Abstract
PCR in bronchoalveolar lavage (BAL) fluid has not been accepted as a diagnostic criterion for invasive pulmonary aspergillosis (IPA). We conducted a systematic review assessing the diagnostic accuracy of PCR in BAL fluid with a direct comparison versus galactomannan (GM) in BAL fluid. We included prospective and retrospective cohort and case-control studies. Studies were included if they used the EORTC/MSG consensus definition criteria of IPA and assessed ≥80% of patients at risk for IPA. Two reviewers abstracted data independently. Risk of bias was assessed using QUADAS-2. Summary sensitivity and specificity values were estimated using a bivariate model and reported with a 95% confidence interval (CI). Nineteen studies published between 1993 and 2012 were included. The summary sensitivity and specificity values (CIs) for diagnosis of proven or probable IPA were 90.2% (77.2 to 96.1%) and 96.4% (93.3 to 98.1%), respectively. In nine cohort studies strictly adherent to the 2002 or 2008 EORTC/MSG criteria for reference standard definitions, the summary sensitivity and specificity values (CIs) were 77.2% (62 to 87.6%) and 93.5% (90.6 to 95.6%), respectively. Antifungal treatment before bronchoscopy significantly reduced sensitivity. The diagnostic performance of PCR was similar to that of GM in BAL fluid using an optical density index cutoff of 0.5. If either PCR or GM in BAL fluid defined a positive result, the pooled sensitivity was higher than that of GM alone, with similar specificity. We conclude that the diagnostic performance of PCR in BAL fluid is good and comparable to that of GM in BAL fluid. Performing both tests results in optimal sensitivity with no loss of specificity. Results are dependent on the reference standard definitions.
INTRODUCTION
Invasive aspergillosis (IA) is an opportunistic fungal infection, affecting mainly patients with severe and prolonged neutropenia (3). Early diagnosis of invasive pulmonary aspergillosis (IPA) is notoriously difficult, and few diagnostic tools are available (16). Between 1978 and 1992, the percentage of patients in whom postmortem-proven IPA was suspected or confirmed antemortem improved only from 9% to 32% (17). Histological confirmation is difficult to obtain in patients at risk for IA due to concomitant severe thrombocytopenia. Efforts should be made to facilitate the diagnosis, as early therapy probably reduces mortality (9, 43).
PCR in bronchoalveolar lavage (BAL) fluid has not been incorporated into the diagnostic criteria of IPA because of the lack of standardization of extraction and amplification protocols, differences in DNA targets, and differences in the methods used to examine the amplified products (5, 11). We systematically reviewed all studies assessing PCR in BAL fluid for the diagnosis of IPA. We also compared and assessed the value of PCR compared to and combined with galactomannan (GM) in BAL fluid.
(Presented in part at the 22nd European Congress of Clinical Microbiology and Infectious Diseases, London, United Kingdom, 2012.)
MATERIALS AND METHODS
Inclusion criteria.
We included prospective and retrospective cohort studies and case-control studies. Participants (both cases and controls) were patients at risk for IA as defined by the host criteria in the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) 2002 definitions for IA (3). Thus, case-control studies in which controls were healthy people or people otherwise not at risk for IA were excluded. In studies that also included patients not at risk for IA, we extracted data only for patients at risk. If this was not possible, we excluded studies with >20% of patients who did not have host criteria. We excluded patients in whom HIV was the only underlying immune deficiency, since this is not the patient population of interest in our review.
The index test was PCR for Aspergillus spp. performed on BAL fluid or bronchial secretions. Any PCR test, including standard, real-time, nested, multiplex, or other PCR and targeting all or specific Aspergillus spp., was acceptable. We used the sample taken at the time closest to the onset of infection. When reported, we used only PCR tests taken within 14 days of infection onset and excluded further samples. We also extracted data on GM in BAL fluid in studies reporting both tests. The optical density index (ODI) cutoff values to define GM positivity were documented. The target condition was IPA. In the primary analysis, we considered only proven and probable IPA (cases classified as possible IPA were excluded). In a secondary analysis, we also considered possible IPA as a disease. The reference standard was the consensus definition of IPA of the EORTC/MSG Consensus Group. We accepted the definitions published in 2002 and 2008 (3, 11). We accepted studies conducted before the publication of these guidelines but using similar criteria (2, 14). We documented the adherence of the study methods to the definitions of the reference standard used in the study and assessed its effect on results. In studies that provided a full description of all patients with suspected IA, we reapplied the 2008 definitions if possible.
Search strategy.
We searched PubMed with the phrase “(bronchoalveolar lavage OR BAL OR sputum OR respiratory) AND (PCR OR real-time OR RT-PCR OR nested-PCR OR PCR) AND (aspergil*).” Search updates were reviewed until June 2012. In addition, we searched the European Congress of Clinical Microbiology and Infectious Diseases and Interscience Conference on Antimicrobial Agents and Chemotherapy between the years 2004 and 2011 using only the key words for aspergillus or aspergillosis. We scanned the references of all included studies and reviews cited in included studies. No language, date, or publication restrictions were used.
Data collection and risk of bias assessment.
Two reviewers independently selected studies for inclusion and extracted all data from the studies. A risk-of-bias assessment was conducted using the QUADAS-2 tool (45). The tool tailored for our review is described in Table S1 in the supplemental material.
