located at the edge of the spoke-like connection important for the L-type and R-type switching (28). This place corresponds to the “corner” of the bi-stable subunits (scheme A) in Calladine's mechanical model (7). Generation of noncanonical helical forms is therefore reasonable (see Fig. S4 in the supplemental material). Among these noncanonical helices, L-curly flagella have been observed in a mutant of Salmonella wichita (25), a marine bacterium Idiomaria loihiensis (40), and some polar flagellated bacterial species (14). However, the swimming behavior of none of them has been carefully studied.
The transformation of the w-coil flagella to the normal form occurred at higher rotation speed than that of the R-normal flagella. The slightly curved flagella of the M123 mutant were the most difficult to transform mechanically. The mutated flagella in a bundle spontaneously revert to the stationary form at low rotation speed that lead to pseudotumbles in runs. During a real tumble, polymorphic transition between canonical forms was interrupted by reversion to the noncanonical forms. Thus far, the only known bacterium that relays on flagellar polymorphic transition to swim is the monotrichous R. sphaeroides (33). Although its flagella does not change the handedness after transformation like these E. coli mutants (1), the principle is similar: the morphology of stationary flagella is not critical for motility, as far as they can deform into the normal helical shape upon drag load, the cell is motile.
These mutations accumulated in the long-term laboratory cultivation. Laboratory events shed light on natural processes just like that domestication reveal the principle of natural selection in Darwin's evolution theory. At the beginning of the molecular biology era, phages were widely used in genetics to mediate recombination (34). Right-handed flagella are inherently resistant to flagellotropic phage (39) because their surface grooves prevent the phage particle from sliding along the filament when rotation initiates. A small pH variation of the media might have played an unconsciously significant role in the selection of strain LMG194. Wobbling of the cell body and the spontaneous deformation of the mutant flagella might also influence long-range hydrodynamic interaction among bacteria, which would have a profound effect in the crowd liquid broth (4). Nevertheless, this research demonstrated that the equilibrium morphologies of bacterial flagella are not critical for their functionality, and evolution would favor their diversification. We expect that such a phenomenon common in the microbial world. Some different helical forms have been observed on different bacteria (14), it would be interesting to see which one is functional and how they work.
Supplementary Material
ACKNOWLEDGMENTS
We thank Howard C. Berg, Keiichi Namba, Christopher R. Calladine, and Fumio Hayashi for helpful discussion and insightful comments. We also thank John S. Parkinson and Jonathan R. Beckwith for providing strains and Keiichi Namba for providing the protein coordinates.
This research was supported by the Max-Planck Society, and W.W. was also supported by the MPG-CAS Joint Doctoral Promotion Program.
Footnotes
Published ahead of print 24 August 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Armitage JP, Macnab RM. 1987. Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J. Bacteriol. 169: 514– 518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Armstrong JB, Adler J, Dahl MM. 1967. Nonchemotactic mutants of Escherichia coli. J. Bacteriol. 93: 390– 398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bachmann BJ. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36: 525– 557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baskaran A, Marchetti MC. 2009. Statistical mechanics and hydrodynamics of bacterial suspensions. Proc. Natl. Acad. Sci. U. S. A. 106: 15567– 15572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berg HC. 2003. Escherichia coli in motion. Springer-Verlag, New York, NY [Google Scholar]
- 6. Berg HC, Anderson RA. 1973. Bacteria swim by rotating their flagellar filaments. Nature 245: 380– 382 [DOI] [PubMed] [Google Scholar]
- 7. Calladine CR. 1978. Change of waveform in bacterial flagella: role of mechanics at molecular level. J. Mol. Biol. 118: 457– 479 [Google Scholar]
- 8. Calladine CR. 1975. Construction of bacterial flagella. Nature 255: 121– 124 [DOI] [PubMed] [Google Scholar]
- 9. Casadaban MJ, Cohen SN. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138: 179– 207 [DOI] [PubMed] [Google Scholar]
- 10. Chattopadhyay S, Moldovan R, Yeung C, Wu XL. 2006. Swimming efficiency of bacterium Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 103: 13712– 13717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Darnton NC, Berg HC. 2007. Force-extension measurements on bacterial flagella: triggering polymorphic transformations. Biophys. J. 92: 2230– 2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Darnton NC, Turner L, Rojevsky S, Berg HC. 2007. On torque and tumbling in swimming Escherichia coli. J. Bacteriol. 189: 1756– 1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Doetsch RN, Sjoblad RD. 1980. Flagellar structure and function in eubacteria. Annu. Rev. Microbiol. 34: 69– 108 [DOI] [PubMed] [Google Scholar]
- 14. Fujii M, Shibata S, Aizawa SI. 2008. Polar, peritrichous, and lateral flagella belong to three distinguishable flagellar families. J. Mol. Biol. 379: 273– 283 [DOI] [PubMed] [Google Scholar]
