Abstract
Clostridium difficile causes one of the leading nosocomial infections in developed countries, and therapeutic choices are limited. Some strains of C. difficile produce phage tail-like particles upon induction of the SOS response. These particles have bactericidal activity against other C. difficile strains and can therefore be classified as bacteriocins, similar to the R-type pyocins of Pseudomonas aeruginosa. These R-type bacteriocin particles, which have been purified from different strains, each have a different C. difficile-killing spectrum, with no one bacteriocin killing all C. difficile isolates tested. We have identified the genetic locus of these “diffocins” (open reading frames 1359 to 1376) and have found them to be common among the species. The entire diffocin genetic locus of more than 20 kb was cloned and expressed in Bacillus subtilis, and this resulted in production of bactericidal particles. One of the interesting features of these particles is a very large structural protein of ∼200 kDa, the product of gene 1374. This large protein determines the killing spectrum of the particles and is likely the receptor-binding protein. Diffocins may provide an alternate bactericidal agent to prevent or treat infections and to decolonize individuals who are asymptomatic carriers.
INTRODUCTION
Clostridium difficile, a now-notorious nosocomial pathogen, is a Gram-positive, spore-forming, obligate anaerobe. It can innocuously colonize the intestinal tract of mammals. However, upon expansion, often induced by the administration of antibiotics that damage the intestinal microbiota, C. difficile can cause Clostridium difficile infection (CDI), with symptoms ranging from diarrhea to pseudomembraneous colitis and toxic megacolon. C. difficile has emerged as the leading cause of nosocomial infections in developed countries (1, 23, 34). Elderly patients and those on sustained antibiotic treatment are particularly vulnerable. Among the virulence factors of this pathogen are the potent exotoxins TcdA and TcdB (39). Strains exhibiting increased toxin production, such as North American pulse field type 1 (NAP1; also known as BI, or ribotype 027), have emerged and can rapidly spread in hospital settings (19, 28, 41). Therapeutic options are currently limited to treatment with nonspecific antibiotics, such as vancomycin, metronidazole, or fidaxomicin (20, 35, 36), as well as transplantation of healthy feces (4); additional therapies are needed. Furthermore, no means of prophylaxis is available except for care facility and personal hygiene, such as thorough hand washing by those persons exposed to the spores, especially hospital staff. Studies showing that phage tail-like R-type bacteriocins can be used orally and parentally to treat bacterial infections (6, 12, 21, 29, 30) have prompted us to identify similar entities specific for C. difficile for possibile use in the treatment or prevention of CDI.
High-molecular-weight or phage tail-like bacteriocins are found throughout the Eubacteria domain. The best-studied examples are the R-type pyocins of Pseudomonas aeruginosa (for a review, see reference 22). However, similar entities have been found in other Gram-negative (5, 33) as well as Gram-positive (7, 13, 37, 46) bacteria. Typically, these phage tail-like bacteriocins are produced in response to SOS induction and accumulate within the cell. At some programmed time point, the cells lyse, releasing the particles into the surrounding tissue. Two general types of phage tail-like bacteriocins have been identified. R-type bacteriocins resemble the structures of the tail apparatus of Myoviridae phages (contractile, nonflexible tails), while the F-type bacteriocins of P. aeruginosa morphologically and genetically resemble those of Siphoviridae phages (noncontractile, flexible tails) (26). Because of their genetic relationship and structural similarities to temperate phages, it has often been speculated that they share a common ancestry.
R-type bacteriocins kill target cells by first using a receptor-binding protein, such as a tail fiber, to attach to a bacterial surface receptor. Attachment is followed by sheath contraction and insertion of the core through the envelope of the target bacterium. The core penetration results in a rapid depolarization of the cell membrane potential and prompt cell death (38). Contact with a single R-type bacteriocin particle can result in cell death, accounting for the potent bactericidal activity. The natural targets of R-type bacteriocins are typically other strains of the same bacterial species that produce the particles, although rare coincidental killing of other species by R-type pyocins has been noted (2, 3, 8, 24, 25). As a general rule, strains that produce these particles are resistant to their own bacteriocin. R-type bacteriocins must provide a selective advantage to their producers by killing competing, closely related bacteria. However, this activity is ecologically complex: bacteria that produce R-type bacteriocins must lyse to release the particles. But since those bacteria and their offspring, which inherit the R-type bacteriocin genes, are resistant to their own particular bacteriocin, the selective advantage must be the one conveyed to the direct kinship, a form of bacterial altruism.
We describe here R-type phage tail-like bacteriocins, “diffocins,” that are encoded in the genomes of many strains of C. difficile. Upon induction of the SOS response, some strains produce diffocin particles, which we have shown to have bactericidal activity against other C. difficile isolates. The large genetic locus encoding diffocins has been identified and cloned from C. difficile, and when it is expressed in Bacillus subtilis, it produces active diffocins. Furthermore, we have identified the putative receptor-recognizing, spectrum-determining protein for diffocins by showing that switching this one protein switches the bactericidal specificity of the diffocin.
MATERIALS AND METHODS
Growth of C. difficile.
C. difficile strains were grown under anaerobic conditions in an atmosphere of 5% hydrogen, 5% CO2, 90% nitrogen. All culture media were reduced by anaerobic incubation for at least 24 h prior to use. Cultures were maintained for a short term on C. difficile isolation agar plates (BBL). Liquid cultures were grown in brucella broth (Difco). The incubation temperature was 37°C.
Induction of diffocins.
Diffocins were induced from C. difficile strains by growing cultures anaerobically (typically, 20-ml volumes) in brucella broth to an optical density at 600 nm (OD600) of 0.1 to 0.3/cm, followed by addition of mitomycin C to a final concentration of 3 μg/ml. Cultures were then incubated at 37°C overnight. Diffocins were concentrated and purified by first centrifuging the induced/lysed culture at 5,000 × g to remove cellular debris. The supernatant was collected as a crude lysate. The crude lysates were then passed through a 0.45-μm cellulose acetate filter and centrifuged in a Beckman L8-M ultracentrifuge with a Ti60 rotor at 90,000 × g to pellet the diffocins. The pellets containing diffocins were resuspended in a 1/100 volume of 10 mM Tris-HCl (pH 7.5), 50 mM NaCl.
