Abstract
Transcription factors play key roles in the formation of a multilayered cerebral cortex consisting of neurons and glial cells. Krüppel-like factor 4 (KLF4) is expressed in neural stem cells and controls axonal regeneration. Its dysregulation leads to hydrocephalus in postnatal mouse brains. Here, we further show that KLF4 regulates neurogenesis and radial migration of neurons in the developing cerebral cortex. Neural progenitors with constitutive expression of KLF4 fail to migrate and develop into mature neurons but, rather, form cells with a glial identity. Notably, the JAK-STAT pathway is altered by KLF4, with increased phosphorylation of STAT3 at tyrosine 705. Blocking STAT3 activation with a dominant negative form can rescue the migration defect induced by constitutive KLF4 expression. Furthermore, downregulation of endogenous KLF4 significantly promotes radial migration and the transition of newly born migrating neurons from multipolar to bipolar morphology. Together, these results suggest that precise regulation of KLF4 expression is critical to neuronal differentiation and migration during the formation of a cerebral cortex.
INTRODUCTION
The mammalian brain is composed of numerous neurons and glia cells, all of which are derived from neural stem cells (NSCs) that reside in the ventricular zone (VZ) during neural development (22, 40). These NSCs exhibit a radial glial morphology and extend their long processes to the pial surface. Through asymmetric division, NSCs first give rise to neuronal cell types during the neurogenic phase and then to glial cells during the later gliogenic phase (22).
Newly generated cortical neurons migrate along radial glia fibers away from the VZ over substantial distances and settle in defined cortical layers. These cortical layers, mainly layers II to VI, are generated in an inside-out manner. Neurons born earlier occupy the deeper layers, whereas later-generated neurons pass through existing layers to form more superficial layers (23). The migrating neurons are highly polarized in the direction of their movement. Upon birth, they first go through a transient multipolar shape. Then, they change to a bipolar morphology with a leading process in the direction of radial migration and a trailing process (23). The molecular mechanisms regulating neuronal polarity and radial migration are still not fully understood. Interestingly, our current study indicates that Krüppel-like factor 4 (KLF4) plays a role in these processes.
KLF4 is a zinc finger-containing transcription factor that regulates multiple biological functions, including proliferation and differentiation. Germ line deletion of Klf4 results in a skin barrier defect, which leads to postnatal lethality due to severe dehydration (34). Mice with this mutation also show impaired differentiation of goblet cells in the colon (20). Depending on the cellular context, KLF4 may serve as either a tumor suppressor or an oncogene (11, 12, 14), likely by inhibiting Wnt/β-catenin signaling (9, 43) or p53 function (31). Interestingly, KLF4 plays a critical role in maintaining self-renewal of embryonic stem cells (ESCs) (16, 19, 42) and is also one of the original four factors that reprogram somatic cells into induced pluripotent stem cells (iPSCs) (39).
In the nervous system, Moore et al. reported that KLF4 acts as a transcriptional repressor of axonal growth in regenerating retinal ganglion cells (27). Previously, we showed that KLF4 is expressed in NSCs. Its dysregulation in transgenic mice leads to disrupted ventricular cilia and hydrocephalus (30). To have a better understanding of the role of KLF4 in NSCs and in their proliferation and differentiation in vivo, we carried out gain-of-function and loss-of-function studies by in utero electroporation in the developing mouse neocortex.
MATERIALS AND METHODS
Animals.
Wild-type C57BL/6 mice were purchased from the Jackson Laboratory. Wild-type ICR mice were purchased from the Harlan Laboratory. All mice were housed under a 12-h light/dark cycle and had ad libitum access to food and water in a controlled animal facility. Experimental protocols were approved by the Institutional Animal Care and Use Committee at the University of Texas (UT) Southwestern Medical Center.
Plasmids, shRNAs, and lentivirus production.
A cDNA encoding mouse KLF4 was amplified by PCR and inserted into the vector pCAG-IRES-eGFP (where IRES is internal ribosome entry site and eGFP is enhanced green fluorescent protein) or pCAG-IRES-tomato at the ClaI and XhoI restriction sites. A cDNA encoding a dominant negative form of STAT3 (dnSTAT3), in which the tyrosine residue at position 705 was mutated to phenylalanine (Y705F) by site-directed mutagenesis, was subcloned into the vector pCAG-IRES-eGFP at the SalI site. For short hairpin RNA (shRNA)-mediated knockdown experiments, two pairs of synthetic oligonucleotides were individually cloned into the pSuper vector, in which the shRNA is under the control of a human H1 promoter. The sense strands that have cloning sites for the two shRNA constructs are GAT CCC CAC CTA TAC CAA GAG TTC TCA TTT CAA GAG AAT GAG AAC TCT TGG TAT AGG TTT TTT A and GAT CCC CGC GTG AGG AAC TCT CTC ACA TTT CAA GAG AAT GTG AGA GAG TTC CTC ACG CTT TTT A. To make lentivirus, Klf4 cDNA was inserted into the lentiviral vector CS-CDF-CG-PRE, which also expresses GFP under the control of an IRES. Lentiviruses were produced as previously described (25).
In utero electroporation.
In utero electroporation was conducted according to previously published methods (32, 41). Briefly, a simple laparotomy was performed on wild-type ICR pregnant females at 14.5 days of gestation under anesthesia. While the embryos were still in the uterus, 1.5 μl of a mixture of plasmid DNA and Fast Green (final concentrations of 4 mg/ml and 2 mg/ml, respectively) was directly injected into the lateral ventricles of the embryonic forebrain by using a glass micropipette. Five electric pulses at 35 V with a duration of 50 ms per pulse at 950-ms intervals were applied through the uterus by using an electroporator (CUY21; Nepagene). During this procedure, the uterus was kept wet with warm saline. After the electroporation, the uterus was repositioned carefully into the abdominal cavity. Warm saline was filled into the cavity to replenish the abdominal fluids. The abdominal wall and the skin were separately sutured. For coelectroporation, the plasmids (6 mg/ml each) were mixed at a 1:1 ratio.
Western blot analysis and quantitative PCR (qPCR).