Data analysis.
We assessed the sensitivity, specificity, and diagnostic odds ratio (DOR) of PCR in BAL fluid. We compared the test performance of GM and PCR and that of GM alone to that in the situation in which either PCR or GM positivity defines a positive test result. To avoid incorporation bias, we excluded from this analysis patients in whom the microbiological criterion to define IPA was the GM test (including only proven or culture-positive IPA cases). GM samples taken together with PCR or within 14 days of infection onset were used. Only direct test comparisons (paired analysis) were performed. Studies were included only once in the analysis (except when comparing PCR methods), selecting the more advanced PCR method. We used the bivariate model for data summary. Parameter estimates from the model were used to obtain hierarchical summary receiver operating curves (HSROC).
The main covariates assessed were the quality of the reference standard in the study (classified as fully adherent to the 2002 or 2008 EORTC/MSG criteria versus incomplete [i.e., not fully adherent] [studies to which the EORTC/MCG criteria could be reapplied by using individual patient data reported in them were classified as fully adherent]), the antimold antifungal treatment at the time of BAL (segregated by studies in which 50% or more of the patients were treated versus studies in which less than 50% of the patients were treated), the timing of PCR testing after onset of infection (segregated by more than 14 days versus within 14 days), and type of PCR (nested and real-time versus other). Other covariates examined included the study design, inclusion criteria (patients with host factor alone or host and clinical criteria at the time of BAL), use of the 18S rRNA primer versus other primers, other PCR method characteristics, the Aspergillus spp. targeted by the PCR (all versus selective), and the QUADAS-2 domains (45). Covariates were included in the bivariate model, and the P value for the difference in the likelihood ratio tests for the model with and without the covariate is reported.
Analyses were conducted using SAS 9.2 (SAS Institute Inc., Cary, NC) and Review Manager 5.1 (10).
RESULTS
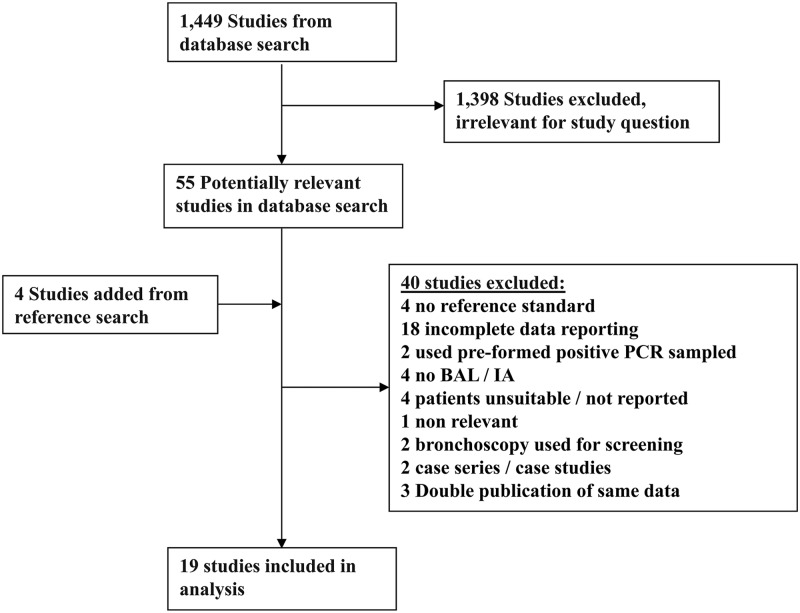
The search identified 1,449 references, of which 55 were selected for full-text review (Fig. 1). Forty studies were excluded (see Table S2 in the supplemental material), and four were added through reference searching. A total of 19 studies published between 1993 and 2011 were included (6, 7, 15, 19–22, 24, 27–33, 38, 40, 42). All studies reported on the diagnostic accuracy of PCR in BAL fluid, and 10 studies also reported on GM in BAL fluid (15, 19, 24, 27, 28, 30, 31, 32, 33, 40).
Fig 1.
Study flow chart.
Altogether, 1,585 patients at risk for IA were included, of whom 319 (20.1%) had proven/probable IPA and 169 (10.5%) had possible IPA. The median percentages of patients with hematological malignancy and hematopoietic stem cell transplantation (HSCT) were 75% and 23%, respectively. BAL cultures were positive in 180/319 (56.4%) of patients with proven/probable IPA. Eight studies reported the number of patients treated with antimold antifungals at the time of bronchoscopy, and in three of these, more than 50% of patients were treated. Other study characteristics are presented in Table S3 in the supplemental material.
The QUADAS-2 risk-of-bias assessment and applicability criteria are shown in Table S4 in the supplemental material. Only three were case-control studies, and including these, nine studies were at high risk of bias regarding patient selection. Concerns regarding classification, interpretation, and applicability of the reference standard were present in 11 studies. The interval between suspicion of IPA and performance of BAL was reported as 14 days or fewer in six studies.
