Abstract
The right and left nares of healthy adults (n = 251) were swabbed separately to determine carriage of Staphylococcus aureus in each nostril. Carriers were significantly more likely to carry S. aureus in one nostril than in both. Of those carrying S. aureus in both nostrils, 20% carried genetically distinct strains in each. Nostrils belonging to a single individual should not be assumed to be homogenous with respect to carriage of S. aureus.
TEXT
Staphylococcus aureus is the leading cause of nosocomial infections in the United States (8) and is increasingly associated with community-acquired infections as well (4, 15). Between 25% and 40% of healthy adults asymptomatically carry S. aureus (5, 11). A common site of S. aureus carriage in asymptomatic carriers is the anterior nares (9), and most screening protocols involve swabbing the nostrils. However, both hospital and surveillance protocols vary in their recommendations as to whether both nostrils should be sampled, and those that do recommend the sampling of both nostrils typically recommend the use of a single swab to do so. (See reference 19 for an example of a clinical protocol that swabs both nostrils using a single swab; see references 1, 2, and 6 for examples of surveillance studies doing so.) Implicit in such protocols is the idea that nostrils are homogenous with respect to the bacterial populations they carry and that it is therefore legitimate to treat two separate nostrils as if they were a single body site. This study tested that hypothesis by sampling right and left nostrils separately for S. aureus carriage.
(This work was presented in part as poster 2155 at the 111th General Meeting of the American Society for Microbiology on 24 May 2011 in New Orleans, LA.)
In this study, the right and left nostrils of healthy adult volunteers (n = 251) were swabbed separately with sterile polyester swabs (Fisher Scientific, Pittsburgh, PA), and swabs were immediately plated on mannitol salt agar (Becton, Dickinson Diagnostic Systems, Sparks, MD). Volunteers ranged in age from 18 years old to 70 years old, and 58% (n = 146) were women. Volunteers were recruited from the community via advertising in northwestern Iowa and northeastern Nebraska. Volunteers were not compensated for participation. Volunteers were excluded from this study if they reported ever having been diagnosed with a methicillin-resistant S. aureus (MRSA) infection, had been treated for any infection in the last 2 months, or were immunocompromised in any way. Sample collection began in November 2007 and continued until May 2010. Samples were defined as culture positive for S. aureus if the bacteria were mannitol-fermenting, salt-tolerant, catalase- and coagulase-positive, DNase-positive, Gram-positive staphylococci. Methicillin resistance was determined using the Kirby-Bauer technique utilizing oxacillin disks (Remel, Lenexa, KS) and Clinical and Laboratory Standards Institute (9th edition) zone of inhibition breakpoints (3). At least two isolates per study participant were tested for methicillin resistance. S. aureus cultures were preserved in 10% glycerol-90% tryptic soy broth at −70°C. This sample collection protocol and study was approved by the Human Subjects Committee of the Institutional Research Review Board at Morningside College.
In this population, 123 isolates were recovered from 88 (35%) healthy adult volunteers (n = 251) who carried S. aureus in their nostrils, broadly consistent with the findings of previous studies that have treated both nostrils as a single body site. However, 60% (n = 53) of these volunteers were culture positive for S. aureus in only one nostril. Proportions of S. aureus carriers carrying bacteria in both nostrils versus only one nostril, and proportions of single-nostril carriers carrying S. aureus in right versus left nostrils were compared using a test of population proportions, testing the hypotheses that the probability of carrying S. aureus in both nostrils was less than the probability of carrying S. aureus in one nostril and that the probability of carrying S. aureus in the left nostril was equal to the probability of carrying S. aureus in the right nostril. Nasal carriers of S. aureus were significantly more likely to carry in one nostril than in both (test of population proportions, P = 0.0015). For single-nostril carriers, neither the right nostril nor the left nostril was more likely to be an individual's site of S. aureus colonization (test of population proportions, P = 0.39). All MRSA carriers (n = 8; 3.2% of study population) carried MRSA in only one nostril, although three of these individuals carried a methicillin-susceptible S. aureus (MSSA) strain in their other nostril.
Agarose plugs containing S. aureus isolates were prepared using standard pulsed-field gel electrophoresis (PFGE) techniques (12). The plugs were digested with SmaI for 12 to 18 h at 24°C. PFGE was run for 21 h at 6.0 V/centimeter with an initial switch time of 5 min and a final switch time of 40 min on a contour-clamped homogenous electric field (CHEF) apparatus (Bio-Rad, Hercules, CA). PFGE plugs were prepared in this way for all 123 isolates in this study, and PFGE running conditions were identical for all isolates across all gels run. S. aureus NCTC 8325 was used as an internal control. Gels were stained with ethidium bromide and visualized with a GelDoc system (Bio-Rad, Hercules, CA). PFGE banding patterns were analyzed using BioNumerics software.
