Abstract
The Tat pathway transports folded proteins across the bacterial cytoplasmic membrane and is a major route of protein export in the Streptomyces genus of bacteria. In this study, we have examined the localization of Tat components in the model organism Streptomyces coelicolor by constructing enhanced green fluorescent protein (eGFP) and mCherry fusions with the TatA, TatB, and TatC proteins. All three components colocalized dynamically in the vegetative hyphae, with foci of each tagged protein being prominent at the tips of emerging germ tubes and of the vegetative hyphae, suggesting that this may be a primary site of Tat secretion. Time-lapse imaging revealed that localization of the Tat components was highly dynamic during tip growth and again demonstrated a strong preference for apical sites in growing hyphae. During aerial hypha formation, TatA-eGFP and TatB-eGFP fusions relocalized to prespore compartments, indicating repositioning of Tat components during the Streptomyces life cycle.
INTRODUCTION
In bacteria, the general secretory (Sec) and twin arginine protein translocation (Tat) pathways operate in parallel to transport proteins across the cytoplasmic membrane. In most bacteria, Sec is the predominant route of protein export. Proteins are targeted to the Sec machinery by the presence of N-terminal signal peptides and are extruded across the membrane in an unfolded conformation (14). Proteins are also targeted to the Tat pathway by N-terminal signal peptides, which in this case contain a conserved twin-arginine motif (3, 4). The major difference between the Sec and Tat pathways is that the Tat machinery transports folded proteins (reviewed in references 18 and 37). Protein transport by the Tat pathway is powered solely by the proton motive force (7, 56).
The Tat machineries of Gram-negative bacteria and Gram-positive actinobacteria are composed of the TatA, TatB, and TatC proteins (5, 20, 40, 42, 50), while those of low G+C Gram-positive bacteria require only TatA and TatC (25, 26). TatB and TatC form a membrane-bound complex that binds Tat substrate proteins through their twin-arginine signal peptides (6, 8). TatA is the most highly produced of all of the Tat components (23), and many copies of this monotopic membrane protein are believed to cluster around a substrate-bound TatBC complex to bring about the transport of the folded protein across the membrane (9, 30, 34).
Although the Tat pathway is generally seen as a relatively minor route of protein traffic, bacteria of the Streptomyces genus exceptionally seem to encode large numbers of Tat substrates (13). Streptomycetes are mycelial organisms that undergo complex morphological differentiation (17) and are of prime importance in industry and medicine since they produce a large number of commercially important secondary metabolites, enzymes, and other secreted protein products (21). Proteomic analyses of Streptomyces coelicolor and the plant-pathogenic species Streptomyces scabies indicate that a diverse array of hydrolytic enzymes utilize the Tat export pathway, as do substrate binding proteins of ABC transporters which are subsequently lipid modified (27, 47, 51, 53). The Streptomyces Tat machinery comprises the TatA, TatB, and TatC proteins, which are functional homologues of similar proteins found in Gram-negative bacteria such as Escherichia coli (20, 43, 44, 51). Inactivation of the Tat pathway in Streptomyces spp. results in pleiotropic phenotypes, including impaired morphological differentiation, a retarded growth rate, and increased permeability of the cell envelope (27, 44, 51). Given the importance of the Tat pathway to the biology of Streptomyces, in this study, we have examined the localization of the Tat transport machinery during the life cycle of S. coelicolor by constructing individual C-terminal fusions to enhanced green fluorescent protein (eGFP) and mCherry, which are commonly used as reporters for protein localization in Streptomyces (16, 39, 55). Our studies show that the three Tat components are highly dynamic and that they frequently associate with the tips of vegetative hyphae.
MATERIALS AND METHODS
Bacterial growth conditions.
E. coli strains were routinely grown in Luria-Bertani medium and supplemented with arabinose as necessary. S. coelicolor strains were grown on soya flour mannitol agar, Difco nutrient broth agar (BD Diagnostics), and a 50:50 mixture of tryptone soya broth (TSB; Oxoid) and yeast extract-malt extract (YEME) agar. Liquid cultures were grown in Difco nutrient broth or a 50:50 mixture of TSB and YEME. All of the growth medium recipes used are those of Kieser et al. (29). For agarase assays, strains were cultured on MM medium (per liter, 10 g agar, 1 g (NH4)2SO4, 0.5 g K2HPO4 · 7H2O, 0.2 g Mg2SO4 · 7H2O, 0.01 g FeSO4 · 7H2O). The bacteria inoculated onto the plates were grown for 5 days at 30°C, after which they were stained with Lugol's solution (Sigma). For fluorescence microscopy of germinating spores and young hyphae (up to 18 h), spores were incubated in liquid culture in germination medium (29). Samples from liquid cultures were spotted onto a 1.5% agarose bed on a glass microscope slide before microscopy analysis. Images of older vegetative hyphae (26 h onward) and aerial hyphae/spore chains (collected at 44 or 72 h) were collected from samples that had been inoculated at the acute-angle junction of coverslips inserted at a 45° angle in minimal medium agar plates containing 1% mannitol (29).
Strain and plasmid construction.
The strains and plasmids used in this study are shown in Table 1. Strains producing TatA-eGFP and TatC-eGFP fusion constructs from the native chromosomal location were assembled in a similar manner by using the PCR targeting approach of Gust et al. (19). Briefly, an eGFP-aac3(IV)-oriT cassette (conferring apramycin resistance) was amplified with primer pairs tatA-link-egfp fw (5′-CTCCCGTCCGGTCACCGAGCCGACGGACACGACCAAGCGCCTGCCGGGCCCGGAGCTG-3) and tatA-link-egfp rv (5′-CTTGTTGCGGGCAGGCTTCAGCAACCCACGTTCCCATCTCATATGTGTAGGCTGGAGCTGCTTC-3′) or tatC-link-egfp fw (5′-GGCCACGAAGGACCGGGTCAACGGCTACGACGACGTGACCCTGCCGGGCCCGGAGCTG-3′) and tatC-link-egfp rv (5′-ATCATTATCGGGATCGGCCGTGTGGGGGTTCCGTGGGGGCCATATGTGTAGGCTGGAGCT GCTTC-3′) and inserted downstream of tatA or tatC, respectively, in kanamycin resistance (Kanr)-marked cosmid DNA I41 (2). This approach includes an LPGPELPGPE linker sequence between the end of the Tat protein and eGFP, an arrangement which has been observed to improve the folding and activity of eGFP fusions (39). The resultant constructs were transferred separately to S. coelicolor M145 or FM145 by intergeneric conjugation. Apramycin-resistant exconjugants were screened for the loss of kanamycin resistance, indicating the double-crossover allelic exchange of the tatA or tatC locus, and the strains were designated BRO1 (M145 tatA::tatA-eGFP), BRO2 (M145 tatC::tatC-eGFP), BRO3 (FM145 tatA::tatA-eGFP), and BRO4 (FM145 tatC::tatC-eGFP).