Assay of diffocin activity.
Diffocin bactericidal activity was assayed by a spot method. Log-phase target bacteria (100 μl) were added to 5 ml of molten, tempered, reduced 0.5% brucella overlay agar and poured onto a 1.5% brucella agar plate. After the overlay agar set, 3- to 5-μl diffocin samples were spotted onto the agar and allowed to dry. The plates were then incubated overnight at 37°C. Killing was observed by a clearing of bacterial growth at the spot where the diffocin was applied. Killing activity was also assessed in a survival titration assay based on the method described for R-type pyocins (29, 30). Bactericidal events based on the survival titration assay are expressed as killing units.
Electron microscopy.
Lysates containing phage tail-like particles were deposited onto 400-mesh Formvar/carbon-coated copper grids (Cedarlane Laboratories, Burlington, ON, Canada) and negatively stained with 2% uranyl acetate (Cedarlane Laboratories) as described before (9). The grids were observed at 60 kV with a Hitachi H-7500 transmission electron microscope equipped with a 16-megapixel AMT TR160 digital camera controlled by using the AMT software (Advanced Microscopy Techniques).
Identification of diffocin proteins.
To identify diffocin proteins, purified samples were analyzed on 10% SDS-PAGE (Invitrogen) followed by silver staining (IBI Scientific Silver Bullit). Bands were excised, digested in-gel with trypsin, and subjected to tandem mass spectrometry analysis, which was conducted at the University of California Davis Proteomics Core.
Sequencing the genomes of C. difficile strains CD4 and CD16.
Genomic DNA was extracted from C. difficile strains CD4 and CD16 (E.Z.N.A., Omega Biotech) and then subjected to whole-genome draft sequencing using 454 pyrosequencing technology according to the manufacturer's Titanium protocol (Roche, Branford, CT). Briefly, genomic DNA was nebulized into 300- to 800-nucleotide fragments by using nitrogen gas, and then specialized 454 adaptors were ligated to either end of the DNA fragments for use in downstream purification, clonal amplification, and sequencing steps. Roughly 273,000 and 672,000 reads were generated for strain CD16 and strain CD4, respectively. Reads were assembled de novo using Newbler software (454; Roche). Signal processing and assembly were carried out off-rig on a Linux cluster of 10 nodes connected via a gigabit ethernet; each node contained eight 64-bit processing cores running at 2.3 GHz with 8 GB of RAM.
Cloning and expression of the diffocin gene clusters in Bacillus subtilis.
A detailed description of the cloning strategy for the diffocin gene fragments as well as the genetic modifications to B. subtilis are provided in the supplemental material. Also included in the supplemental material are the primer/oligonucleotide sequences (see Table S2) used in making these constructs.
Transformation into B. subtilis.
Transformation was accomplished by the single-step method using MC medium (16).
RESULTS
C. difficile strains produce R-type bacteriocins.
After exposure to mitomycin C, many strains of C. difficile have been found by electron microscopy to produce particles with phage tail-like morphology (9, 27, 32) (Fig. 1). However, even in the best producer strains, the particles were released in low quantities in lysates, quantities too low for antibacterial activity to be detected using typical assays developed for this class of bacteriocins (30). To overcome this limitation and test for bactericidal activity, we made lysates of two producer strains, CD4 and CD16, by growing cells to early log phase and inducing them with mitomycin C. Lysates were then concentrated, and the diffocin particles were purified by high-speed centrifugation (see Materials and Methods). These samples were shown by electron microscopy to contain concentrated diffocin particles; Fig. 1A shows diffocins isolated from CD4. Bactericidal activity in the purified, concentrated material was assayed by a spot test method whereby samples were applied to an overlay lawn of the target bacteria. Since most R-type bacteriocins target nonself strains of the same producer species, the tested target strains consisted of a panel of C. difficile isolates (Table 1). After overnight incubation, sharp-bordered clear zones on the lawn where the material was spotted indicated bactericidal activity. Figure 2A shows a typical spot assay of diffocin 4 spotted onto a lawn of a susceptible C. difficile isolate (19137). Figure 2B is an image of a plate that shows the results of a liquid culture survival titration assay. Typical yields, based on the survival assay, were ∼1 × 109 killing units/ml of culture volume. Diffocin 4 and diffocin 16 demonstrated different bactericidal spectra on clinical isolates of C. difficile (Table 1), with some isolates being insensitive to both.
Fig 1.
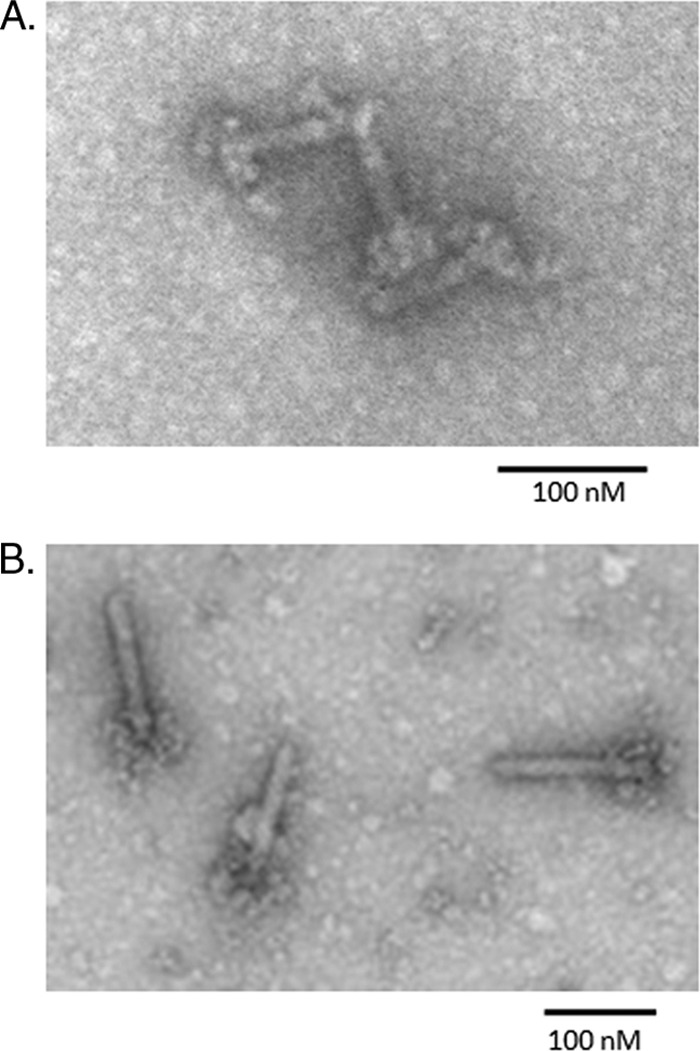
Electron micrograph of negatively stained purified diffocin particles. (A) Diffocins isolated from CD4. (B) Diffocins isolated from strain ATCC 43593. Note the large flower-like appendages, which are likely the receptor-binding protein.