Details of the PCR techniques are presented in Table S5 in the supplemental material. Standard PCR was used in six studies (with or without enzyme-linked immunosorbent assay [ELISA]), real-time PCR was used in 11, and nested PCR was used in four (one study tested both real-time and nested PCR [32]). Twelve studies used primers targeting the 18S rRNA gene, five used mitochondrial DNA primers, and three used the alkaline protease gene (one study used both mitochondrial DNA and alkaline protease genes [29]). DNA extraction was usually performed by the use of either a QIAamp kit (n = 6) or phenol-chloroform protocols (n = 9). An internal/inhibition control and a contamination control were each reported in 15 studies.
Performance of PCR.
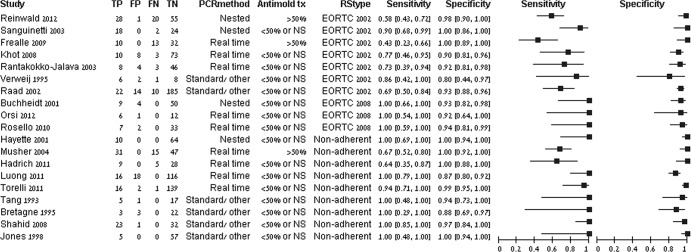
The coupled forest plots for PCR to diagnose proven/probable IPA are presented in Fig. 2. Specificity was uniformly high. Sensitivity was more variable. By visual inspection, results were affected by the reference standard and by use of antifungal treatment at the time of bronchoscopy.
Fig 2.
Sensitivity and specificity of PCR for the detection of proven or probable IPA. The reference standard (RStype) was classified as the 2002 or 2008 EORTC/MSG criteria (3, 11) or as nonadherent to both definitions. Antimold treatment at the time of bronchoscopy (Antimold tx) was classified as >50% of patients or less than 50% or not stated (NS). Studies are sorted by reference standard type and PCR method. Values in brackets for sensitivity and specificity are 95% CIs. TP, true positive; FP, false positive; FN, false negative; TN, true negative.
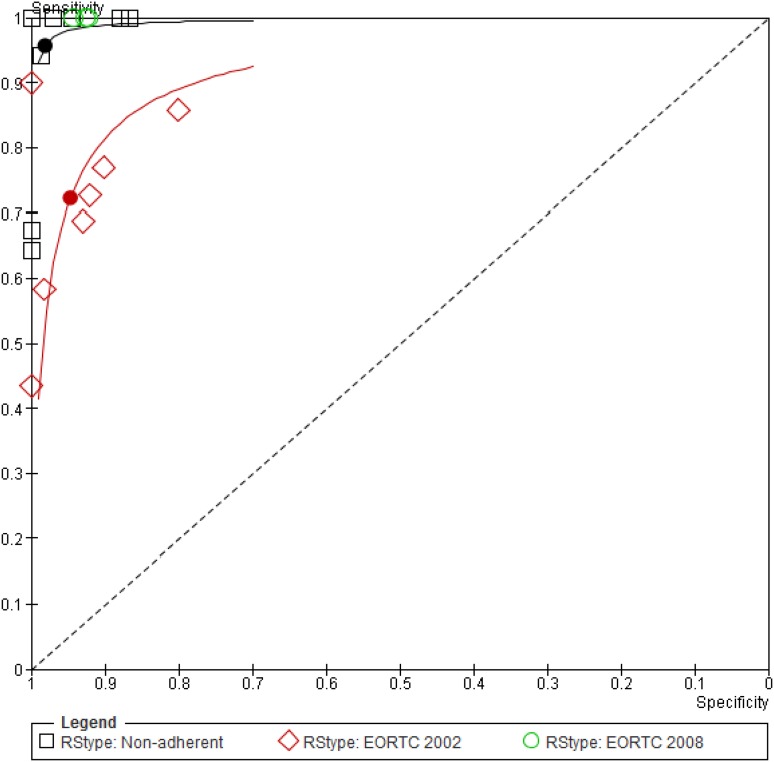
The summary sensitivity and specificity values of the bivariate model were 90.2% (95% CI, 77.2 to 96.1%) and 96.4% (95% CI, 93.3 to 98.1%), respectively, in 19 studies (see Fig. S1 in the supplemental material). The DOR was 243 (95% CI, 81 to 726). Examination of the main covariates showed that results were affected by the reference standard definition. Three studies used the 2008 EORTC/MSG criteria, and all had 100% sensitivity and high specificity (92 to 94%). Studies using and adhering to the 2002 EORTC/MSG criteria had significantly lower sensitivity than studies classified as nonadherent to both sets of criteria (P = 0.003) (Fig. 3 and Table 1). Similar results were observed when assessing the QUADAS-2 reference standard risk-of-bias item. In the three studies reporting the use of mold-active antifungals by more than 50% of patients undergoing BAL, the sensitivity (58%; 95% CI, 44 to 70.9%) was significantly lower than that in other studies (93.1%; 95% CI, 82.4 to 97.5%) (P = 0.002). There were no statistically significant differences in the performance of real-time, nested, and other PCRs. Visual inspection of SROC by the levels of other covariates did not show an effect of other study methods, the PCR amplification method, the inclusion criteria, or the probability of IA in the study. In a subset of cohort studies, adhering to acceptable reference standard definitions (2002 or 2008 criteria), the pooled sensitivity was 77.2% (95% CI, 62 to 87.6%) while specificity remained high (Table 1).
Fig 3.