Table 1.
Strains and plasmids used in this study
| Strain | Description | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | F− ϕ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Laboratory stock |
| BW25113/pIJ790 | K-12 derivative; ΔaraBAD ΔrhaBAD/λ-Red (gam bet exo) cat araC rep101(Ts) | 19 |
| ET12567/pUZ8002 | dam-13::Tn9 dcm cat tet hsdM hsdR zjj-201::Tn10/tra neo RP4 | 36 |
| S. coelicolor | ||
| M145 | SCP1− SCP2− | 2 |
| FM145 | Derivative of M145 with reduced autofluorescence | 55 |
| TP5 | M145 ΔtatB | 51 |
| TP6 | TP5 harboring pTDW134 | This work |
| TP7 | TP5 harboring pTDW135 | This work |
| BRO1 | M145 tatA::tatA-egfp aac3(IV)-oriT | This work |
| BRO2 | M145 tatC::tatC-egfp aac3(IV)-oriT | This work |
| BRO3 | FM145 tatA::tatA-egfp aac3(IV)-oriT | This work |
| BRO4 | FM145 tatC::tatC-egfp aac3(IV)-oriT | This work |
| BRO5 | BRO3 harboring pTDW136 | This work |
| BRO6 | BRO4 harboring pTDW136 | This work |
| BRO7 | BRO3 harboring pTDW137 | This work |
| BRO8 | FM145 harboring pTDW135 | This work |
| Plasmids | ||
| pTDW134 | pIJ8660 tatBp-tatBstop-egfp | This work |
| pTDW135 | pIJ8660 tatBp-tatB-egfp | This work |
| pTDW136 | pIJ6902 tatB-mcherry | This work |
| pTDW137 | pIJ6902 tatC-mcherry | This work |
Two tatB-expressing constructs were assembled in a similar manner, one of which produces transcriptionally coupled TatB and eGFP as separate proteins (pTDW134) and the other of which produces them as a fusion (pTDW135). To construct pTDW134, DNA encoding tatB along with 200 bp of upstream DNA was amplified with oligonucleotides tatbpromf (5′-GGCGCGGGATCCGACGGCAAGGAGACGAAG and tatbcodcompr (5′-GGCGCGGGATCCTCAGGTGGCGTCCATGTCGAAG-3′), the PCR product was digested with BamHI and cloned into pIJ8660 (45). The clones were assessed to ensure that the tatB gene and the gene for eGFP were expressed in the same orientation, and the resultant plasmid was designated pTDW134. To construct pTDW135, the tatB gene was amplified with tatbpromf and tatbcodr (5′-GGCGCGGGATCCCATATGGGTGGCGTCCATGTCGAAG-3′); the resulting PCR product was digested with BamHI and cloned into BamHI-digested pBluescript. The cloned products were then excised by digestion with BamHI and NdeI and cloned into similarly digested pIJ8660 to give pTDW135. These two constructs were then integrated into the chromosome of S. coelicolor strain TP5 (M145 ΔtatB [51]) at the ϕC31 site to give strains TP6 (produces TatB and eGFP as separate proteins) and TP7 (produces a TatB-eGFP fusion).
To construct plasmids producing C-terminal mCherry fusions with TatB and TatC, the S. coelicolor tatB gene was amplified with the primer pair tatBNdeI_fw (5′-GAGCTTCATATGGTGTTCAATGACATAGGCG-3′) and tatBXbaI_rv (5′-GTACGTCTAGAGGTGGCGTCCATGTCGAAG-3′) and the tatC gene was amplified with primer pair tatCNdeI_fw (5′-GCACGTCATATGCTGAAGCCTGCCCGCAAC-3′) and tatCXbaI_rv (5′-GTGCATCTAGAGGTCACGTCGTCGTAGCCG-3′). The amplified genes were digested with NdeI and XbaI and cloned into similarly digested pIJ6902 (22) to give pIJ6902-TatB and pIJ6902-TatC, respectively. The gene coding for mCherry was amplified with primers mCherryXbaI_fw (5′-CTACATTCTAGAGTGAGCAAGGGCGAGGAG-3′) and mCherryKpnI_rv (5′-GGCTAGGTACCTTACTTGTACAGCTCGTCC-3′), digested with XbaI and KpnI, and cloned into similarly digested pIJ6902-TatB and pIJ6902-TatC to give pTDW136 (pIJ6902-TatB-mCherry) and pTDW137 (pIJ6902-TatC-mCherry), respectively. Plasmid pTDW136 was integrated into the chromosomes of S. coelicolor strains BRO3 and BRO4 at the ϕC31 site to give strains BRO5 and BRO6, respectively. Plasmid pTDW137 was similarly integrated into the chromosome of strain BRO3 to give strain BRO7.
Protein methods.
To assess the subcellular localization of fusion proteins, suspensions of S. coelicolor hyphae were sonicated and subsequently separated into cytoplasmic and membrane fractions according to reference 47. Fluorescent proteins were analyzed following SDS-PAGE (12% acrylamide; samples were not boiled prior to separation) by scanning with a phosphorimager (FujiFilm FLA-5100) equipped with a 473-nm laser and a local binary pattern filter as described previously (39). Protein concentration was determined by the method of Lowry et al. (31).
Microscopy.
Samples were analyzed using a Zeiss Axio Imager M1 or Axioskop A1 equipped with a 150×/1.35 or 100×/1.35 objective, respectively, a reflector (38 HE for eGFP or 63 HE for mCherry), and a condenser contrast of DIC3, and images were captured using an 5-megapixel camera. Images were examined using the Zeiss AxioVision software. Live imaging experiments were conducted with MM agar plates. Samples were grown on cellophane squares for 12 h and then prepared for imaging as described previously (28). Imaging of time-lapse movies was performed as described previously (54) with 2-min intervals between frames and a 100-ms exposure time (for TatC, the exposure time was 500 ms).
RESULTS
Fluorescent-protein fusions with S. coelicolor Tat components are stable and retain Tat transport activity.
