Abstract
In Saccharomyces cerevisiae, the histone chaperone Rtt106 binds newly synthesized histone proteins and mediates their delivery into chromatin during transcription, replication, and silencing. Rtt106 is also recruited to histone gene regulatory regions by the HIR histone chaperone complex to ensure S-phase-specific expression. Here we showed that this Rtt106:HIR complex included Asf1 and histone proteins. Mutations in Rtt106 that reduced histone binding reduced Rtt106 enrichment at histone genes, leading to their increased transcription. Deletion of the chromatin boundary element Yta7 led to increased Rtt106:H3 binding, increased Rtt106 enrichment at histone gene regulatory regions, and decreased histone gene transcription at the HTA1-HTB1 locus. These results suggested a unique regulatory mechanism in which Rtt106 sensed the level of histone proteins to maintain the proper level of histone gene transcription. The role of these histone chaperones and Yta7 differed markedly among the histone gene loci, including the two H3-H4 histone gene pairs. Defects in silencing in rtt106 mutants could be partially accounted for by Rtt106-mediated changes in histone gene repression. These studies suggested that feedback mediated by histone chaperone complexes plays a pivotal role in regulating histone gene transcription.
INTRODUCTION
Cell cycle-regulated transcription of the canonical histone genes is a hallmark of eukaryotic organisms. During S phase, a coordinated burst of histone gene transcription is required to double the level of histone proteins to package newly replicated DNA into chromatin (13, 22). The tight coupling of both the timing and the level of histone gene expression with DNA synthesis is critical for cell viability. Mutations that perturb DNA replication lead to altered histone gene transcription (3, 23). Conversely, misregulation of the timing and/or level of histone gene expression leads to genomic instability and cell cycle defects (23, 43). Consequently, cells have evolved a complex regulatory mechanism to repress histone gene transcription outside S phase and to promote a precise level of transcription during S phase.
The Saccharomyces cerevisiae genome contains two nonallelic copies of each canonical histone gene which are organized in head-to-head pairs of H2A-H2B (HTA1-HTB1 and HTA2-HTB2) and H3-H4 (HHT1-HHF1 and HHT2-HHF2). Nucleosomes, the fundamental units of chromatin structure, contain two H2A/H2B dimers, one H3/H4 tetramer, and ∼147 bp of DNA wrapped around the outer surface (37). Repression of histone transcription outside S phase is maintained by the HIR H3/H4 histone-protein chaperone complex, referred to here as the HIR complex (Hir1, Hir2, Hir3, and Hpc2). The HIR complex localizes to negative (NEG) sequence elements within the regulatory regions of HTA1-HTB1, HHT1-HHF1, and HHT2-HHF2 (44, 45, 59). Although HTA2-HTB2 transcription is S-phase specific, its promoter does not contain NEG elements, and the factors involved in its HIR-independent repression are unknown (45). A recent study implicated two additional H3/H4 chaperones, Rtt106 and Asf1, in maintaining histone gene repression at the HIR-regulated loci (16). Rtt106 is recruited to histone gene regulatory regions in a HIR-dependent manner (16), and it contributes to transcriptional repression through the direct recruitment of the RSC chromatin-remodeling complex outside S phase (15, 42). Mutations that disrupt Rtt106:H3 binding lead to reduced Rtt106 enrichment at histone gene promoters (32, 51), suggesting that the Rtt106:HIR interaction might be mediated by histone proteins. Asf1 copurifies with the HIR complex (21), and in asf1Δ cells, Rtt106 localization is reduced, leading to inappropriate histone expression outside S phase (51, 53). Although these data suggest that Asf1 plays a direct role in histone gene regulation, whether Asf1 localizes to histone gene promoters as a member of the HIR:Rtt106 regulatory complex was unclear.
During late G1, histone gene repression is alleviated by the dissociation of the RSC complex and by the Rtt106-dependent recruitment of the SWI/SNF chromatin-remodeling complex (8, 15). SWI/SNF promotes active transcription, perhaps by exposing the upstream activating sequences (UAS) to the cell cycle-regulated transcription factors Spt10 and SBF (9, 12, 58). Additional activating factors include Rtt109, a histone acetyltransferase that acetylates newly synthesized histone H3 proteins at lysine 56 (H3K56ac) (10, 24, 48, 55), and Yta7, an AAA-ATPase protein that maintains certain chromatin boundaries (16, 20, 31, 54). The H3K56 acetylation is S-phase specific (40) and is required to overcome chaperone-mediated histone gene repression through an unknown mechanism (16). Yta7 blocks the apparent spreading of Rtt106 and RSC into the histone gene open reading frames and facilitates efficient promoter escape and elongation by RNA polymerase II (16, 31). In rtt109Δ and yta7Δ cells, HTA1 transcription is reduced (16). The transcriptional effects caused by these mutations at the other histone loci were unknown.
The H3/H4 chaperones that regulate histone gene transcription also facilitate nucleosome turnover throughout the genome (6, 7). During S phase, newly synthesized H3/H4 histones are bound by Asf1 and presented to Rtt109 for acetylation at H3K56 (1). These new histones are then delivered into chromatin by the CAF-1 and Rtt106 histone chaperones through a direct interaction with the replication machinery (5, 33, 50, 61). Rtt106 also facilitates replication-independent nucleosome turnover, along with the HIR complex and Asf1 during both transcription initiation and elongation (21, 27, 47). Additionally, CAF-1 performs an overlapping function with Asf1, Rtt106, and the HIR complex to maintain silent chromatin (25, 26, 30, 49, 56). The involvement of histone chaperones in both the assembly of histone proteins into chromatin and the regulation of histone gene transcription suggests that the two processes might be linked (13, 51). Thus, histone chaperones might facilitate a negative-feedback loop to allow cells to adjust histone gene transcription as a function of free histone protein levels.
In this study, we expanded our understanding of histone gene regulation by establishing the nature of the holocomplex at histone regulatory regions. Our data provided evidence that Asf1 acted directly at histone promoters to mediate repression. Additionally, we observed a relationship between Rtt106's physical interaction with histone proteins, its localization at histone promoters, and histone gene transcription. These results suggested that the histone binding activity of Rtt106 might facilitate negative-feedback regulation of histone gene transcription. Our data revealed an unexpected diversity of responses among the distinct histone gene pairs to changes in the composition of the regulatory complex, suggesting the existence of locus-specific mechanisms controlling histone gene transcription. Moreover, our data reveal a new distinction between the two H3-H4 histone gene pairs and implicate changes in histone gene transcription as a foundation for Rtt106's role in silencing.
MATERIALS AND METHODS
Yeast strains, plasmids, and culture.
All yeast strains were derived from S. cerevisiae W303 (Table 1). Gene deletions were generated by one-step integration of knockout cassettes (36) followed by PCR analysis of the 5′ and 3′ ends of the targeted gene. A C-terminal 3× FLAG tag (17) was integrated in frame with the targeted genes by one-step integration and verified by PCR. Strains containing mutant rtt106 S80E, R86A, and T265E plasmids or the hmr-a1Δ::URA3 reporter were previously described (60, 61).
Table 1.