HSROC for PCR in the diagnosis of proven or probable IPA by reference standard. Nineteen studies are shown in the ROC space. HSROC and summary points are shown for 7 studies adhering to the 2002 EORTC/MSG definitions (red) with relatively low sensitivity, 3 studies adhering to the 2008 definition (green), all with 100% sensitivity and specificity ranging between 92 to 94%, and 9 nonadherent studies (black) with excellent sensitivity and specificity (see values in Table 1). Nonadherent studies did not strictly adhere to the EORTC/MSG 2002 or 2008 definition (3, 11).
Table 1.
PCR in BAL fluid for the diagnosis of proven/probable IPA
| Study subset (no. of studies) | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | DOR (95% CI) | NPV/PPV (%) for IPA prevalence ofa: |
|
|---|---|---|---|---|---|
| 15% | 10% | ||||
| All (19) | 90.2 (77.2–96.1) | 96.4 (93.3–98.1) | 243 (81–726) | 97.7/81.6 | 97.8/73.6 |
| 2008 criteria, adherent (3) | 100 | 92–94 | Not assessedb | ||
| 2002 criteria, adherent (7) | 72.2 (51.5–86.5) | 94.7 (84.1–98.4) | 46 (22–96) | 93.5/70.6 | 93.9/60.2 |
| Nonadherent (9) | 95.7 (79.3–99.2) | 98.1 (94.3–99.4) | 1,157 (222–6,022) | 99/89.9 | 99/84.8 |
| Adherent, cohort studies (9) | 77.2 (62–87.6) | 93.5 (90.6–95.6) | 49 (24–97) | 94.6/67.7 | 94.9/56.9 |
| Antimold antifungals (3) | 58 (44–70.9) | 93.1 (82.4–97.5) | 702 (6–79,323) | 90.4/59.7 | 90.9/48.3 |
NPV, negative predictive value; PPV, positive predictive value. NPVs and PPVs were calculated for baseline prevalences of IPA of 15% and 10%. In cohort studies included in the current review, the median prevalence of IPA among high-risk patients undergoing BAL was 15%.
The bivariate model was not able to be calculated for the 3 studies using the 2008 EORTC/MSG criteria. All reported 100% sensitivity, and the specificity ranged between 92 and 94%.
Only seven studies reported on diagnostic performance when proven/probable and possible IPA per EORTC/MSG definitions were considered cases. There was large heterogeneity in sensitivity, and the summary sensitivity and specificity values of the bivariate model were 80.1% (95% CI, 50.6 to 94%) and 94.5% (95% CI, 88.4 to 97.5%), respectively.
PCR versus GM.
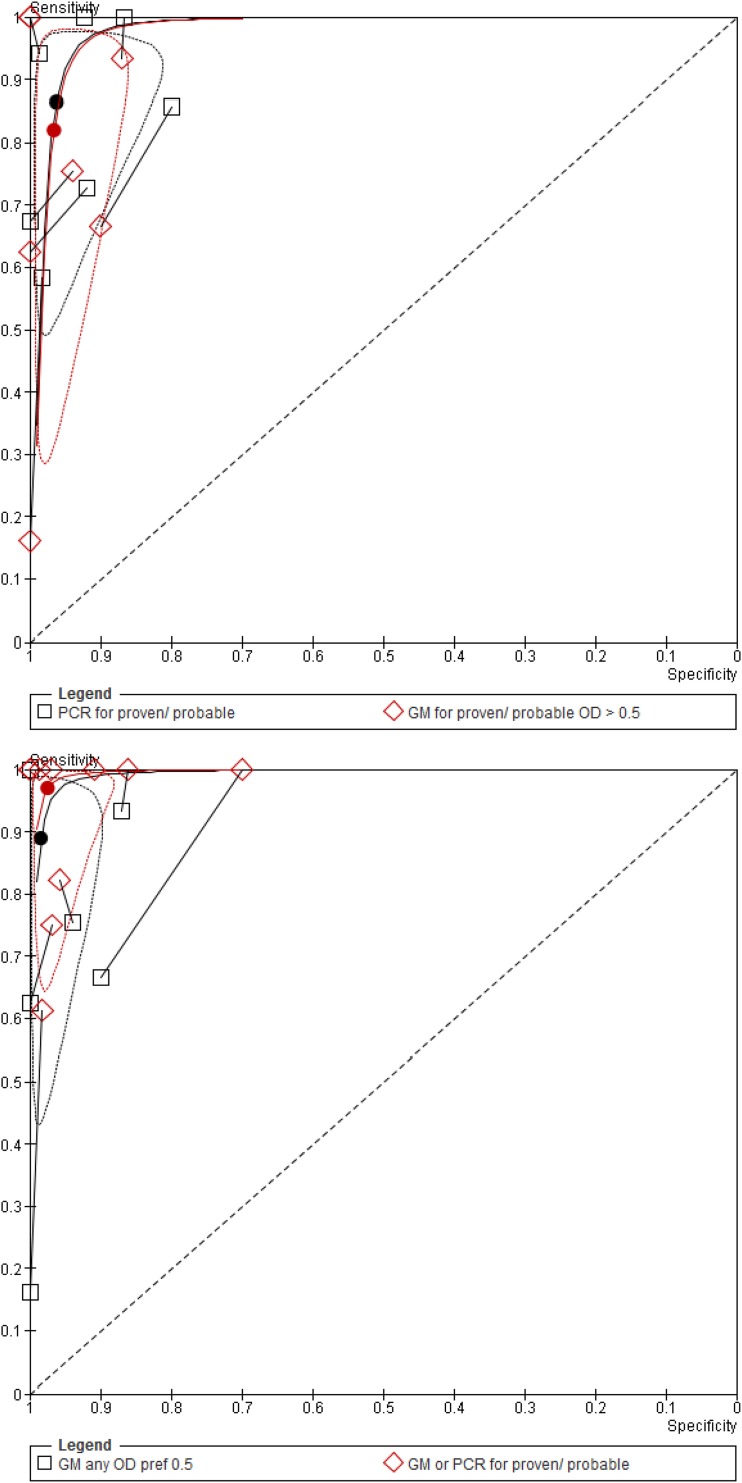
In the direct comparison of PCR and GM, PCR in BAL fluid had higher sensitivity than GM with no loss of specificity. The difference was not statistically significant for the comparison with a GM ODI cutoff of 0.5 and was statistically significant for the comparison with a GM ODI cutoff of 1.0 (Table 2 and Fig. 4). When both tests were performed in BAL fluid and the result was considered positive if any test was positive or negative if both were negative, the pooled sensitivity was 97% (95% CI, 83 to 99.5%), significantly better than that of GM alone in this set of studies (89% [95% CI, 63 to 97.5%]), with no loss in specificity (P = 0.001) (Table 2). In absolute numbers, this improved sensitivity corresponded to 21/150 (14%) of patients with IPA who had a positive PCR in BAL fluid with a negative GM (10 studies).
Table 2.
Direct comparisons of PCR and GM
| Comparison (no. of studies) | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | DOR (95% CI) | P value |