Table 1.
Bactericidal activities of diffocins against clinical isolates of C. difficile
| Strain (ribotype) | Activity against the strain |
||
|---|---|---|---|
| Diffocin 4 | Diffocin 16 | Diffocin 43593 | |
| 19099 (002) | − | + | − |
| 19103 (001) | + | − | − |
| 19104 (027) | − | − | + |
| 19135 (001) | + | − | + |
| 19137 (015) | + | − | − |
| 19142 (NA) | − | − | − |
| 19145 (153) | − | + | − |
| 19155 (NA) | − | − | − |
Fig 2.
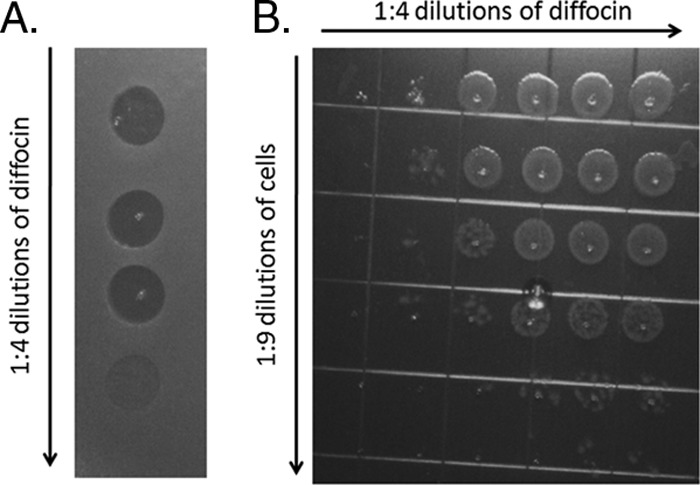
Bactericidal activity of diffocin 4 on strain 19137. (A) Lawn spot assay, showing zones of clearing due to the bactericidal activity of diffocin. (B) Liquid survival titration assay. A fixed number of target cells were incubated with dilutions of the diffocin preparation. The cells were then diluted and spotted onto an agar plate.
C. difficile ATCC strain 43593 was also shown to produce diffocins upon mitomycin exposure (Fig. 1B). This diffocin displayed yet another killing spectrum (Table 1). Of note is that its targets include a ribotype 027 strain, which is among the most prevalent strains in North America (19). This diffocin was further tested against a panel of 12 other 027 strains, all of which were sensitive (data not shown).
No bactericidal activity was detected by spot assay of diffocins 4, 16, or 43593 against a panel of various other species, including Clostridium acetobutylicum, Clostridium sordelli, Clostridium sporogenes, Clostridium biofermentens, Bifidobacterium breve, Bacteroides fragilis, Listeria ivanovii, Listeria monocytogenes, Listeria innocua, Bacillus subtilis, or Escherichia coli (see Table S1 in the supplemental material).
Identification of the diffocin genetic locus.
Purified diffocin particles isolated from strain CD4 were analyzed by SDS-PAGE and silver staining (Fig. 3). Two individual bands, of ∼200 kDa and ∼40 kDa, were each excised and analyzed by mass spectrometry. Peptides from the 40-kDa band were found to match the predicted products from two separate C. difficile open reading frames encoded in several of the C. difficile strains for which the genomes have been sequenced, including reference strain 630. The first of these was ORF 1363, which encodes a 39,192-Da phage-like sheath protein. The second predominant polypeptide in this band corresponded to ORF 1371, a 39,565-Da phage-like baseplate protein. Since these proteins were coincidentally very close in molecular mass, they migrated as the same SDS-PAGE band. The 200-kDa band yielded a dominant polypeptide that corresponded to ORF 1374 of strain 630, a gene that is downstream (3′) of the two other identified phage-like ORFs. A third band of ∼60 kDa was also analyzed and matched a GroEL-like protein (ORF 0194); it was likely a particulate impurity.
Fig 3.
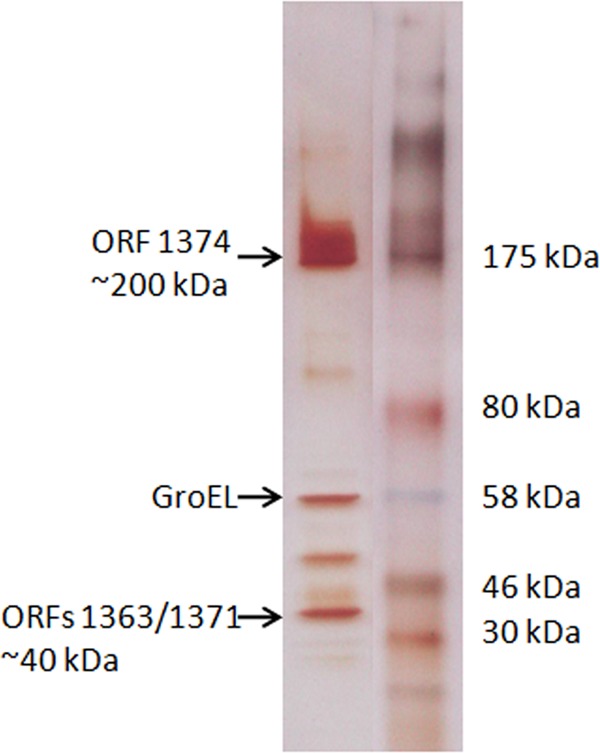
Silver stained SDS-PAGE gel of purified diffocin particles from CD4 (left lane). The arrows indicate bands that were excised for mass spectrometric analysis. The molecular mass markers (right lane) were pasted from a different lane of the same gel.