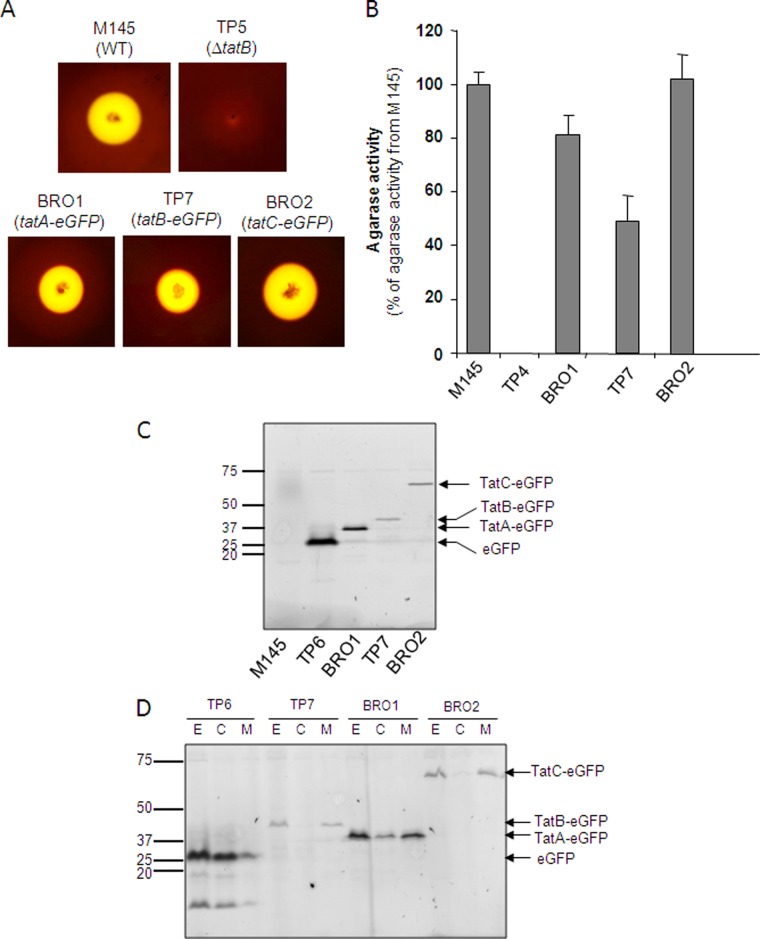
We first assessed whether the S. coelicolor strains that produced eGFP-tagged variants of TatA, TatB, or TatC in place of the native protein retained Tat transport activity. Some functionality of the Tat fusion proteins was suggested by the fact that each strain which harbored a replacement of the native tat gene with a construct producing an eGFP fusion protein showed wild-type growth and differentiation on laboratory growth medium. To obtain a more quantitative assessment of Tat transport activity in these strains, we assessed the activity of the Tat substrate agarase. Agarase secretion results in the breakdown of agar around S. coelicolor colonies, which can be visualized following staining with Lugol's solution. As shown in Fig. 1A, strains producing TatA-eGFP (BRO1), TatB-eGFP (TP7), or TatC-eGFP (BRO2) produced strong halos of agarase activity. Quantification of agarase activity by measurement of zones of clearing around replicates (Fig. 1B) (51, 52) indicated that the level of agarase secretion by the strains producing TatA-eGFP or TatC-eGFP was similar to that of the wild type. Since the tatA and tatC genes appear to be organized as a transcription unit, the observation that the construct producing TatA-eGFP from the native chromosomal location has good Tat transport activity gives a clear indication that the transcription/translation of tatC was not affected in this strain. While the TatB-eGFP fusion also clearly retained function, the total exported agarase activity was lower than that of the wild-type strain (Fig. 1B). The reason for the lower level of agarase activity in the strain producing the TatB-eGFP fusion is not clear; it may reflect a difference between the levels of expression of the single-copy tatB-eGFP fusion produced at a heterologous chromosomal location and that produced at the native site. Alternatively, fusion of GFP to the C terminus of TatB may reduce its functionality.
Fig 1.
Fusions of eGFP to the TatA, TatB, and TatC proteins of S. coelicolor are stable and do not abolish Tat transport activity. (A, B) Semiquantitative analysis of extracellular agarase activity mediated by S. coelicolor strains. The indicated strains were cultured on MM medium for 5 days at 30°C, after which they were stained with Lugol's solution. In panel A, zones of clearing around colonies after staining with Lugol's solution are shown, while panel B, shows relative agarase activity, which was estimated as described previously (51). The error bars represent the standard error of the mean; n = 6 to 9. (C, D) S. coelicolor strains M145, TP6 (M145 ΔtatB ϕC31 tatBp-tatBstop-eGFP), BRO1 (M145 tatA::tatA-eGFP), TP7 (M145 ΔtatB ϕC31 tatBp-tatB-eGFP), and BRO2 (M145 tatC::tatC-eGFP) were cultured aerobically for 24 h at 30°C in a 1:1 mixture of TSB and YEME media. Cell extracts (formed by sonication of hyphal suspension) (panel C) and subcellular fractions fractionated from extract (lanes E) into cytoplasmic (lanes C), and membrane (lanes M) fractions (panel D) were separated by SDS-PAGE (12% acrylamide; samples were not boiled prior to separation), and fluorescent proteins were analyzed by phosphorimaging. Twenty micrograms of total protein was loaded into each lane. The values to the left of panels C and D are molecular sizes in kilodaltons.
To confirm that each of the fusion proteins was stable, we briefly sonicated hyphal suspensions of the strains and separated total proteins by seminative PAGE (i.e., in the presence of SDS but without heat denaturing the samples). The presence of fluorescent proteins was then analyzed with a phosphorimager. As shown in Fig. 1C, each of the Tat-eGFP fusion proteins appeared to be stable and migrated close to the predicted mass. Comparison of the fluorescence intensities of three fusions showed that the TatA-eGFP fusion was present at higher levels than the other two constructs, consistent with the findings that in E. coli, TatA is far more abundant than TatB and TatC (23, 41). Taken together, these observations indicate that fusions of eGFP with TatA, TatB, and TatC are stable and do not inactivate these proteins.
Before we analyzed cells producing these eGFP fusions by fluorescence microscopy, we investigated their subcellular localization. Hyphae from liquid-grown cultures were lysed by sonication, and the extract was separated into cytoplasmic and membrane fractions. As shown in Fig. 1D, each of the Tat-eGFP fusion proteins localized predominantly to the membrane fraction, while free eGFP (produced from strain TP6) was found mainly in the cytoplasmic fraction. We could detect no TatB-eGFP protein in the cytoplasmic fraction, in contrast to TatB from S. lividans, which has been shown to have both membrane and cytoplasmic localizations (10–12). However, we did detect some TatA-eGFP in the cytoplasmic fraction, which has been observed previously for untagged TatA from S. lividans (10–12).