|---|---|---|---|---|
| PCR vs GM with an ODI of >0.5 (7) | 0.088 | |||
| PCR | 86.4 (68.2–95) | 96.2 (90.2–98.6) | 163 (47–560) | |
| GM | 82 (52.7–94.9) | 96.6 (92.2–98.6) | 129 (32–516) | |
| PCR vs GM with an ODI of >1.0 (7) | 0.01 | |||
| PCR | 92.6 (69.9–98.5) | 97.7 (92.6–99.3) | 516 (82–3,248) | |
| GM | 85.1 (62.5–95.1) | 99.7 (97.3–100) | 1,731 (202–14,802) | |
| Any positive result vs GM (10)a | 0.001 | |||
| GM or PCR | 97 (83–99.5) | 97.5 (92.9–99.1) | 1,258 (155–10,215) | |
| GM | 89 (63–97.5) | 98.5 (94.5–99.6) | 516 (74–3,611) |
Comparison between a positive result defined by either PCR or GM positivity when both tests are performed and GM alone. Paired analysis was conducted with all studies reporting on GM in BAL fluid preferably using the results obtained with an ODI cutoff of >0.5.
Fig 4.
Paired comparisons of PCR and GM in BAL fluid. (top) PCR compared to GM with an ODI cutoff of 0.5 in BAL fluid. (bottom) PCR and GM versus GM alone.
DISCUSSION
We examined the accuracy of PCR alone and in comparison with GM in BAL fluid for the diagnosis of IPA among patients at risk for IA. Overall, in 19 included studies, the summary sensitivity and specificity values for the diagnosis of proven/probable IPA were 90.2% and 96.4%. Results were affected by the reference standard used and by adherence to the diagnostic criteria. Studies adhering to the EORTC/MSG 2002 criteria (3) had significantly lower sensitivity than studies classified as nonadherent to both sets of criteria. Three newer studies adhering to the 2008 EORTC/MSG (11) criteria had very high sensitivity and specificity. The QUADAS-2 reference standard risk of bias similarly affected results. In a subset of cohort studies adhering to acceptable reference standard definitions, the summary sensitivity and specificity estimates were 77.2% and 93.5%, respectively. Use of antifungal treatment before BAL reduced the sensitivity of PCR in BAL fluid. In 10 studies reporting on both PCR and GM, the diagnostic performance of PCR in BAL fluid was nonsignificantly better than that of GM in BAL fluid using an ODI cutoff of 0.5 on account of improved sensitivity. When PCR was performed in addition to GM in BAL fluid, the sensitivity of the PCR significantly improved from 89% with GM alone to 97% with GM and PCR, with no loss of specificity.
Using the pooled sensitivity and specificity estimates of our review, the negative and positive predictive values (NPVs and PPVs, respectively) of the test can be calculated using a defined prevalence of disease (1). NPVs for PCR in BAL fluid were high in all analyses, usually above 95%; PPVs ranged between 60 and 80% (Table 1). The values for a GM ODI cutoff of 0.5 in BAL fluid were similar. When both PCR and GM were performed in BAL fluid and either positive result defined a positive test, the NPV was 99% and PPVs were 87% and 81% for disease prevalences of 15% and 10%, respectively. Thus, a negative PCR or GM alone excludes the diagnosis of IPA with a very high probability (≥98%). Performing both tests slightly increases the probability of diagnosing an infection without affecting specificity.