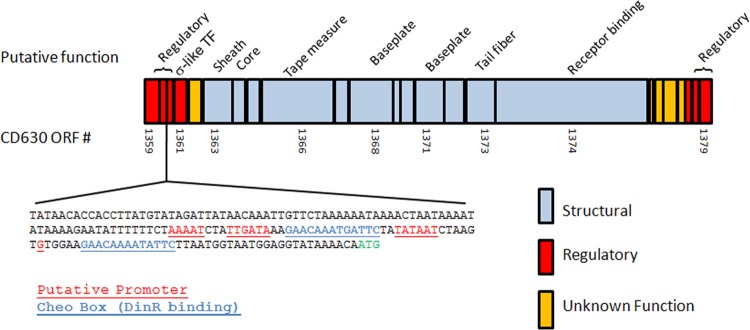
Since these ORFs mapped in very close proximity within the C. difficile genomes, we analyzed the surrounding genomic region. A prophage-like element was found between bases 1574593 and 1596384 of strain 630, which includes ORFs 1359 to 1379 (Fig. 4; Table 2). Several genes encoded in this region are homologues of the components of the tail structure of a typical Myoviridae phage, including sheath, baseplate, tail fiber, and tail length determination proteins. No genes encoding capsid, capsid assembly proteins, or portal proteins were found. Also absent from the locus were any ORFs that encoded putative DNA replication or DNA packaging machinery; this locus is consistent with that of a phage tail-like bacteriocin, for example, the R2 pyocin locus of P. aeruginosa (26). Flanking the structural genes were several ORFs that encoded putative phage-like regulatory elements. Since several ORFs from this locus encode polypeptides identified in diffocin particles, and due to the fact that the gene cluster resembles what one might predict for an R-type bacteriocin, we assigned the diffocin locus to this region. The putative structural genes (ORFs 1363 to 1374) are all encoded on the same DNA strand. Several of these structural proteins display sequence similarities to known C. difficile bacteriophages, including phages ϕ119 and ϕC2 (10, 11) (Table 2), suggesting a common ancestry.
Fig 4.
Map of the genetic locus of the diffocin cluster. ORFs encoding putative structural components are shown in blue, while putative regulatory proteins are shown in red. Those ORFs for which no function can be predicted based on sequence similarity are shown in gold. The intergenic sequence shown between ORFs 1360 and 1360a represents a potential operator region with two strong DinR consensus-binding sites (sequence portions indicated in red). Further annotations of the ORFs are in Table 2.
Table 2.
Open reading frames within the diffocin locia
| ORF | No. of aa residues | Predicted mass (kDa) | Strand sense | Putative function | Comment |
|---|---|---|---|---|---|
| CD1359 | 168 | 19.5 | − | Regulatory | Putative toxin component of toxin/antitoxin system |
| CD1360 | 106 | 12.6 | − | Repressor | Helix-turn-helix XRE family-like proteins |
| CD1360A | 65 | 7.6 | + | Regulatory | Helix-turn-helix XRE family-like proteins |
| CD1361 | 146 | 17.6 | + | Sigma-like factor | DNA-binding domain motif |
| CD1362 | 147 | 17.2 | + | Unknown | |
| CD1363 | 354 | 39.2 | + | Sheath | Similar to many Myoviridae sheath proteins, including C. difficile phages and PBSX |
| CD1364 | 142 | 16 | + | Unknown | |
| CD1365 | 447 | 16.8 | + | Unknown | |
| CD1365A | 55 | 6.4 | + | Unknown | |
| CD1366 | 817 | 89 | + | Tape measure | Determines length of sheath structure |
| CD1367 | 140 | 16.2 | + | Unknown | |
| CD1368 | 509 | 57 | + | Baseplate | Phage cell well hydrolase homology |
| CD1369 | 108 | 12.6 | + | Unknown | |
| CD1370 | 143 | 16.5 | + | Unknown | |
| CD1371 | 350 | 39.6 | + | Baseplate J | Common component of Myoviridae tail structures |
| CD1372 | 232 | 26.4 | + | Unknown | Homology to PBSX XkdT and XkdU |
| CD1373 | 327 | 36.9 | + | Tail fiber | N terminus is similar to Myoviridae phages, including C. difficile phages |
| CD1374 | 1,773 | 197.8 | + | Receptor binding | The most divergent protein among diffocins; spectrum determinant |
| CD1375 | 98 | 11 | + | Unknown | Shortened to C-terminal 39 residues in strain 630 |
| CD1376 | 86 | 9.8 | + | Unknown | |
| CD1377 | 151 | 16.8 | − | Unknown | Not required in B. subtilis |
| CD1378 | 139 | 16.2 | − | Regulatory | Not required for diffocin production in B. subtilis; helix-turn-helix XRE family-like proteins |
| CD1378A | 69 | 8 | + | Regulatory | Not required for diffocin production in B. subtilis; helix-turn-helix XRE family-like proteins |
| CD1378B | 73 | 8.1 | + | Unknown | Not required for diffocin production in B. subtilis |
| CD1379 | 132 | 15.5 | − | Regulatory | Not required for diffocin production in B. subtilis; helix-turn-helix XRE family-like proteins |
ORF numbering is based on the annotation of C. difficile strain 630. Homologues of each ORF are present in strains CD4 and CD16.
Of particular note was ORF 1374. This gene encodes a large polypeptide that is located just downstream of a small ORF with similarity to phage tail fibers (ORF 1373). Electron micrographs of diffocins have revealed a large flower-like structure at one pole of the tubular particle; we hypothesize that this large polypeptide 1374, along with the putative product of ORF 1373, constitutes this flower-like structure (see below).
Notable was the absence of holin or endolysin genes (for a review, see reference 40). These genes are typically carried by phages and are present in some phage tail-like bacteriocin gene clusters, including the R-type pyocin cluster of P. aeruginosa. The products of these genes are involved in the timed lysis of bacterial cells to release the bacteriocin (or phage) particles.