TatB-eGFP localizes at the tips of vegetative hyphae.
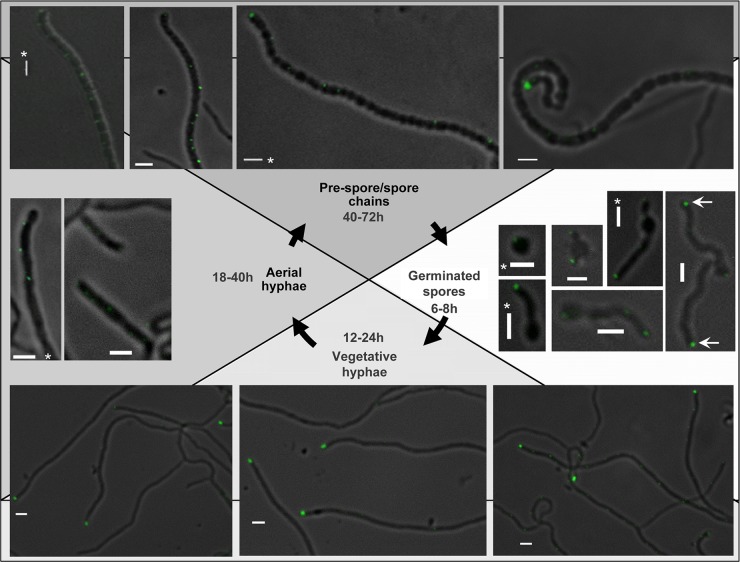
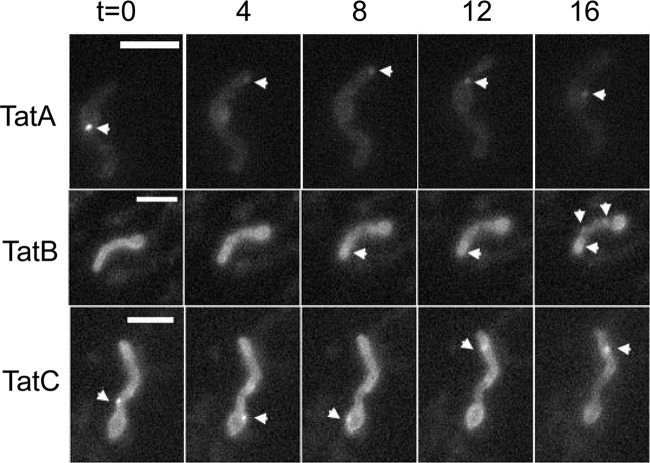
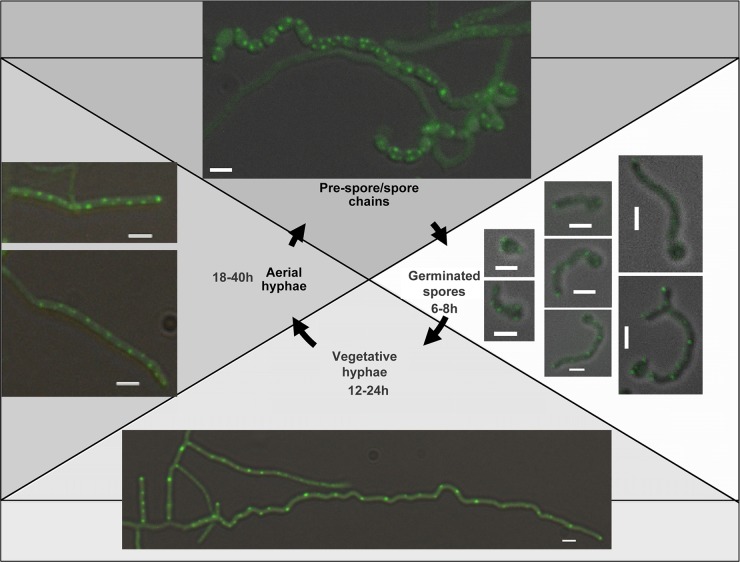
The first fusion that we analyzed by fluorescence microscopy was TatB-eGFP. Images were captured from spores of strain TP7 that had been allowed to germinate in liquid culture for 6 to 8 h. As shown in Fig. 2, after 6 to 8 h and for each germinating spore examined, a strong focus of fluorescence was visible at the hyphal tip, while a second focus was frequently visible farther away from the tip. Where two hyphae emerged from a germinating spore, foci were present at both tips (Fig. 2, arrows). Analysis of 52 germinating hyphae indicated that foci were always present at the tips (Table 2). When germinating spores were observed by time-lapse imaging (54), TatB-eGFP was again revealed to be primarily at the tips of the hyphae, but this localization was highly dynamic throughout the germ tubes, moving to distinctly different sites within seconds (Fig. 3, row 2; see Movie SV1 in the supplemental material).
Fig 2.
Analysis of TatB-eGFP localization in S. coelicolor during growth and development by fluorescence microscopy. Representative images of strain TP7 at different life cycle stages are shown. The fluorescence images were all collected with a 400-ms exposure time, except for those with asterisks, which were collected with a 600-ms exposure time. Scale bars, 2 μm.
Table 2.
Frequency of occurrence of fluorescent foci at the hyphal tips of S. coelicolor producing each of the Tat protein-eGFP fusions
| Time (h) | % (no./total) of tips with foci |
||||
|---|---|---|---|---|---|
| TatA-eGFP |
TatB-eGFP |
TatC-eGFP, vegetative hyphae | |||
| Vegetative hyphae | Aerial hyphae and spore chains | Vegetative hyphae | Aerial hyphae and spore chains | ||
| 6–8 | 87 (65/75) | 100 (52/52) | 98 (163/166) | ||
| 12 | 52 (68/131) | 56 (368/645) | |||
| 24 | 55 (165/300) | 44 (497/1124) | |||
| 44 | 41 (192/467) | —a | 24 (498/2093) | 65 (33/51) | |
| 72 | 32 (84/259) | 53 (26/49) | 22 (316/1452) | 41 (32/79) | |
—, almost no aerial hyphae could be detected at 44 h for this strain.
Fig 3.