Strains used in this studya
| Strain | Genotype | Source or reference | Figure(s) |
|---|---|---|---|
| JRY3009 | MATα ade2-1 his3-11 leu2-3,112 trp1-1 ura3-52 can1-100 | R. Rothstein | Fig. 1C; Fig. 2A, B, and D |
| JRY9253 | MATα rtt106Δ::KanMX [pJR2877 HIS3 RTT106] | 61 | Fig. 1A and B; Fig. 2D and E; Fig. 3A to F |
| JRY9255 | MATα rtt106Δ::KanMX [pJR2879 HIS3 RTT106-3×FLAG::KanMX] | 61 | Fig. 1A and B; Fig. 2D and E; Fig. 3A to F; Fig. 4A to C |
| JRY9262 | MATα rtt106Δ::KanMX hir1Δ::HygMX [pJR2879 HIS3 RTT106-3×FLAG::KanMX] | 61 | Fig. 1A and B; Fig. 3B |
| JRY9258 | MATα rtt106Δ::KanMX [pJR2899 HIS3 rtt106(S80E)-3×FLAG::KanMX] | 61 | Fig. 1A, B, and D; Fig. 3D; Fig. 4B |
| JRY9259 | MATα rtt106Δ::KanMX [pJR2972 HIS3 rtt106(R86A)-3×FLAG::KanMX] | 61 | Fig. 1A, B, and D; Fig. 3D; Fig. 4B |
| JRY9260 | MATα rtt106Δ::KanMX [pJR2978 HIS3 rtt106(T265E)-3×FLAG::KanMX] | 61 | Fig. 1A, B, and D; Fig. 3D; Fig. 4B |
| JRY9401 | MATα rtt106Δ::KanMX asf1Δ::HygMX [pJR2879 HIS3 RTT106-3×FLAG::KanMX] | This study | Fig. 1A and B |
| JRY9266 | MATα rtt106Δ::KanMX rtt109Δ::HygMX [pJR2879 HIS3 RTT106-3×FLAG::KanMX] | 61 | Fig. 1A and B; Fig. 3E; Fig. 4C |
| JRY9402 | MATα rtt106Δ::KanMX [pJR3186 HIS3 rtt106(1-301)-3×FLAG::KanMX] | This study | Fig. 1A, B, and D |
| JRY9268 | MATα rtt106Δ::KanMX [pRS313] | 61 | Fig. 1D; Fig. 4A |
| JRY9403 | MATα HIR1-3×FLAG::KanMX | This study | Fig. 1C; Fig. 2D and E |
| JRY9404 | MATα HIR1-3×FLAG::KanMX rtt106Δ::HygMX | This study | Fig. 1C |
| JRY9405 | MATα HIR1-3×FLAG::KanMX rtt109Δ::HygMX | This study | Fig. 1C |
| JRY9406 | MATα HIR1-3×FLAG::KanMX hst3Δ::HygMX hst4Δ::NatMX | This study | Fig. 1C |
| JRY9407 | MATα HIR1-3×FLAG::KanMX yta7Δ::HygMX | This study | Fig. 2D and E |
| JRY9408 | MATα ASF1-3×FLAG::KanMX | This study | Fig. 2A to E |
| JRY9411 | MATα ASF1-3×FLAG::KanMX rtt106Δ::HygMX | This study | Fig. 2B and C |
| JRY9412 | MATα ASF1-3×FLAG::KanMX hir1Δ::HygMX | This study | Fig. 2B and C |
| JRY9413 | MATα ASF1-3×FLAG::KanMX yta7Δ::HygMX | This study | Fig. 2D and E |
| JRY9414 | MATα rtt106Δ::KanMX yta7Δ::HygMX [pJR2879 HIS3 RTT106-3×FLAG::KanMX] | This study | Fig. 2D and E; Fig. 3A to E; Fig. 4A to C |
| JRY9415 | MATα rtt106Δ::KanMX hir1Δ::HygMX yta7Δ::NatMX [pJR2879 HIS3 RTT106-3×FLAG::KanMX] | This study | Fig. 3A and B |
| JRY9416 | MATα rtt106Δ::KanMX yta7Δ::HygMX [pJR2899 HIS3 rtt106(S80E)-3×FLAG::KanMX] | This study | Fig. 3C and D; Fig. 4B |
| JRY9417 | MATα rtt106Δ::KanMX yta7Δ::HygMX [pJR2972 HIS3 rtt106(R86A)-3×FLAG::KanMX] | This study | Fig. 3C and D; Fig. 4B |
| JRY9418 | MATα rtt106Δ::KanMX yta7Δ::HygMX [pJR2978 HIS3 rtt106(T265E)-3×FLAG::KanMX] | This study | Fig. 3C and D; Fig. 4B |
| JRY9419 | MATα rtt106Δ::KanMX yta7Δ::HygMX rtt109Δ::NatMX [pJR2879 HIS3 RTT106-3×FLAG::KanMX] | This study | Fig. 3C and E; Fig. 4C |
| JRY9420 | MATα rtt106Δ::KanMX yta7Δ::HygMX [pRS313] | This study | Fig. 4A |
| JRY9267 | MATα rtt106Δ::KanMX hst3Δ::HygMX hst4Δ::NatMX [pJR2879 HIS3 RTT106-3×FLAG::KanMX] | 61 | Fig. 3F |
| JRY8883 | MATa hmr-a1Δ::KlURA3 | 60 | Fig. 5A to C |
| JRY9421 | MATa hmr-a1Δ::KlURA3 sir3Δ::KanMX | This study | Fig. 5A to C |
| JRY9422 | MATa hmr-a1Δ::KlURA3 hht1-hhf1Δ::KanMX | This study | Fig. 5A and B |
| JRY9423 | MATa hmr-a1Δ::KlURA3 hht2-hhf2Δ::KanMX | This study | Fig. 5A and B |
| JRY9424 | MATa hmr-a1Δ::KlURA3 rtt106Δ::KanMX | This study | Fig. 5A and B |
| JRY9425 | MATa hmr-a1Δ::KlURA3 cac1Δ::KanMX | This study | Fig. 5A and B |
| JRY9426 | MATa hmr-a1Δ::KlURA3 hir1Δ::KanMX | This study | Fig. 5A and B |
| JRY9427 | MATa hmr-a1Δ::KlURA3 hht1-hhf1Δ::HygMX rtt106Δ::KanMX | This study | Fig. 5A and B |
| JRY9428 | MATa hmr-a1Δ::KlURA3 hht2-hhf2Δ::HygMX rtt106Δ::KanMX | This study | Fig. 5A and B |
| JRY9429 | MATa hmr-a1Δ::KlURA3 hht1-hhf1Δ::KanMX cac1Δ::HygMX | This study | Fig. 5A and B |
| JRY9430 | MATa hmr-a1Δ::KlURA3 hht2-hhf2Δ::KanMX cac1Δ::HygMX | This study | Fig. 5A and B |
| JRY9431 | MATa hmr-a1Δ::KlURA3 hht1-hhf1Δ::KanMX hir1Δ::HygMX | This study | Fig. 5A and B |
| JRY9432 | MATa hmr-a1Δ::KlURA3 hht2-hhf2Δ::KanMX hir1Δ::HygMX | This study | Fig. 5A and B |
| JRY9243 | MATa hmr-a1Δ::KlURA3 rtt106Δ::KanMX cac1Δ::HygMX | 61 | Fig. 5A to C |
| JRY9433 | MATa hmr-a1Δ::KlURA3 rtt106Δ::KanMX cac1Δ::HygMX hht1-hhf1Δ::NatMX | This study | Fig. 5A to C |
| JRY9434 | MATa hmr-a1Δ::KlURA3 rtt106Δ::KanMX cac1Δ::HygMX hht2-hhf2Δ::NatMX | This study | Fig. 5A to C |
KlURA3, Kluyveromyces lactis URA3.
Site-directed mutagenesis was used to delete residues 302 to 455 from RTT106-3×FLAG (pJR2879) to generate rtt106(1-301)-3×FLAG (pJR3186) as described previously (39). All plasmids were introduced into each strain by transformation (18).
Silencing of the hmr-a1Δ::URA3 reporter and sensitivity to genotoxic chemicals were monitored by spotting 5-fold serial dilutions of mid-log-phase cultures onto selective media, as described previously (61).
Yeast whole-cell extract analysis.
Yeast whole-cell extracts were precipitated with 20% trichloroacetic acid. Extracts were solubilized in SDS loading buffer and visualized by SDS-PAGE and immunoblotting using standard procedures. Immunoblots were visualized and quantified with a LI-COR Odyssey imaging system. Anti-Flag M2 antibody (F3165; Sigma) was used to detect FLAG-tagged proteins. Anti-Pgk1 (Invitrogen) was used as a loading control.
Yeast coimmunoprecipitation.
Rtt106-FLAG coimmunoprecipitations (co-IP) were performed as described previously (61). Yeast lysate was incubated with 50 μl of anti-FLAG M2 agarose (Sigma). SDS-PAGE, immunoblotting, and LI-COR imaging were performed using standard procedures. Anti-Flag M2 antibody (F3165; Sigma), anti-H3 (Ab1791), and anti-H3K56ac (07-677; Millipore) were used to detect Rtt106-FLAG and copurifying histone proteins, respectively.
RNA preparation and analysis.
Reverse transcription-quantitative PCR (RT-qPCR) analysis was performed as described previously (61). Total RNA was extracted using an RNeasy minikit (Qiagen). Genomic DNA was digested on the column using RNase-free DNase (Qiagen). cDNA was synthesized using the SuperScript III first-strand synthesis system for RT-PCR kit (Invitrogen) and oligo(dT) primers. qPCR was performed using a DyNAmo HS SYBR green qPCR kit (New England BioLabs) and an Mx3000P machine (Stratagene). Amplification values for each primer set were normalized to the amplification values from ACT1 cDNA. Each background was analyzed in triplicate and for three independent RNA preparations. Statistical comparisons were performed using a two-tailed unpaired t test. Oligonucleotides for qPCR are listed in Table 2.
Table 2.
Oligonucleotides used in this study
| Method and oligonucleotide | Forward | Reverse | Annealing temp (°C) | Source or reference |
|---|---|---|---|---|
| ChIP | ||||
| Chromosome V UTRa | GCA ATC AAC ATC TGA AGA AAA GAA AGT AGT | CAT AAT CTG CGT AAA AAT GGC GTA AAT | 55 | 16 |
| HTA1-HTB1 promoter | ATA GTT AAC GAC CCA ACC GCG T | ACG GGC GTT TCT TCA ACA ACG A | 55 | This study |
| HTA1 5′ ORF | ACG GAT TTG GTT ATT TCT CAG TGA A | ATT CCA AGA CAG CAG TCA AGT AGA C | 55 | 16 |
| HTA1 3′ ORF | CAA AGA AGT CTG CCA AGG CT | AGC AGT TTA GTT CCT TCC GC | 55 | This study |
| HTA1 3′ UTR | AGG TTC ATT GGG CAC TGT TG | ACA GTT CTC CGT GAC AGG AT | 55 | This study |
| HTB1 5′ ORF | CCA CAA ATA AAC CAT ACA CAC | GGA AAT ACC AGT GTC AGG GT | 55 | This study |
| HTB1 3′ ORF | CCA GGT GAA TTG GCT AAG CA | GCA TTC CCT CTA TGA GAC CA | 55 | This study |
| HTB1 3′ UTR | CGA AAC TTC AGA GCA TTG GC | GGG TTC AAT CTC CAA GGC AT | 60 | This study |
| HHT1-HHF1 promoter | ATT TAC CAC CGT ATT CGC GG | AGG TGC AGA GCA AGG AAA TG | 55 | This study |
| HHT1 ORF | GCA ATT AGC TTC TAA GGC TGC CAG | GCA GCC AAG TTG GTA TCT TCA A | 60 | This study |
| HHT1 3′ UTR | GCC TTG TAG GAG GCA AGA TT | CGT ATG CGG CTT CAA GTT GT | 55 | This study |
| HHF1 5′ ORF | CCG CGA ATA CGG TGG TAA AT | TGG CAC CAC CTT TAC CTA GA | 55 | This study |
| HHF1 3′ ORF | TCA TCA GAG ACT CTG TTA CC | GTT ACC GTT TTC TTA GAA TTA G | 55 | 31 |
| HHF1 3′ UTR | ATC TGA GAG CAG GAA GAG CA | GTG TGT CAG CAT CAG AGG TT | 55 | This study |
| HHT2-HHF2 promoter | AAA TGA CCA ACT CCC ATC CG | TTT GTT CTG GTC TGG TCT GC | 55 | This study |
| HTA2-HTB2 promoter | AAT GGT AGC ACG TCG CGT TT | TGA CGG CAA GTG TCT CAC TGT T | 55 | This study |
| RT-qPCR | ||||
| ACT1 | GTC GGT AGA CCA AGA CAC C | GGG TGT TCT TCT GGG GC | 55 | 31 |
| HTA1 | ACG TTA CCA TTG CCC AAG G | GTT TAG TTC CTT CCG CCT TC | 55 | 31 |
| HHT1 | AAT CTT CTG CCA TCG GTG CC | CTA AAA CTG ATG ACA ATC AAC | 55 | 31 |
| HHF1 | TCA TCA GAG ACT CTG TTA CC | GTT ACC GTT TTC TTA GAA TTA G | 55 | 31 |
| HTA2 | TAT TGG GTA ATG TTA CCA TC | GCT TTG TTT CTT TTC AAC TCA G | 55 | 31 |
| hmr-a1::URA3 | CTT CCA AGG GTT CTC TAG CAC ACG | CTG TAC TGC TGA CCC AAT GCA TCG | 55 | 61 |
| HML-α2 | TCC ACA AAT CAC AGA TGA GT | GTT GGC CCT AGA TAA GAA TCC | 55 | This study |
| MATa | TGG ATG ATA TTT GTA GTA TGG CGG A | TCC CTT TGG GCT CTT CTC TT | 55 | 61 |
UTR, untranscribed region; ORF, open reading frame.