The region between ORFs 1360 and 1360a, which are transcribed in opposite directions, represents a potential operator (Fig. 4). A strong promoter consensus sequence upstream of 1360a is present. Also noted were two consensus Cheo (DinR) boxes, which are likely binding sites for DinR, the Gram-positive equivalent of the SOS transcriptional regulator LexA (43, 44).
Sequence analysis of CD4 and CD16.
Our assignment of the diffocin cluster was based on the published sequence of strain 630. We wished to confirm this by identifying the analogous diffocin gene cluster(s) in strains CD4 and CD16 and expressing the clusters in another species that was not anaerobic (see below). To identify the CD4 and CD16 diffocin loci, we obtained draft genome sequences of CD4 and CD16 (see Materials and Methods). Diffocin loci analogous to that of 630 were found in both strains. All of the ORFs shared a high degree of sequence similarity (>98% amino acid sequence identity), with the exception of ORF 1374. Annotations of the diffocin gene cluster(s) are shown in Table 2.
The ORF 1374 is by far the most divergent ORF among the diffocin gene clusters. The ORF 1374 (1,725 amino acids [aa]) of CD16 and that of strain 630 (1,743 aa) are very similar, with 88% identity; however, much of the divergence is in one central region of the protein. The CD4 ORF 1374 (1,773 aa) is 38% identical to both those of CD16 and 630, with only the N-terminal and C-terminal portions being highly similar. A clustal analysis and a dendrogram of the ORF 1374 amino acid sequences are shown in Fig. S1 of the supplemental material. We also amplified and sequenced ORF 1374 from ATCC 43593; it was found to be very similar to that of 630, with 99% amino acid sequence identity (see Fig. S1).
Cloning and expression of diffocin 16 and diffocin 4 in Bacillus subtilis.
The entire diffocin gene cluster from CD16 was cloned and inserted into a pETcoco-based bacterial artificial chromosome (BAC) (see the materials description in the supplemental material). Briefly, the 5′ and 3′ regions of the Bacillus subtilis amyE gene were introduced to flank the entire predicted diffocin gene cluster, ORFs 1359 to 1379, which along with the spectinomycin resistance marker (aad9) was inserted into pETcoco-1 to create pDG488. This plasmid was then transformed into B. subtilis strain BDR123, which has the chloramphenicol resistance marker (cat) inserted into amyE. Recombinants that were chloramphenicol sensitive but spectinomycin resistant were selected, picked, and verified by PCR to contain the entire diffocin 16 locus flanked by amyE sites in the B. subtilis chromosome. This strain was named BDR123-488.
Diffocins were expressed in BDR123-488 by induction with mitomycin C, as for C. difficile. B. subtilis is known to have a functional SOS system, and we suspected that the B. subtilis DinR would bind to the diffocin operator and be cleaved by activated B. subtilis RecA upon induction, resulting in expression of diffocins. Note that B. subtilis encodes a defective prophage, PBSX (14, 45), which is induced by the SOS response and results in lysis of cells and release of defective phage-like PBSX particles into the culture supernatant. We anticipated that because PBSX carries the holin and lysin genes, these would serve to release the recombinant diffocin particles along with PBSX particles (15). Particles collected from these lysates were tested for bactericidal activity against target C. difficile strains. Bactericidal activity was detected with the predicted bactericidal spectrum of diffocin 16 on target strains 19099 and 19145. No activity was seen for strains 19137 and 19135, which are insensitive to diffocin 16. No bactericidal activity against any tested C. difficile isolate was detected from lysates of B. subtilis BDR123, which contains no diffocin cluster.
The three downstream putative regulatory genes, ORFs 1377 to 1379, were deleted from pDG488, creating pDG491. This plasmid was transformed into BDR123 as described above, and appropriate recombinant bacteria were selected. When induced with mitomycin C, this construct (BDR123-491) also produced functional diffocin 16, indicating that these deleted genes are not required for diffocin expression in B. subtilis.
Similarly, we cloned ORFs 1359 to 1376 from strain CD4, creating plasmid pDG580. This BAC was introduced into BDR123, and recombinants were selected. This strain (BDG12) also produced diffocins that exhibited the predicted target specificity of diffocin 4, killing strains 19137 and 19135 but not 19099 or 19145. Thus, diffocin 4 and diffocin 16 can each be expressed in B. subtilis and retain the distinct killing spectra.
While the above expression system produced viable diffocin particles based on spot plate assays, expression levels appeared to be low compared with the natural material produced from C. difficile. To remedy this we pursued two approaches. The first was to delete the PBSX gene cluster from B. subtilis strain BDR11 (see the supplemental material). We suspected that production of these phage-like particles might limit diffocin production. This deletion was accomplished by the marker-free deletion method of Liu et al. (18), by which we created strain BDG9. The second modification was to switch the integrant selective marker from spectinomycin to chloramphenicol, which often results in better protein expression (E. Ferrara, personal communication). A construct was designed to integrate the diffocin 4 cluster with spectinomycin selection into the amyE as a single crossover (pDG589). This construct was transformed into BDG9, and an integrant was isolated and termed BDG21. BDG21 was further modified to switch the spectinomycin resistance marker to chloramphenicol. This was accomplished by creating a simple recombination plasmid, pDG621, which had the chloramphenicol gene flanked by homologous portions of the diffocin cluster and the front portion of amyE. Upon successful transformation and recombination, this process generated BDG45.
BDG45 was grown to an OD600 of 1.0 and induced with mitomycin as described above. After overnight induction, some lysis was observed despite the lack of PBSX and its lysis cassette, although the amount of lysis was considerably less than with the wild type: a 30 to 35% reduction in the OD600 compared to a >90% reduction for PBSX-containing strains. The culture was centrifuged, and the bacterial pellet was treated with Bugbuster to release the cellular contents. The cellular contents were then added to the supernatant, and the total diffocin activity was examined by spot assay. Based on serial 5-fold dilutions, we observed an approximately 125-fold increase in diffocin production above that of the previous construct (BDG21). With these changes in the production strain and purification modifications, we were within the range to measure total killing units (KU) using the survival assay (30). The yield was comparable to, or slightly higher than, that produced from C. difficile, ∼1 × 109 KU/per ml of culture volume.