Leeflang et al. reviewed the accuracy of GM antigen in blood of patients with IPA in 30 studies (23). With an ODI cutoff of 0.5, the summary sensitivity and specificity values were 78% (95% CI, 61 to 89%) and 81% (95% CI, 72 to 88%), respectively. Results were sensitive to the ODI cutoff used and the patient population assessed (sensitivity was lower among patients with unresponsive fever alone). Similar to our review, the use of strict 2002 EORTC criteria as a reference standard was associated with lower sensitivity. Prior antifungal therapy was associated with higher GM sensitivity and lower specificity. Mengoli et al. pooled results from 16 prospective studies that studied the diagnostic value of PCR in whole blood and serum for the diagnosis of IPA (26). Sensitivity and specificity were 75% (95% CI, 54 to 88%) and 87% (95% CI, 78 to 0.93%), respectively, if two consecutive positive samples were required to define positivity. The results for a single test were 88% (95% CI, 75 to 94%) and 75% (95% CI, 63 to 84%), respectively, and other covariates could not be assessed due to the paucity of studies. Thus, although useful as a screening test, blood samples have limited ability to rule in IPA. The evidence from our review and a previous review summarizing all studies that evaluated GM in BAL fluid (18) shows that BAL fluid testing increases diagnostic performance over that of blood testing.
One of the main criticisms against the use of PCR as a criterion in the diagnosis of IPA is the lack of standardization in performance and reporting of the PCR methods (8). In 2010, the European Aspergillus PCR Initiative (EAPCRI) reported on performance and results of PCR in blood in different European centers and provided recommendations based on the results observed (44). The main conclusion was that results are dependent on the extraction procedure rather than the PCR amplification. General recommendations included the use of a large sample volume (>3 ml for blood), bead beating for fungal cell lysis, use of negative and positive extraction controls and an internal control, and the use of DNA elution volumes smaller than 100 μl. These recommendations refer to blood sampling and are mostly nonrelevant to performance of PCR in BAL fluid. Similar guidance is needed to standardize PCR for Aspergillus spp. in BAL fluid. Variable methods of sampling, extraction, and amplification protocols were used in the studies included in our review (see Table S4 in the supplemental material). We did not observe an effect of the type of PCR or other PCR methods on results. However, this is a complex matrix of variables, and the assessment of each separately is probably insufficient. The number of studies included in our review was too small, and reporting was insufficient to assess individually and in combination the large number of variables relating to PCR methods.
The main problem of studies on the diagnosis of IPA is the imperfect reference standard (4, 16, 17, 39). Indeed, adherence to reference standard definitions was the main variable underlying heterogeneity in our analysis. Although we attempted to include only studies using the EORTC/MSG criteria as a reference standard, we found that a large percentage of studies did not perfectly adhere to these criteria, even when declaring their use as a reference standard. Examples included considering PCR in BAL fluid as diagnostic of IPA (incorporation bias that exaggerates sensitivity, as observed in our review), nonavailability of GM testing, mandating positive sputum cultures for proven IPA diagnoses, misclassification of possible IA as probable, or addition of clinical criteria (response to therapy, survival) to the diagnostic algorithm. Whether the overall results or the results of the adherent studies are more relevant is debatable, because the existing reference standard is imperfect. We therefore present both results. Our analysis regarding previous antimold treatment was limited by the paucity of studies reporting these data. The number of patients that were treated with antifungals prior to BAL as treatment or prophylaxis is probably much higher than that reported in our review. Results were heterogeneous, and the paucity of studies did not allow for assessment of multiple covariates. Our systematic review adds to previous systematic reviews (37, 41) since we included six new studies published after 2009 (19, 24, 28, 31, 40) (Roselló et al., presented at the European Congress of Clinical Microbiology and Infectious Diseases, 2011), we addressed the clinically relevant situation in which GM testing in BAL fluid is available, and heterogeneity was investigated using the recommended methodology (12). We excluded studies assessing healthy patients or patients that are otherwise not at risk for IA or that were not able to be separated from patients at risk (13, 34), studies with PCR sampling of pretested positive samples (35), and studies with an invalid reference standard for which we were not able to apply consensus definitions (25, 36).
In summary, we show good sensitivity and specificity of PCR in BAL fluid for the detection of IPA in patients at risk. The NPV of the test, given a plausible range of disease prevalence and subgroup analyses, was consistently 95% or higher. PPVs were lower, ranging between 60 and 80%. The diagnostic performance of PCR in BAL fluid was comparable to that of GM in BAL fluid, and the addition of PCR to GM resulted in optimal sensitivity and PPV values without a significant loss in specificity. BAL without biopsy can be safely performed in most patients at risk for IA. These results favor the use of PCR in BAL fluid for patients with suspected IPA and point to the combination of PCR and GM as the optimal testing strategy. Standards for PCR performance in BAL fluid are needed.
Supplementary Material
ACKNOWLEDGMENTS
All authors declare no conflict of interest.
This study was supported in part by a grant from the Rothschild-Caesarea Foundation, Optimal antibiotic treatment of moderate to severe infections: integration of PCR technology and medical informatics/artificial intelligence.