Cells were lysed in a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.5% sodium deoxycholate, 1% NP-40, and protease inhibitors (Roche). Protein samples were then separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore). The membranes were sequentially blotted with the corresponding primary and secondary antibodies and processed for enhanced chemiluminescence (ECL) detection (Amersham). The following primary antibodies were used: KLF4 (rabbit, 1:1,000; Santa Cruz), glial fibrillary acidic protein (GFAP; mouse, 1:2,000; Sigma-Aldrich), cyclin-dependent kinase 5 (Cdk5; rabbit, 1:1,000; Santa Cruz), Janus kinase 2 (JAK2; rabbit, 1:1,000; Cell Signaling), signal transducer and activator of transcription 3 (STAT3; mouse, 1:1,000; Cell Signaling), phospho-STAT3 (pSTAT3; rabbit, 1:1,000; Cell Signaling), phospho-glycogen synthase kinase 3α/β (pGSK3α/β; rabbit, 1:1,000; Cell Signaling), and β-actin (mouse, 1:8,000; Sigma-Aldrich).
Total RNAs were isolated from independent samples in triplicate using the TRIzol method (Invitrogen). After conversion into cDNAs using a SuperScript First-Strand cDNA Synthesis kit (Invitrogen), qPCR was carried out with an IQ SYBR green Supermix kit (Bio-Rad) and performed in a 384-well plate using an ABI 7900HT instrument. The PCR program consisted of 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 58°C. Primer quality was checked by dissociation curves. The relative expression of each gene was analyzed by the ΔΔCT method (where CT is threshold cycle) using Hprt as a housekeeping gene.
Primary NSC culture and lentiviral transduction.
Mouse NSCs were isolated from forebrains at embryonic day 13.5 (E13.5) by microdissection and physical dissociation. They were cultured as neurospheres in Dulbecco's modified Eagle's medium (DMEM)–F-12 medium supplemented with 1 mM l-glutamine, 1× N2 supplement, 20 ng/ml epidermal growth factor (EGF), and 20 ng/ml fibroblast growth factor 2 (FGF2). Neuronal or glial differentiation was initiated with, respectively, a cocktail of 5 μM forskolin (FSK) and 1 μM retinoic acid (RA) or with 50 ng/ml leukemia inhibitory factor (LIF) in the above medium lacking EGF and FGF2. For lentiviral transduction, NSCs were plated at a density of 2 × 106 cells per 6-cm dish. All cells were cultured at 37°C under a humidified atmosphere of 5% CO2.
Immunohistochemistry and imaging.
Embryonic brains were extracted at the ages indicated below and in the figure legends and fixed with 4% paraformaldehyde. Postnatal brains were extracted and fixed in 4% paraformaldehyde after transcardial perfusion. Brains were further postfixed overnight and then cryoprotected with 30% sucrose in phosphate-buffered saline (PBS) at 4°C. Coronal sections were cut at 16-μm thickness with a Cryostat (Leica) and mounted onto Superfrost Plus microscope slides (Fisher). For immunostaining, sections were washed with PBS and blocked with 5% bovine serum albumin (BSA) in PBS. After overnight incubation with primary antibodies, diluted in the blocking solution, with gentle agitation at 4°C, sections were washed and incubated with corresponding Alexa-conjugated secondary antibodies (1:1,000; Molecular Probes). Nuclei were counterstained with Hoechst 33342. The following primary antibodies were used: GFP (chick, 1:500; AVES), GFAP (mouse, 1:1,000; Sigma-Aldrich), proliferating cell nuclear antigen (PCNA; mouse, 1:200; Santa Cruz), NeuN (mouse, 1:500; Chemicon), Sox2 (rabbit, 1:500; Chemicon), phospho-STAT3 (pSTAT3; rabbit, 1:200; Cell Signaling), chondroitin sulfate proteoglycan 4 (CSPG4/NG2; rabbit, 1:500; Millipore), and glutamine synthetase (GS; mouse, 1:200; Millipore). Fluorescent images were acquired on a Zeiss LSM510 META confocal system or Olympus BX51 microscope equipped with a Hamamatsu Orca charge-coupled-device (CCD) camera. The morphology of migrating neurons in the cortex was traced by using Neurolucida, version 9.0, software (MBF Bioscience).
Statistical analysis.
Data are expressed as means ± the standard deviations. Statistical significance was determined using an unpaired Student's t test. A P value of <0.05 was considered significant.
RESULTS
Downregulation of KLF4 is essential for normal neurogenesis.
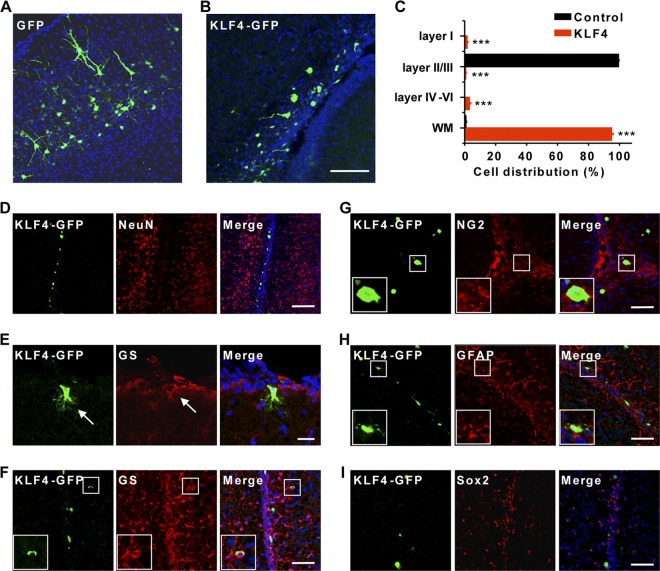
KLF4 is expressed in NSCs but drastically is downregulated in differentiated neurons (30). To investigate the role of such KLF4 downregulation during neural development in vivo, we electroporated KLF4-IRES-GFP (KLF4-GFP) or a control GFP reporter under the constitutive CAG promoter into the ventricular zone at E14.5. Cell fate was examined at postnatal day 7 (P7), which was 2 weeks postelectroporation. In the control GFP-electroporated brains, most of the labeled cells reached cortical layers II and III (II/III) and exhibited a pyramidal-like neuronal morphology with multiple dendrites and a single axon (Fig. 1A). In sharp contrast, the majority of cells with constitutive expression of KLF4 were not detected in the cortical plate but, rather, were found along the fiber tracts within the white matter (Fig. 1B). To quantify this difference, we analyzed the distribution of transfected cells in each brain region, including layer I, layers II/III, layers IV to VI, and the white matter. In control brains, 99.23% of GFP-labeled cells migrated into layers II/III with only 0.77% of cells located in the white matter. In contrast, 94.76% of KLF4-expressing cells were located in the white matter, with the remaining cells dispersed from layer I to layer VI (Fig. 1C).