The product of ORF 1374 is the bactericidal spectrum determinant.
ORF 1374 encodes a large polypeptide (∼200 kDa) that was shown by mass spectrometry to be part of the purified diffocin structure. When comparing the gene clusters of diffocins 4 and diffocin 16, most of the gene products, particularly those that were predicted to be structural components, were nearly identical at the amino acid level. The major difference between the two clusters was ORF 1374. For this and other reasons discussed below, we speculated that this gene product confers the target specificity of the diffocins. To test this, we completely replaced ORF 1374 of diffocin 4 with that of diffocin 16 in the integration vector (see the supplemental material) to create pDG603. pDG603 was then used to make a B. subtilis recombinant (BDG55), from which the diffocins were expressed as described above. The resulting hybrid diffocin particles, termed diffocin 4-16, had bactericidal activities against strains 19099 and 19145, both of which are sensitive to natural diffocin 16. Diffocin 4-16 did not kill 19135 or 19137; both are sensitive to diffocin 4 (Fig. 5 shows spot plate assays of diffocins 4 and 4-16 on these strains). The observed bactericidal specificities of these chimeric diffocin particles after replacement of just this one ORF demonstrated that target specificity is determined by the product of ORF 1374, the putative receptor-binding protein.
Fig 5.
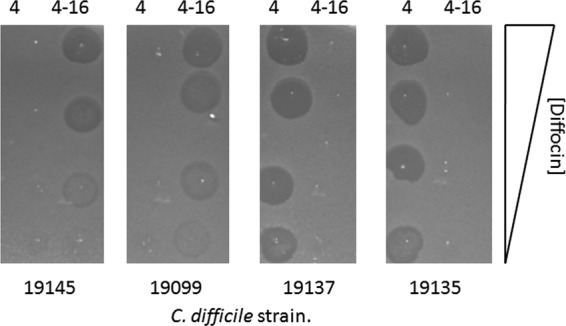
Spot plate assays of diffocin 4 and the hybrid diffocin 4-16 on target strains 19145, 19099, 19137, and 19135. Diffocin 4 was produced in B. subtilis BDG45, and the hybrid diffocin 4-16 was produced in BDG55. The diffocins were purified, and serial 5-fold dilutions of diffocins were spotted onto lawns of target bacteria, with diffocin 4 on the left and diffocin 4-16 on the right of each lawn. The plates were incubated overnight at 37°C before imaging.
DISCUSSION
We have shown that some strains of Clostridium difficile produce R-type bacteriocins, termed diffocins, which have bactericidal activity against other strains of the same species. The genetic locus that encodes these structures has been identified, cloned, and expressed in B. subtilis, an aerobic organism that is used to produce food and pharmaceutical products. This is apparently the first such entity to have been genetically and functionally identified and described for this species. Phage tail-like bacteriocins have been identified in C. botulinum; however, no functional or genetic characterizations have been published (7, 13).
Like the R-type pyocins of Pseudomonas aeruginosa, the bactericidal activities of diffocins appear to be specific to subsets of strains within the species. The spectrum determinant, presumably the receptor-binding protein of the particle, is the product of ORF 1374.
The diffocin locus has been located between ORFs 1359 and 1376, based on identification of structural genes, examination of the surrounding region for similarities to other phage tail-like ORFs, including regulatory genes, and the fact that this locus can be transferred to a different organism and produce bactericidal particles. We originally annotated ORFs 1377 to 1379 as part of the locus; however, production of active diffocins in B. subtilis does not require these ORFs. These small ORFs, which show some similarities to phage-like regulatory proteins, could be evolutionary remnants; however, they may play some regulatory role in C. difficile. Several other small ORFs that also have sequence similarities to regulatory proteins have been noted upstream; it is also unclear what role they play. A potential operator region was identified between the transcriptionally divergent ORFs 1360 and 1360a. This region contains two potential DinR-binding (functionally equivalent to LexA of E. coli) sequences flanking a strong promoter consensus that would likely initiate transcription of ORFs 1360a, 1361, 1362, and 1363 (the latter being the first identifiable structural gene), and also structural genes further downstream. The most likely regulatory scenario is that DinR binds these Cheo boxes between the ORF 1360 and 1360a sites and represses transcription, possibly in both directions; upon cleavage of DinR by activated RecA, the operator is derepressed. However, the presence of two potential lambda Cro-like repressors (ORFs 1360 and 1360a) and a putative sigma factor (ORF 1361) suggests that transcription is fine-tuned by diffocin-specific regulatory proteins. A considerable amount of work will be needed to elucidate the regulatory switch. In any event, when introduced into B. subtilis, this operon appears to be regulated in a similar manner, suggesting that the DinR proteins are functionally interchangeable between C. difficile and B. subtilis.
Noticeably absent from the diffocin loci are ORFs encoding cell lysis functions (i.e., a lysis cassette). This is somewhat surprising, since lysis is required to release particles from the cells. It appears that most, if not all, C. difficile strains do carry prophages that carry lysis genes that could be induced to release diffocins. Upon induction of the SOS response, there may be a genetic switch that in some instances enables production of phages and under other conditions, diffocins.
A unique feature of diffocins is the large protein encoded by ORF 1374, which we have shown to be the spectrum determinant that likely binds to a target cell surface receptor. Typically, Myoviridae phage tails and related bacteriocins possess tail fibers that act as receptor-binding proteins. These homotrimeric tail fiber structures are attached to the baseplate and are likely to be present at 6 copies per particle (17). This is the case for the Myoviridae phages of C. difficile, including ϕ119 and ϕC2 (10, 11), which likely share a common ancestry with diffocins. However, diffocins do not encode a full-length tail fiber. Instead, ORF 1373 encodes a truncated homologue of just the N terminus of the phage ϕCD119 and ϕCD2 tail fibers. In fact, there is even significant sequence similarity between ORF 1373 and the N terminus of the tail fiber protein of the P. aeruginosa R2 pyocin (Prf15), the region shown to be involved in baseplate binding (31, 42). We believe that the N terminus of the product of ORF 1373 retains its baseplate-binding function, but rather than acting as a receptor-binding protein, it interacts with the product of ORF 1374, which fulfills the latter role. The large flower-like appendages seen in the diffocin electron micrographs could be explained by such a structure. Also notable is the fact that the gene products of ORFs 1374 of various C. difficile isolates have diverged considerably more than the other ORFs in the diffocin clusters. This would be expected, considering that receptor-binding proteins would have diverged to target diverse receptors in order to kill diverse strains. Future work is needed to identify the molecular targets or receptors for diffocin binding on the surfaces of target bacteria.