Footnotes
Published ahead of print 5 September 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Altman DG, Bland JM. 1994. Diagnostic tests 2: predictive values. BMJ 309:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ascioglu M, De Pauw B, Bennett JE. Analysis of definitions used in clinical research on invasive fungal infections (IFI): consensus proposal for new, standardized definitions, abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., San Francisco, CA.: 1999. p. 1639. [Google Scholar]
- 3.Ascioglu S, et al. 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect. Dis. 34:7–14 [DOI] [PubMed] [Google Scholar]
- 4.Bodey G, et al. 1992. Fungal infections in cancer patients: an international autopsy survey. Eur. J. Clin. Microbiol. Infect. Dis. 11:99–109 [DOI] [PubMed] [Google Scholar]
- 5.Bretagne S, Costa JM. 2005. Towards a molecular diagnosis of invasive aspergillosis and disseminated candidosis. FEMS Immunol. Med. Microbiol. 45:361–368 [DOI] [PubMed] [Google Scholar]
- 6.Bretagne S, et al. 1995. Detection of Aspergillus species DNA in bronchoalveolar lavage samples by competitive PCR. J. Clin. Microbiol. 33:1164–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchheidt D, et al. 2001. Detection of Aspergillus species in blood and bronchoalveolar lavage samples from immunocompromised patients by means of 2-step polymerase chain reaction: clinical results. Clin. Infect. Dis. 33:428–435 [DOI] [PubMed] [Google Scholar]
- 8.Bustin SA, et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622 [DOI] [PubMed] [Google Scholar]
- 9.Caillot D, et al. 1997. Improved management of invasive pulmonary aspergillosis in neutropenic patients using early thoracic computed tomographic scan and surgery. J. Clin. Oncol. 15:139–147 [DOI] [PubMed] [Google Scholar]
- 10.The Cochrane Collaboration 2011. Review Manager (RevMan) version 5.1. The Nordic Cochrane Centre, The Cochrane Collaboration, Copenhagen, Denmark [Google Scholar]
- 11.De Pauw B, et al. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 46:1813–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diagnostic Test Accuracy Working Group Handbook for DTA Reviews. The Cochrane Collaboration; http://srdta.cochrane.org/handbook-dta-reviews Accessed 5 September 2012 [Google Scholar]
- 13.Einsele H, et al. 1998. Prediction of invasive pulmonary aspergillosis from colonisation of lower respiratory tract before marrow transplantation. Lancet 352:1443. [DOI] [PubMed] [Google Scholar]
- 14.EORTC International Antimicrobial Therapy Cooperative Group 1989. Empiric antifungal therapy in febrile granulocytopenic patients. Am. J. Med. 86:668–672 [DOI] [PubMed] [Google Scholar]
- 15.Frealle E, et al. 2009. Diagnosis of invasive aspergillosis using bronchoalveolar lavage in haematology patients: influence of bronchoalveolar lavage human DNA content on real-time PCR performance. Eur. J. Clin. Microbiol. Infect. Dis. 28:223–232 [DOI] [PubMed] [Google Scholar]
- 16.Girmenia C, et al. 27 September 2011, posting date. New category of probable invasive pulmonary aspergillosis in haematological patients. Clin. Microbiol. Infect. doi:10.1111/j.1469-0691.2011.03685.x. [DOI] [PubMed] [Google Scholar]
- 17.Groll AH, et al. 1996. Trends in the postmortem epidemiology of invasive fungal infections at a university hospital. J. Infect. 33:23–32 [DOI] [PubMed] [Google Scholar]
- 18.Guo YL, et al. 2010. Accuracy of BAL galactomannan in diagnosing invasive aspergillosis: a bivariate metaanalysis and systematic review. Chest 138:817–824 [DOI] [PubMed] [Google Scholar]
- 19.Hadrich I, et al. 2011. Comparison of PCR-ELISA and real-time PCR for invasive aspergillosis diagnosis in patients with hematological malignancies. Med. Mycol. 49:489–494 [DOI] [PubMed] [Google Scholar]
- 20.Hayette MP, et al. 2001. Detection of Aspergillus species DNA by PCR in bronchoalveolar lavage fluid. J. Clin. Microbiol. 39:2338–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones ME, et al. 1998. PCR-ELISA for the early diagnosis of invasive pulmonary aspergillus infection in neutropenic patients. J. Clin. Pathol. 51:652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khot PD, Ko DL, Hackman RC, Fredricks DN. 2008. Development and optimization of quantitative PCR for the diagnosis of invasive aspergillosis with bronchoalveolar lavage fluid. BMC Infect. Dis. 8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leeflang MM, et al. 2008. Galactomannan detection for invasive aspergillosis in immunocompromized patients. Cochrane Database Syst. Rev. 2008:CD007394 doi:10.1002/14651858.CD007394 [DOI] [PubMed] [Google Scholar]
- 24.Luong ML, et al. 2011. Comparison of an Aspergillus real-time polymerase chain reaction assay with galactomannan testing of bronchoalveolar lavage fluid for the diagnosis of invasive pulmonary aspergillosis in lung transplant recipients. Clin. Infect. Dis. 52:1218–1226 [DOI] [PubMed] [Google Scholar]
- 25.Melchers WJ, et al. 1994. General primer-mediated PCR for detection of Aspergillus species. J. Clin. Microbiol. 32:1710–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mengoli C, Cruciani M, Barnes RA, Loeffler J, Donnelly JP. 2009. Use of PCR for diagnosis of invasive aspergillosis: systematic review and meta-analysis. Lancet Infect. Dis. 9:89–96 [DOI] [PubMed] [Google Scholar]