Chromatin immunoprecipitation.
Chromatin immunoprecipitations (ChIP) were performed as described previously (61). A total of 50 optical-density-at-600-nm (OD600) units of log-phase yeast culture were cross-linked with 1% formaldehyde for 30 min at room temperature. Chromatin was sonicated to ∼500 bp. Rtt106-FLAG, Hir1-FLAG, and Asf1-FLAG were immunoprecipitated using anti-FLAG M2 agarose (Sigma). Precipitated DNA fragments were analyzed by qPCR as described above. Amplification values for all primer sets were normalized to a previously described reference locus within an untranscribed region on chromosome V (16). Samples were analyzed in triplicate and for at least three independent chromatin preps. Statistical comparisons were performed using a two-tailed unpaired t test. Oligonucleotides for qPCR are listed in Table 2.
RESULTS
Rtt106:H3 binding was required for histone gene repression.
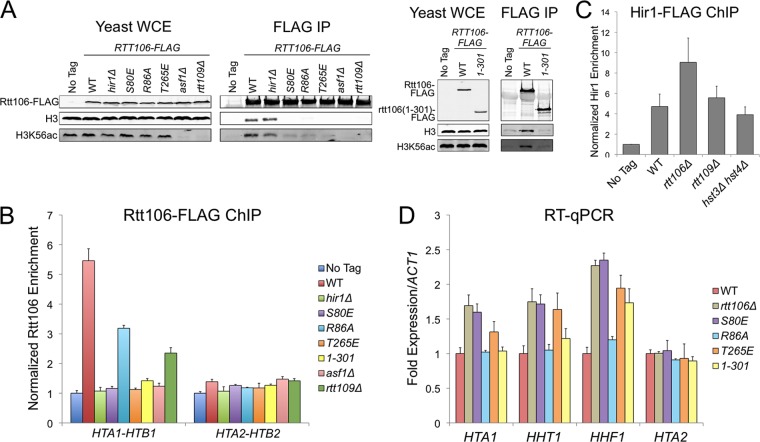
Throughout the cell cycle, the HIR complex recruits Rtt106 to histone gene regulatory regions, where together they facilitate transcriptional repression outside S phase and transcriptional activation during S phase (13, 22). Although Rtt106 coimmunoprecipitates (co-IP) with the HIR complex, it is unclear whether this interaction is direct or involves intermediate proteins (16). Notably, the deletion of RTT109 or ASF1, which prevents H3K56 acetylation and consequently decreases Rtt106:H3 binding, reduces Rtt106 enrichment at histone promoters (Fig. 1A and B) (32, 33, 51) without reducing HIR localization (Fig. 1C) (16). Therefore, the HIR:Rtt106 interaction might be bridged by additional proteins, with histones H3/H4 being likely candidates. To test whether Rtt106:H3 binding directly recruited Rtt106 to histone gene promoters, we monitored the localization of three Rtt106 point mutants that we previously discovered disrupt the histone binding surface along its double pleckstrin homology (PH) domain (Rtt106 S80E, R86A, and T265E) (61). S80E and R86A disrupt the charge of a basic patch within the N-terminal PH domain, and T265E alters the charge on a loop connecting two β strands within the C-terminal PH domain. Consistent with the histone-bridging hypothesis, each Rtt106 mutant protein had reduced H3 binding and reduced enrichment at the HTA1-HTB1 promoter (Fig. 1A and B). The lower levels of enrichment for Rtt106 S80E and T265E compared to that for R86A suggested that R86A had subtle changes in histone binding that were detectable by ChIP but not by earlier co-IP experiments. Our previous work found a similar increase in the severity of replication and silencing defects associated with the S80E and T265E alleles compared to those associated with R86A, further suggesting that Rtt106 R86A is partially functional (61). Like most histone chaperones, Rtt106 contains an acidic C-terminal tail. Although the tail of Rtt106 is not necessary for histone binding in vitro (33, 34, 52), the tail was necessary for histone binding in vivo and for HTA1-HTB1 promoter recruitment (Fig. 1A and B). Together these results suggested that HIR recruited Rtt106 to histone gene promoters through a histone protein bridge.
Fig 1.
Rtt106:H3 binding directly regulated histone gene transcription. (A) Mutants that disrupted Rtt106:H3 binding. Rtt106-FLAG was immunoprecipitated (IP) from yeast whole-cell extract (WCE) of the indicated mutant strains with anti-FLAG resin. Copurifying proteins were detected by immunoblotting with antibodies against FLAG, H3, and H3K56ac. WT, wild type. (B) ChIP analysis of Rtt106-FLAG at the HTA1-HTB1 and HTA2-HTB2 promoters. Values were obtained with qPCR and were normalized to a previously described negative control within an untranscribed region of chromosome V. No-tag ratios were normalized to 1. Error bars here and elsewhere represent standard deviations from the mean (n ≥ 3). (C) ChIP analysis of Hir1-FLAG at the HTA1-HTB1 promoter. Data were analyzed as described for panel B. (D) RT-qPCR analysis of histone transcripts in rtt106 mutant strains. Transcript values were normalized to ACT1 mRNA and adjusted to the WT value, which was normalized to 1. rtt106Δ, S80E, and T265E each had significantly increased HTA1, HHT1, and HHF1 transcription compared to the WT (P < 0.01). R86A and rtt106(1-301) each had significantly increased HHF1 transcription compared to the WT (P < 0.01).
Similar trends in defective recruitment of Rtt106 were observed at the HHT1-HHF1 and HHT2-HHF2 promoters (data not shown). However, the overall enrichment signal at the two H3-H4 gene pairs was reduced compared to that at HTA1-HTB1. Currently, it is unclear whether the smaller dynamic range of wild-type Rtt106 enrichment at the H3-H4 promoters, compared to that at H2A-H2B, reflected a biological distinction or an experimental limitation of ChIP at the H3-H4 promoter sequences. The HTA2-HTB2 locus, which is not subject to regulation by the HIR complex (45), exhibited minimal Rtt106 enrichment above background (Fig. 1B).
To promote S-phase-specific transcription, Rtt106 directly recruits the chromatin remodeling enzymes RSC and SWI/SNF to generate repressive and active chromatin architectures, respectively (8, 15, 42). In rtt106Δ strains, histone genes are transcribed outside S phase, leading to an increase in total histone mRNA in asynchronously growing cultures (16). Each rtt106 mutant strain, in which Rtt106 failed to localize to the promoter, had increased histone mRNA levels (Fig. 1D). HTA1 and HHT1 transcription in rtt106 R86A and rtt106(1-301) strains was similar to that in the wild type, suggesting that a threshold amount of Rtt106 was able to maintain repression. In the absence of Rtt106, the extent of transcriptional derepression was greater at HHF1 than at HTA1 and HHT1, suggesting nonidentical regulatory mechanisms at each histone gene pair. The direct inverse relationship between the extent of Rtt106:H3 binding and the level of histone gene expression suggested a potential Rtt106-mediated negative-feedback loop, as discussed below.
Asf1 localized to histone gene regulatory regions with Rtt106 and the HIR complex.