Dynamic localization of TatA-eGFP (BRO3), TatB-eGFP (BRO8), and TatC-eGFP (BRO4) at the tips of germinating spores. Images were taken from Movies SV5, SV1, and SV3 in the supplemental material, respectively, and represent 4-min time intervals. Images were collected using a Zeiss Observer with a Hamamatsu C9100-02 electron-multiplying charge-coupled device camera. Exposure times were 100 ms for TatA and TatB and 500 ms for TatC movies. For all constructs, setting of minimum/maximum intensity was used to show similar fluorescence intensities. Scale bars, 2 μm. The arrowheads indicate examples of the fluorescent protein fusions of the respective Tat components.
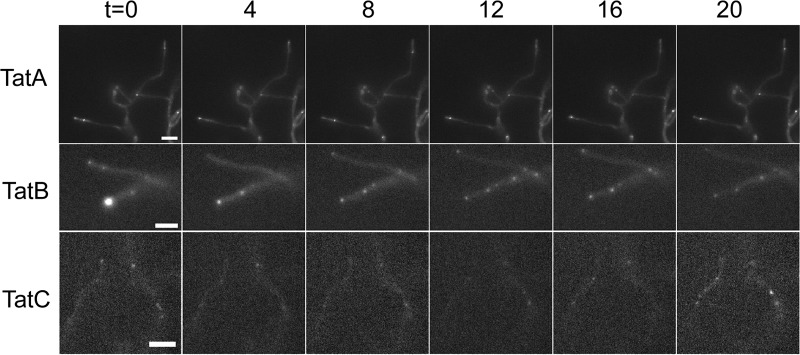
When vegetative hyphae were observed (12 to 24 h postgermination; Fig. 2, bottom panels), bright fluorescence was still visible at the tips but the frequency of tip localization had declined, with only around half of the hyphae examined showing this localization pattern (Table 2). In addition, further, less intense fluorescent foci could be seen at positions away from the tip. The same dynamic localization of TatB observed in germinating spores was also found in vegetative hyphae by time-lapse imaging, i.e., with foci of fluorescence moving through the hyphae on a time scale of seconds (Fig. 4, row 2; see Movie SV2 in the supplemental material). This indicates that the TatB-eGFP fusion is highly mobile. It should be noted that an E. coli TatA-yellow fluorescent protein (YFP) fusion was also shown to be highly mobile (30).
Fig 4.
Dynamic localization of TatA-eGFP, TatB-eGFP, and TatC-eGFP in vegetative hyphae. The images shown were taken from Movies SV6, SV2, and SV4 in the supplemental material, respectively, and represent 4-min time intervals. The strains and image settings are the same as those in Fig. 3. Scale bars, 2 μm.
Interestingly, when aerial hyphae were observed (Fig. 2, left), regularly spaced, punctate fluorescence could be seen that seemed to coincide with the positioning of prespore compartments. In addition, some of the aerial hyphae examined also showed a brighter fluorescent focus at the tip. Thus, it appears that the TatB-eGFP fusion is highly mobile in substrate hyphae of S. coelicolor and that it relocalizes to prespores during aerial growth.
TatC-eGFP colocalizes with TatB during vegetative growth.
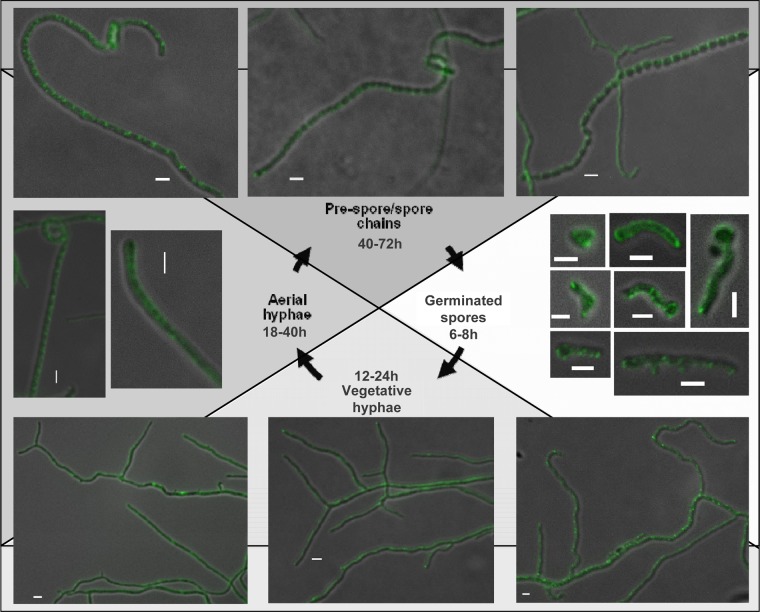
In E. coli, TatB forms a stoichiometric complex with TatC that functions as a receptor for substrates (6, 46). The fact that the S. coelicolor TatB and TatC proteins are each able to functionally substitute for their E. coli counterparts (20) suggests that the S. coelicolor TatB and TatC proteins similarly function together. We therefore anticipated that we should see similar localization of the TatB and TatC fusions. However, the fluorescence of the TatC-eGFP fusion in living cells was not as bright as that of TatB-eGFP, requiring a longer exposure time (600 ms), which produced significant background autofluorescence. Nevertheless foci could be seen at or close to the tips of emerging hyphae in germinating spores (Fig. 5, right). For this reason, the TatC-eGFP construct was introduced into strain FM145, a low-autofluorescent derivative of M145 (55), allowing longer exposure times (of up to 5 s). This revealed localizations of TatC in both germinating spores and growing hyphae of BRO4 (FM145 tatC::tatC-eGFP) similar to those seen in BRO2 (compare Fig. 5 with Fig. 3 and 4; see Movies SV3 and SV4 in the supplemental material). The intensity of the fluorescent foci of the TatC-eGFP fusion at the hyphal tips was less pronounced than that of TatB-eGFP, and other equally bright foci could be seen at regularly spaced intervals along the vegetative hyphae (Fig. 5, bottom). Examination of aerial hyphae and prespores (Fig. 5, right and top) again indicated regularly spaced, punctate fluorescence apparently coinciding with prespore compartments. We also noted that, similar to TatB-eGFP, TatC-eGFP was also highly mobile, moving through the hyphae on a time scale of seconds (Fig. 4, bottom row; see Movie SV4 in the supplemental material).
Fig 5.
Localization of TatC-eGFP during growth and development of S. coelicolor. Representative images of germinating spores (after 6 to 8 h of germination) of S. coelicolor strain BRO2 are shown. Representative images of strain BRO4 at other life cycle stages are also shown. Fluorescence images were collected with a 600-ms exposure time for germinating spores and 5 s for other stages. Scale bars, 2 μm.