- 27.Musher B, et al. 2004. Aspergillus galactomannan enzyme immunoassay and quantitative PCR for diagnosis of invasive aspergillosis with bronchoalveolar lavage fluid. J. Clin. Microbiol. 42:5517–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orsi CF, et al. 2012. Performance of 2 commercial real-time polymerase chain reaction assays for the detection of Aspergillus and pneumocystis DNA in bronchoalveolar lavage fluid samples from critical care patients. Diagn. Microbiol. Infect. Dis. 73:138–143 [DOI] [PubMed] [Google Scholar]
- 29.Raad I, et al. 2002. Diagnosis of invasive pulmonary aspergillosis using polymerase chain reaction-based detection of aspergillus in BAL. Chest 121:1171–1176 [DOI] [PubMed] [Google Scholar]
- 30.Rantakokko-Jalava K, et al. 2003. Semiquantitative detection by real-time PCR of Aspergillus fumigatus in bronchoalveolar lavage fluids and tissue biopsy specimens from patients with invasive aspergillosis. J. Clin. Microbiol. 41:4304–4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reinwald M, et al. 2012. Therapy with antifungals decreases the diagnostic performance of PCR for diagnosing invasive aspergillosis in bronchoalveolar lavage samples of patients with haematological malignancies. J. Antimicrob. Chemother. 67:2260–2267 [DOI] [PubMed] [Google Scholar]
- 31a.Roselló E, et al. 2011. Evaluation of MycAssay Aspergillus molecular diagnostic test and automated EZ1 DNA extraction for detecting Aspergillus spp. DNA in bronchoalveolar samples of patients with risk factors for pulmonary invasive aspergillosis, abstr P1942. 21st Eur. Congress Clin. Microbiol. Infect. Dis., Milano, Italy. http://www.poster-submission.com/search/select/26 [Google Scholar]
- 32.Sanguinetti M, et al. 2003. Comparison of real-time PCR, conventional PCR, and galactomannan antigen detection by enzyme-linked immunosorbent assay using bronchoalveolar lavage fluid samples from hematology patients for diagnosis of invasive pulmonary aspergillosis. J. Clin. Microbiol. 41:3922–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shahid M, Malik A, Bhargava R. 2008. Bronchogenic carcinoma and secondary aspergillosis—common yet unexplored: evaluation of the role of bronchoalveolar lavage-polymerase chain reaction and some nonvalidated serologic methods to establish early diagnosis. Cancer 113:547–558 [DOI] [PubMed] [Google Scholar]
- 34.Skladny H, et al. 1999. Specific detection of Aspergillus species in blood and bronchoalveolar lavage samples of immunocompromised patients by two-step PCR. J. Clin. Microbiol. 37:3865–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spiess B, et al. 2003. Development of a LightCycler PCR assay for detection and quantification of Aspergillus fumigatus DNA in clinical samples from neutropenic patients. J. Clin. Microbiol. 41:1811–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spreadbury C, Holden D, Aufauvre-Brown A, Bainbridge B, Cohen J. 1993. Detection of Aspergillus fumigatus by polymerase chain reaction. J. Clin. Microbiol. 31:615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun W, et al. 2011. Evaluation of PCR on bronchoalveolar lavage fluid for diagnosis of invasive aspergillosis: a bivariate metaanalysis and systematic review. PLoS One 6:e28467 doi:10.1371/journal.pone.0028467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang CM, Holden DW, Aufauvre-Brown A, Cohen J. 1993. The detection of Aspergillus spp. by the polymerase chain reaction and its evaluation in bronchoalveolar lavage fluid. Am. Rev. Respir. Dis. 148:1313–1317 [DOI] [PubMed] [Google Scholar]
- 39.Tarrand JJ, et al. 2003. Diagnosis of invasive septate mold infections. A correlation of microbiological culture and histologic or cytologic examination. Am. J. Clin. Pathol. 119:854–858 [DOI] [PubMed] [Google Scholar]
- 40.Torelli R, et al. 2011. Diagnosis of invasive aspergillosis by a commercial real-time PCR assay for Aspergillus DNA in bronchoalveolar lavage fluid samples from high-risk patients compared to a galactomannan enzyme immunoassay. J. Clin. Microbiol. 49:4273–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuon FF. 2007. A systematic literature review on the diagnosis of invasive aspergillosis using polymerase chain reaction (PCR) from bronchoalveolar lavage clinical samples. Rev. Iberoam. Micol. 24:89–94 [PubMed] [Google Scholar]
- 42.Verweij PE, et al. 1995. Comparison of antigen detection and PCR assay using bronchoalveolar lavage fluid for diagnosing invasive pulmonary aspergillosis in patients receiving treatment for hematological malignancies. J. Clin. Microbiol. 33:3150–3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Eiff M, et al. 1995. Pulmonary aspergillosis: early diagnosis improves survival. Respiration 62:341–347 [DOI] [PubMed] [Google Scholar]
- 44.White PL, et al. 2010. Aspergillus PCR: one step closer to standardization. J. Clin. Microbiol. 48:1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whiting PF, et al. 2011. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 155:529–536 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.