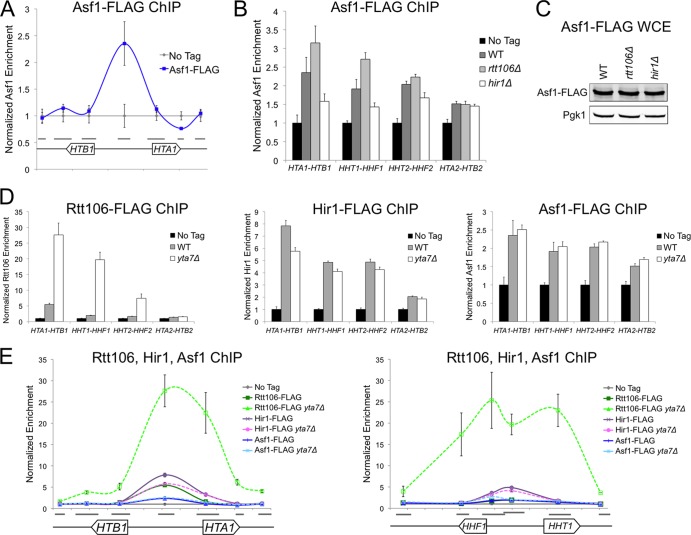
Because Rtt106 enrichment is reduced and histone transcription is increased in asf1Δ cells, it has been suggested that Asf1 itself localizes to histone promoters and recruits Rtt106 (51). However, since Rtt106:H3 binding was necessary for Rtt106 recruitment to the promoter (Fig. 1A) and the affinity of Rtt106 for H3 is enhanced by the Asf1-dependent acetylation of H3K56 (33), Rtt106 mislocalization in asf1Δ cells could result from reduced Rtt106:H3 binding due to the absence of H3K56ac. To test whether Asf1 directly regulated histone gene transcription, we evaluated Asf1's enrichment in the regulatory region of the HTA1-HTB1 gene pair and at the other histone gene loci. Intriguingly, like Rtt106 and the HIR complex (16, 21), Asf1 localized to the center of the HTA1-HTB1 promoter (Fig. 2A). This localization profile was consistent with Asf1's known direct physical interaction with HIR and its indirect interaction with Rtt106 , which is thought to be mediated by H3/H4 binding (21, 32). Together, these findings supported a previous suggestion that the HIR complex, localized to the center of the HTA1-HTB1 regulatory region, recruits Asf1, which in turn recruits Rtt106 through an H3/H4 bridge (51).
Fig 2.
Asf1 localized with Rtt106 and the HIR complex at histone gene regulatory regions. (A) ChIP analysis of Asf1-FLAG at the HTA1-HTB1 locus (chromosomal coordinates from 913.9 to 916.3 kb). Gray bars indicate the sequences amplified by each qPCR primer set. qPCR values were normalized as described for Fig. 1B. (B) ChIP analysis of Asf1-FLAG at the center of the HTA1-HTB1, HHT1-HHF1, HHT2-HHF2, and HTA2-HTB2 promoters in the indicated mutant strains. WT, wild type. (C) Yeast whole-cell extract (WCE) immunoblotted with anti-FLAG and anti-Pgk1 (loading control) antibodies. (D) ChIP analysis of Rtt106-FLAG (left), Hir1-FLAG (center), and Asf1-FLAG (right) as described for panel B. (E) ChIP analysis of Rtt106-FLAG, Hir1-FLAG, and Asf1-FLAG at the HTA1-HTB1 (left) and HHT1-HHF1 (right, chromosomal coordinates 254.6 to 257.1 kb) loci, as described for panel A.
Consistent with this proposed order of recruitment, Asf1 enrichment was Rtt106 independent and was reduced in the absence of Hir1 (Fig. 2B). The cellular levels of Asf1 in rtt106Δ, hir1Δ, and wild-type cells were similar (Fig. 2C). Therefore, changes in Asf1 enrichment were not due to altered protein levels. Both Asf1 and Hir1 exhibited increased promoter enrichment in rtt106Δ cells (Fig. 2B and Fig. 1C). Therefore, the absence of Rtt106 may have caused greater exposure of the Hir1 and Asf1 epitopes, leading to the appearance of increased enrichment. Alternatively, since histone gene derepression is greater in hir1Δ cells than in rtt106Δ cells (16), the increased enrichment of HIR and Asf1 in rtt106Δ cells may partially compensate for the absence of Rtt106. Nevertheless, this similar trend in Hir1 and Asf1 enrichment strongly suggested that Asf1 played a direct role at histone gene promoters and was recruited prior to Rtt106.
Similarly to Rtt106 and the HIR complex (Fig. 1B and below), Asf1 exhibited a low level of enrichment at the HTA2-HTB2 promoter (Fig. 2B). Since HTA2-HTB2 does not contain the negative regulatory sequence elements that recruit the HIR complex at the other three histone gene loci, this low-level enrichment of histone chaperones was enigmatic. The enrichment of Hir1, Rtt106, and Asf1 at HTA2-HTB2 was unchanged in mutant backgrounds that altered chaperone recruitment at the three HIR-dependent histone gene loci (Fig. 2B and D). Additionally, HTA2 transcription was similarly unaffected in such mutants compared to that of loci containing negative regulatory sequence elements (Fig. 1D) (45). Therefore, if these low-level enrichments reflected a true histone chaperone complex at HTA2-HTB2, the complex was clearly subject to distinct regulation.
Next we tested whether Asf1 localization was restricted to the center of histone regulatory sequences by the AAA-ATPase chromatin boundary protein Yta7, which regulates the localization of Rtt106. In yta7Δ cells, Rtt106 has increased enrichment at the promoter and in the 5′ end of the histone open reading frames (ORFs) (Fig. 2D and E) (16). Subsequent increased recruitment of RSC throughout the region is thought to create a transcriptionally refractive chromatin environment (31). Unlike Rtt106, the HIR complex exhibits similar levels of enrichment between yta7Δ and wild-type cells (Fig. 2D and E) (16). Like the HIR complex and unlike Rtt106, in the absence of Yta7, Asf1 enrichment was similar to its enrichment in wild-type cells (Fig. 2D and E). Therefore, the HIR complex and Asf1 were nucleating factors that recruited Rtt106 to the regulatory region of histone genes. In the absence of the Yta7-mediated boundary, only Rtt106 and RSC showed increased enrichment in both histone gene promoters and ORFs to repress transcription (31). In the absence of Yta7, Rtt106 enrichment was more extensive at HHT1-HHF1 than at HTA1-HTB1, suggesting a locus-specific variation of this regulation (Fig. 2E).
Yta7 in Rtt106:H3 binding and histone gene localization.
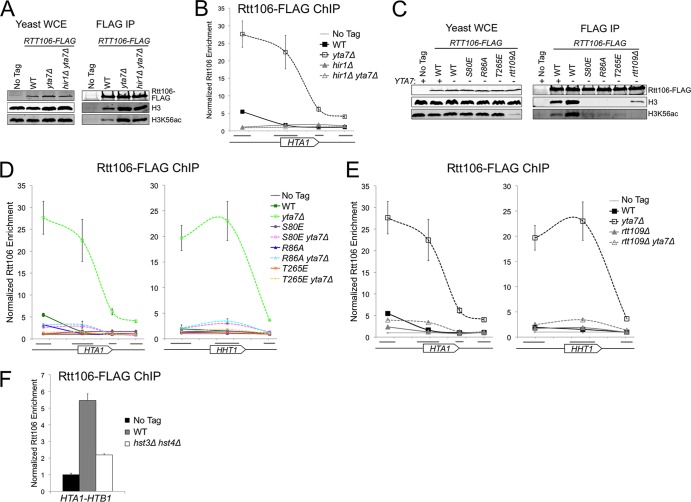
The relationship between Rtt106's histone binding activity and its recruitment to histone promoters suggested that Rtt106 plays a pivotal role in a negative-feedback loop for histone gene regulation. Reduced Rtt106:H3 binding led to reduced Rtt106 enrichment at the promoter and increased histone gene transcription (Fig. 1). Therefore, Rtt106 may act as a sensor to fine-tune histone transcription as a function of histone protein levels. A recent study reported an increased level of chromatin-bound H3 in yta7Δ cells (35). The negative-feedback model predicted that the increased promoter enrichment of Rtt106 and the reduced histone transcription observed in yta7Δ cells might depend on Rtt106 “sensing” these elevated histone protein levels. Indeed, co-IP experiments revealed an increased amount of H3 bound to Rtt106 in yta7Δ cells compared to wild-type cells (Fig. 3A). This increase was maintained in hir1Δ yta7Δ double mutants, indicating that the elevated Rtt106:H3 binding in yta7Δ single mutants occurred prior to Rtt106 recruitment to histone gene promoters (Fig. 3A and B). Eliminating Rtt106's ability to “sense” histone proteins, either by mutating its histone binding surface (S80E, R86A, or T265E) or by depleting H3K56ac (rtt109Δ), suppressed both the elevated Rtt106:H3 binding and the increased Rtt106 enrichment at histone promoters in yta7Δ cells (Fig. 3C to E). This suppression indicated that elevated enrichment of Rtt106 in the yta7Δ mutant relied upon the previously characterized Rtt106:H3 binding surface, implying that the additional Rtt106 was bound to H3/H4. Moreover, in the presence of excess histone protein, increased Rtt106:H3 binding was necessary for Rtt106 recruitment and subsequent repression of histone gene transcription.
Fig 3.
Elevated Rtt106:H3 binding in yta7Δ cells was necessary for increased Rtt106 enrichment at histone genes. (A) Rtt106-FLAG immunoprecipitated from the indicated mutant strains as in Fig. 1A. WT, wild type. (B) ChIP analysis of Rtt106-FLAG at the HTA1 locus, as described in Fig. 2E (chromosomal coordinates from 915.0 to 916.3 kb). (C) Rtt106-FLAG immunoprecipitated from yeast whole-cell extract (WCE) as in panel A. (D and E) ChIP analysis of Rtt106-FLAG at the HTA1 (left) and HHT1 (right, chromosomal coordinates 255.8 to 257.1 kb) loci, as in panel B. (F) ChIP analysis of Rtt106-FLAG at the HTA1-HTB1 promoter, as in Fig. 1B.
Monitoring Rtt106 localization in yta7Δ cells revealed a previously unappreciated distinction between the severity of the S80E and T265E mutations, which had similar phenotypes in wild-type cells (Fig. 1) (61). A small but significant amount of both Rtt106 S80E and R86A spread into histone gene ORFs in yta7Δ cells compared to each mutant's localization in wild-type cells (P < 0.05 at HTA1, P < 0.01 at HHT1), whereas Rtt106 T265E localization in yta7Δ was similar to that of the wild type (Fig. 3D). These results suggested that mutations within Rtt106's C-terminal loop (T265E) led to stronger H3 binding defects than mutations within the N-terminal basic patch (S80E and R86A).