Of 35 dual-nostril carriers, 28 had S. aureus strains with 100% identical PFGE patterns in each nostril, and two more dual-nostril carriers had at least 90% PFGE identity (i.e., isolates varying by only one band) between the isolates in each nostril. Five dual-nostril carriers harbored strains that had less than 50% PFGE identity between the isolates in each nostril. Thus, for these five dual-nostril carriers, isolates in a single individual's right and left nostrils were more distantly related to each other than hospital-associated MRSA strain USA100 is related to community-associated MRSA strain USA300, two distinctly different lineages (18) (Fig. 1 and Table 1).
Fig 1.
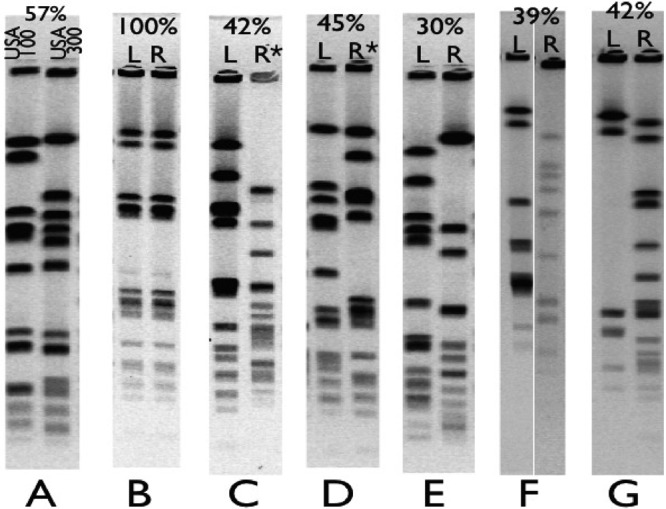
Comparison of PFGE patterns of S. aureus found in separate nostrils of the same individual. Five individuals (C through G) carried unrelated S. aureus strains in their right and left nostrils. For each pair of lanes, the percentage shows the relatedness per BioNumerics. L and R refer to whether the sample was found in the left (L) or right (R) nostril; an asterisk indicates that the isolate is methicillin resistant. (A) Reference strains USA100 and USA300 with 57% PFGE identity; (B) paired nostril isolates with 100% PFGE identity; (C) paired nostril isolates with 42% PFGE identity, where the left isolate is susceptible to methicillin and the right isolate is MRSA; (D) paired nostril isolates with 45% PFGE identity where the left isolate is susceptible to methicillin and the right isolate is MRSA; (E) paired nostril isolates with 30% PFGE identity where both isolates are MSSA; (F) paired nostril isolates with 39% PFGE identity where both isolates are MSSA; (G) paired nostril isolates with 42% PFGE identity where both isolates are MSSA.
Table 1.
Colonization of separate nostrils of healthy volunteers by distinct S. aureus strainsa
| S. aureus and colonization of nostrils | No. of volunteers (no. of isolates with characteristic) unless specified otherwise |
|---|---|
| Total S. aureus | |
| Colonized in both nostrils | |
| 100% PFGE identity between strains | 28 (56) |
| <95% PFGE identity between strains | 7 (14) |
| Colonized in only one nostril | |
| Only the right nostril colonized | 26 (26) |
| Only the left nostril colonized | 27 (27) |
| Total no. of volunteers colonized (total no. of isolates) | 88 (123) |
| MRSA (mecA positive) | |
| Colonized in both nostrils | |
| MRSA in both nostrils | 0 (0) |
| MRSA in one nostril and MSSA in the other nostril | 3 (3) |
| Colonized in only one nostril | |
| Only the right nostril colonized | 1 (1) |
| Only the left nostril colonized | 4 (4) |
| Total no. of volunteers colonized (total no. of MRSA isolates) | 8 (8) |
| pvl-positive S. aureus | |
| Colonized in both nostrils | |
| pvl-positive strain in both nostrils | 2 (4) |
| pvl-positive strain in one nostril and pvl-negative strain in the other nostril | 1 (1) |
| Colonized in only one nostril | |
| Only the right nostril colonized | 2 (2) |
| Only the left nostril colonized | 3 (3) |
| Total no. of volunteers colonized (total no. of pvl-positive isolates) | 8 (10) |
There were a total of 251 healthy volunteers in this study.
The prevalence of the Panton-Valentine leukocidin (pvl) and methicillin resistance cassette A (mecA) genes in this population was determined using the PCR technique using previously described primers (see references 10 and 14, respectively). DNA was extracted from overnight cultures of S. aureus using the Puregene yeast/bacterial kit (Qiagen, Valencia, CA). All amplifications used a Phusion PCR kit (New England BioLabs, Ipswich, MA). Analysis of the short-sequence repeat region of the protein A gene (spa typing) of the isolates was conducted as previously described using Ridom spa-type software (16, 17). For spa typing, PCR products were purified using a QiaQuick kit (Qiagen, Valencia, CA) and sequenced by the University of Iowa Carver College of Medicine DNA Core Facility.