Fig 1.
Neurogenesis is sensitive to constitutive expression of KLF4. (A and B) Representative coronal sections taken from P7 brains, which were electroporated at E14.5 with plasmids constitutively expressing GFP or KLF4-GFP, are shown. Most KLF4-expressing cells were identified along the corpus callosum, whereas control GFP-expressing cells migrated and settled in layers II/III. Nuclei were counterstained with Hoechst (blue). Scale bar, 100 μm. (C) Quantification of cellular distribution, including layer I, layers II/III, layers IV to VI, and white matter (WM) of cerebral cortices at P7. n = 3; ***, P < 0.001. (D to I) Immunohistochemistry analysis of KLF4-expressing cells at P7. They are neither neurons (NeuN+) (D) nor stem cells (Sox2+) (I). In contrast, they express markers for glial cells, such as GS (E and F), NG2 (G), and GFAP (H). Magnified views of cells in boxed regions are also shown. Arrows indicate a KLF4-expressing cell colabeled with the glial marker GS in layer I (E). Scale bars, 100 μm (D), 20 μm (E), and 50 μm (F to I).
The identity of the electroporated cells was analyzed by immunohistochemistry. Unlike the control GFP-expressing cells that showed neuronal morphology, none of the KLF4-expressing cells stained positive for NeuN, a marker for mature neurons (Fig. 1D). In addition, these cells had a round cell body that rarely extended any neuron-like processes. In fact, all of the KLF4-expressing cells in layer I showed glial morphology with the expression of GS, a marker for astrocytes (Fig. 1E). The cells in the white matter also showed expression of glia markers, such as GS, GFAP, and NG2, although they rarely had any processes (Fig. 1F to H). Since KLF4 is expressed in NSCs and plays a critical role in cellular reprogramming (30, 39), we examined whether constitutive expression of KLF4 kept these cells in a stem cell-like state. However, none of them stained positive for Sox2, a marker for NSCs (Fig. 1I). Therefore, we conclude that neuronal differentiation requires downregulation of endogenous KLF4.
Modulating the JAK-STAT pathway by KLF4 in neural progenitors.
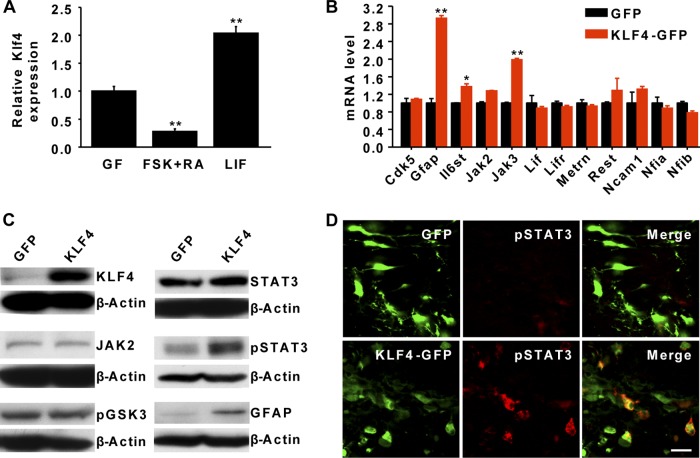
Our observation that constitutive expression of KLF4 in NSCs kept them in a glia-like fate led us to examine the role of KLF4 in gliogenic pathways. NSCs cultured in vitro can differentiate into neurons or glial cells, depending on the induction signals. Neuronal fate is induced by treatment with forskolin (FSK) and retinoic acid (RA) (33), whereas glial differentiation can be initiated by cytokines such as leukemia inhibitory factor (LIF), interleukin-6 (IL-6) family proteins, ciliary neurotrophic factor (CNTF), and cardiotrophin-1 (CT-1) (26). We first examined KLF4 expression under these culture conditions and found that it was drastically downregulated by FSK/RA treatment but enhanced by LIF signaling (Fig. 2A).
Fig 2.
Regulation of the JAK-STAT3 pathway by KLF4. (A) qPCR analysis of endogenous KLF4 expression in NSCs or under differentiation conditions. KLF4 is induced by LIF but downregulated by FSK and RA treatment. n = 3; **, P < 0.01. (B) qPCR analysis of gene expression in NSCs 36 h posttransduction with lentiviruses expressing either GFP or KLF4-GFP. n = 3; *, P < 0.05; **, P < 0.01. Lifr, leukemia inhibitory factor receptor; Metrn, meteorin; Rest, RE1-silencing transcription factor; Ncam1, neural cell adhesion molecule 1; Nfia, nuclear factor I/A; Nfib, nuclear factor I/B. (C) Western blot analysis. The expression of GFAP and pSTAT3 was induced by KLF4 in NSCs 3 days after lentiviral transduction. β-Actin was used as a loading control. (D) Confocal images taken from coronal brain sections after staining for GFP (green) and pSTAT3 (red). Brains were electroporated at E14.5 and examined at E17.5. Strong pSTAT3 staining was detected in KLF4-expressing but not control GFP-expressing cells. Scale bar, 20 μm.