Surprisingly, unlike in rtt109Δ single mutants (Fig. 1A), a low level of Rtt106:H3 binding was detectable by co-IP in rtt109Δ yta7Δ cells (Fig. 3C). Additionally, compared to rtt109Δ single mutants, a small amount of Rtt106 spread into histone gene ORFs in rtt109Δ yta7Δ double mutants (P < 0.01)(Fig. 3E). Therefore, in the absence of Yta7, the acetylation of H3K56 by Rtt109 was no longer required for detectable Rtt106:H3 binding. These results were consistent with in vitro binding experiments, which show that although the acetylation of H3 at K56 increases the affinity of Rtt106 for H3 by ∼15- to 20-fold, Rtt106:H3 binding is still detectable with recombinant, unacetylated H3/H4 (Kd [dissociation constant], ∼1 μM) (52). Therefore, it is possible that in rtt109Δ cells a low level of Rtt106:H3 binding that was below the level of detection by co-IP occurred, whereas in yta7Δ cells the increased cellular concentration of histones allowed the visualization of Rtt106's weak in vivo interaction with H3 proteins lacking acetylation at K56. This idea was supported by the Rtt106 enrichment at the HTA1-HTB1 promoter in rtt109Δ which, though substantially reduced with respect to the wild type, was nevertheless higher than the enrichment of Rtt106 carrying the S80E and T265E point mutations along the Rtt106:H3 interaction surface (Fig. 1B).
The correlation between increased Rtt106:H3 binding and Rtt106 enrichment at histone gene promoters in yta7Δ cells predicted that additional mutant backgrounds with elevated Rtt106:H3 would lead to similarly increased Rtt106 enrichment within the regulatory region of histone genes. Cells lacking the H3K56ac-specific histone deacetylases Hst3 and Hst4 have increased Rtt106:H3 binding, presumably due to increased levels of H3K56ac (4, 33). However, unlike in yta7Δ cells, Rtt106 enrichment was reduced at the regulatory region of HTA1-HTB1 in hst3Δ hst4Δ cells (Fig. 3F). Therefore, elevated Rtt106:H3 binding does not always lead to increased Rtt106 localization at the histone gene loci, as discussed further below.
Feedback regulation of histone gene transcription by Yta7 and Rtt106.
According to the negative-feedback model, changes in Rtt106 enrichment at histone loci in yta7Δ cells should lead to changes in transcription. However, the effect of yta7Δ on the level of histone transcripts is somewhat controversial. Gradolatto et al. reported precocious increases in histone mRNA levels prior to S phase in yta7Δ cells, though the trend was dependent upon the type of synchronizing agent and was observed only at a subset of histone gene loci (19). In contrast, Fillingham et al. reported reduced total HTA1 mRNA in yta7Δ cells, whereas Lombardi et al. reported that the HHT1 mRNA level in yta7Δ cells is similar to the level in wild-type cells (16, 35). Here, we recapitulated that in yta7Δ cells total HTA1 transcripts were significantly reduced, whereas HHT1 and HHF1 mRNA levels were indistinguishable from those in the wild type (Fig. 4A). The repression of HTA1 expression in yta7Δ cells was suppressed in an isogenic yta7Δ rtt106Δ double mutant, suggesting that Rtt106 spreading caused the HTA1 repression (Fig. 4A). In summary, enhanced Rtt106 enrichment at HTA1-HTB1 correlated with reduced transcription at that locus whereas enhanced enrichment at HHT1-HHF1 did not, implying the existence of additional feedback mechanisms operating at HHT1-HHF1 to ensure proper transcriptional regulation.
Fig 4.
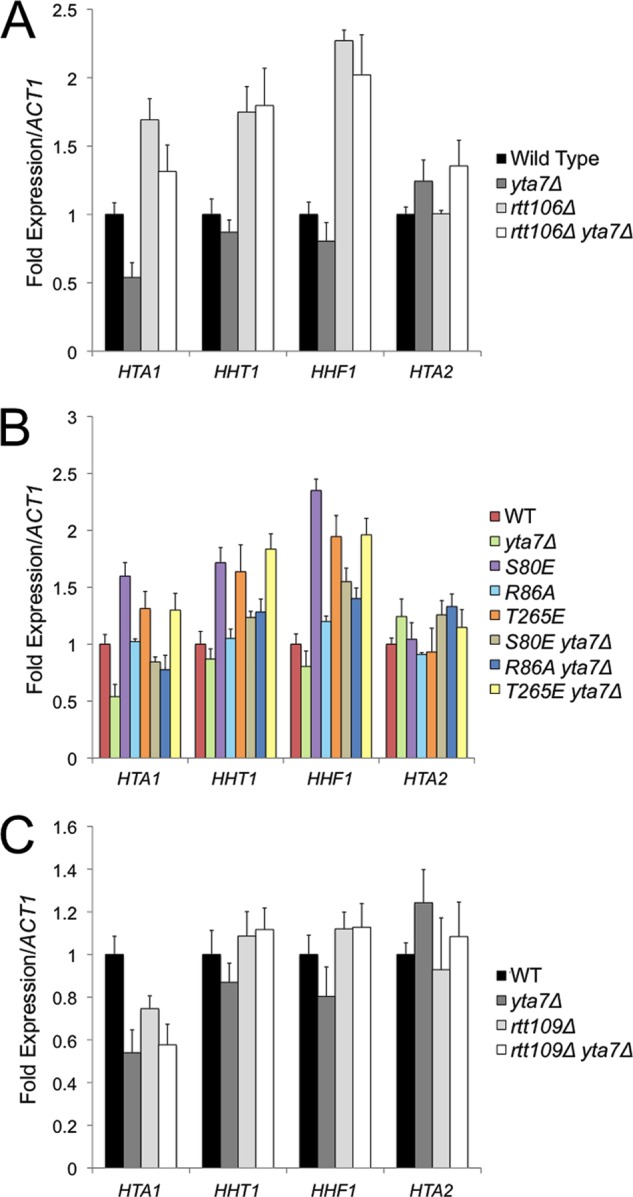
Increased Rtt106 enrichment at histone gene loci repressed HTA1 transcription. RT-qPCR of histone gene mRNA levels in the indicated strains as described for Fig. 1D. HTA1 transcription was significantly reduced in yta7Δ and rtt109Δ strains compared to that in the wild type (WT) (P < 0.01).
Changes in HTA1, HHT1, and HHF1 transcript levels mirrored the changes in Rtt106 S80E, R86A, and T265E localization between wild-type and yta7Δ cells. Rtt106 T265E localization was unchanged in yta7Δ cells (Fig. 3D), and histone transcript levels were similar in T265E and T265E yta7Δ mutants (Fig. 4B). Although Rtt106 S80E and R86A were strongly reduced in their affinity for H3 compared to wild-type Rtt106, these mutants displayed some spreading in the absence of Yta7 compared to the promoter-specific localization of Rtt106 in wild-type cells (Fig. 3D). This partial formation of a repressive complex at the promoter led to reduced HTA1, HHT1, and HHF1 transcripts in S80E yta7Δ and R86A yta7Δ compared to T265E yta7Δ and rtt106Δ yta7Δ, where no spreading occurred (Fig. 4B). Therefore, the enhanced dynamic range of Rtt106 enrichment at histone gene regulatory regions in yta7Δ cells allowed further recognition of functionally significant differences in the localization of Rtt106 mutants.
Previous studies classified Rtt109 along with Yta7 as an activator of HTA1 transcription (16). However, as in yta7Δ, we observed transcriptional repression in rtt109Δ cells only at HTA1; HHT1 and HHF1 mRNA levels were similar to those of the wild type (Fig. 4C). Rtt106 enrichment was partially reduced at the HTA1-HTB1 promoter in rtt109Δ cells (Fig. 1B) presumably due to the reduced level of H3K56ac, the preferred histone ligand of Rtt106. Nevertheless, HTA1 transcript levels are reduced due to the lack of Rtt109's activator function, which is required to overcome histone chaperone-mediated repression (see below) (Fig. 4C) (16). As expected, HTA1 transcription was reduced in rtt109Δ yta7Δ cells due to both the absence of the Rtt109 activator and the small but significant amount of Rtt106 enrichment within the HTA1 ORF (Fig. 4C and Fig. 3E).
Relationship between Rtt106's roles in replication, silencing, and histone gene transcription.
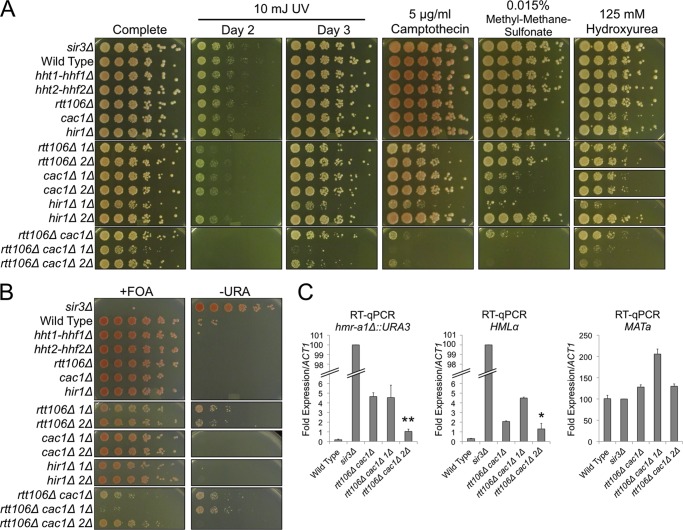
In addition to the cell cycle regulation of histone gene transcription, Rtt106 mutations also exhibit genetic interactions with mutations in the CAF-1 histone chaperone complex during replication and silencing (25, 26, 33). The direct physical interaction of Rtt106 with CAF-1 suggests that Rtt106's role in silencing and replication is direct (25). Nevertheless, it was also possible that rtt106Δ cac1Δ phenotypes regarding replication and silencing were at least partially due to inappropriate histone gene transcription throughout the cell cycle. By this hypothesis, a reduction in histone gene dosage might suppress the silencing defects and genotoxic chemical sensitivities observed in rtt106Δ cac1Δ cells. To test this hypothesis, we monitored growth on media containing chemicals that induce DNA damage during S phase and on media that measure silencing of an HMRa1 reporter (hmr-a1Δ::URA3) in rtt106Δ cac1Δ cells lacking one of the two H3-H4 histone gene pairs (hht1-hhf1Δ or hht2-hhf2Δ).