Strain 630 encodes a locus that we used to first identify the diffocin genes. Given that its ORF 1374 is nearly identical to that of ATCC strain 43593, we might predict it to have a similar bactericidal spectrum. However, we have not been able to induce active diffocin particles from strain 630 by using mitomycin C. It is possible that the diffocin gene cluster is defective in this strain. Induction of the SOS response in 630 does induce prophages (10); perhaps there is a subtle regulatory balance between diffocin induction versus phage induction. During preparation of the manuscript, several whole-genome and draft genome sequences of C. difficile have been released, most of which contained a diffocin or diffocin-like gene cluster. It appears that these particles are widespread among the species. In addition, a recent study involving a collection of ribotype 027 C. difficile isolates found that they produced phage tail-like particles that morphologically resembled diffocins similar to our 027 diffocin producer, CD16 (27). Further work is needed to determine if diffocins from all 027 strains have identical killing spectra.
CDI has become a leading nosocomial infectious disease. Treatment of full-blown infection requires extensive use of antibiotics, such as metronidazole, vancomycin, or fidaxomycin, all of which are often successful therapies. However, the relapse rates are high and increase with each recurrence. Alternative therapies and management are needed. Particularly attractive is the potential prophylactic use of these specifically targeted bactericidal diffocins for prevention of colonization or for decolonization of asymptomatic human carriers, particularly individuals scheduled for antibiotic-mediated insults to their intestinal microbiota. Oral and parenteral R-type bacteriocins have been demonstrated to be effective prophylactics and therapeutics in animal models (6, 12, 21, 29, 30). We expect that orally administered diffocins could similarly be used to treat or prevent CDI. These agents might also be used to suppress the C. difficile bloom and consequential colitis in high-risk carrier patients and perhaps to prevent relapse, all without causing unintended collateral damage to the intestinal microbiota. At least one diffocin, the one produced by 43593, kills 027 isolates and could be used itself in locations where this ribotype is dominant. However, other ribotypes are becoming more prevalent, and a broader-spectrum diffocin will be needed; our work in identifying the receptor-binding protein is the first step in potentially being able to broaden the spectrum of diffocins produced by C. difficile by using genetic manipulation.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Rudner for supplying strains and plasmids. We also thank David Rudner, Linc Sonenshein, and Eugenio Ferrara for helpful advice with the Bacillus constructs.
The NMRC portion of this work was supported by the Transformational Medical Technologies Program under contract TMTI_IB06RSQ002 through the Defense Threat Reduction Agency of the U.S. Department of Defense.
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, or the U.S. Government.
Footnotes
Published ahead of print 14 September 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Ananthakrishnan AN. 2011. Clostridium difficile infection; epidemiology, risk factors, and management. Nat. Rev. Gastroenterol. Hepatol. 8:17–26 [DOI] [PubMed] [Google Scholar]
- 2. Blackwell CC, Law JA. 1981. Typing of non-serogroupable Neisseria meningitidis by means of sensitivity to R-type pyocins of Pseudomonas aeruginosa. J. Infect. 3:370–378 [DOI] [PubMed] [Google Scholar]
- 3. Blackwell CC, Winstanley FP, Telfer Brunton WA. 1982. Sensitivity of thermophilic campylobacters to R-type pyocins of Pseudomonas aeruginosa. J. Med. Microbiol. 15:247–251 [DOI] [PubMed] [Google Scholar]
- 4. Brandt LJ, et al. 2012. Long-term follow-up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am. J. Gastroenterol. 107:1079–1087 [DOI] [PubMed] [Google Scholar]
- 5. Coetzee HL, De Klerk HC, Coetzee JN, Smit JA. 1968. Bacteriophage tail-like particles associated with intra-species killing of Proteus vulgaris. J. Gen. Virol. 2:29–36 [DOI] [PubMed] [Google Scholar]
- 6. Damasko C, et al. 2005. Studies of efficacy of enterocoliticin, a phage-tail like bacteriocin, as antimicrobial agent against Yersinia enterocolitica serotype O3 in a cell culture system and in mice. J. Vet. Med. 52:171–179 [DOI] [PubMed] [Google Scholar]
- 7. Ellison JS, Kautter JA. 1970. Purification and some properties of two botocins. J. Bacteriol. 104:19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Filiatrault MJ, Munson RS, Jr, Campagnari AA. 2001. Genetic analysis of a pyocin-resistant lipooligosaccharide (LOS) mutant of Haemophilus ducreyi: restoration of full-length LOS restores pyocin sensitivity. J. Bacteriol. 183:5756–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fortier L-C, Moineau S. 2007. Morphological and genetic diversity of temperate phages in Clostridium difficile. Appl. Environ. Microbiol. 73:7358–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goh S, Ong PF, Song KP, Riley TV, Chang BJ. 2007. The complete genome sequence of Clostridium difficile phage ϕC2 and comparisons to ϕCD119 and inducible prophages of 630. Microbiology 153:676–685 [DOI] [PubMed] [Google Scholar]
- 11. Govind R, Fralick JA, Rolfe RD. 2006. Genomic organization and molecular characterization of Clostridium difficile bacteriophage ϕCD119. J. Bacteriol. 188:2568–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haas H, Sacks T, Saltz N. 1974. Protective effect of pyocin against lethal Pseudomonas aeruginosa infections in mice. J. Infect. Dis. 129:470–472 [DOI] [PubMed] [Google Scholar]
- 13. Kautter DA, Harmon SM, Lynt RK, Jr, Lilly T. 1966. Antagonistic effect on Clostridium botulinum type E by organisms resembling it. Appl. Microbiol. 14:616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krogh S, O'Reilly M, Nolan N, Devine KM. 1996. The phage-like element PBSX and part of the skin element, which are resident at different locations on the Bacillus subtilis chromosome, are highly homologous. Microbiology 142:2031–2040 [DOI] [PubMed] [Google Scholar]