One downstream target of LIF signaling is the JAK-STAT pathway, which plays a critical role during gliogenesis (4, 28). Interestingly, several genes in this pathway may also be direct targets of KLF4, based on chromatin immunoprecipitation data in ESCs (21). We examined gene expression by qPCR using RNA samples from cultured NSCs that were transduced with lentiviruses expressing either GFP or KLF4-IRES-GFP. Ectopic KLF4 further enhanced the expression of the cytokine receptor IL6ST (IL-6 signal transducer) and JAK3, a tyrosine kinase that phosphorylates and activates STATs (Fig. 2B). Indeed, phosphorylation at tyrosine 705 (Y705) of STAT3 (pSTAT3), which is a key effector of the JAK-STAT pathway during gliogenesis (4, 17), was significantly increased though STAT3 expression was not altered (Fig. 2B and C). Furthermore, the expression of GFAP, a marker for astrocytes, was increased more than 2-fold (Fig. 2B). We also examined the activation status of STAT3 in vivo by immunohistochemistry and confocal microscopy using an antibody that specifically recognizes pSTAT3. Embryonic brains were electroporated at E14.5 and examined by E17.5. pSTAT3 was hardly detectable in control GFP-electroporated cells during this neurogenic stage. In contrast, enhancing KLF4 expression induced robust activation of STAT3, indicated by strong staining of pSTAT3 (Fig. 2D). Similarly, pSTAT3 was also much enhanced in the VZ of transgenic mice, with inducible expression of KLF4 in the NSCs (30).
Radial migration in the cerebral cortex requires downregulation of KLF4.
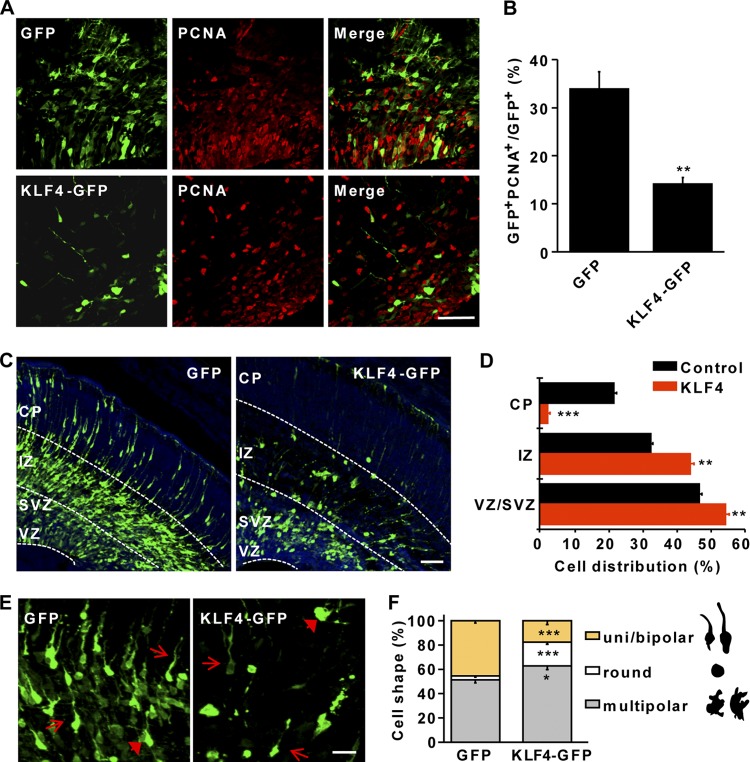
In mouse ESCs, KLF4 is a downstream target of the LIF-JAK-STAT3 pathway (16, 29). In the absence of LIF, overexpression of KLF4 is sufficient to maintain embryonic stem cell pluripotency. Our finding that KLF4 also enhances activation of STAT3 suggests that KLF4 and the JAK-STAT pathway form a feed-forward loop. Such a result suggests that ectopic KLF4 in NSCs may keep them in a constant self-renewal state. We examined this possibility by immunohistochemistry on E17.5 brain sections that were electroporated at E14.5 with a plasmid constitutively expressing KLF4. Proliferating cells were then detected by staining for endogenous expression of PCNA, a marker for cell proliferation. During this neurogenic period, one-third of control GFP-expressing cells continued to proliferate. However, only 15% of cells with ectopic KLF4 stained positive for PCNA, indicating that overexpression of KLF4 partially inhibited proliferation of neural progenitors (Fig. 3A and B). This result suggests that KLF4 plays a context-dependent role in cell proliferation even though it is still involved in the JAK-STAT pathway in cells such as ESCs.
Fig 3.
Radial neuronal migration requires downregulation of KLF4. (A and B) Constitutive KLF4 expression inhibits proliferation of neural progenitors in vivo. Brains were electroporated at E14.5 and examined at E17.5. Proliferating cells were labeled by PCNA staining (red signal in panel A) and quantified by counting PCNA+ GFP+ cells in ventricular zone/subventricular zone. n = 5; **, P < 0.01. Scale bar, 50 μm. (C and D) Radial neuronal migration is impaired by constitutive KLF4 expression. E14.5 mouse embryos were electroporated in utero with GFP or KLF4-GFP plasmids and examined at E17.5. Coronal sections were counterstained with Hoechst (blue signal). Very few KLF4-expressing cells migrated to the cortical plate (CP). n = 5; **, P < 0.01; ***, P < 0.001. Scale bar, 100 μm. (E and F) Multipolar-to-bipolar transition of migrating neurons is compromised by KLF4. Images were taken from the intermediate zone. Brains were electroporated at E14.5 and examined at E17.5. Arrows and arrowheads show cells with uni- or bipolar and multipolar morphologies, respectively. n = 5; *, P < 0.05; **, P < 0.01. Scale bar, 20 μm. IZ, intermediate zone; VZ, ventricular zone; SVZ, subventricular zone.
One striking feature of KLF4-overexpressing cells was that most of them remained in the ventricular zone/subventricular zone (VZ/SVZ), indicating a neuronal migration defect. By dividing the cortex into three regions—the VZ/SVZ, the intermediate zone (IZ), and the cortical plate (CP)—we quantified the distribution of electroporated cells. We found that there was a greater than 7-fold decrease of cells with ectopic KLF4 that migrated to the CP region compared to levels in the control GFP-electroporated cells (Fig. 3C and D). Furthermore, constitutive expression of KLF4 also resulted in more cells with a round or multipolar morphology and a much lower number of cells with bipolar processes (Fig. 3E and F). It should be noted that such a migration defect was unique to KLF4 function since no phenotype could be identified in cells overexpressing KLF5, another transcription factor belonging to the Krüppel-like family (24).