In the presence of genotoxic agents, a reduction in H3-H4 dosage did not suppress the growth defects of rtt106Δ cac1Δ cells, suggesting that increased histone gene transcription was not the primary cause of these chemical sensitivities (Fig. 5A). During these growth assays, we were surprised to discover a synthetic growth defect phenotype between rtt106Δ and hht1-hhf1Δ but not hht2-hhf2Δ (Fig. 5A; most evident in “day 2” panel). This distinction between H3-H4 gene copies was unexpected; the two loci encode identical H3 and H4 proteins and are thought to be functionally equivalent (9). To tease apart whether the synthetic sickness was due to Rtt106's role in histone gene transcription or replication-coupled nucleosome turnover, we tested whether mutants of the HIR or CAF-1 complex had similar synthetic interactions with hht1-hhf1Δ. The cac1Δ hht1-hhf1Δ and cac1Δ hht2-hhf2Δ double mutants both phenocopied the cac1Δ single mutant, indicating that the two H3-H4 gene pairs were equivalent with respect to replication-coupled chromatin assembly. In contrast, like rtt106Δ, hir1Δ hht1-hhf1Δ had a strong synergistic growth defect compared to that of hir1Δ hht2-hhf2Δ (Fig. 5A). The increased severity of the hir1Δ hht1-hhf1Δ growth defects compared to that of rtt106Δ hht1-hhf1Δ was consistent with the larger fold increase in histone gene transcription in hir1Δ cells compared to that in rtt106Δ cells (16). These results unveiled a previously unappreciated distinction between histone chaperone-mediated repression at HHT1-HHF1 and that at HHT2-HHF2.
Fig 5.
Reduced H3-H4 dosage partially suppressed rtt106Δ cac1Δ silencing phenotypes. (A) Five-fold serial dilutions of yeast culture were spotted onto media containing the indicated genotoxic agents (1Δ, hht1-hhf1Δ; 2Δ, hht2-hhf2Δ). (B) Silencing of the hmr-a1Δ::URA3 reporter was monitored by spotting 5-fold serial dilutions of yeast culture onto medium containing 5-fluoroorotic acid (+FOA), a counterselection for URA3 expression, and onto medium lacking uracil (−URA), a selection for URA3 expression. (C) RT-qPCR analysis of hmr-a1Δ::URA3, HMLα, and MATa. Transcript values were normalized to ACT1 mRNA and adjusted to the sir3Δ value, which was normalized to 100. Transcription was significantly reduced in rtt106Δ cac1Δ hht2-hhf2Δ cells compared to that in rtt106Δ cac1Δ cells (*, P < 0.05; **, P < 0.01).
Intriguingly, unlike the sensitivities to DNA-damaging agents, knocking out hht2-hhf2Δ partially suppressed the silencing defects in rtt106Δ cac1Δ cells (Fig. 5B, bottom panels). The absence of suppression in rtt106Δ cac1Δ hht1-hhf1Δ cells further highlighted differences between the two copies of H3-H4. Direct mRNA measurements revealed that rtt106Δ cac1Δ hht2-hhf2Δ cells partially suppressed the rtt106Δ cac1Δ silencing defects at both the hmr-a1Δ::URA3 reporter and endogenous HMLα (Fig. 5C). In contrast, transcription at the MATa locus in rtt106Δ cac1Δ hht2-hhf2Δ cells was similar to that in the wild type (Fig. 5C). Therefore, the enhanced silencing at hmr-a1Δ::URA3 and HMLα in rtt106Δ cac1Δ hht2-hhf2Δ cells compared to that in rtt106Δ cac1Δ cells was not due to a general transcription defect. Together, these results suggested a partial overlap of Rtt106's roles in regulating histone gene transcription and maintaining repression at the silent mating-type loci.
DISCUSSION
In this study, we evaluated the composition and function of protein complexes at the negative regulatory regions of histone gene pairs. Our data suggested that in wild-type cells, the HIR complex at the center of the bidirectional promoters recruited Asf1, which in turn recruited Rtt106 via a histone protein bridge (Fig. 6A, left). Rtt106 directly recruits either the RSC or SWI/SNF chromatin remodeling complex to facilitate transcriptional repression or activation, respectively (8, 15, 42). We observed a direct relationship between the level of Rtt106:H3 binding and Rtt106 enrichment at histone promoters and an inverse relationship between Rtt106 enrichment and transcriptional output. rtt106 mutants that reduced Rtt106:H3 binding had reduced Rtt106 enrichment at histone promoters and increased transcription (Fig. 6A, center). Conversely, yta7Δ mutants exhibited increased Rtt106:H3 binding, increased Rtt106 enrichment at histone promoters, and reduced transcription (Fig. 6A, right). The relationship between Rtt106's physical interaction with histone proteins and the level of histone gene transcription suggested that Rtt106 acted as a sensor to facilitate a negative-feedback loop to regulate histone levels.
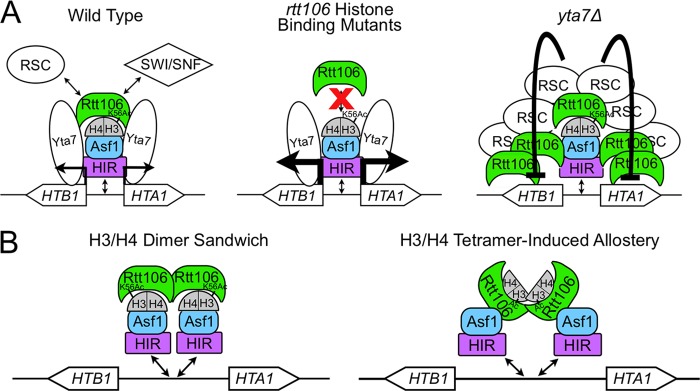
Fig 6.
Model for negative-feedback regulation of histone gene transcription. (A) In wild-type cells (left), HIR, Asf1, and Rtt106 histone chaperones along with Yta7 localize to histone gene regulatory regions. Rtt106 directly recruits RSC or SWI/SNF chromatin remodeling enzymes to facilitate S-phase-specific histone gene transcription. rtt106 mutants that can no longer bind histone H3 (center) have reduced Rtt106 enrichment at histone regulatory regions and increased histone transcription. In yta7Δ cells (right), Rtt106:H3 binding was increased, leading to elevated Rtt106 enrichment throughout histone gene loci and reduced transcription. (B) Two models for the histone-mediated interaction between Rtt106 and Asf1.
The regulatory complex of histone gene pairs: a sandwich of histone proteins and chaperones.
Previous work established that the HIR complex localizes and recruits Rtt106 to a negative regulatory element in the promoters of three of the four histone gene pairs (16, 44, 45, 59). We found that Asf1, another H3/H4 chaperone previously shown to interact with both the HIR complex and Rtt106 (21, 32), also localized to the promoter regions of all HIR-regulated histone gene pairs (Fig. 2A and B). Recruitment of Asf1 was HIR dependent and Rtt106 independent (Fig. 2B). In contrast, the recruitment of Rtt106 was both Asf1 dependent and HIR dependent (Fig. 1B) (16, 51). These results suggested that a HIR:Asf1:Rtt106 regulatory complex formed at histone promoters to maintain S-phase-specific expression.
The involvement of three different H3/H4 chaperones at histone gene regulatory regions implied that histone proteins themselves might be critical components of the complexes. Indeed, we found that mutations in RTT106 that reduced the affinity of Rtt106 for histone H3 reduced the enrichment of Rtt106 at histone promoters (Fig. 1A and B) and led to increased histone expression (Fig. 1D). Because HIR and Asf1 enrichment at these control regions was Rtt106 independent (Fig. 1C and Fig. 2B) (16), these data implied that a histone protein bridge mediated the interaction between Asf1 and Rtt106. The histone dependence of this chaperone-chaperone interaction was unique; Rtt106:CAF-1, Asf1:CAF-1, and Asf1:HIR interactions are direct and independent of chaperone-histone binding (21, 25, 57). This additional layer of complexity suggested a new mechanism for chaperone-mediated interactions. Previous studies established that Asf1 fails to co-IP with Rtt106 in hir1Δ cells (32). Therefore, the Asf1:H3/H4:Rtt106 interaction likely occurs at histone gene promoters. At this point, we have not resolved whether the HIR complex dependence of this interaction is due to sequential recruitment of Asf1 followed by Rtt106 or whether the HIR complex stabilizes the Asf1:H3/H4:Rtt106 interaction, possibly by making direct contacts with Rtt106.