The prevalence of pvl in this population of isolates was 8%. The prevalence of mecA in this population of isolates was 6.5%. One dual-nostril carrier harbored a pvl-positive strain in one nostril and a pvl-negative strain in the other nostril, and three dual-nostril carriers harbored a mecA-positive strain in one nostril and a mecA-negative strain in the other nostril. All MRSA isolates carried the mecA gene (Table 1). Both isolates from a single individual with identical PFGE patterns also had matching spa types, with spa type 012 being the most common (15% of isolates).
On the basis of these data, nostrils belonging to a single individual should not be assumed to be homogenous with respect to carriage of S. aureus.
This study used culture positivity to define S. aureus carriage, rather than the PCR-based techniques that are becoming more common in surveillance studies. PCR detection of S. aureus is more sensitive than culture, though less specific (7). For this reason, this study may underestimate the rate of S. aureus nasal carriage, although the overall rates of S. aureus carriage reported in this study are in agreement with those reported by others (5, 11). It is unlikely that this potential to underestimate S. aureus carriage rates in general explains the disparity in proportions of single-nostril versus dual-nostril carriers shown here or the genetic differences observed among isolates from separate nostrils in the same individual.
This study sampled each volunteer's nares only once, rather than repeatedly over time. This study therefore cannot distinguish between intermittent and persistent carriers of S. aureus (13). Future studies could examine whether dual-nostril carriers are more likely to be persistent carriers than are single-nostril carriers and whether intermittent carriers are more likely to harbor genetically distinct S. aureus strains in each nostril, as well as to help confirm the results reported here.
Researchers conducting surveillance studies and clinicians screening patients for S. aureus should be aware of the potential for a single individual's nostrils not to be homogenous in terms of bacterial carriage and should consider swabbing nostrils separately when testing for these bacteria.
ACKNOWLEDGMENTS
Funding for the genetic techniques used in this research was generously provided by the FUTURE in Biomedicine program through the University of Iowa Carver College of Medicine.
We thank the labs of Daniel Diekema and Alexander Horswill, both of the University of Iowa, for allowing use of their equipment and for technical support.
Footnotes
Published ahead of print 22 August 2012
REFERENCES
- 1.Andrews JI, Fleener DK, Messer SA, Kroeger JS, Diekema DJ. 2009. Screening for Staphylococcus aureus carriage in pregnancy: usefulness of novel sampling and culture strategies. Am. J. Obstet. Gynecol. 201:396.e1-396.e5 doi:10.1016/j.ajog.2009.06.062 [DOI] [PubMed] [Google Scholar]
- 2.Bode LGM, et al. 2010. Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N. Engl. J. Med. 362:9–17 [DOI] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute 2006. Performance standards for antimicrobial disk susceptibility tests; approved standard— 9th ed CLSI document M2-A9, vol 26, p 1 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 4.Davis SL, et al. 2007. Epidemiology and outcomes of community-associated methicillin-resistant Staphylococcus aureus infection. J. Clin. Microbiol. 45:1705–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorwitz RJ, et al. 2008. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001-2004. J. Infect. Dis. 197:1226–1234 [DOI] [PubMed] [Google Scholar]
- 6.Hamdan-Partida A, Sainz-Espunes T, Bustos-Martinez J. 2010. Characterization and persistence of Staphylococcus aureus strains isolated from the anterior nares and throats of healthy carriers in a Mexican community. J. Clin. Microbiol. 48:1701–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong L, Wu K, Fan D. 1 June 2008. Universal surveillance for MRSA: polymerase chain reaction tests vs. culture-based assays. Comment on A. Robicsek et al., Ann. Intern. Med. 148:409-418, 2008. Available from: http://www.annals.org/content/148/6/409.abstr/reply#annintmed_el_7716618347349 [Google Scholar]
- 8.Klein E, Smith DL, Laxminarayan R. 2007. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. Emerg. Infect. Dis. 13:1840–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kluytmans J, van Belkum A, Verbrugh H. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10:505–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lina G, et al. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128–1132 [DOI] [PubMed] [Google Scholar]
- 11.Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 12.McDougal LK, et al. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nouwen JL, et al. 2004. Predicting the Staphylococcus aureus nasal carrier state: derivation and validation of a “culture rule.” Clin. Infect. Dis. 39:806–811 [DOI] [PubMed] [Google Scholar]
- 14.Oliveira DC, de Lencastre H. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ray TG, Suaya JA, Baxter R. 2012. Trends and characteristics of culture-confirmed Staphylococcus aureus infections in a large U.S. integrated health care organization. J. Clin. Microbiol. 50:1950–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shopsin B, et al. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strommenger B, et al. 2006. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J. Clin. Microbiol. 44:2533–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenover FC, et al. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44:108–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.University of Iowa Department of Pathology 2012. Laboratory services handbook. Staphylococcus aureus (MRSA/MSSA) by PCR. University of Iowa Department of Pathology, Iowa City, IA: http://www.healthcare.uiowa.edu/path_handbook/handbook/test2800.html [Google Scholar]