Rescuing migration defect by a dominant negative form of STAT3.
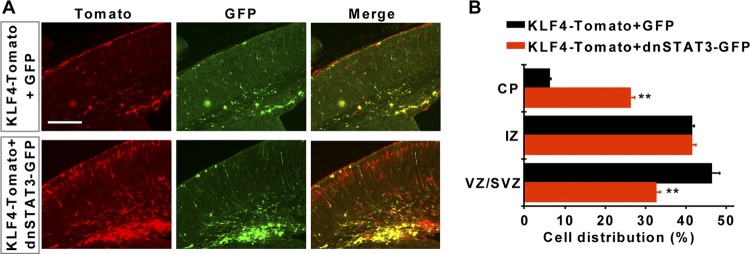
The radial migration defect of KLF4-expressing cells may be due to KLF4-induced superactivation of STAT3. To examine this possibility, we used a dominant negative form of STAT3 (dnSTAT3) in which the Y705 residue was mutated to phenylalanine, thereby preventing its phosphorylation and activation (7, 13). dnSTAT3-IRES-GFP or a control IRES-GFP was then coelectroporated with KLF4-IRES-Tomato into E14.5 forebrains. We examined distribution of electroporated cells within the cerebral cortex at E18.5. Coexpression of dnSTAT3 but not GFP significantly rescued radial migration of cells with ectopic KLF4, as indicated by strikingly more cells being localized in the cortical plate (Fig. 4A and B). This result suggests that overactivation of STAT3 indeed plays a role in the KLF4-induced radial migration defect.
Fig 4.
Dominant negative STAT3 rescues KLF4-induced migration defect. Brains were electroporated at E14.5 and examined at E18.5. Coexpression of a dominant negative form of STAT3 (dnSTAT3) with KLF4 significantly improved cellular migration (red signal) to the cortical plate (CP). IZ, intermediate zone; VZ/SVZ, ventricular zone/subventricular zone. n = 3; **, P < 0.01. Scale bar, 200 μm.
Downregulation of KLF4 enhances radial migration.
To examine the endogenous role of KLF4 in neural progenitors, we carried out knockdown experiments using short hairpin RNA (shRNA) under the control of a human H1 promoter. Two shRNAs were confirmed to effectively silence KLF4 expression by cotransfection with a Flag epitope-tagged KLF4 in HEK293 cells (Fig. 5A). We also coelectroporated these shRNAs with KLF4 into E14.5 brains. When brains were examined at E17.5, coexpression of shRNA with KLF4 resulted in significantly more cells that migrated to the cortical plate (Fig. 5B). Additionally, shRNA expression also rescued the morphological defect caused by KLF4 overexpression, with more cells showing neuronal processes. Such results indicated that these shRNAs could indeed abolish KLF4 function.
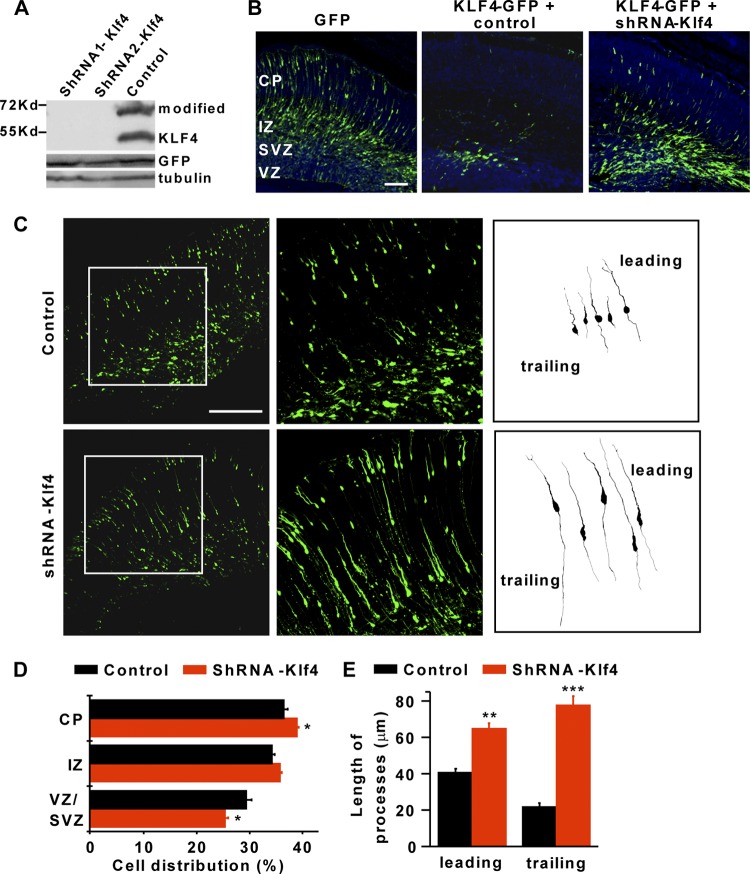
Fig 5.
Knockdown of KLF4 enhances radial migration of cortical neurons. (A) Two shRNAs against KLF4 (shRNA-Klf4) efficiently silenced the expression of transfected KLF4 in HEK293 cells by Western blot analysis. GFP and β-tubulin were used as loading controls. A modified form of KLF4 with a higher molecular mass is also indicated. (B) shRNA-Klf4 partially rescued the neuronal migration defect induced by constitutive KLF4. Brains were coelectroporated with KLF4-GFP and shRNA-Klf4 or a control at E14.5 and analyzed at E17.5. Scale bar, 100 μm. (C to E) Enhanced radial neuronal migration by knocking down endogenous KLF4. Brains were electroporated at E14.5 and analyzed at E18.5. A GFP marker driven by the constitutive CAG promoter was coelectroporated to label transfected cells. Representative traces of migrating neurons with leading and trailing processes are shown at the far right in panel C. (D and E) Quantification of regional distribution and processes of migrating neurons. CP, cortical plate; IZ, intermediate zone; VZ/SVZ, ventricular zone/subventricular zone. n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Scale bar, 200 μm.