Since Asf1:H3 binding occludes the tetramerization surface of H3/H4 (2, 11, 41), the proposed Rtt106:H3/H4:Asf1 interaction must utilize H3/H4 dimers. Although Rtt106 was recently shown to bind H3/H4 tetramers in vivo, the possibility of Rtt106 binding H3/H4 dimers has not been excluded (14). Additionally, Rtt106 itself was recently shown to function as a dimer (52). Together, these results suggest that the regulatory complex at histone gene promoters contains one Rtt106 dimer bound to two individual H3/H4 dimers each bound by Asf1 (Fig. 6B, left). Asf1's direct interaction with each H3/H4 dimer and the HIR complex would block H3/H4 tetramerization and promote Rtt106 recruitment to histone promoters, respectively. Alternatively, Rtt106 binding to H3/H4 tetramers might induce a conformational change in Rtt106 required for a direct Rtt106:Asf1 interaction (Fig. 6B, right).
On the spreading of, and repression by, Rtt106.
One of the most surprising findings in this study was the lack of a consistent mechanistic link between Rtt106 spreading from the regulatory regions into the coding regions of histone gene pairs and the repression of those genes. In wild-type cells, Yta7 restricts Rtt106 localization to the center of the regulatory region (16, 31), perhaps analogously to the way in which Yta7 acts as a boundary to the spreading of silenced chromatin (20, 28, 54). Our data at the HTA1-HTB1 gene pair confirmed earlier results: in the absence of Yta7, increased Rtt106 enrichment within the HTA1-HTB1 locus led to reduced HTA1 transcription (Fig. 2E and 4A) (16). However, this paradigm was not shared across all histone gene loci. Rtt106 enrichment in yta7Δ cells was less extensive within the HTB1 ORF than within the HTA1 ORF (Fig. 2E). This asymmetric localization was consistent with previous findings that Rtt106 has no effect on HTB1 transcription in otherwise wild-type cells (16). Additionally, we found even more extensive spreading of Rtt106 from the regulatory region of HHT1-HHF1 into the coding sequences in yta7Δ cells, yet HHT1 and HHF1 expression in yta7Δ was similar to that in wild-type cells (Fig. 2E and 4A). Therefore, repression was not an obligate consequence of increased Rtt106 localization. Additional factors, such as SWI/SNF versus RSC recruitment, might be the source of these transcriptional differences. These results highlight the importance of investigating transcriptional effects at all histone gene pairs, unlike many studies that focused only on the HTA1 locus.
The potential for histone chaperones spreading along chromatin represents a new mechanism for transcriptional regulation. In the case of Rtt106, its binding preference for H3K56ac over unmodified H3 suggests that spreading may be dependent on the presence of H3K56ac, which is an S-phase-specific modification (40). Indeed, the spreading of Rtt106 in yta7Δ mutants was largely suppressed in yta7Δ rtt109Δ double mutants, which lack the H3K56 acetyltransferase (Fig. 3D). Curiously, in wild-type cells Rtt106 enrichment at histone gene promoters is consistent throughout the cell cycle, whereas Yta7 enrichment is S-phase specific (16, 31). Therefore, either Rtt106 spreading is also S-phase specific or Rtt106 spreading outside S phase is restricted by Yta7-independent factors. These possibilities will be distinguished by monitoring Rtt106 enrichment in synchronized yta7Δ cells. Recent studies suggest that Rtt106 might form higher-order oligomeric structures in vitro (14, 52), which could also facilitate the increased enrichment in the absence of Yta7. Future studies will address whether, as in yta7Δ cells, artificial overexpression of histone proteins leads to increased Rtt106:H3 binding, increased Rtt106 localization at endogenous histone gene regulatory regions, and reduced histone gene transcription.
The altered Rtt106 localization that we observed in yta7Δ cells was consistent with results of previous studies that proposed that Rtt106 spreads into histone ORFs (16). However, the increased Rtt106 ChIP signal in yta7Δ cells might reflect a change in the dynamics of Rtt106's association with the promoter rather than an increase in the total amount of Rtt106 protein bound to each locus. Additionally, since the histone genes are small (∼400 bp), the limitations of conventional ChIP make rigorous assessment of changes in Rtt106 localization below ∼500 bp difficult. Future studies will address whether the altered Rtt106 localization represents true spreading or a distinct mechanism of recruitment that is antagonized by Yta7.
In yta7Δ cells, the enhanced Rtt106:H3 binding may have resulted from the increased levels of chromatin-bound H3 (35). Although Rtt106:H3 binding is H3K56ac dependent (33) and the ratio of H3K56ac to H3 was unchanged in yta7Δ cells (Fig. 3A), a contribution by other posttranslational modifications to Rtt106:H3 binding remains possible. Intriguingly, a cocrystal structure of Rtt106 and acetylhistamine revealed two acetylhistamine-binding sites, one on each PH domain of Rtt106 (52) (Protein Data Bank [PDB] identification [ID] code 3TW1). If these two acetylhistamine binding sites reveal binding sites also capable of binding acetyllysine, then Rtt106:H3 binding might be influenced by a second acetyl modification elevated in yta7Δ cells.
Role of H3K56ac in histone gene transcription.
Currently, the role of H3K56ac in the regulation of histone gene transcription is paradoxical. In wild-type cells, H3K56ac is highly enriched at histone gene promoters and is thought to induce histone turnover and active transcription (29). Cells lacking the histone gene activators Spt10 or Rtt109 have reduced H3K56ac within the promoter and are transcriptionally repressed (16, 29, 58). Therefore, H3K56ac appears to be necessary for histone gene activation. However, asf1Δ cells, which lack H3K56ac (Fig. 1A) (1), have elevated histone gene transcription compared to that of wild-type cells (16, 53), suggesting that H3K56 acetylation is not required for high-level expression. Unlike rtt109Δ cells in which Rtt106 enrichment at the HTA1-HTB1 promoter is partially reduced, Rtt106 is reduced to background levels in asf1Δ cells due to Asf1's direct role in Rtt106 recruitment (Fig. 1B). Therefore, H3K56ac may only be necessary for transcription in the presence of histone chaperones. Additionally, in hir1Δ and rtt106Δ cells, histone promoters are depleted of histone protein and transcription is elevated compared to that of the wild type (16, 45), further suggesting that H3K56ac is not necessary for active transcription. These results suggest that H3K56ac is necessary to overcome the histone chaperone-mediated repression at histone promoters, perhaps by promoting nucleosome turnover to expose the DNA to activators. Monitoring chaperone-dependent histone incorporation within the histone gene regulatory regions throughout the cell cycle will further define the relationship between chaperone-mediated nucleosome assembly and histone gene transcription.
In addition to acting locally at the histone gene regulatory regions, H3K56ac might antagonize chaperone-mediated repression at a distance by titrating chaperones away from histone promoters and into other areas of the genome. Consistent with this hypothesis, although Rtt106:H3 binding was increased in hst3Δ hst4Δ cells (33), which have elevated levels of H3K56ac, Rtt106 enrichment was partially reduced at histone gene promoters (Fig. 3F). According to a simple negative-feedback model, increased Rtt106:H3 binding should lead to increased Rtt106 localization and reduced histone gene transcription. However, nearly 100% of H3 proteins are acetylated at H3K56 in hst3Δ hst4Δ cells (4), which could titrate Rtt106 away from the histone gene loci and into other areas of the genome. Additionally, excess H3K56ac in this double mutant leads to elevated DNA damage, which might titrate chaperones to sites of repair (4, 38, 46). Thus, the mechanism of regulation appeared to involve a fine-tuning of affinities and concentrations among histone proteins and their chaperones.
Relationship between histone gene expression, replication, and silencing.
In addition to mediating H3K56ac delivery into chromatin (33, 61), the role of Rtt106 in histone gene regulation may promote DNA replication and silencing, most evident in cells lacking CAF-1. A reduction in H3 and H4 gene dosage partially suppressed the silencing defects but not the genotoxic chemical sensitivities associated with rtt106Δ cac1Δ cells (Fig. 5). These results were consistent with findings that overexpression of the histone genes enhanced the telomeric silencing defects observed in cac mutants, further suggesting that in the absence of CAF-1, silencing was sensitive to changes in histone levels (30). Additionally, rtt106Δ rtt109Δ mutants partially suppress the histone transcription defects associated with each single mutant (16), and rtt109Δ rtt106Δ cac1Δ mutants partially suppress rtt106Δ cac1Δ silencing phenotypes (61). Together, these results suggest that Rtt106's role in histone gene regulation may at least partially account for the silencing defects associated with rtt106Δ.
Analyzing changes in histone gene dosage revealed a genetic interaction between the replication-independent histone chaperones RTT106 and HIR1 and one of the two loci encoding H3 and H4 (HHT1-HHF1) (Fig. 5A). This distinction between the two loci encoding H3 and H4 suggested that Rtt106 and Hir1 might perform unique functions at HHT1-HHF1 compared to HHT2-HHF2. Although, we did not detect differences in the level of chaperone enrichment at these two loci in wild-type cells (Fig. 2D), differences in Rtt106:H3 binding and Rtt106 enrichment at histone gene regulatory regions in hht1-hhf1Δ and hht2-hhf2Δ cells may have contributed to the observed phenotypes. Another possibility was that in hht1-hhf1Δ cells, the increased transcription of HHT2-HHF2 was detrimental in the absence of Rtt106 or Hir1. In rtt106Δ and hir1Δ cells, the fold increase of HHT and HHF transcription is larger at the HHT1-HHF1 locus than at HHT2-HHF2 (16). Additionally, each H3-H4 locus produces a slightly different ratio of HHT to HHF transcripts. Because all H3 and H4 proteins are thought to exist as either an H3/H4 dimer or tetramer, altering the amount of each histone type can be detrimental to the cell. Therefore, in the absence of replication-independent histone chaperones, an altered HHT/HHF ratio might lead to increased sensitivity to DNA-damaging agents. Importantly, strains used to analyze histone point mutants historically have both endogenous copies of H3 and H4 knocked out and HHT2-HHF2 expressed from a plasmid. These strains are effectively hht1-hhf1Δ, and therefore results should be interpreted with caution when analyzing replication-independent histone chaperone phenotypes.