- 15. Krogh S, Jørgensen ST, Devine KM. 1998. Lysis genes of the Bacillus subtilis defective prophage PBSX. J. Bacteriol. 180:2110–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kunst F, Msadek T, Rapopot G. 1994. Signal transduction network controlling degradative enzyme synthesis and competence in Bacillus subtilis, p 1–20 In Piggot PJ, Moran CP, Jr, Youngman P. (ed), Regulation of bacterial differentiation. American Society for Microbiology, Washington, DC [Google Scholar]
- 17. Leiman PG, Scheider MM. 2012. Contractile tail machines of bacteriophages. Adv. Exp. Med. Biol. 726:93–114 [DOI] [PubMed] [Google Scholar]
- 18. Liu S, Endo K, Ara K, Ozaki K, Ogasawara N. 2008. Introduction of marker-free deletions in Bacillus subtilis using AraR repressor and the ara promoter. Microbiology 154:2562–2570 [DOI] [PubMed] [Google Scholar]
- 19. Loo VG, et al. 2005. A predominantly clonal multiinstitutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442–2449 [DOI] [PubMed] [Google Scholar]
- 20. Louie TJ, et al. 2011. Fidaxomicin verses vancomycin for Clostridium difficile infection. N. Engl. J. Med. 364:422–431 [DOI] [PubMed] [Google Scholar]
- 21. Merrikin DJ, Terry CS. 1972. Use of pyocin 78-C2 in the treatment of Pseudomonas aeruginosa infection in mice. Appl. Microbiol. 23:164–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499–510 [DOI] [PubMed] [Google Scholar]
- 23. Miller BA, Chen LF, Sexton DJ, Anderson DJ. 2011. Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals. Infect. Control. Hosp. Epidemiol. 32:387–390 [DOI] [PubMed] [Google Scholar]
- 24. Morse SA, Vaughan P, Johnson D, Iglewski BH. 1976. Inhibition of Neisseria gonorrhoeae by a bacteriocin from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 10:354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morse SA, Jones BV, Lysko PG. 1980. Pyocin inhibition of Neisseria gonorrhoeae: mechanism of action. Antimicrob. Agents Chemother. 18:416–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakayama K, et al. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38:213–231 [DOI] [PubMed] [Google Scholar]
- 27. Nale JY, et al. 2012. Diverse temperate bacteriophage carriage in Clostridium difficile 027 strains. PLoS One 7:e37263 doi:10.1371/journal.pone.0037263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pepin J, et al. 2005. Increasing risk of relapse after treatment of Clostridium difficile colitis in Quebec, Canada. Clin. Infect. Dis. 40:1591–1597 [DOI] [PubMed] [Google Scholar]
- 29. Ritchie JM, et al. 2011. An E. coli O157-specific engineered pyocin prevents and ameliorates infection by E. coli O157:H7 in an animal model of diarrheal disease. Antimicrob. Agents Chemother. 55:5469–5474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scholl D, Martin DW., Jr 2008. Antibacterial efficacy of R-type pyocins towards Pseudomonas aeruginosa in a mouse peritonitis model. Antimicrob. Agents Chemother. 52:1647–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Scholl D, et al. 2009. An engineered R-type pyocin is a highly specific and sensitive bactericidal agent for the foodborne pathogen, E. coli O157:H7. Antimicrob. Agents Chemother. 53:3074–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shan J, et al. 15 June 2012. Prophage carriage and diversity within clinically relevant strains of Clostridium difficile. Appl. Environ. Microbiol. 78:6027–6034 [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Strauch E, et al. 2001. Characterization of enterocoliticin, a phage tail-like bacteriocin, and its effect on pathogenic Yersinia enterocolitica strains. Appl. Environ. Microbiol. 67:5634–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sunenshine RH, McDonald CL. 2006. Clostridium difficile-associated disease: new challenges from an established pathogen. Cleve. Clin. J. Med. 73:187–197 [DOI] [PubMed] [Google Scholar]
- 35. Surawicz CM, Alexander J. 2011. Treatment of refractory and recurrent Clostridium difficile infection. Nat. Rev. Gastroenterol. Hepatol. 8:330–339 [DOI] [PubMed] [Google Scholar]
- 36. Tannock GW, et al. 2010. A new macrocyclic antibiotic, fidaxomicin (OPT-80), causes less alteration to the bowel microbiota of Clostridium difficile-infected patients than does vancomycin. Microbiology 156:3354–3359 [DOI] [PubMed] [Google Scholar]
- 37. Thompson NE, Pattee PA. 1981. Genetic transformation in Staphylococcus aureus: demonstration of a competence-conferring factor of bacteriophage origin in bacteriophage 80a lysates. J. Bacteriol. 148:294–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Uratani Y, Hoshino T. 1984. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J. Bacteriol. 157:632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Voth DE, Ballard JD. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18:247–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang IN, Smith DL, Young R. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799–825 [DOI] [PubMed] [Google Scholar]
- 41. Warny M, et al. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084 [DOI] [PubMed] [Google Scholar]
- 42. Williams S, Gebhart D, Martin DW, Scholl D. 2008. Re-targeting R-type pyocins to generate novel bactericidal protein complexes. Appl. Environ. Microbiol. 74:3868–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Winterling KW, Levine AS, Yasbin RE, Woodgate R. 1997. Characterization of DinR, the Bacillus subtilis SOS repressor. J. Bacteriol. 179:1698–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Winterling KE, et al. 1998. The Bacillus subtilis DinR binding site: redefinition of the consensus sequence. J. Bacteriol. 180:2201–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wood HE, Dawson MT, Devine KM, McConnell DJ. 1990. Characterization of PBSX, a defective prophage of Bacillus subtilis. 172:2667–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zink R, Loessner MJ, Schere S. 1995. Characterization of cryptic prophages (monocins) in Listeria and sequence analysis of a holin/endolysin gene. Microbiology 141:2577–2584 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.