We next performed in utero electroporation with an shRNA targeting Klf4 (shRNA-Klf4) or a control at E14.5. A coelectroporated GFP marker under the constitutive CAG promoter was used to identify transfected cells at E18.5. Consistent with a role of KLF4 in radial migration, its knockdown by shRNA led to a 7% increase of cells in the cortical plate and a corresponding decrease in the VZ/SVZ (Fig. 5C and D). Interestingly, downregulation of endogenous KLF4 by shRNA also resulted in cells with much longer leading and trailing processes (an increase of 65% and 300%, respectively) (Fig. 5E). This phenotype was specific since cells electroporated with shRNAs against KLF5 behaved similarly to control cells. Together, these results suggest that the expression level of KLF4 is critical to normal cellular behaviors during neural development.
KLF4 regulates multipolar-to-bipolar transition of migrating neurons.
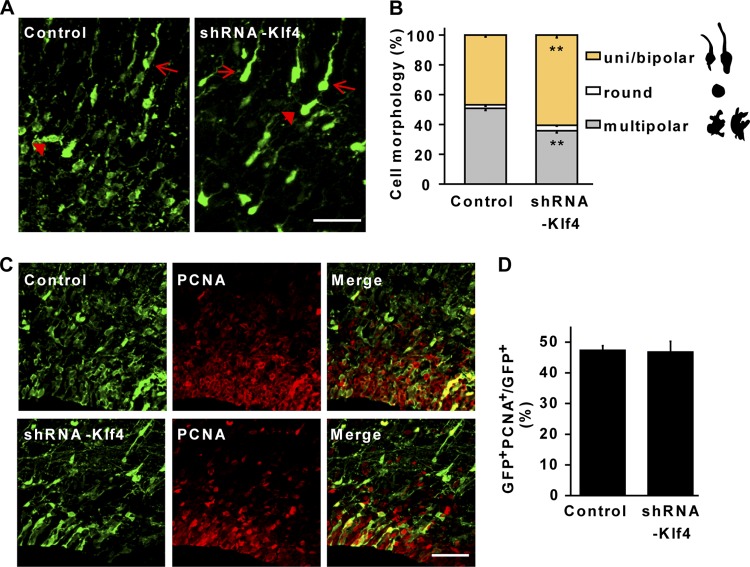
Newly born migrating neurons become transiently multipolar in the SVZ/IZ before converting to a highly polarized morphology with leading and trailing processes (37). We examined in detail the morphology of cells with KLF4 downregulation. Cells in the VZ were electroporated with shRNA-Klf4 or a control GFP and examined 4 days later. Quantitative analysis of transfected cells in the IZ showed that downregulation of KLF4 led to a 25% increase of cells becoming uni- or bipolar and a corresponding decrease of cells with a multipolar morphology (Fig. 6A and B). This early morphological transition may be caused by a role of KLF4 in cell proliferation, which was analyzed by immunohistochemistry. However, no difference was identified in the number of PCNA-positive cells in the brains with KLF4 downregulation (Fig. 6C and D). This result suggests that KLF4 has a direct role in governing the morphological change of migrating neurons.
Fig 6.
Downregulation of KLF4 promotes multipolar-to-bipolar transitions of migrating neurons. (A) Representative confocal images showing morphological changes induced by knocking down endogenous KLF4 by shRNA. Arrows and arrowheads mark cells with bipolar and multipolar morphologies, respectively. Scale bar, 40 μm. (B) Quantification of neurons with bipolar, round, or multipolar morphology in the intermediate zone. n = 3; **, P < 0.01. (C and D) Knockdown of KLF4 has no effect on cell proliferation, which was quantified by staining for PCNA. Brains were electroporated at E14.5 and examined at E18.5 (n = 3). Scale bar, 50 μm.
Knocking down KLF4 has no long-term effect on neurons.
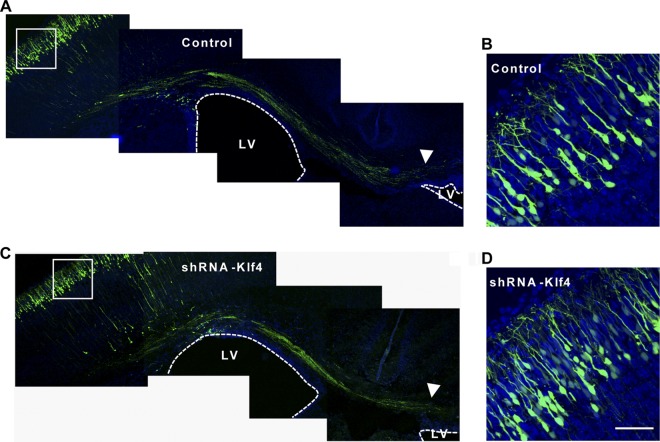
To determine the long-term effect of KLF4 downregulation on the final morphology and position of fully differentiated neurons, we performed in utero electroporation with a plasmid expressing shRNA-Klf4 or a control at E14.5 and analyzed the brains at P3. Similar to controls, KLF4-downregulated neurons were positioned at layers II/III, most of which exhibited the typical pyramidal morphology (Fig. 7). During the first postnatal week of the developing cortex, the leading process gives rise to the apical dendrite while the trailing process becomes an axon when the migrating cell body translocates to its final destination (2). We found that the apical dendrites that extended toward the pial surface, as well as the axons, showed morphologies that were similar for the KLF4-silenced neurons and their controls (Fig. 7B and D). Multiple axons formed bundles in the IZ and elongated tangentially along the corpus callosum. Some of these traveled across the midline of the brain and terminated close to the lateral ventricle (LV) of the contralateral hemisphere (Fig. 7A and C). Since KLF4 is normally downregulated in differentiated neurons, it may not be unexpected that shRNA-mediated knockdown of KLF4 did not produce long-term effects on the behavior of neurons. Alternatively, cells with KLF4 knockdown may eventually be rescued in a cell-nonautonomous manner by surrounding wild-type cells since only a small fraction of cells in the LV were initially transfected by in utero electroporation.