- 15. Garstecki P, Tierno P, Weibel DB, Sagués F, Whitesides GM. 2009. Propulsion of flexible polymer structures in a rotating magnetic field. J. Phys. Condens. Matter 21: 204110. [DOI] [PubMed] [Google Scholar]
- 16. Gonzalez-Beltran C, Burge RE. 1974. Subunit arrangements in bacterial flagella. J. Mol. Biol. 88: 711– 716 [DOI] [PubMed] [Google Scholar]
- 17. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177: 4121– 4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hayashi F, Tomaru H, Oosawa K. 2010. The analysis of the flagellar filaments of the Salmonella pseudorevertants. Kobunshi Ronbunshu 67: 666– 678 [Google Scholar]
- 19. Hedblom ML, Adler J. 1980. Genetic and biochemical properties of Escherichia coli mutants with defects in serine chemotaxis. J. Bacteriol. 144: 1048– 1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kamiya R, Asakura S, Wakabayashi K, Namba K. 1979. Transition of bacterial flagella from helical to straight forms with different subunit arrangements. J. Mol. Biol. 131: 725– 742 [DOI] [PubMed] [Google Scholar]
- 21. Kanto S, Okino H, Aizawa S-I, Yamaguchi S. 1991. Amino acids responsible for flagellar shape are distributed in terminal regions of flagellin. J. Mol. Biol. 219: 471– 480 [DOI] [PubMed] [Google Scholar]
- 22. Kim M, Bird JC, Van Parys AJ, Breuer KS, Powers TR. 2003. A macroscopic scale model of bacterial flagellar bundling. Proc. Natl. Acad. Sci. U. S. A. 100: 15481– 15485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kitao A, et al. 2006. Switch interactions control energy frustration and multiple flagellar filament structures. Proc. Natl. Acad. Sci. U. S. A. 103: 4894– 4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kondoh H, Yanagida M. 1975. Structure of straight flagellar filaments from a mutant of Escherichia coli. J. Mol. Biol. 96: 641– 652 [DOI] [PubMed] [Google Scholar]
- 25. Leifson E. 1951. Staining, shape, and arrangement of bacterial flagella. J. Bacteriol. 62: 377– 389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Link AJ, Phillips D, Church GM. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179: 6228– 6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Macnab RM. 1977. Bacterial flagella rotating in bundles: a study in helical geometry. Proc. Natl. Acad. Sci. U. S. A. 74: 221– 225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maki-Yonekura S, Yonekura K, Namba K. 2010. Conformational change of flagellin for polymorphic supercoiling of the flagellar filament. Nat. Struct. Mol. Biol. 17: 417– 422 [DOI] [PubMed] [Google Scholar]
- 29. Martinez RJ, Ichiki AT, Lundh NP, Tronick SR. 1968. A single amino acid substitution responsible for altered flagellar morphology. J. Mol. Biol. 34: 559– 564 [DOI] [PubMed] [Google Scholar]
- 30. Namba K, Vonderviszt F. 1997. Molecular architecture of bacterial flagellum. Q. Rev. Biophys. 30: 1– 65 [DOI] [PubMed] [Google Scholar]
- 31. Pedretti A, Villa L, Vistoli G. 2004. VEGA: an open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J. Comput. Aided Mol. Des. 18: 167– 173 [DOI] [PubMed] [Google Scholar]
- 32. Ping L. 2010. The asymmetric flagellar distribution and motility of Escherichia coli. J. Mol. Biol. 397: 906– 916 [DOI] [PubMed] [Google Scholar]
- 33. Ping L. 2012. Cell orientation of swimming bacteria: from theoretical simulation to experimental evaluation. Sci. China Life Sci. 55: 202– 209 [DOI] [PubMed] [Google Scholar]
- 34. Ping L. 2009. Pea, fly, germ, and us: a brief history of genetics. World Scientific Publishing, Ltd, Singapore, Philippines [Google Scholar]
- 35. Ping L, Mavridou DAI, Emberly E, Westermann M, Ferguson SJ. 2012. Vital dye reaction and granule localization in periplasm of Escherichia coli. PLoS One 7: e38427 doi:10.1371/journal.pone.0038427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ping L, Vogel H, Boland W. 2008. Cloning of prokaryotic genes by a universal degenerate primer PCR. FEMS Microbiol. Lett. 287: 192– 198 [DOI] [PubMed] [Google Scholar]
- 37. Purcell EM. 1997. The efficiency of propulsion by a rotating flagellum. Proc. Natl. Acad. Sci. U. S. A. 94: 11307– 11311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Samatey FA, et al. 2001. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410: 331– 337 [DOI] [PubMed] [Google Scholar]
- 39. Samuel ADT, et al. 1999. Flagellar determinants of bacterial sensitivity to χ-phage. Proc. Natl. Acad. Sci. U. S. A. 96: 9863– 9866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shibata S, Alam M, Aizawa SI. 2005. Flagellar filaments of the deep-sea bacteria Idiomarina loihiensis belong to a family different from those of Salmonella typhimurium. J. Mol. Biol. 352: 510– 516 [DOI] [PubMed] [Google Scholar]
- 41. Shimada K, Kamiya R, Asakura S. 1975. Left-handed to right-handed helix conversion in Salmonella flagella. Nature 254: 332– 334 [DOI] [PubMed] [Google Scholar]
- 42. Strauch KL, Beckwith J. 1988. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc. Natl. Acad. Sci. U. S. A. 85: 1576– 1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Turner L, Ryu WS, Berg HC. 2000. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182: 2793– 2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamashita I, et al. 1998. Structure and switching of bacterial flagellar filaments studied by X-ray fiber diffraction. Nat. Struct. Mol. Biol. 5: 125– 132 [DOI] [PubMed] [Google Scholar]
- 45. Yonekura K, Maki-Yonekura S, Namba K. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424: 643– 650 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.