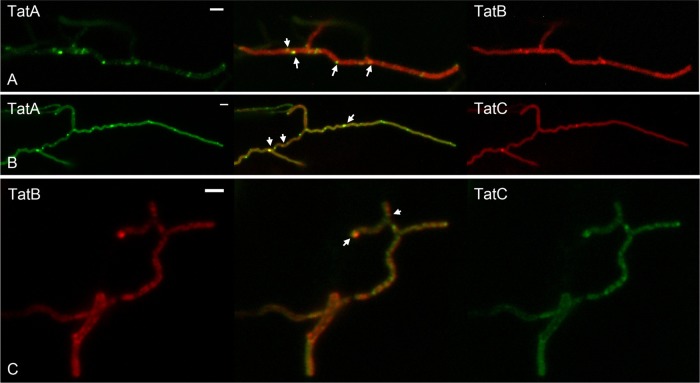
On the basis of 166 images of hyphae emerging from germinating spores of strain BRO2, the frequency of tip-localized fluorescence was estimated at 98% (Table 2). Since foci of TatB-eGFP were also found at the hyphal tips at a high frequency, this suggests that the two proteins colocalize. To investigate this further, we constructed a fusion of mCherry to the C terminus of TatB encoded by plasmid pIJ6902 and incorporated a single copy into strain BRO4, which produces TatC-enhanced GFP from the native chromosomal location. Since TatB-mCherry expression in this new strain, BRO6, depended on the leakiness of the uninduced tipA promoter, it accumulated at lower levels than TatB–eGFP which was expressed from the tatB promoter. Therefore, imaging of TatB-mCherry required long exposure times (on the order of 5 to 10 s). Nonetheless, analysis of the strain producing both the TatC-eGFP and TatB-mCherry fusion proteins showed mostly colocalization of the two proteins in vegetative hyphae (see Fig. 7C).
Fig 7.
Colocalization of TatA-eGFP and TatB-mCherry (strain BRO5) (A), TatA-eGFP and TatC-mCherry (strain BRO7) (B), and TatB-mCherry and TatC-eGFP (strain BRO6) (C) in each case in vegetative hyphae of S. coelicolor FM145. The arrows indicate colocalizing foci. Images were acquired with an Axioskop A1 and an Mrc5 camera with 2-s (for eGFP) and 10-s (for mCherry) exposure times. Scale bars, 2 μm.
Colocalization of TatA-eGFP with other Tat components in vegetative hyphae.
Finally we examined the behavior of TatA-eGFP. As shown in Fig. 6, this fusion appeared to be much brighter than the TatB- or TatC-eGFP fusion protein and showed a more dispersed localization, but in all cases at the periphery of hyphae, consistent with membrane localization. In germinating hyphae, fluorescence could be seen throughout the membrane, appearing in some cases as continuous fluorescence and in other instances as more punctate spots (Fig. 6, right). A similar pattern of mixed punctate and dispersed fluorescence has also been seen in a TatA-YFP protein produced at native levels in E. coli, with punctate fluorescence observed only in the presence of the other Tat components (30). In almost all instances, a strong fluorescent focus was also visible at the germling tips (Table 2). In older hyphae, fluorescent foci were visible throughout, including in the tip region (Fig. 6, bottom). Like the TatC-eGFP fusion, the TatA-eGFP fusion appeared to be reasonably evenly distributed over the hyphal length. In aerial hyphae and prespores, again distribution of the TatA fusion protein was along the entire length, with foci coinciding with each prespore compartment (Fig. 6, left and top; see Movie SV5 in the supplemental material). Time-lapse imaging of TatA-eGFP in vegetative hyphae again showed dynamic localization of the fusion. Interestingly, it appeared in some cases that foci of fluorescence assembled from more dispersedly fluorescent regions (Fig. 4, top row; see Movie SV6 in the supplemental material). This may potentially correspond to the assembly of dispersed monomers or small oligomers of TatA into large assemblies, as implied by other studies (30).
Fig 6.
Localization of TatA-eGFP during growth and development of S. coelicolor. Representative images of strain BRO1 at different life cycle stages are shown. All fluorescence images were collected with a 600-ms exposure time. Scale bars, 2 μm.
To ascertain whether TatA colocalizes with the other Tat components, we constructed dually tagged strains that coproduced either TatB-mCherry or TatC-mCherry under the control of the tipA promoter with natively encoded TatA-eGFP (strains BRO5 and BRO7, respectively). Very long exposure times (5 to 10 s) were required to visualize mCherry, making colocalization studies difficult. However, it appears that in vegetative hyphae, all of the foci of TatB-mCherry colocalize with foci of TatA-eGFP (Fig. 7A). While all of the TatC-mCherry foci also colocalized with TatA-eGFP foci, some of the TatA-eGFP appeared to localize independently of TatC-mCherry (Fig. 7B).
DISCUSSION
In this study, we have used fluorescent protein fusions to determine the subcellular localization of the S. coelicolor Tat components throughout the complex life cycle of this organism. We clearly observed foci of eGFP-tagged TatA, TatB, and TatC at the tips of germinating hyphae. As the vegetative hyphae aged, the proportion with foci of TatA-eGFP and TatB-eGFP at the hyphal tips decreased, although even at 24 h postgermination, almost 50% of the hyphae we examined had tip-localized foci of TatA and TatB. Colocalization experiments showed that TatA-eGFP foci colocalized with TatB-mCherry and TatC-mCherry foci and that, likewise, TatC-eGFP foci also colocalized with TatB-mCherry foci. It is likely that these colocalizing Tat foci represent active Tat transport complexes since it is known from studies with other organisms that TatA associates only transiently with TatBC during the protein transport step (1, 34).
The localization of Tat proteins at the tips of vegetative hyphae suggests that one of the major sites of protein secretion by the Tat pathway is at or near the hyphal tips, analogous to tip-dependent secretion by fungi (15). However, Tat complexes also assembled at other sites along the hyphal wall, suggesting that tip secretion is not the only route. Interestingly, during time-lapse imaging, TatA and TatB appeared to be highly dynamically localized and TatA foci were frequently seen to move away from the tips and settle at a position approximately 2 μm below the apical site. However, since TatA is relatively highly abundant, we cannot rule out the possibility that some TatA remains at the tips, because of the very short exposure times. TatB was always visible at the tips of vegetative hyphae, but in time-lapse imaging, several foci were observed farther away from the tip, which appeared to retrace back to the tip during growth. We are currently analyzing the dynamics of the Tat components and their colocalization in time and space.