Fig 7.
Knocking down KLF4 has no long-term effect on differentiated neurons. Representative images of coronal brain sections, which were electroporated at E14.5 and examined at P3, are shown. A pCAG-EGFP plasmid was coelectroporated to label transfected cells. Sections were counterstained with Hoechst (blue signal). Arrowhead marks axonal bundles that crossed the midline. Images in panels B and D are higher-magnification views of boxed regions in panels A and C, respectively. Scale bar, 50 μm.
DISCUSSION
Precise cellular differentiation and migration are critical to the development of a mammalian cerebral cortex. Our studies showed that these developmental processes require downregulation of KLF4. Importantly, we provided evidence that KLF4 interacts with the JAK-STAT pathway by enhancing phosphorylation of STAT3 in a cell-autonomous manner in neural progenitors. These data add new insights into the molecular mechanism by which the behavior of NSCs and migrating neurons is transcriptionally controlled during brain development.
Increasing evidence suggests that KLF4 plays a context-dependent role in controlling cellular behavior. Overexpression of KLF4 is sufficient to maintain ESC pluripotency in the absence of LIF (16). It is also one of the key factors required for the induction of iPSCs (38). Consistent with our previous observations in transgenic mice, however, our current study using in utero electroporation showed that overexpression of KLF4 inhibits proliferation of NSCs. Such an inhibitory role of KLF4 on cell proliferation was also described under several other cellular contexts (35, 44). Mechanistically, KLF4 inhibits cell cycle progression (i) by suppressing the expression of cell cycle-promoting genes, such as cyclin D1 and cyclin B1, and (ii) by recruiting p53 to activate the expression of p21Cip1/Waf1, a cyclin-dependent kinase (CDK) inhibitor (44). It should be noted that downregulation of KLF4 in the neural system has no significant effect on cell proliferation. Such a result may be due to redundant functions of other KLF family members since several of them are highly expressed in NSCs (30).
In accordance with an inhibitory role of KLF4 in controlling axonal regeneration of cultured retinal ganglion cells or cortical neurons (3, 27), we revealed that enhanced expression of KLF4 in vivo significantly abolished neurite outgrowth and radial migration of developing neurons. Conversely, its downregulation by shRNA drastically enhanced the polarity of migrating neurons by increasing the length of leading and trailing processes. This role of KLF4 seems to oppose the function of several genes that promote neuronal migration, such as neurogenin 2 (Neurog2) (18), CDK5 (1), and semaphorin-3A (5). It will be interesting to examine whether KLF4 genetically interacts with these factors in the future. Since cytoskeletal dynamics play a critical role in neurite outgrowth and during radial neuronal migration (8), KLF4 may transcriptionally regulate the expression of genes involved in the formation of the cytoskeleton in developing neurons. Supporting this hypothesis is the finding that KLF4 directly controls keratins, a family of intermediate filaments associated with cellular differentiation and cytoskeletal organization (6). It should be noted, however, that knockdown of KLF4 in vivo has no long-term effect on the final position or morphology of mature neurons. This result indicates that developing neurons, as well as regenerating neurons after damage or in culture (27), are more sensitive than mature neurons to the reduced expression level of KLF4.
KLF4 expression is directly activated by JAK-STAT3 signaling in response to LIF treatment in ESCs (29). This pathway is important for both ESC self-renewal and maintenance of pluripotency (15, 29, 42). Similarly, our current study showed that KLF4 is induced in cultured NSCs by LIF. Interestingly, we also found that overexpression of KLF4 can further enhance activation of STAT3 by increasing its phosphorylation at Y705. However, instead of promoting self-renewal of NSCs, overexpression of KLF4 inhibits their proliferation and induces the expression of GFAP. At a later stage, cells with constitutive expression of KLF4 express markers of glial cells, such as GS, GFAP, and NG2. These observations may not be unexpected since activation of JAK-STAT3 signaling in NSCs has been previously shown to promote gliogenesis (4, 17, 28).
During early neural development, gliogenesis is suppressed by neurogenic factors such as neurogenin 1 and 2 (Neurog1 and Neurog 2). In addition to promoting neuronal differentiation, neurogenins also repress glial differentiation by inhibiting JAK-STAT3 signaling. Such inhibition is accomplished both by reducing STAT3 phosphorylation and by sequestering the CBP/p300-Smad1 complex away from STAT3 (36). By enhancing activation of STAT3, KLF4 may oppose the neurogenic functions of neurogenins. Moreover, KLF4 was shown to directly bind to cofactor CBP/p300 (10), thereby reducing its availability to neurogenins and further tipping the balance toward gliogenesis. Interestingly, emerging evidence also links neurogenesis to molecular machinery that controls migration (1). Future experiments are warranted to determine the detailed interactions between these factors and signaling pathways that govern the behavior of NSCs and migrating neurons.
ACKNOWLEDGMENTS
We thank members of the Zhang laboratory for discussions and reagents.
C.-L.Z. is a W. W. Caruth, Jr., Scholar in Biomedical Research. This work was supported by the Whitehall Foundation Award (2009-12-05), the Welch Foundation Award (I-1724), the Ellison Medical Foundation Award (AG-NS-0753-11), and NIH grants (1DP2OD006484 and R01NS070981 to C.-L.Z.).