Concluding remarks on histone chaperone-mediated negative feedback.
Because all newly synthesized H3 molecules are acetylated on K56 (4), and Rtt106:H3 binding is K56ac dependent (33), our results suggest that Rtt106 may act as a sensor to regulate histone gene transcription as a function of new histone protein levels. This feedback structure positions the HIR:Asf1:H3/H4:Rtt106 interaction as a critical node in regulating histone transcription. Future analysis of the relationship between RNA polymerase II loading and histone chaperone occupancy at the histone gene loci throughout the cell cycle will deepen our understanding of this complex regulatory mechanism. Since histone gene promoters are sites of rapid histone turnover (29), monitoring the colocalization of Rtt106, Asf1, and the HIR complex at other sites in the genome with similar histone dynamics will likely identify genes encoding proteins that are critical for chromatin assembly and thus are regulated by similar feedback loops.
ACKNOWLEDGMENTS
We thank Barbara Meyer, Robert Tjian, and Robert Fischer for advice and support. We thank Laura Lombardi for critical discussions and technical support, along with Anne Dodson and James Fraser for review of the manuscript.
This research was supported by a National Science Foundation graduate research fellowship (to R.M.Z.) and a National Institutes of Health research grant (GM31105 to J.R.).
Footnotes
Published ahead of print 20 August 2012
REFERENCES
- 1. Adkins MW, Carson JJ, English CM, Ramey CJ, Tyler JK. 2007. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J. Biol. Chem. 282: 1334–1340 [DOI] [PubMed] [Google Scholar]
- 2. Antczak AJ, Tsubota T, Kaufman PD, Berger JM. 2006. Structure of the yeast histone H3-ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics. BMC Struct. Biol. 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonner WM, Wu RS, Panusz HT, Muneses C. 1988. Kinetics of accumulation and depletion of soluble newly synthesized histone in the reciprocal regulation of histone and DNA synthesis. Biochemistry 27: 6542–6550 [DOI] [PubMed] [Google Scholar]
- 4. Celic I, et al. 2006. The sirtuins Hst3 and Hst4p preserve genome integrity by controlling histone H3 lysine 56 deacetylation. Curr. Biol. 16: 1280–1289 [DOI] [PubMed] [Google Scholar]
- 5. Clemente-Ruiz M, Gonzalez-Prieto R, Prado F. 2011. Histone H3K56 acetylation, CAF1, and Rtt106 coordinate nucleosome assembly and stability of advancing replication forks. PLoS Genet. 7: e1002376 doi:10.1371/journal.pgen.1002376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Das C, Tyler JK, Churchill ME. 2010. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem. Sci. 35: 476–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Koning L, Corpet A, Haber JE, Almouzni G. 2007. Histone chaperones: an escort network regulating histone traffic. Nat. Struct. Mol. Biol. 14: 997–1007 [DOI] [PubMed] [Google Scholar]
- 8. Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley MA. 1999. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol. Cell 4: 75–83 [DOI] [PubMed] [Google Scholar]
- 9. Dollard C, Ricupero-Hovasse SL, Natsoulis G, Boeke JD, Winston F. 1994. SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 5223–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Driscoll R, Hudson A, Jackson SP. 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315: 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. 2006. Structural basis for the histone chaperone activity of Asf1. Cell 127: 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eriksson PR, Ganguli D, Clark DJ. 2011. Spt10 and Swi4 control the timing of histone H2A/H2B gene activation in budding yeast. Mol. Cell. Biol. 31: 557–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eriksson PR, Ganguli D, Nagarajavel V, Clark DJ. 2012. Regulation of histone gene expression in budding yeast. Genetics 191: 7–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fazly A, et al. 2012. Histone chaperone Rtt106 promotes nucleosome formation using (H3–H4)2 tetramers. J. Biol. Chem. 287: 10753–10760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferreira ME, Flaherty K, Prochasson P. 2011. The Saccharomyces cerevisiae histone chaperone Rtt106 mediates the cell cycle recruitment of SWI/SNF and RSC to the HIR-dependent histone genes. PLoS One 6: e21113 doi:10.1371/journal.pone.0021113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fillingham J, et al. 2009. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell 35: 340–351 [DOI] [PubMed] [Google Scholar]
- 17. Gelbart ME, Rechsteiner T, Richmond TJ, Tsukiyama T. 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21: 2098–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gietz RD, Woods RA. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96 [DOI] [PubMed] [Google Scholar]
- 19. Gradolatto A, et al. 2008. Saccharomyces cerevisiae Yta7 regulates histone gene expression. Genetics 179: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gradolatto A, et al. 2009. A noncanonical bromodomain in the AAA ATPase protein Yta7 directs chromosomal positioning and barrier chromatin activity. Mol. Cell. Biol. 29: 4604–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Green EM, et al. 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15: 2044–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gunjan A, Paik J, Verreault A. 2005. Regulation of histone synthesis and nucleosome assembly. Biochimie 87: 625–635 [DOI] [PubMed] [Google Scholar]
- 23. Gunjan A, Verreault A. 2003. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115: 537–549 [DOI] [PubMed] [Google Scholar]
- 24. Han J, et al. 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315: 653–655 [DOI] [PubMed] [Google Scholar]
- 25. Huang S, et al. 2005. Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc. Natl. Acad. Sci. U. S. A. 102: 13410–13415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang S, Zhou H, Tarara J, Zhang Z. 2007. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J. 26: 2274–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Imbeault D, Gamar L, Rufiange A, Paquet E, Nourani A. 2008. The Rtt106 histone chaperone is functionally linked to transcription elongation and is involved in the regulation of spurious transcription from cryptic promoters in yeast. J. Biol. Chem. 283: 27350–27354 [DOI] [PubMed] [Google Scholar]
- 28. Jambunathan N, et al. 2005. Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics 171: 913–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaplan T, et al. 2008. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 4: e1000270 doi:10.1371/journal.pgen.1000270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaufman PD, Cohen JL, Osley MA. 1998. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 18: 4793–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kurat CF, et al. 2011. Restriction of histone gene transcription to S phase by phosphorylation of a chromatin boundary protein. Genes Dev. 25: 2489–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lambert JP, et al. 2010. Defining the budding yeast chromatin-associated interactome. Mol. Syst. Biol. 6: 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li Q, et al. 2008. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134: 244–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Y, et al. 2010. Structural analysis of Rtt106p reveals a DNA binding role required for heterochromatin silencing. J. Biol. Chem. 285: 4251–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lombardi LM, Ellahi A, Rine J. 2011. Direct regulation of nucleosome density by the conserved AAA-ATPase Yta7. Proc. Natl. Acad. Sci. U. S. A. 108: E1302–E1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Longtine MS, et al. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- 37. Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- 38. Maas NL, Miller KM, DeFazio LG, Toczyski DP. 2006. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell 23: 109–119 [DOI] [PubMed] [Google Scholar]
- 39. Makarova O, Kamberov E, Margolis B. 2000. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques 29: 970–972 [DOI] [PubMed] [Google Scholar]
- 40. Masumoto H, Hawke D, Kobayashi R, Verreault A. 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436: 294–298 [DOI] [PubMed] [Google Scholar]
- 41. Natsume R, et al. 2007. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 446: 338–341 [DOI] [PubMed] [Google Scholar]
- 42. Ng HH, Robert F, Young RA, Struhl K. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 16: 806–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Osley MA. 1991. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 60: 827–861 [DOI] [PubMed] [Google Scholar]
- 44. Osley MA, Gould J, Kim S, Kane MY, Hereford L. 1986. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell 45: 537–544 [DOI] [PubMed] [Google Scholar]
- 45. Osley MA, Lycan D. 1987. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol. Cell. Biol. 7: 4204–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pan X, et al. 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081 [DOI] [PubMed] [Google Scholar]
- 47. Prochasson P, Florens L, Swanson SK, Washburn MP, Workman JL. 2005. The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 19: 2534–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A. 2006. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J. Biol. Chem. 281: 37270–37274 [DOI] [PubMed] [Google Scholar]
- 49. Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11: 463–473 [DOI] [PubMed] [Google Scholar]
- 50. Shibahara K, Stillman B. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96: 575–585 [DOI] [PubMed] [Google Scholar]
- 51. Silva AC, et al. 2012. The replication-independent histone H3–H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. J. Biol. Chem. 287: 1709–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Su D, et al. 2012. Structural basis for recognition of H3K56-acetylated histone H3–H4 by the chaperone Rtt106. Nature 483: 104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sutton A, Bucaria J, Osley MA, Sternglanz R. 2001. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics 158: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tackett AJ, et al. 2005. Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 169: 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tsubota T, et al. 2007. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell 25: 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tyler JK, et al. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402: 555–560 [DOI] [PubMed] [Google Scholar]
- 57. Tyler JK, et al. 2001. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Cell. Biol. 21: 6574–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xu F, Zhang K, Grunstein M. 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121: 375–385 [DOI] [PubMed] [Google Scholar]
- 59. Xu H, Kim UJ, Schuster T, Grunstein M. 1992. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 5249–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zill OA, Scannell D, Teytelman L, Rine J. 2010. Co-evolution of transcriptional silencing proteins and the DNA elements specifying their assembly. PLoS Biol. 8: e1000550 doi:10.1371/journal.pbio.1000550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zunder RM, Antczak AJ, Berger JM, Rine J. 2012. Two surfaces on the histone chaperone Rtt106 mediate histone binding, replication, and silencing. Proc. Natl. Acad. Sci. U. S. A. 109: E144–E153 [DOI] [PMC free article] [PubMed] [Google Scholar]