We noted that the tip-localized fluorescent foci appeared to be brighter for TatB-eGFP than for TatA- or TatC-eGFP, suggesting that, for reasons that are currently unclear, there is more TatB than TatA or TatC at the tip. We also noted that the frequency and brightness of tip-localized foci appeared to increase when cells had stopped growing, for example, when embedded in agarose prior to imaging. This might suggest that energy is required to move the fluorescent proteins away from the tip. It should also be noted that we attempted to investigate whether we could use an agarase signal peptide-eGFP fusion to image Tat-dependent secretion; however, we found that, as previously noted for Bacillus subtilis (33), eGFP could not be exported in a fluorescent form by the Streptomyces Tat pathway (D. Widdick, J. K. Fyans, and T. Palmer, unpublished data).
During aerial hypha formation, there was relocalization of the Tat-eGFP fusion proteins such that foci of fluorescence appeared to coincide with prespore compartments. This would serve to ensure that Tat components are partitioned to each spore during sporulation. Partitioning of protein clusters has attracted some attention in recent years, with proteins of the MinD/ParA family being increasingly implicated in protein segregation (32, 48). These protein-partitioning ParA proteins are often termed “orphan” ParA proteins to reflect the fact that they do not function with ParB partner proteins. It is interesting that in actinobacteria, including Streptomyces, while tatA and tatC are found together at the same chromosomal location, tatB is found at a conserved location elsewhere on the chromosome. The chromosomal neighborhood of tatB (SCO5150) includes genes predicted to code for a σE-type extracytoplasmic function sigma factor (SCO5147), a HtrA/DegP protease (SCO5149), and of particular interest, an orphan ParA protein (SCO5152) (38). It would be interesting to ascertain whether any of these genes, in particular, SCO5152, plays a major role in the localization of Tat components.
In previous studies, it was shown that the SsgA protein, which is an actinomycete-specific protein that acts as an activator of cell wall remodeling, e.g., during septum formation, branching, and germination (24, 35), is required for proper secretion of the Tat substrate tyrosinase (49). Deletion of ssgA strongly enhanced the expression of the tat genes, most likely as a compensation effect (35). During germination and active growth, both SsgA and the Tat proteins localize to the apical sites and both show a dynamic localization pattern. Strains of S. coelicolor overexpressing SsgA show strong fragmentation of the mycelia during fermentation and displayed 3-fold-increased secretion of Tat substrates (49). Data presented in this work suggest that this increased secretion may be explained by the fact that Tat complexes preferentially localize to apical sites, which increase dramatically when hyphae fragment. The localization of the protein secretion machinery is highly relevant in strain improvement approaches.
In conclusion, fluorescence and time-lapse imaging showed that components of the Tat export pathway dynamically localize throughout the Streptomyces life cycle, actively changing during each growth phase. It will be interesting to ascertain whether other protein secretion machineries show similar dynamic localization and to determine the localization of Tat substrates during translocation in S. coelicolor during the same developmental time course.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by the Biotechnology and Biological Sciences Research Council through grants BB/F002947/1 to T.P. and by a VICI grant from the Netherlands Technology Foundation STW to G.P.V.
We are grateful to Dagmara Jakimowicz for providing cosmid H24parB-egfp-aac3(IV), which was used as the template to amplify the eGFP-aac3(IV)-oriT cassette. We thank colleagues at the John Innes Centre for helpful discussion.
Footnotes
Published ahead of print 21 September 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alami M, et al. 2003. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell 12:937–946 [DOI] [PubMed] [Google Scholar]
- 2. Bentley SD, et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147 [DOI] [PubMed] [Google Scholar]
- 3. Berks BC. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22:393–404 [DOI] [PubMed] [Google Scholar]
- 4. Berks BC, Sargent F, Palmer T. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260–274 [DOI] [PubMed] [Google Scholar]
- 5. Bogsch EG, et al. 1998. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273:18003–18006 [DOI] [PubMed] [Google Scholar]
- 6. Bolhuis A, Mathers JE, Thomas JD, Barrett CM, Robinson C. 2001. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J. Biol. Chem. 276:20213–20219 [DOI] [PubMed] [Google Scholar]
- 7. Cline K, Ettinger WF, Theg SM. 1992. Protein-specific energy requirements for protein transport across or into thylakoid membranes. Two lumenal proteins are transported in the absence of ATP. J. Biol. Chem. 267:2688–2696 [PubMed] [Google Scholar]
- 8. Cline K, Mori H. 2001. Thylakoid DeltapH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J. Cell Biol. 154:719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dabney-Smith C, Mori H, Cline K. 2006. Oligomers of Tha4 organize at the thylakoid Tat translocase during protein transport. J. Biol. Chem. 281:5476–5483 [DOI] [PubMed] [Google Scholar]
- 10. De Keersmaeker S, et al. 2005. Functional analysis of TatA and TatB in Streptomyces lividans. Biochem. Biophys. Res. Commun. 335:973–982 [DOI] [PubMed] [Google Scholar]
- 11. De Keersmaeker S, et al. 2005. Structural organization of the twin-arginine translocation system in Streptomyces lividans. FEBS Lett. 579:797–802 [DOI] [PubMed] [Google Scholar]
- 12. De Keersmaeker S, Vrancken K, Van Mellaert L, Anne J, Geukens N. 2007. The Tat pathway in Streptomyces lividans: interaction of Tat subunits and their role in translocation. Microbiology 153:1087–1094 [DOI] [PubMed] [Google Scholar]
- 13. Dilks K, Rose RW, Hartmann E, Pohlschroder M. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 185:1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Driessen AJ, Nouwen N. 2008. Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77:643–667 [DOI] [PubMed] [Google Scholar]
- 15. Fischer R, Zekert N, Takeshita N. 2008. Polarized growth in fungi—interplay between the cytoskeleton, positional markers and membrane domains. Mol. Microbiol. 68:813–826 [DOI] [PubMed] [Google Scholar]
- 16. Flärdh K. 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 49:1523–1536 [DOI] [PubMed] [Google Scholar]
- 17. Flärdh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7:36–49 [DOI] [PubMed] [Google Scholar]