Footnotes
Published ahead of print 20 August 2012
REFERENCES
- 1. Ayala R, Shu T, Tsai LH. 2007. Trekking across the brain: the journey of neuronal migration. Cell 128: 29– 43 [DOI] [PubMed] [Google Scholar]
- 2. Barnes AP, Polleux F. 2009. Establishment of axon-dendrite polarity in developing neurons. Annu. Rev. Neurosci. 32: 347– 381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blackmore MG, et al. 2010. High content screening of cortical neurons identifies novel regulators of axon growth. Mol. Cell Neurosci. 44: 43– 54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bonni A, et al. 1997. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science 278: 477– 483 [DOI] [PubMed] [Google Scholar]
- 5. Chen G, et al. 2008. Semaphorin-3A guides radial migration of cortical neurons during development. Nat. Neurosci. 11: 36– 44 [DOI] [PubMed] [Google Scholar]
- 6. Chen X, Whitney EM, Gao SY, Yang VW. 2003. Transcriptional profiling of Kruppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J. Mol. Biol. 326: 665– 677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Darnell JE, Jr, Kerr IM, Stark GR. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264: 1415– 1421 [DOI] [PubMed] [Google Scholar]
- 8. Dehmelt L, Halpain S. 2004. Actin and microtubules in neurite initiation: are MAPs the missing link? J. Neurobiol 58: 18– 33 [DOI] [PubMed] [Google Scholar]
- 9. Evans PM, Chen X, Zhang W, Liu C. 2010. KLF4 interacts with beta-catenin/TCF4 and blocks p300/CBP recruitment by beta-catenin. Mol. Cell. Biol. 30: 372– 381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Evans PM, et al. 2007. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J. Biol. Chem. 282: 33994– 34002 [DOI] [PubMed] [Google Scholar]
- 11. Foster KW, et al. 2000. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 60: 6488– 6495 [PubMed] [Google Scholar]
- 12. Foster KW, et al. 2005. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene 24: 1491– 1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Funamoto M, et al. 2000. Signal transducer and activator of transcription 3 is required for glycoprotein 130-mediated induction of vascular endothelial growth factor in cardiac myocytes. J. Biol. Chem. 275: 10561– 10566 [DOI] [PubMed] [Google Scholar]
- 14. Ghaleb AM, et al. 2007. Haploinsufficiency of Kruppel-like factor 4 promotes adenomatous polyposis coli dependent intestinal tumorigenesis. Cancer Res. 67: 7147– 7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo G, et al. 2009. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136: 1063– 1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hall J, et al. 2009. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell 5: 597– 609 [DOI] [PubMed] [Google Scholar]
- 17. He F, et al. 2005. A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat. Neurosci. 8: 616– 625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heng JI, et al. 2008. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature 455: 114– 118 [DOI] [PubMed] [Google Scholar]
- 19. Jiang J, et al. 2008. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 10: 353– 360 [DOI] [PubMed] [Google Scholar]
- 20. Katz JP, et al. 2002. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129: 2619– 2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim J, Chu J, Shen X, Wang J, Orkin SH. 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132: 1049– 1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kriegstein A, Alvarez-Buylla A. 2009. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32: 149– 184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marin O, Valiente M, Ge X, Tsai LH. 2010. Guiding neuronal cell migrations. Cold Spring Harb. Perspect. Biol. 2: a001834 doi:10.1101/cshperspect.a001834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol. Rev. 90: 1337– 1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mei J, Bachoo R, Zhang CL. 2011. MicroRNA-146a inhibits glioma development by targeting Notch1. Mol. Cell. Biol. 31: 3584– 3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miller FD, Gauthier AS. 2007. Timing is everything: making neurons versus glia in the developing cortex. Neuron 54: 357– 369 [DOI] [PubMed] [Google Scholar]
- 27. Moore DL, et al. 2009. KLF family members regulate intrinsic axon regeneration ability. Science 326: 298– 301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakashima K, et al. 1999. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science 284: 479– 482 [DOI] [PubMed] [Google Scholar]
- 29. Niwa H, Ogawa K, Shimosato D, Adachi K. 2009. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460: 118– 122 [DOI] [PubMed] [Google Scholar]
- 30. Qin S, Liu M, Niu W, Zhang CL. 2011. Dysregulation of Kruppel-like factor 4 during brain development leads to hydrocephalus in mice. Proc. Natl. Acad. Sci. U. S. A. 108: 21117– 21121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rowland BD, Bernards R, Peeper DS. 2005. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat. Cell Biol. 7: 1074– 1082 [DOI] [PubMed] [Google Scholar]
- 32. Saito T. 2006. In vivo electroporation in the embryonic mouse central nervous system. Nat. Protoc. 1: 1552– 1558 [DOI] [PubMed] [Google Scholar]
- 33. Schneider JW, et al. 2008. Small-molecule activation of neuronal cell fate. Nat. Chem. Biol. 4: 408– 410 [DOI] [PubMed] [Google Scholar]
- 34. Segre JA, Bauer C, Fuchs E. 1999. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 22: 356– 360 [DOI] [PubMed] [Google Scholar]
- 35. Shie JL, Chen ZY, Fu M, Pestell RG, Tseng CC. 2000. Gut-enriched Kruppel-like factor represses cyclin D1 promoter activity through Sp1 motif. Nucleic Acids Res. 28: 2969– 2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun Y, et al. 2001. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell 104: 365– 376 [DOI] [PubMed] [Google Scholar]
- 37. Tabata H, Nakajima K. 2003. Multipolar migration: the third mode of radial neuronal migration in the developing cerebral cortex. J. Neurosci. 23: 9996– 10001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takahashi K, et al. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861– 872 [DOI] [PubMed] [Google Scholar]
- 39. Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663– 676 [DOI] [PubMed] [Google Scholar]
- 40. Temple S. 2001. The development of neural stem cells. Nature 414: 112– 117 [DOI] [PubMed] [Google Scholar]
- 41. Walantus W, Castaneda D, Elias L, Kriegstein A. 2007. In utero intraventricular injection and electroporation of E15 mouse embryos. J. Vis. Exp. 6: e239 doi:10.3791/239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang P, Andrianakos R, Yang Y, Liu C, Lu W. 2010. Kruppel-like factor 4 (Klf4) prevents embryonic stem (ES) cell differentiation by regulating Nanog gene expression. J. Biol. Chem. 285: 9180– 9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang W, et al. 2006. Novel cross talk of Kruppel-like factor 4 and beta-catenin regulates normal intestinal homeostasis and tumor repression. Mol. Cell. Biol. 26: 2055– 2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang W, et al. 2000. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J. Biol. Chem. 275: 18391– 18398 [DOI] [PMC free article] [PubMed] [Google Scholar]