- 18. Fröbel J, Rose P, Muller M. 2012. Twin-arginine-dependent translocation of folded proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:1029–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hicks MG, et al. 2006. Formation of functional Tat translocases from heterologous components. BMC Microbiol. 6:64 doi:10.1186/1471-2180-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horinouchi S. 2007. Mining and polishing of the treasure trove in the bacterial genus Streptomyces. Biosci. Biotechnol. Biochem. 71:283–299 [DOI] [PubMed] [Google Scholar]
- 22. Huang J, et al. 2005. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 58:1276–1287 [DOI] [PubMed] [Google Scholar]
- 23. Jack RL, Sargent F, Berks BC, Sawers G, Palmer T. 2001. Constitutive expression of Escherichia coli tat genes indicates an important role for the twin-arginine translocase during aerobic and anaerobic growth. J. Bacteriol. 183:1801–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jakimowicz D, van Wezel GP. 2012. Cell division and DNA segregation in Streptomyces: how to build a septum in the middle of nowhere? Mol. Microbiol. 85:393–404 [DOI] [PubMed] [Google Scholar]
- 25. Jongbloed JD, et al. 2004. Two minimal Tat translocases in Bacillus. Mol. Microbiol. 54:1319–1325 [DOI] [PubMed] [Google Scholar]
- 26. Jongbloed JD, van der Ploeg R, van Dijl JM. 2006. Bifunctional TatA subunits in minimal Tat protein translocases. Trends Microbiol. 14:2–4 [DOI] [PubMed] [Google Scholar]
- 27. Joshi MV, et al. 2010. The twin arginine protein transport pathway exports multiple virulence proteins in the plant pathogen Streptomyces scabies. Mol. Microbiol. 77:252–271 [DOI] [PubMed] [Google Scholar]
- 28. Jyothikumar V, Tilley EJ, Wali R, Herron PR. 2008. Time-lapse microscopy of Streptomyces coelicolor growth and sporulation. Appl. Environ. Microbiol. 74:6774–6781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 30. Leake MC, et al. 2008. Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc. Natl. Acad. Sci. U. S. A. 105:15376–15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 32. Lutkenhaus J. 2012. The ParA/MinD family puts things in their place. Trends Microbiol. 20:411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meissner D, Vollstedt A, van Dijl JM, Freudl R. 2007. Comparative analysis of twin-arginine (Tat)-dependent protein secretion of a heterologous model protein (GFP) in three different Gram-positive bacteria. Appl. Microbiol. Biotechnol. 76:633–642 [DOI] [PubMed] [Google Scholar]
- 34. Mori H, Cline K. 2002. A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid [Delta]pH/Tat translocase. J. Cell Biol. 157:205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Noens EE, et al. 2007. Loss of the controlled localization of growth stage-specific cell-wall synthesis pleiotropically affects developmental gene expression in an ssgA mutant of Streptomyces coelicolor. Mol. Microbiol. 64:1244–1259 [DOI] [PubMed] [Google Scholar]
- 36. Paget MS, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. 1999. Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Palmer T, Berks BC. 2012. The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 10:483–496 [DOI] [PubMed] [Google Scholar]
- 38. Palmer T, Hutchings MI. 2010. Protein secretion in Streptomyces, p 87–104. In Dyson PJ. (ed), Streptomyces molecular biology and biotechnology. Horizon Press, Norwich, United Kingdom [Google Scholar]
- 39. Ruban-Ośmialowska B, Jakimowicz D, Smulczyk-Krawczyszyn A, Chater KF, Zakrzewska-Czerwinska J. 2006. Replisome localization in vegetative and aerial hyphae of Streptomyces coelicolor. J. Bacteriol. 188:7311–7316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sargent F, et al. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sargent F, et al. 2001. Purified components of the Escherichia coli Tat protein transport system form a double-layered ring structure. Eur. J. Biochem. 268:3361–3367 [DOI] [PubMed] [Google Scholar]
- 42. Sargent F, Stanley NR, Berks BC, Palmer T. 1999. Sec-independent protein translocation in Escherichia coli. A distinct and pivotal role for the TatB protein. J. Biol. Chem. 274:36073–36082 [DOI] [PubMed] [Google Scholar]
- 43. Schaerlaekens K, et al. 2001. Twin-arginine translocation pathway in Streptomyces lividans. J. Bacteriol. 183:6727–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schaerlaekens K, Van Mellaert L, Lammertyn E, Geukens N, Anne J. 2004. The importance of the Tat-dependent protein secretion pathway in Streptomyces as revealed by phenotypic changes in tat deletion mutants and genome analysis. Microbiology 150:21–31 [DOI] [PubMed] [Google Scholar]
- 45. Sun J, Kelemen GH, Fernandez-Abalos JM, Bibb MJ. 1999. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145:2221–2227 [DOI] [PubMed] [Google Scholar]
- 46. Tarry MJ, et al. 2009. Structural analysis of substrate binding by the TatBC component of the twin-arginine protein transport system. Proc. Natl. Acad. Sci. U. S. A. 106:13284–13289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thompson BJ, et al. 2010. Investigating lipoprotein biogenesis and function in the model Gram-positive bacterium Streptomyces coelicolor. Mol. Microbiol. 77:943–957 [DOI] [PubMed] [Google Scholar]
- 48. Thompson SR, Wadhams GH, Armitage JP. 2006. The positioning of cytoplasmic protein clusters in bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:8209–8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van Wezel GP, et al. 2006. Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. Appl. Environ. Microbiol. 72:5283–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weiner JH, et al. 1998. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell 93:93–101 [DOI] [PubMed] [Google Scholar]
- 51. Widdick DA, et al. 2006. The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proc. Natl. Acad. Sci. U. S. A. 103:17927–17932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Widdick DA, Eijlander RT, van Dijl JM, Kuipers OP, Palmer T. 2008. A facile reporter system for the experimental identification of twin-arginine translocation (Tat) signal peptides from all kingdoms of life. J. Mol. Biol. 375:595–603 [DOI] [PubMed] [Google Scholar]
- 53. Widdick DA, et al. 2011. Dissecting the complete lipoprotein biogenesis pathway in Streptomyces scabies. Mol. Microbiol. 80:1395–1412 [DOI] [PubMed] [Google Scholar]
- 54. Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. 2011. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 25:89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Willemse J, van Wezel GP. 2009. Imaging of Streptomyces coelicolor A3(2) with reduced autofluorescence reveals a novel stage of FtsZ localization. PLoS One 4:e4242 doi:10.1371/journal.pone.0004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yahr TL, Wickner WT. 2001. Functional reconstitution of bacterial Tat translocation in vitro. EMBO J. 20:2472–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.